Abstract
Transcripts derived from select clades of transposable elements are among the first to appear in early mouse and human embryos, indicating transposable elements and the mechanisms that regulate their activity are fundamental to the establishment of the founding mammalian lineages. However, the mechanisms by which these parasitic sequences are involved in directing the developmental program are still poorly characterized. Transposable elements are regulated through epigenetic means, where combinatorial patterns of DNA methylation and histone 3 lysine 9 trimethylation (3K9me3) suppress their transcription. From studies in rodents, SET Domain Bifurcated 1 (SETDB1) has emerged as the core methyltransferase responsible for marking transposable elements with H3K9me3, and temporally regulating their transcriptional activity. SetDB1 loss of function studies in mice reveal that while extraembryonic tissues do not require this methyltransferase, establishment of the embryo proper fails without it. As the bovine embryo initiates the processes of epigenetic programming earlier in the preimplantation phase, we sought to determine if suppressing SETDB1 would block the formation of the inner cell mass. We report here that bovine SETDB1 transcripts are present throughout preimplantation development, and RNAi-based depletion blocks embryo growth at the morula stage of development. While we did not observe alterations in global histone methylation or transposable element transcription, we did observe increased global levels of H3K27 acetylation; an epigenetic mark associated with active enhancers. Our observations suggest SETDB1 might interact with the epigenetic machinery controlling enhancer function, and that suppression of this methyltransferase may disrupt the bovine developmental program.
Keywords: SETDB1, Transposable Elements, Preimplantation Development, Enhancer, Lineage Specification, Inner Cell Mass, Epigenetic, Developmental Program
1.0 Introduction
Preimplantation development represents a critical window in the process of epigenetic programming as during the first few cleavage divisions, a major reorganization of chromatin structure occurs establishing the patterns of gene expression necessary to direct growth and differentiation of the mammalian embryo. This programming phase is one of the key events in the formation of the embryonic and extraembryonic cellular lineages [1,2]. Defects in the capacity of the embryo to properly establish this epigenetic program result in developmental failure or onset of disease due to inappropriate patterns of gene expression [3–5].
For the past 25 years, the preimplantation kinetics of DNA methylation have been the subject of intense study, while the dynamics of post-translational histone methylation have remained poorly defined [6,7]. This is particularly true of the histone methylation machinery and processes hypothesized to demarcate the genome into regions of euchromatin as well as both constitutive and facultative heterochromatin. Within this realm, lysine nine of histone 3 (H3K9) is a core target of multiple epigenetic modifiers implicated in controlling both gene transcription as well as supra-nucleosomal chromatin structure. For example, acetylated lysine 9 has been identified within the 5′ regions of transcriptionally active genes, while methylation at this specific residue has been linked to the nucleation of both transcriptional silencing and pericentric heterochromatin [8–11]. Interestingly, trimethylation (me3) of H3K9 is enriched within the regulatory regions and bodies of various families of transposable elements [12,13], and regulation of these parasitic nucleic acid sequences directly influence stem cell pluripotency in both primates and rodents [14–17]. Thus, understanding the mechanisms responsible for establishing and maintaining H3K9 epigenetic post-translational marks is essential to better defining the processes of developmental programming and preimplantation lineage specification.
Six mammalian histone methyltransferases with demonstrated H3K9 catalytic activity have been identified, including EHMT1 (Glp), EHMT2 (G9a), SUV39H1, SUV39H2, SETDB1, (Eset) and SETDB2 [18]. SUV39H1 and SUV39H2 are responsible for the dynamics of pericentric heterochromatin formation [8] while EHMT1, EHMT2, and SETDB1 appear to regulate both protein coding genes and transposable elements [19–21]. SETDB2 remains poorly characterized and may be involved in immune function [10]. Interestingly, while EHMT2 appears necessary to maintain DNA methylation at transposable elements, SETDB1 appears to be the core methyltransferase responsible for marking these sequences with H3K9me3 and temporally regulating their transcriptional activity [20,21]. Given the importance of transposable element sequences in regulating the pluripotency transcriptional program [14–17], better understanding the dynamics of SETDB1 during preimplantation development may be crucial to deciphering the processes directing specification of the embryonic lineage.
In the mouse, SetDB1 transcripts (and possibly the maternal SETDB1 protein) are present in mature oocytes. Zygotic transcription of SetDB1 begins at the blastocyst stage, and is essential for the survival of the mouse inner cell mass (ICM). In contrast, the extraembryonic tissues do not appear to require this methyltransferase as at day 6.5 of gestation, null embryos display a fully developed trophoblast and ectoplacental cone, while the embryo proper shows a complete lack of development [22]. Given the established link between ES cell pluripotency and transposable elements, we sought to determine if suppressing maternal SetDB1 transcripts would disrupt blastocyst formation by blocking the formation of an ICM. In mice, the transition from zygote to blastocyst takes 3.5 days with zygotic transcription initiating at the late 1-cell to 2-cell stage [23]. While an RNA interference (RNAi)-based approach could be employed to deplete SetDB1 mRNAs, stores of maternal SETDB1 are hypothesized to be present, and depending on the half-life of the protein, may allow the formation of the ICM to proceed unperturbed [22]. In contrast, during bovine embryogenesis, the transition from zygote to blastocyst takes between seven to eight days to occur, with zygotic transcription beginning at the 8-cell stage [24]. This greater length of gestational time should allow opportunity to interrogate the consequences of SETDB1 loss of function on both specification of the ICM and blastocyst formation. We report here that RNAi-based depletion of SETDB1 transcripts blocks bovine preimplantation development at the morula stage and increases global levels of H3K27 acetylation, an epigenetic mark associated with active enhancers. Our results add one more piece of data to suggest SetDB1 is essential for the earliest events in lineage specification.
2.0 Materials and Methods
2.1 Embryo Production and siRNA Injection
Bovine oocytes were collected at a commercial abattoir (DeSoto Bioscience, Seymour, TN, USA) and shipped in a MOFA metal bead incubator (MOFA Global, Verona, WI. USA) at 38.5 °C overnight in sealed sterile vials containing 5% CO2 in air equilibrated Medium 199 with Earles salts (Invitrogen, Life Technologies Inc., Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 1% Penicillin/Streptomycin (Invitrogen), 0.2 mM sodium pyruvate, 2 mM l-Glutamine (Sigma Chemical Co., St. Louis, MO, USA) and 5.0 μg/ml of Folltropin (Vetoquinol, Pullman, WA, USA). The oocytes were matured in this medium for 22 to 24 h. Matured oocytes were washed twice in warm Tyrode-lactate (TL) Hepes (Vetoquinol) supplemented with 50 μg/μL Gentamicin (Invitrogen) while being handled on a 38.5 °C stage warmer (Minitube). In vitro fertilization was conducted using a 2 h pre-equilibrated modified Tyrode-lactate medium supplemented with 250 μM sodium pyruvate, 1% P/S, 6 mg·mL -1 BSA Fatty Acid free (Sigma), 20 μM Penicillamine, 10 μM Hypotaurine, and 10 μg/mL Heparin (Sigma) in 38.5 °C, 5% CO2 a humidified air incubator. Frozen semen was thawed at 35 °C for 1 min, then separated by centrifugation at 200 × g for 20 min in a density gradient medium (Isolate®, Irvine Scientific, Santa Ana, CA, USA) 50% upper/90% lower. Supernatant was removed, sperm pellet re-suspended in 2 mL Modified Tyrodes medium and centrifuged at 200 × g for 10 min to wash. The sperm pellet was removed and placed into a warm 0.65 mL microtube (VWR Scientific, Pittsburg, PA, USA) prior to bulk fertilizing in Nunclon® 4-well multi-dishes (VWR) containing up to 50 matured oocytes per well at a concentration of 1.0 × 106 sperm/mL. Sixteen to eighteen hours post-insemination, oocytes were cleaned of cumulus cells by a 2 min vortex in 45 μL TL Hepes in 0.65 mL microtube (VWR), washed in TL Hepes, and then randomly assigned to three different treatment groups: non-injected controls (CNTL), non-targeting siRNA injected controls (siNULL), and injection with siRNA targeting SETDB1 (siSETDB1).
Three short interfering RNA (siRNA) sequences were designed to bovine SETDB1: SETDB1 siRNA 545 (5′ CCGUGAAGCUAUGGCUGCCUUAAGA SETDB1 3′), siRNA 2799 (5′ CCUGAUGACCGAAACAGGAUGUCAA 3′), and SETDB1 siRNA 3225 (5′ GACGACAGCUCAGAUGACAACUUCU 3′). The sequences shown correspond to the sense strand and the numbers indicate the targeted position within the bovine SETDB1 (NM_001191388.2) transcript. The control siRNA (Invitrogen, Cat # AM 4621) was labeled with a fluorescent Cy3 tag. Of the three SETDB1 siRNAs 545 and 3225 were the most effective and a 50%/50% mixture of each was utilized for all subsequent experiments. 20 μm of both the control and SETDB1 siRNAs were mixed with green fluorescent dextran in 1 x Tris-EDTA, and approximately 100 pL injected cytoplasmically. Post-injection, fluorescent embryos were separated and cultured in bovine evolve medium (Zenith Biotech, ZEBV-100) supplemented with 4 mg·mL−1 of BSA (Probumin, Millipore, 810014) and cultured until the blastocyst stage. Cleavage rates were recorded on Day 2 and viable embryos were separated from nonviable embryos. Embryos were monitored daily for morphological progression and blastocyst rates recorded on Day 8 post-IVF.
2.2 RNA isolation and Reverse Transcription
RNA was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA. Cat # 74104) with the following modifications. Pools of ten bovine MII oocytes or embryos (2, 4–8, 16-cell, morula and blastocyst stage) were washed twice in DPBS and snap frozen in 10ul of RLT lysis buffer. Samples were thawed on ice and brought to a volume of 100ul by adding additional lysis buffer. 100ul of 80% ethanol was added, samples mixed by pipetting, the total 200μl volume transferred to an RNeasy spin column, and centrifuged at 8,000g for 30 seconds. Columns were washed with 350μl of buffer RW1, then again with 500μl of 80% ethanol, and after centrifugation, allowed to air dry for 5 minutes at room temperature. RNA was eluted by adding 20μl RNase free water to the column, allowing it to sit at room temperature for 1 minute, and then centrifuging at 8000g for 1 minute. Isolated RNA was treated with amplification grade DNaseI (Sigma, St. Louis, MO. Cat# AMPD1) by adding 1μl of DNA and 2μl of buffer to the 20μl volume of isolated RNA. Samples were brought to room temperature for 15 minutes and then placed on ice. Reverse transcription reactions were conducted using the SuperScriptII kit (Invitrogen, Carlsbad, CA. Cat# 18064-071) by combining 1 μl random hexamer oligonucleotides (Invitrogen, Carlsbad, CA. Cat# 48190011), 1 μl 10 mM dNTP (Invitrogen, Carlsbad, CA. Cat# 18427-013), and 23 μl of DNAse treated RNA. This mixture was brought to 70°C for 5 minutes then cooled to room temperature. A total of 5μl of SuperScriptII reaction buffer, 3μl of dithiothreitol, and 1μl of SuperScriptII were then added, and the reaction mixture brought to 42°C for 50 minutes, 45°C for 20 minutes, 50°C for 15 minutes, then 70°C for five minutes. Samples were stored at −80°C until use.
2.3 Real-Time PCR Amplification
Primer sequences not acquired from previous publications were designed using the NetPrimer software tool (www.premierbiosoft.com/netprimer). Real-time PCR analysis of mRNA levels was carried out using the DyNAmo Flash SYBR Green qPCR Mastermix (Fisher Scientific, Pittsburgh PA. Cat # F-415L) following the manufacturer’s instructions. Reactions were performed on a StepOnePlus Real Time PCR system (Applied Biosystems, Foster City CA.). Quantitative measurements of SETDB1 transcript levels were obtained using the primer sequences bSETDB1 Fwd (5′ GCTGAGACACCAAACGTCAAAA 3′) and bSETDB1 Rev (5′ ACATAGGAAGCATAGCCATCATCA 3′) followed by normalization of these readings to the geometric mean to three reference genes, as previously described [25]. The three reference genes selected were GAPDH, YWHAZ & SDHA based on previously published data [26]. Primer sequences used were: Gapdh (Fwd 5′-CTGCCCGTTCGACAGATAG-3′, Rev 5′-CTCCGACCTTCACCATCTTG-3′) Ywhaz (Fwd 5′-CTGAACTCCCCTGAGAAAGC-3′ Rev 5′-CCTTCTCCTGCTTCAGCTTC-3′) Sdha (Fwd 5′-ACCTGATGCTTTGTGCTCTG-3′ Rev 5′-TCGTACTCGTCAACCCTCTC-3′). Primer sequences used in the analysis of bovine transposable elements were LINE1 (Fwd 5′ CCCAGGTCCAGACGGCTT 3′ Rev 5′ GGGTGATGGTGGCCTCATAGA 3′), BOV-B (Fwd 5′ CAAGTGCGCCCGAGAGGA 3′ Rev 5′ GCCACCACCCCTGACCTC 3′) bERV g4 LTR (Fwd 5′ GAGCTGTGATGCGCCGAG 3′ Rev 5′ CGGGAGCTTCGCTGAGTTG 3′), bERV g4 Pol (Fwd 5′ CCAGGGGTCCCAGGAGGT 3′ Rev 5′ GGCAGGCTGAGGCGTGAC 3′) [27], and bERV b3 LTR (Fwd 5′ GTTCGCCGTTTCCAAGTCC 3′ Rev 5′ CTCTTTCTGCAGGGCTCGC 3′) [28]. Relative gene expression levels from each sample were calculated in triplicate using the ΔΔCT method described previously [29]. Values from these calculations were transferred into the statistical analysis program Graph Pad for analysis.
2.4 Immunocytochemistry
2.4.1 Analysis of 5-methylcytosine and 5-hydroxy-methylcytosine
IVF embryos at the morula stage (148 hours post-fertilization) were removed from culture, washed twice with 1 x PBS and fixed with 99% pure methanol at −20°C for 5 minutes. Embryos were washed in PBS and permeabilized for 30 minutes at room temperature with 0.2% Triton X-100 in PBS and treated with 3 M HCl diluted in ddH2O with 0.1% Triton X-100 to denature the DNA and allow binding of the primary antibody to methylated/hydroxymethylated DNA. The HCl was removed once the zona was dissolved (10–15 minutes) and a 100 mM Trizma Hydrochloride buffer, pH 8.5, was added for 30 minutes to neutralize the HCl. Embryos were incubated in blocking buffer with 3.0% BSA (Sigma Cat# 9022) and 0.1% Triton X-100 in PBS for four hours at room temperature. Primary antibodies listed in Table 1 were diluted 1:200 in blocking buffer and embryos incubated in 500μl drops for one hour at 37°C. Embryos were then washed through blocking buffer three times for thirty minutes each. Alexa 488 (Invitrogen) and Alexa 594 (Invitrogen) secondary antibodies were diluted 1:200 in PBS with 0.1% Tween-20 and embryos incubated in this solution for one hour at 37°C with no light (remaining steps were done without light). This secondary antibody was washed three times with PBS with 0.1% Tween-20 for five minutes each at 37°C. RNase A was diluted to 25 μg/ml in PBS and incubated for 20 minutes at 37°C. Hoechst (Sigma Cat# 33342) was diluted 25 μg/ml in PBS, added to the embryos and incubated for 15 minutes at 37°C. Embryos were washed three times with PBS with 0.1% Tween-20 for thirty minutes each at 37°C. Cover slips were mounted with a 50/50 solution of anti-fade and glycerol on glass slides and sealed with clear fingernail polish.
Table 1.
Antibodies utilized in immunocytochemical analysis.
| Type | Target | Antibody | Source (Cat #) |
|---|---|---|---|
| Primary | 5mc | 5-Methylcytidine Mouse mAb | Epigentek (33D3) |
| Primary | 5hmc | 5-Hydroxymethylcytosine Rabbit pAb | Active Motif (39791) |
| Primary | H3K9me3 | Histone 3 trimethyl Lys9 Rabbit pAb | Active Motif (39161) |
| Primary | H3K27me3 | Histone 3 trimethyl lys27 Mouse mAb | Abcam (ab6002) |
| Primary | H3K4me3 | Histone 3 trimethyl Lys3 Rabbit pAb | Abcam (ab6000) |
| Primary | H3K9me2 | Histone 3 dimethyl Lys9 Rabbit pAb | Active Motif (39375) |
| Primary | H3K27ac | Histone 3 acetyl Lys 27 Rabbit pAb | Abcam (ab4729) |
| Secondary | Rabbit IgG | Alexa Flour 488 Goat Anti-Rabbit IgG | Invitrogen (A-11008) |
| Secondary | Mouse IgG | Alexa Flour 594 Goat Anti-Mouse IgG | Invitrogen (A-11005) |
2.4.2 Analysis of post-translational histone modifications
Morula embryos were fixed by placing embryos in cold methanol for a minimum of 1 minute, and stored in PBS-0.1% Tween 20 (PBS-Tw) at 4°C until further use. Embryos were permeabilized with 1% Triton X-100 in PBS (PBS-Tr) for 1 hour at room temperature and washed thoroughly through PBS with 0.1% Tween-20 over 15 minute intervals. Following the permeabilization and wash, embryos were blocked in fresh blocking buffer (10 mg/mL BSA, 2% v/v goat serum and 11.25 mg/mL glycine in 1X PBS) overnight in order to prevent any nonspecific antibody binding. Embryos were labeled with the appropriate primary antibody, at room temperature for one hour, washed through fresh blocking buffer again and labeled with the corresponding secondary antibody. Primary and secondary antibodies are listed in Table 1 and were all diluted in PBS with 0.1% Tween-20. (Same method as before for DNA staining with Hoechst (Sigma Cat# 33342) as the 5mc and 5mc protocol).
2.4.3 Digital Microscopy
Digital imaging microscopy was performed in the Texas A&M University College of Veterinary Medicine & Biomedical Sciences Image Analysis Laboratory at the Zeiss stallion digital imaging work station. A z-series was taken from individual embryos with 5 images per embryo with various distances depending on their shape and size. Each section contained an image of the antibody of interest (5-methylcytosine, H3K9 me2, me3 etc.) and one for the Hoechst nuclear stain. Mean intensity measurements of the embryo images were taken using NIS Elements 3.0 (Nikon New York, USA) software where the images of samples with no primary antibody were set as the baseline. Antibody stain intensity measurements were divided by the intensity of the Hoechst stained nuclei to give a ratio for each image plane in an embryo.
2.5 Statistical Analysis
For analysis of gene expression, the measured cycle threshold (CT) experimental values for each transcript were compiled and normalized to the geometric mean of the reference genes listed above. Normalized expression levels were calculated using the Delta Delta CT method described previously (Schmittgen and Livak, 2008). Values from these calculations were transferred into the statistical analysis program GraphPad (GraphPad Software, Inc., La Jolla, CA) and an analysis of variance (ANOVA) was run to assay differences between experimental treatments. For determination of differences in embryo development rates, Chi-Square analysis was performed between three independent groups of embryos. For image analysis, ratios for each embryo were averaged to obtain a mean ratio for each treatment group and staining combination. Using these means, a one-way ANOVA model was constructed in order to determine significant differences between treatment groups for each staining. For all experiments, statistical significance was set at alpha = 0.05. For comparisons with p-values < 0.05, we applied Tukey’s HSD analysis and have marked statistically significant differences with the lowercase letters.
3.0 Results
3.1 Bovine SETDB1 expression and siRNA mediated depletion in preimplantation embryos
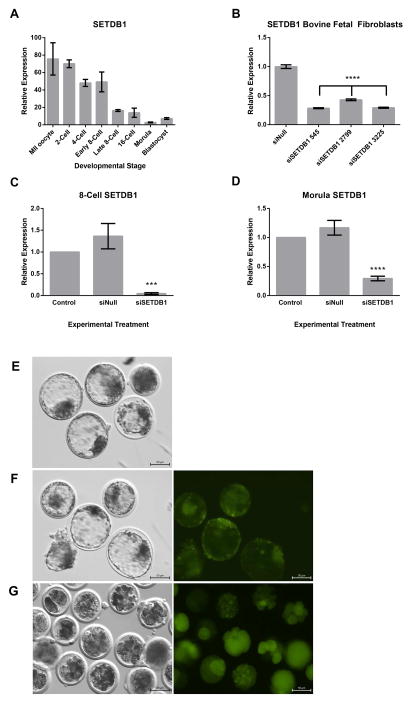
Bovine SETDB1 is located on chromosome 3, and encodes a 1290 amino acid protein with 96% conservation to the human homologue. We began our investigation by examining the mRNA transcript profile of SETDB1 during bovine preimplantation development using quantitative reverse transcription polymerase chain reaction (qRT-PCR). SETDB1 transcripts are abundant in the MII oocyte and maintain high abundance until the early 8-cell stage (74–75 hours post-fertilization) (Fig 1a). These high levels begin to precipitously decline during the transition to the late 8-cell stage (94–96 hours) and remained low, yet consistently detectable through the blastocyst stage (172 to 180 hours) (Fig 1a). To examine the consequences of SETDB1 loss of function on bovine preimplantation development, we designed and tested three short interfering RNAs (siRNAs) to bovine SETDB1, and validated their capacity to deplete SETDB1 transcripts using transient transfection assays in bovine embryonic fibroblasts (Fig 1b). These analyses identified two siRNAs (545 and 3225) as the most effective. In previous studies examining DNMT1 suppression, we identified an additive effect of two siRNAs when used in combination [30]. Therefore, all further experiments utilized an equimolar ratio of siRNAs 545 and 3225. To evaluate the impact of SETDB1 suppression during embryogenesis, the siRNA mixture was injected into bovine zygotes 16–18 hours post-fertilization (siSETDB1). For comparison, a separate group was injected with a non-targeting siRNA (siNull) described previously [30], and a third group was left untreated (Control).
Fig 1.
Bovine SETDB1 expression and siRNA mediated depletion in bovine preimplantation embryos. A) Measurements of transcripts encoding bovine SETDB1 during embryogenesis. Levels of SETDB1 mRNAs were measured in bovine oocytes and at various stages of preimplantation development using q-RT-PCR. B) Validation of siRNAs targeting SETDB1 using transient transfection into bovine fetal fibroblasts. SETDB1 transcript levels were quantified using q-RT-PCR. C–D) Measurement of SETDB1 transcripts in bovine 8-cell (C) and morula stage embryos (D) from the Control, siNull and siSETDB1 treatment groups using q-RT-PCR. For A–D, samples were normalized to the geometric mean of GAPDH, YWHAZ & SDHA and graphed using the 2−ΔΔ CT method [52]. Graphs are representative of four independent experiments. Error bars represent the SEM, *** denotes p < 0.001 and **** p < 0.0001. E–G) Developmental arrest of siSETDB1 treated embryos. Confocal images of expanded blastocyst stage noninjected Control (E), and siNull embryos (F), as well as arrested morula stage siSETDB1 injected embryos (G). 20x images displaying the injected green fluorescent dextran dye are displayed on the right while bright field images are displayed on the left.
Due to the lack of bovine-specific antibodies to SETDB1, we utilized qPCR to examine the effect siSETDB1 treatments. To verify that injection of the siRNAs was depleting the target mRNA, 8-cell (70–72 hours) and morula (146 to 148 hours) stage embryos were collected, RNA isolated and levels of SetDB1 transcripts measured using qRT-PCR. As can be seen in Figure 2c–d, SETDB1 siRNAs injected 16–18 hours post-fertilization produced a nearly complete depletion at the 8-cell stage, with robust suppression persisting into the morula stage of development, as compared to siNULL injected embryos (p < 0.01). We next examined the impact of siSETDB1 treatment on embryo viability, as measured by development to the blastocyst stage. Embryo cleavage rates between all experimental groups were similar, and while the injection process did produce a measurable, yet expected decline in embryo viability (25% for siNull vs 44% for Control), 0 out of 361 embryos injected with siRNAs targeting SETDB1 survived to the blastocyst stage (Table 2). As can be seen in Figures 1e–g, Control and siNull experimental treatments yielded viable expanded blastocysts on day 8 while siSETDB1 treated embryos were only viable until day 6 (146 to 148 hours) of cultured development, producing degenerate embryos and some poor quality morula past this point.
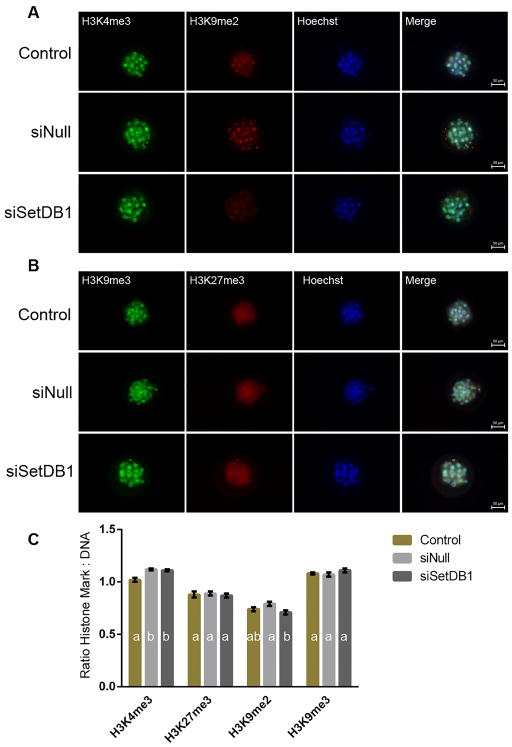
Fig 2.
Analysis of post-translational histone modifications in siSETDB1 injected embryos. Representative confocal images of viable morula-stage embryos co-stained with (A) H3K4me3 (green)/H3K9me2 (red)/Hoechst (blue), as well as (B) H3K9me3 (green)/H3K27 (red)/Hoechst (blue). Images are at 20 × magnification. C) Average fluorescence intensity ratios were calculated for each embryo by dividing the intensity of the specific target (H3K4me3, H3K9me2, H3K9me3 and H3K27me3) by the intensity of DNA stain (Hoechst). Ratios for each post-translational modification from each embryo were then averaged to obtain a mean ratio for each treatment group and staining combination. Using these means, a one-way ANOVA model was constructed in order to determine differences between treatment groups for each staining. Bars displaying different letters were considered to be significantly different (p < 0.05). Error bars represent the SEM.
Table 2.
Embryo development rates for Control, siNull and siSETDB1 injected embryos. The number of embryos either cleaving to the 2-cell stage, or developing to the blastocyst stage were counted and used to generate a one-way ANOVA model in order to determine differences between treatment groups. Groups displaying different superscript letters were considered to be different (p < 0.05).
| Treatment | n | Cleaved | % Cleavage | Blastocysts | % Blastocysts |
|---|---|---|---|---|---|
| CNTL | 189 | 162 | 85.7 ± 2.2%a | 85 | 44.9 ± 4.9%a |
| NULL | 283 | 222 | 78.4 ± 6.9%a | 73 | 25.7 ± 6.0%b |
| siSETDB1 | 361 | 308 | 85.3 ± 2.8%a | 0 | 0%c |
3.2 Analysis of DNA and Histone 3 Lysine 9 methylation in siSETDB1 embryos
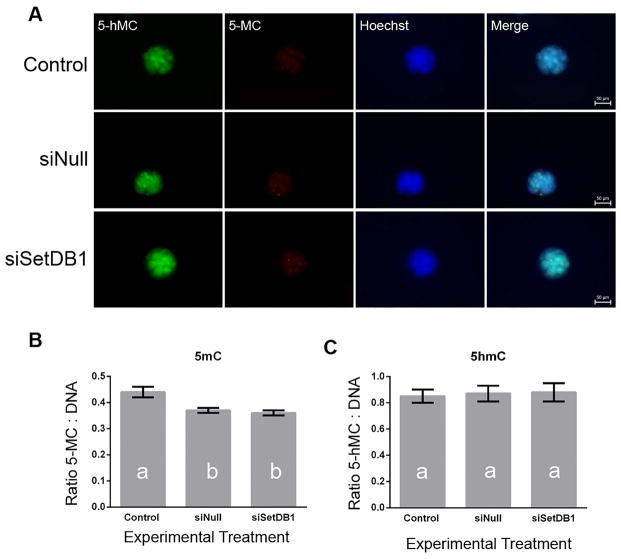
While late-stage, compacting morula from the siSETDB1 treatments were fragmented and of very poor quality, early morula stage embryos (146 to 148 hours post fertilization) from this treatment group appeared morphologically normal. We therefore concentrated on this developmental time-point in our subsequent investigations. In order to determine the cause of the observed lethality, we measured potential alterations to the developing epigenome. SETDB1 catalyzes the conversion of dimethylated H3K9 into the trimethylated form [31]. Thus, we began by measuring global levels of dimethylated and trimethylated H3K9. For this analysis, we employed immunocytochemical analysis, and focused our efforts on the morula stage of development (146 to 148 hours post fertilization), which was last viable point of development for the siSetDb1 treatment group. Consistent with observations reported in mouse SETDB1 null embryos and ES cells [22], analysis of both K3K9 me2 and H3K9 me3 failed to identify a measurable difference across all treatment groups (Fig 2a–c). Published evidence exists to suggest interplay between H3K9me and methylation of both histone 3 lysine 4 (H3K4) and lysine 27 (H3K27) [32,33]. Therefore embryo treatments were examined to determine if SETDB1 suppression altered global levels of any of these post-translational histone modifications. As can be seen in Figure 2, no alterations could be detected. Given the established link between DNA methylation and both histone methylating enzymes [34], as well as the H3K9me3 mark itself [35], we evaluated all embryo treatment groups for measurable changes in both global 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). While the injection procedure produced a significant reduction in global levels of 5mC, no significant differences were observed between the siNull and siSETDB1 treatments (Fig 3). These results indicate the injection procedure impacted levels of 5mC, but suppression of SETDB1 did not specifically impact this epigenetic mark.
Fig 3.
Analysis of global levels of 5-methyl-cytosine and 5-hydroxy-methyl-cytosine in siSETDB1 injected embryos. A) Representative confocal images of morula-stage embryos co-stained with antibodies recognizing 5-hydroxy-methyl-cytosine (green)/5-methyl-cytosine (red)/the DNA stain Hoechst (blue). Images are at 20 × magnification. B) Average fluorescence intensity ratios were calculated for each embryo by dividing the intensity of the specific target (5-hMC or 5-MC) by the intensity of DNA stain (Hoechst). Ratios from each embryo were then averaged to obtain a mean ratio for each treatment group. Using these means, a one-way ANOVA model was developed in order to examine differences between treatment groups. Bars displaying different letters were considered to be different (p < 0.05). Error bars represent the SEM.
3.3 Analysis of transposable element transcriptional activity in siSetDB1 embryos
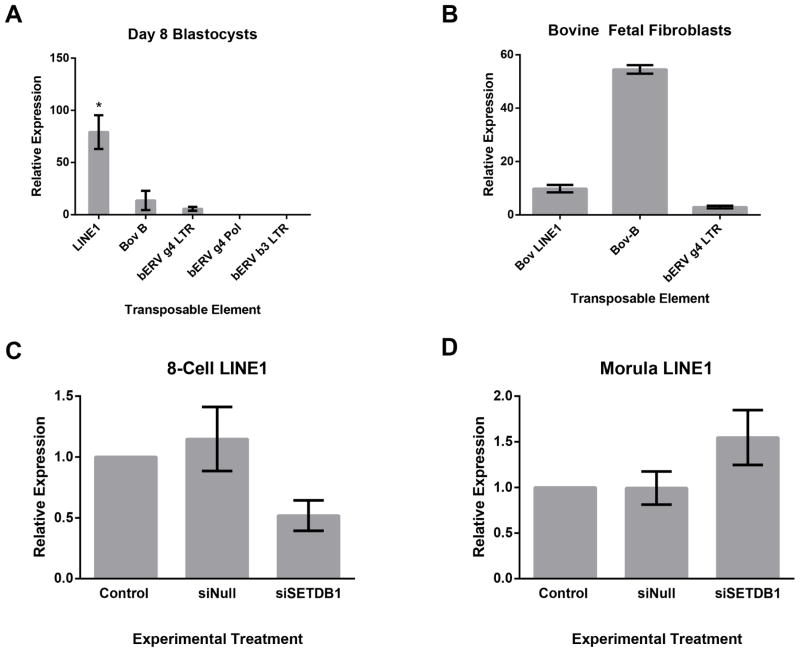
Given the importance of transposable elements in regulating the pluripotency transcriptional program of primates and rodents [14–17], as well as the role of SETDB1 in their control, we examined siSETDB1 embryos for altered patterns of transposable element transcription. Transposable elements account for more than 44% of the cattle genome and are suspected to play an equally prominent role in the pluripotency program of the bovine embryo [36,37]. To characterize potential alterations in transposable element transcriptional activity arising as a consequence of SETDB1 loss of function, we sought to first identify candidate RNAs encoding characterized bovine endogenous retrotransposons and transposons transcribed in preimplantation embryos. To this end, qRT-PCR was utilized to examine transcript levels of the Long Terminal Repeat (LTR) containing endogenous retrovirus-beta3 (bERV-b3 - Accession EF030818) [28], and endogenous retrovirus gamma4 (bERV-g4 - Accession EU697940), which are the two largest families of endogenous retroviruses identified in cattle [27]. Additionally, we also examined the abundance of both the BOV-B and LINE1 family of non-LTR containing transposable elements [36]. Interestingly, despite the fact that all primer sets could repeatably detect transposable element transcription in cDNA derived from bovine fetal fibroblasts, only transcripts encoding the LINE1 family of transposable elements were consistently detected in day-8 blastocysts (Fig 4a–b). While transcripts derived from the BOV-V, and bERV g4 families were detected in embryos, their signals were just above our detection threshold; and similar to other reports, measurements were not consistent enough for rigorous repeated quantification [38]. We therefore analyzed the bovine LINE1 family of transposable elements in siSETDB1 treated embryos. RNA was collected from 8-cell and morula stage embryos from each treatment group for analysis by qPCR. As can be seen in Figures 4c–d, compared to the two control groups, siSETDB1 embryos displayed a modest reduction in LINE1 activity at the 8-cell stage (p=0.052), and no significant differences at the morula stage of development.
Fig 4.
Analysis of transposable element activity in siSETDB1 injected embryos. A) Measurements of transcripts derived from the indicated clades of bovine transposable elements were measured in bovine blastocysts using q-RT-PCR. B) Measurements of transcripts derived from the indicated clades of bovine transposable elements measured in bovine fetal fibroblasts using q-RT-PCR. C–D) Measurements of bovine LINE1 transcripts in bovine 8-cell (C) and morula stage embryos (D) from Control, siNull and siSETDB1 treatment groups using q-RT-PCR. For A–D, samples were normalized to the geometric mean of GAPDH, YWHAZ & SDHA and graphed using the 2−ΔΔ CT method [52]. Graphs are representative of four independent experiments. Error bars represent the SEM, * denotes p < 0.05.
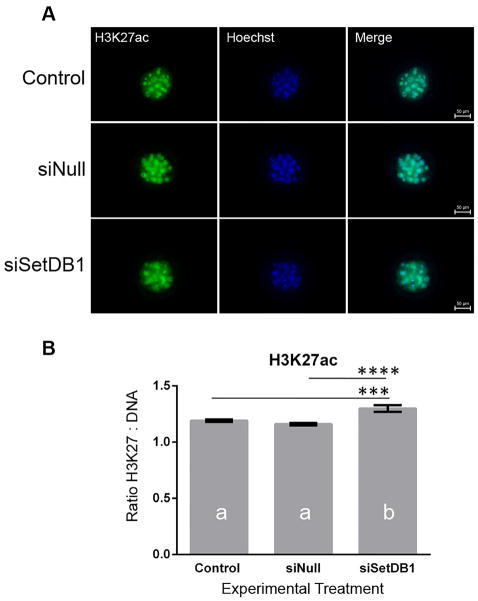
3.4 Analysis of Histone 3 lysine 27 acetylation in siSETDB1 embryos
In addition to serving as alternative promoters, studies in both humans and rodents have suggested that during development, the gene bodies of transposable elements have the capacity to serve as tissue-specific enhancers [14]. In these studies, separate families of transposable elements displayed tissue-specific patterns of epigenetic marks typically associated with enhancer function, and these chromatin modifications correlate with cell-specific patterns of gene expression [39,40]. Using immunocytochemical analysis, we examined embryo treatment groups for alterations in histone 3 lysine 27 acetylation (H3K27ac), a mark associated with transcriptionally active enhancers [41]. As can be seen in Figure 5, suppression of SETDB1 during bovine preimplantation development produced a significant increase in H3K27ac compared to both the uninjected control and the siNull treatment groups (p<0.01). These results suggest that suppression of SetDB1 during early bovine embryonic development changes the global abundance of a key epigenetic mark characterizing transcriptionally active enhancers.
Fig 5.
Analysis of global levels of H3K27 acetylation in siSETDB1 injected embryos. A) Representative confocal images of morula-stage embryos co-stained with antibodies recognizing H3K27ac (green) and the Hoechst DNA stain (blue). Images are at 20 × magnification. B) Average fluorescence intensity ratios were calculated for each embryo by dividing the staining intensity for H3K27ac by the intensity of DNA stain Hoechst. Ratios from each embryo were then averaged to obtain a mean ratio for each treatment group. Using these means, a one-way ANOVA model was constructed in order to determine differences between treatment groups. Bars displaying different letters were considered to be different (p < 0.05). Using a post-hoc Tukeys test, the differences between the siSetDB1 and Control and siNull groups were (p < 0.001). Error bars represent the SEM.
4.0 Discussion
Early studies of placental tissues using electron microscopy revealed an abundance of endogenous type C retroviral particles in syncytial trophoblasts, suggesting robust retroviral activity is present within the tissues of the developing embryo [42,43]. Since these initial observations, we have come to realize that transcripts and proteins encoded by transposable elements are not only abundant in developing ova and early embryos, but are also essential for preimplantation development to proceed [40,44,45]. In 2012, Macfarlan and colleagues demonstrated that many transcripts expressed immediately after murine embryonic genome activation are chimeric ones initiated from long terminal repeats derived from endogenous retroviruses [15]. Specifically, one core family of mouse transposable element, the MuERV-L family, have embedded themselves within the regulatory regions of key genes (Gata4 and Tead4) directing the specification of primitive endoderm and trophectoderm, respectively. Importantly, transcripts driven from these retroviral elements randomly become reactivated within small subpopulations of cultured ES cells, pushing them into a privileged, totipotent state where they are able to contribute to both the extraembryonic and embryonic lineages. A similar circumstance exists in humans where the primate-specific endogenous retrovirus HERVH mediates the transcriptional program essential for naive pluripotency [17]. In summary, it appears co-option of transposable elements by cellular genes provides a mechanism to coordinately link the expression of key factors required for both specification of the trophectoderm and embryonic pluripotency. Thus, understanding the dynamics of how transposable elements are controlled during preimplantation development is key to deciphering the specification of the embryonic lineage.
The mechanisms regulating the transcription of transposable elements appear predominantly epigenetic, and largely mediated through combinatorial patterns of DNA methylation and H3K9me3 [12,13,46]. Previous work from our laboratory has suggested that inactivation of the maintenance methyltransferase DNMT1 during bovine preimplantation development results in arrest of embryonic development at the 8-cell to 16-cell transition [30]. Thus, one core element in the epigenetic control of bovine transposable elements appears firmly required for preimplantation development to proceed. In the current study, we focused on the developmental consequences of inactivating the histone methyltransferase responsible for marking transposable elements with H3K9me3 [20,21]. Previous studies in rodents have suggested SETDB1 null embryos can progress through preimplantation development with no observable defects. However, as mouse development occurs over a comparatively short time-frame, the strong possibility exists that a maternal stock of SetDB1 protein present in the oocyte is sufficient to properly regulate transposable elements, and enable specification of the cell types within the mouse blastocyst [22]. Bovine embryogenesis requires a greater amount of time, which we suspected would allow the opportunity to utilize RNAi to interrogate the consequences of SETDB1 loss of function on both specification of the ICM and blastocyst formation. In our experiments, we observed blastocyst development in our non-injected control embryos as well as in our siNull injected controls, while in contrast no siSETDB1 embryos were able to progress past the early morula stage of development. These results suggest that in contrast to the mouse, even a partial suppression of bovine SETDB1 using RNAi results in developmental arrest.
Studies in mouse ES cells have shown that deletion of the SetDB1 interacting protein KAP1 produced a modest increase in LINE1 transcripts, while LTR containing endogenous retroviral elements exhibited 15–50 fold increase in expression [21,47]. Thus, in our experiments we sought to determine if loss of SETDB1 produced a measurable impact upon the transcription of transposable elements. Previous studies utilizing transcriptional profiling of retrotransposon families in pools of oocytes and embryos, have shown that most families of bovine transposable elements identified to date (BtERVF2-I, ERV2-1-I_BT, ERV2-2-I_BT and ERV2-3-I_BT) have either extremely low expression and/or high variation, which hinders reliable quantification [38]. We therefore focused our analysis on the most abundant families of transposable elements in the bovine genome, (LINE1, BOV-B, bERV-b3 and bERV-g4) in hopes these would be quantifiable. Unexpectedly, levels of BOV-B, bERV-b3 and bERV-g4 fell either at or below our detection threshold, and while fluctuations in LINE1 activity were observed, none of the measured changes attained statistical significance (p=0.056). Admittedly, the number of bovine transposable elements examined in our assays are but a small subset of the total, and importantly, neither the crucial MuERV-L nor HERVH families are the most abundant transposable elements in mice and humans respectively [17,22]. Thus it is possible that other families of transposable elements have altered transcriptional profiles and that these changes are in some way tied to the block in development at the morula stage.
In addition to driving the production of chimeric transcripts, several recent studies have begun to correlate tissue-specific differences in the DNA methylation profiles of LTR-containing transposable elements with enhancer activity [14,39,40]. These studies suggest the bodies and regulatory regions of transposable elements are themselves able to recruit epigenetic marks associated with cell-specific enhancer function. Indeed, a strong correlation exists between transposable element sequences and both embryonic and placental patterns of histone H3 lysine 4 monomethylation (H3K4me1) and H3K27ac, which are chromatin modifications defining enhancers [39,40]. Similar to the studies in mice [22], we were unable to detect global alterations in H3K9me2 or H3K9me3 in bovine siSETDB1 embryos. We acknowledge that ICC analysis is very crude, and that perhaps the signal from pericentric and constitutive heterochromatin masked any changes in H3K9me3 at transposable elements.. Additionally, it is possible that in the bovine embryo, other methyltransferases are able to compensate for loss of SETDB1. We were however, able to detect an increase in global levels of H3K27ac. This epigenetic mark distinguishes active enhancers from inactive or poised enhancer elements containing H3K4me1 alone, and acquiring this mark is a key step in the process of enhancer activation [41]. In porcine embryos, H3K27ac decreases from the earliest pronuclear stage to the 8-cell stage, followed by re-acetylation from the morula stage onwards [48]. The correlate timing of this increase with the events accompanying the first cell lineage specification suggest H3K27ac deposition may be a key aspect of the embryonic developmental program. Our observations of increased levels of H3K27ac suggest that suppression of SETDB1 in bovine embryos possibly impacts the global dynamics of enhancer activity. However, the ICC analysis provides a global measure of H3K27ac, and does not imply these changes localize to functional regions of the genome. We speculate that perhaps SETDB1 is required to oppose the actions of CBP-P300 [49], and that SETDB1 suppression potentially allows key loci to become prematurely activated. Inappropriate patterns of enhancer activity could simultaneously engage multiple differentiation programs, leading to the developmental arrest observed at the morula stage. However, it is possible alterations in this post-translational histone mark are the consequence of chromatin fragmentation arising as a result of decreased viability [1]. Additional studies are required to discern which regions of the genome become hyperactylated as a consequence of SETDB1 inactivation.
5.0 Conclusions
During mouse preimplantation development, a major reorganization of chromatin structure occurs where patterns of DNA methylation are lost, and then subsequently reestablished post-implantation [50]. The bovine genome also undergoes a period of remethylation but in contrast, the processes of epigenetic programming initiates earlier in the preimplantation phase (at the 8-cell stage) [51]. It is therefore possible that the differing sensitivities of bovine and mouse embryos to SETDB1 loss of function is solely tied to the timing of epigenome reestablishment. Transcripts derived from select clades of transposable elements are among the first to appear in early mouse and human embryos. These elements also seem to drive the expression of key pluripotency associated transcription factors, indicating transposable elements and the epigenetic mechanisms that regulate their activity are fundamental to the establishment of the stem cell identify of the ICM. Our studies suggest SETDB1 may be a key player required for the initiation of lineage specification and the proper establishment of the bovine embryonic epigenome.
Highlights.
Suppression of bovine SETDB1 is embryonic lethal, producing a block at the morula stage of development, just prior to lineage specification.
SETDB1 loss of function is associated with a global increase in Histone 3 Lysine 27 acetylation, an epigenetic mark associated with enhancer function.
Loss of SETDB1 does not impact global levels of DNA methylation or trimethylation of histone 3 lysine 9.
Acknowledgments
This project was supported by NIH NICHD grants 1R21HD055631-01A2 and 5R01HD058969-02. KJV was supported by NIH NIAAA grant 1R21AA022484, and MS and JHP were supported by NIH ORIP grant 8R24OD011188-02.
Footnotes
Author Contributions
MCG, MEW, and CRL designed the experiments, supervised their completion and wrote the manuscript. MS, GLW, MP, and JHP conducted the experiments focusing on embryology while GLW, KJV and MP conducted the analysis of gene expression. GLW and KJV conducted the statistical analysis under the supervision of MCG and CRL.
Conflicts of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael C. Golding, Email: mgolding@cvm.tamu.edu.
Matthew Snyder, Email: msnyder@cvm.tamu.edu.
Gayle L. Williamson, Email: gwillimason@cvm.tamu.edu.
Kylee J. Veazey, Email: kveazey@cvm.tamu.edu.
Michael Peoples, Email: mpeoples@cvm.tamu.edu.
Jane H. Pryor, Email: jpryor@cvm.tamu.edu.
Mark E. Westhusin, Email: mwesthusin@cvm.tamu.edu.
Charles R. Long, Email: clong@cvm.tamu.edu.
References
- 1.Beaujean N. Histone post-translational modifications in preimplantation mouse embryos and their role in nuclear architecture. Mol Reprod Dev. 2014;81:100–12. doi: 10.1002/mrd.22268. [DOI] [PubMed] [Google Scholar]
- 2.Paul S, Knott JG. Epigenetic control of cell fate in mouse blastocysts: the role of covalent histone modifications and chromatin remodeling. Mol Reprod Dev. 2014;81:171–82. doi: 10.1002/mrd.22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5:401–8. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 4.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald WA, Mann MR. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol Reprod Dev. 2014;81:126–40. doi: 10.1002/mrd.22220. [DOI] [PubMed] [Google Scholar]
- 6.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 7.Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–28. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 9.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, et al. Central role of Drosophila SU(VAR)3–9 in histone H3•K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schliehe C, Flynn EK, Vilagos B, Richson U, Swaminathan S, Bosnjak B, et al. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat Immunol. 2015;16:67–74. doi: 10.1038/ni.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–62. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–4. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 15.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012 doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–66. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–9. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 18.Völkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 22.Dodge JE, Kang Y, Beppu H, Lei H, Li E. Histone H3-K9 Methyltransferase ESET Is Essential for Early Development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latham KE, Schultz RM. Embryonic genome activation. Front Biosci. 2001;6:D748–59. doi: 10.2741/latham. [DOI] [PubMed] [Google Scholar]
- 24.Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- 25.Veazey KJ, Golding MC. Selection of stable reference genes for quantitative rt-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS One. 2011;6:e27592. doi: 10.1371/journal.pone.0027592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol. 2005;5:27. doi: 10.1186/1471-213X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao R, Park K, Oh Y, Kim J, Park C. Structural characterization of the genome of BERV gamma4, the most abundant endogenous retrovirus family in cattle. Mol Cells. 2008;26:404–8. [PubMed] [Google Scholar]
- 28.Xiao R, Kim J, Choi H, Park K, Lee H, Park C. Characterization of the bovine endogenous retrovirus beta3 genome. Mol Cells. 2008;25:142–7. [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Golding MC, Williamson GL, Stroud TK, Westhusin ME, Long CR. Examination of DNA methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol Reprod Dev. 2011;78:306–17. doi: 10.1002/mrd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12:475–87. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SY, Paylor SW, Magnuson T, Schumacher A. Juxtaposed Polycomb complexes co-regulate vertebral identity. Development. 2006;133:4957–68. doi: 10.1242/dev.02677. [DOI] [PubMed] [Google Scholar]
- 34.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A. 2011;108:5718–23. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–60. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adelson DL, Raison JM, Edgar RC. Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc Natl Acad Sci U S A. 2009;106:12855–60. doi: 10.1073/pnas.0901282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao R, Park K, Lee H, Kim J, Park C. Identification and classification of endogenous retroviruses in cattle. J Virol. 2008;82:582–7. doi: 10.1128/JVI.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Goossens K, Van Poucke M, Forier K, Braeckmans K, Van Soom A, et al. High oxygen tension increases global methylation in bovine 4-cell embryos and blastocysts but does not affect general retrotransposon expression. Reprod Fertil Dev. 2014 doi: 10.1071/RD14133. [DOI] [PubMed] [Google Scholar]
- 39.Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013;45:836–41. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–9. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalter SS, Helmke RJ, Panigel M, Heberling RL, Felsburg PJ, Axelrod LR. Observations of apparent C-type particles in baboon (Papio cynocephalus) placentas. Science. 1973;179:1332–3. doi: 10.1126/science.179.4080.1332. [DOI] [PubMed] [Google Scholar]
- 43.Gross L, Schidlovsky G, Feldman D, Dreyfuss Y, Moore LA. C-type virus particles in placenta of normal healthy Sprague-Dawley rats. Proc Natl Acad Sci U S A. 1975;72:3240–4. doi: 10.1073/pnas.72.8.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brûlet P, Kaghad M, Xu YS, Croissant O, Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci U S A. 1983;80:5641–5. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev. 2006;73:279–87. doi: 10.1002/mrd.20423. [DOI] [PubMed] [Google Scholar]
- 46.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends in genetics. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 47.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 48.Zhou N, Cao Z, Wu R, Liu X, Tao J, Chen Z, et al. Dynamic changes of histone H3 lysine 27 acetylation in pre-implantational pig embryos derived from somatic cell nuclear transfer. Anim Reprod Sci. 2014;148:153–63. doi: 10.1016/j.anireprosci.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP- mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–41. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 51.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–8. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]