Summary
Scaffold proteins are ubiquitous chaperones that bind to proteins and facilitate the physical interaction of the components of signal transduction pathways or multi-enzymatic complexes. In this study, we used a biochemical approach to dissect the molecular mechanism of a membrane-associated scaffold protein, FloT, a flotillin-homolog protein that is localized in Functional Membrane Microdomains of the bacterium Bacillus subtilis. This study provides unambiguous evidence that FloT physically binds to and interacts with the membrane-bound sensor kinase KinC. This sensor kinase activates biofilm formation in B. subtilis in response to the presence of the self-produced signal surfactin. Furthermore, we have characterized the mechanism by which the interaction of FloT with KinC benefits the activity of KinC. Two separate and synergistic effects constitute this mechanism: first, the scaffold activity of FloT promotes more efficient self-interaction of KinC and facilitates dimerization into its active form. Second, the selective binding of FloT to KinC prevents the occurrence of unspecific aggregation between KinC and other proteins that may generate dead-end intermediates that could titrate the activity of KinC. Flotillin proteins appear to play an important role in prokaryotes in promoting effective binding of signaling proteins with their correct protein partners.
Keywords: Flotillin, scaffold activity, kinase, KinC, Bacillus subtilis
Introduction
Spatio-temporal organization of proteins in membranes and organelles promotes interaction specificity and optimizes the efficiency of biological reactions (DeLoache & Dueber, 2013; Diekmann & Pereira-Leal, 2013). An interesting question is how scaffold proteins assemble interacting components (Good et al., 2011). Scaffold proteins are chaperones that bind to proteins and facilitate the physical interaction of the components of signal transduction pathways or multi-enzymatic cascades (Bauer & Pelkmans, 2006; Chapman & Asthagiri, 2009; Good et al., 2011). This type of proteins is present in all kingdoms of life but has traditionally been studied in eukaryotes (Good et al., 2011). For instance, the scaffold protein Ste5 mediates essential steps in the three-tiered mating MAPK signaling cascade in yeast (Chapman & Asthagiri, 2009). Likewise, the scaffold protein JIP1 tethers c-Jun N-terminal kinase (JNK) in the MAPK signaling pathway in human cells (Dickens et al., 1997).
Membrane-bound flotillins are part of the family of scaffold proteins. Flotillins preferentially localize in the lipid rafts of eukaryotic cells (Babuke & Tikkanen, 2007; Morrow & Parton, 2005; Otto & Nichols, 2011; Stuermer, 2011; Zhao et al., 2011), in which a large number of proteins related to signal transduction and membrane trafficking concentrate (Simons & Ikonen, 1997). Thus, it is believed that the scaffold activity of flotillin may recruit proteins that must be localized in lipid rafts to be active and may facilitate their interaction and oligomerization (Babuke & Tikkanen, 2007; Morrow & Parton, 2005; Otto & Nichols, 2011; Stuermer, 2011; Zhao et al., 2011). Consequently, flotillins play a central role in the organization of the multi-component protein reactions that are harbored in the lipid rafts (Babuke & Tikkanen, 2007; Stuermer, 2011; Zhao et al., 2011) and their perturbation affects the functionality of numerous raft-associated signal transduction pathways (Bodrikov et al., 2011; Chen et al., 2006; Hattori et al., 2006; Marin et al., 2013; Schneider et al., 2008).
We recently showed that bacteria organize many signal transduction cascades and multi-protein reactions in Functional Membrane Microdomains (FMMs) (Lopez & Kolter, 2010b) that are functionally and structurally similar to the lipid rafts of eukaryotic cells (Simons & Ikonen, 1997). FMMs are discrete membrane regions enriched in polyisoprenoid lipids and bacterial flotillin proteins (Tavernarakis et al., 1999). Bacterial flotillins seem to play a similar role to eukaryotic flotillins, acting as scaffold proteins and favoring the recruitment of FMM-associated proteins to promote interactions and oligomerization (Good et al., 2011; Langhorst et al., 2005). The bacterial model Bacillus subtilis is currently the best-established cellular system to explore the biological significance of FMMs in prokaryotic membranes (Bach & Bramkamp, 2013; Dempwolff et al., 2012a; Donovan & Bramkamp, 2009; Lopez & Kolter, 2010b; Mielich-Süss et al., 2013; Yepes et al., 2012). The FMMs of B. subtilis contain two different flotillin-like proteins, FloT and FloA, similar to the eukaryotic flotillins FLO-1 and FLO-2 (Bramkamp & Lopez, 2015; Stuermer & Plattner, 2005; Zhao et al., 2011). Flotillins, including those found in B. subtilis, present a membrane-anchoring N-terminal region (Bach & Bramkamp, 2015) and the so-called PHB domain (prohibitin domain), which appears important for the functionality of flotillins although its precise role is still unknown (Bach & Bramkamp, 2015; Tavernarakis et al., 1999). FloT was the first flotillin-like protein discovered in bacteria (Tavernarakis et al., 1999) and further experimental studies demonstrated its association with FMMs in Bacillus halodurans (Zhang et al., 2005). Moreover, FloT had a heterogeneous distribution in discrete puncta across the bacterial membrane and was found to influence sporulation in B. subtilis, as cells lacking FloT showed reduced sporulation efficiency (Donovan & Bramkamp, 2009). FloT was co-localized with FloA in FMMs, which also clustered other proteins related to signal transduction and cell–cell communication (Lopez & Kolter, 2010b). Consequently, a flotillin-defective B. subtilis strain showed severe impairments in biofilm formation, sporulation, activation of natural competence and motility, suggesting that the scaffold activity of flotillin was important for the correct functionality of many signaling transduction pathways that are associated with FMMs (Bach & Bramkamp, 2013; Dempwolff et al., 2012a; Donovan & Bramkamp, 2009; Lopez & Kolter, 2010b; Mielich-Süss et al., 2013; Yepes et al., 2012).
One of the first signaling transduction pathways found in association with the FMMs of B. subtilis was the route to biofilm formation that is triggered by the membrane-bound sensor kinase KinC. KinC is one of the five sensor kinases (KinA–E) that phosphorylate the master regulator Spo0A responsible for biofilm formation in B. subtilis, by transferring a phosphoryl-group to Spo0A via a Spo0F/Spo0B phosphorelay system (Jiang et al., 2000; LeDeaux et al., 1995). The activation of the KinA–E kinases is driven by the action of specific signals of unknown nature (Lopez & Kolter, 2010a; McLoon et al., 2011; Shemesh & Chai, 2013). However, it was recently discovered that KinC responds to particular type of membrane damage that is caused by a battery of small molecules, including the self-produced molecule surfactin (Lopez et al., 2009; Lopez et al., 2010). The presence of surfactin activates KinC, and phosphorylates Spo0A with subsequent activation of the signaling pathway for biofilm formation (Lopez et al., 2009; Lopez et al., 2010). In order to sense surfactin, KinC must be localized in the FMMs of B. subtilis where it co-localizes with FloT and FloA (Lopez & Kolter, 2010b). Moreover, deletion of floT and floA genes results in mis-localization of KinC and also abrogates KinC activity, which prevents the flotillin-defective strain from expressing matrix genes and forming a biofilm in response to the signal surfactin (Lopez & Kolter, 2010b).
The study of scaffold proteins, and more precisely bacterial flotillins, is relatively new and there are numerous questions that need to be answered to fully understand their biological significance. One of the most interesting questions in relation to prokaryotic flotillins is how these proteins benefit the interaction of specific protein components. It is currently believed that scaffold proteins coordinate the physical assembly of protein interaction partners (Good et al., 2011) thereby increasing local concentrations of these proteins and thus, the likelihood of interaction of components (Lingwood & Simons, 2010; Michel & Bakovic, 2007). However, the role of bacterial flotillins over their interacting partners, such as KinC, is yet to be elucidated.
In this study, we used a biochemical approach to provide unambiguous evidence for the influence of the scaffold activity of FloT on the interaction of KinC and other membrane-bound sensor kinases of B. subtilis. First, we used protein–protein interaction experiments to demonstrate that FloT strongly binds to KinC and to KinD to a lesser extent. Second, we showed that the interaction of FloT with KinC promoted more efficient oligomerization of KinC and therefore, the assembly of the active dimeric form of this kinase. At the same time, FloT inhibited non-specific aggregation of KinC with other kinases and prevented the titration of the activity of this kinase via formation of dead-end intermediates. In summary, flotillins facilitated specific interaction of signaling proteins and prevented the formation of non-active intermediates.
Material and methods
Strains, media and culture conditions
All bacterial strains used in this study are listed in table S1 of the supplementary material. Bacilllus subtilis NCIB3610 was used in all experiments unless otherwise stated (Branda et al., 2001). Escherichia coli strain BTH101 was used in all experiments unless otherwise stated (Karimova et al., 1998). E. coli strain DH5α (Reusch et al., 1986) was used for cloning purposes. Cells were usually propagated in Luria-Bertani (LB) medium containing ampicillin (100 μg/ml), kanamycin (50 μg/ml) or gentamicin (2 and 10 μg/ml) when required.
Generation of labeled strains of Bacillus subtilis
Translational fusions PhpFloT-His6 and Php-KinC-CFP were constructed by long flanking homology PCR and subsequently cloned into the plasmids pKM003 and pDR183, respectively. Php is an IPTG-inducible promoter. Primers are listed in table S2 of the supplementary material. Plasmids are listed in table S3 of the supplementary material. These plasmids allowed the integration of the constructs into the bacterial genome at the lacA and amyE loci, respectively. The translational fusions were expressed under the control of an IPTG-inducible promoter (Britton et al., 2002; Erwin et al., 2005; Nakano et al., 2003). Linearized vectors were added to B. subtilis 168 cells grown in competence-inducing conditions. The constructs were integrated into the bacterial genome at the amyE and lacA loci. Double recombination occurred at the amyE locus when using the plasmid pKM003 or the lacA locus when using the plasmid pDR183. Cells were plated on corresponding selective media and colonies were checked for integration of constructed fusions by colony PCR. SPP1 phage transduction was used to transfer constructs from B. subtilis 168 to wild-type NCIB 3610, according to (Yasbin & Young, 1974).
Cell fractionation
Cell fractionation was performed as described by (Yepes et al., 2014). Samples of 100 ml of cultures were harvested and cells were lysed in 25 ml SMM buffer (1 M sucrose, 0.04 M maleic acid, 0.04 M MgCl2 [pH 6.5]) and supplemented with lysozyme (10 μg/ml) prior to sonication (four series of 12 pulses, power output 0.7 and cycle 50%). After cell disruption, a centrifugation step (11 000 × g for 10 min at 4 °C) separated the cell debris (pellet) from the total cell extract fraction. We collected the total cell extract fraction and purified the membrane fraction using ultracentrifugation (100 000 × g for 1 h at 4 °C). Then the membrane fraction was precipitated and solubilized in 3 ml of Tris buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% DDM [n-dodecyl β-d-maltoside], 1 mM phenylmethylsulfonyl fluoride [PMSF]).
Pull-down analysis
Pull-down assays were performed using Ni-NTA resin (Qiagen) and samples were kept at 4 °C throughout the assays. The membrane fractions of the different mutants were resuspended in 7 ml of binding buffer (50 mM Tris-HCl, 500 mM NaCl, 10% glycerol [v/v], 20 mM imidazole, 1% Tween [pH 8]). Proteins (100 mg membrane/strain) were bound to 200 μl of the resin at 4 °C overnight. In order to remove unspecific binding, the resin was washed twice with wash buffer A (50 mM Tris-HCl, 500 mM NaCl, 10% glycerol [v/v], 20 mM imidazole [pH 8]) and twice with wash buffer B (same as wash buffer A but with 50 mM imidazole). His-tagged proteins were eluted using elution buffer that contained a high concentration of imidazole (50 mM Tris-HCl, 500 mM NaCl, 10% glycerol [v/v], 500 mM imidazole). Proteins from the eluted fractions were precipitated by adding trichloroacetic acid 10% to the sample. The protein sample was resuspended in Tris buffer (50 mM Tris-HCl).
Western blot analysis and immunodetection
Immunoblotting was carried out as previously described (Koch et al., 2014). Total protein (80 μg) was separated using 12% SDS polyacrylamide gel. Proteins were transferred from the gel to a PVDF membrane using semi-dry blotting for 1.5 h. After blotting, the membrane was blocked with 10% skim milk for 1 h and probed with 1:4,000 diluted anti-CFP tag antibody (Living Colors) to detect the presence of KinC-CFP (CFP is cyan fluorescence protein). Proteins were detected after incubation with the secondary antibody anti-rabbit IgG-HRP (BioRad) diluted 1:20,000, using a chemiluminescent substrate kit (Thermo Scientific). Chemiluminescence was recorded with the Illumination System ImageQuant LAS4000 (General Electric).
Bacterial two-hybrid analysis
To perform bacterial two-hybrid (BTH) analysis, the coding sequences of floT and the sensor kinases were amplified from the B. subtilis NCIB3610 genome and cloned in frame into the BTH expression vectors. Sanger sequencing was used to verify that resultant colonies contained the plasmids (Primers are listed in table S2 of supplementary material). floT was cloned into the pKNT25 plasmid whereas the sensor kinases were cloned into the pUT18 plasmid (EuroMedex) (Karimova et al., 1998). Pairwise combinations of plasmids that expressed FloT and a sensor kinase were cotransformed in E. coli BTH101 strain, which harbors a lacZ gene under the control of a cAMP-inducible promoter. Upon interaction, the T25 and T18 catalytic domains of the adenylate cyclase formed an active enzyme leading to the production of cAMP and hence to the expression of the reporter (Karimova et al., 1998). Positive cells turn blue in the presence of X-gal. Protein interaction assays were performed following the protocol previously described by (Karimova et al., 1998). Plates were incubated for 48 h at 30 °C. pKT25-zip and pUT18C-zip served as positive controls, and the empty vectors pKNT25 and pUT18C were negative controls. For quantitative measurements, β-galactosidase activity was determined. The transformants were grown for 48 h at 30 °C in LB medium supplemented with ampicillin (100 μg/ml) and kanamycin (50 μg/ml). Optical density at 600 nm was determined before cells were permeabilized using chloroform and 0.01% SDS. β-galactosidase activity was measured according to (Miller, 1972) and results are represented in Miller Units.
Bacterial three-hybrid analysis (B3H assay)
To assay the scaffold activity of FloT, the kinase genes kinB, kinC and kinD were PCR-amplified and cloned into the plasmids pKNT25 and pUT18 (Karimova et al., 1998). Cells were plated in LB medium with 100 μg/ml ampicillin and 50 μg/ml kanamycin and Sanger sequencing verified that resultant colonies contained the plasmids. These strains were used to perform protein–protein interaction assays following the protocol of Karimova et al. (Karimova et al., 1998) to determine the interaction efficiency among sensor kinases. Moreover, the strains were subsequently used to propagate pSEVA modulable plasmids (Silva-Rocha et al., 2013) that produced different levels of FloT. We specifically used pSEVA-621 (2 μg/ml gentamicin), pSEVA-631 (10 μg/ml gentamicin) and pSEVA-641 (10 μg/ml gentamicin) plasmids to produce FloT at different concentrations. These plasmids contain distinct replication origins (RK2, pBR101 and pRO1600, respectively) and propagate in E. coli at low, medium and high copy numbers, respectively (Silva-Rocha et al., 2013). This generates low, medium and high concentration of FloA and FloT in the in BTH E. coli strains in which the plasmids were propagated. Experiments that required the propagation of pSEVA vectors were performed in LB medium with 100 μg/ml ampicillin, 50 μg/ml kanamycin and 2-10 μg/ml gentamicin. Miller Units were quantified to monitor the efficiency of protein interactions, as described in (Miller, 1972).
Native-PAGE
Native PAGE was performed under non-denaturing conditions adapted from standard protocols (Life Technologies). DDM (1%) solubilized membrane samples were equilibrated in Tris-Glycine Native Sample Buffer and run on a 8% polyacrylamide gel. Electrophoresis was performed under non-denaturing conditions using a Tris-Glycine buffered system and run for 1h at 150 V, followed by 1h at 250 V.
Blue native-PAGE
Strains were streaked on LB agar and used to inoculate 100 ml liquid MSgg medium supplemented with 1 mM IPTG. Cultures were incubated overnight at 37 °C with agitation (200 rpm). Cells were collected by centrifugation and membrane fractions were isolated as described above. Membranes were dissolved in 1% DDM and prepared for BN-PAGE according to the manufacturer’s protocol (Life Technologies). Briefly, membranes were dissolved in supplied BN-PAGE buffer containing 1% DDM for 30 minutes on ice. Undissolved material was removed by centrifugation at 20 000 × g at 4°C for 30 minutes. Dissolved membrane proteins were supplemented with Coomassie G-250 and mounted on a 4-20% BN-PAGE gradient gel and run for 1h at 150 V, followed by 1h at 250 V.
Immunoblotting
Native gels were used for standard immunoblotting procedures without further processing. To unfold native proteins and expose hydrophobic sites after blotting, PVDF membranes were fixed with 8 % acetic acid, air-dried and re-wetted with methanol. CFP-tagged KinC was detected using a polyclonal antibody against CFP diluted 1:5000 (Living colors).
Biofilm formation and sporulation assays
A detailed scheme for the procedure to biofilm formation is shown in supplemental figure S3. Briefly, the strains were grown overnight in LB agar. The next day, serial passaging of cells were performed in LB medium. Bacteria grow in exponential phase for many generations to reduce the levels Spo0A that remains activated from stationary phase cells that grew overnight. Cells with lowly activated Spo0A background were used to inculate MSgg medium. LB cultures were washed and cells dispersed in fresh MSgg medium to a OD600 = 1.0. 20 μl of the cell dispersion were used to inoculate 1 ml of MSgg medium that was placed in 24-well plates and incubated at 30°C overnight. To perform the sporulation assay, MSgg cultures were normalized in OD600. Vegetative cells were killed by incubating samples at 80°C for 30 min. Serial dilutions were plated and colony forming units were examined.
Fluorescence microscopy
1 ml of MSgg culture was pelleted, cells were resuspended in 500 μl paraformaldehyde (4%) and incubated 7 min at room temperature to effect fixation. Samples were washed in PBS buffer and mounted Samples were finally mounted on microscope slides with thin agarose pads (0.8% agarose in PBS). Images were taken on a Leica DMI6000B inverted microscope equipped with a Leica CRT6000 illumination system, a HCX PL APO oil immersion objective with 100 × 1.47 magnification, a Leica DFC630FX color camera and an environment control system. BP480/40 excitation filter and BP527/30 emission filter were used to detect GFP by applying excitation times between 100 and 200 ms, while transmitted light images were taken at 36 ms exposure. Leica Application Suite Advanced Fluorescence V3.7 was used to process raw data and fluorescence signals were deconvoluted using AutoQuant software (MediaCybernetics). Further processing of images and calculation of Pearson’s correlation coefficient were performed using ImageJ.
Results
FloT physically interacts with KinC in B. subtilis
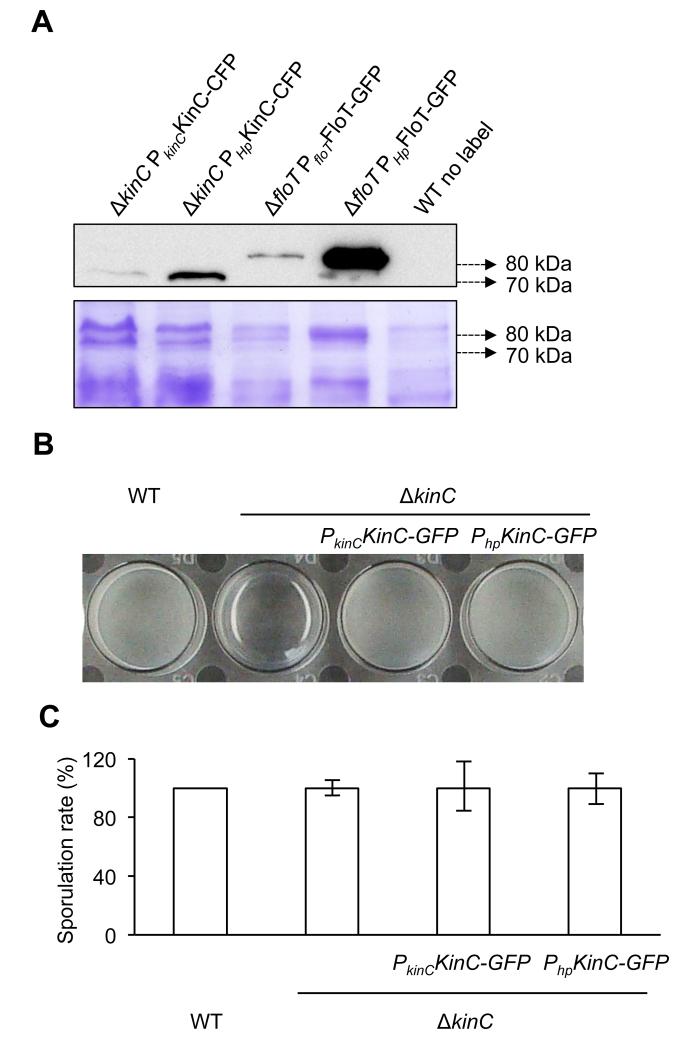
To explore the interaction affinity that exists between FloT and the membrane-bound sensor kinase KinC of B. subtilis, we attempted to co-purify FloT with KinC directly from the cell extracts of B. subtilis using a pull-down assay. To do this, we constructed a double-labeled strain of B. subtilis that expressed IPTG-inducible His6-tagged variant of FloT and a CFP-tagged variant of KinC (FloT-His6 KinC-CFP double-labeled strain). Single-labeled strains harboring FloT-His6 and KinC-CFP constructs were used as control strains. We first tested whether the expression levels of FloT and KinC in the constructed strains are within a physiological range. To do this, we used western blot analysis to compare the relative expression level of FloT-His6 and KinC-CFP under the control of their natural promoters and ITPG-inducible promoters. Using ΔkinC mutant as genetic background, our results showed that the IPTG-inducible promoter expressed KinC-CFP approximately ten-fold higher than wildtype expression levels (Fig. 1a). Similarly, the western blot analyses performed in the ΔfloT mutant showed approximately ten-fold expression FloT-His6 when expressed under the control of an IPTG-inducible promoter in comparison to the wild type promoter (Fig. 1a). It is known that higher expression levels of FloT does not affect the physiology of B. subtilis (Mielich-Süss et al., 2013). Yet, we run additional experiments to determine whether the higher expression levels of KinC affects to the physiology of this bacterium. We performed biofilm formation and sporulation assays of the strain that expressed wildtype levels of KinC-CFP and higher levels of KinC-CFP. After overnight incubation in MSgg medium, the wild type strain was able to form a pellicle on the top of the culture that was not detected in the KinC mutant (Branda et al., 2001; Lopez et al., 2009) (Fig. 1b). When the ΔkinC mutant was complemented with a copy of KinC expressed under the control of its own promoter, we observed recovery of pellicle formation to levels comparable to the wildtype strain (Fig. 1b). Furthermore, the ΔkinC mutant complemented with a copy of KinC expressed via IPTG-inducible promoter showed recovery of pellicle formation similar to the levels of the wildtype strain and the ΔkinC mutant complemented with the wildtype promoter of KinC-CFP (Fig. 1b). In addition to these results, we did not detect any alteration in the sporulation rate in all the strains tested (Fig. 1c). Altogether, these results suggest that the higher expression level KinC and FloT in our system does not alter the physiology of B. subtilis.
Figure 1. Overexpression of physiologically relevant levels of KinC.
(a) Immunoblot detection of native and induced levels of KinC-CFP (left lanes) and FloT-GFP (right lanes) in B. subtilis. Native promoters are represented by PkinC and PfloT, respectively. IPTG-inducible promoter is represented by Php. Right lane is unlabeled wildtype strain (WT) that served as negative control. SDS-PAGE is shown as the loading control. (b) Pellicle formation assay in different genetic backgrounds. KinC-deficient strain is unable to form pellicles. Complementation of the ΔkinC mutant with KinC-GFP translational fusion recovered the ability to form pellicles to wild type levels. Expression of KinC-GFP under the native or IPTG-induced promoter does not affect pellicle formation. Pellicle formation assay was performed in MSgg medium. Cultures were allowed to grow at 30°C overnight. A more detailed protocol for the pellicle formation assay is described in figure S4. (c) Sporulation rate in ΔkinC mutant and complemented strains is similar to WT levels. Cultures were grown in MSgg medium at 30°C overnight.
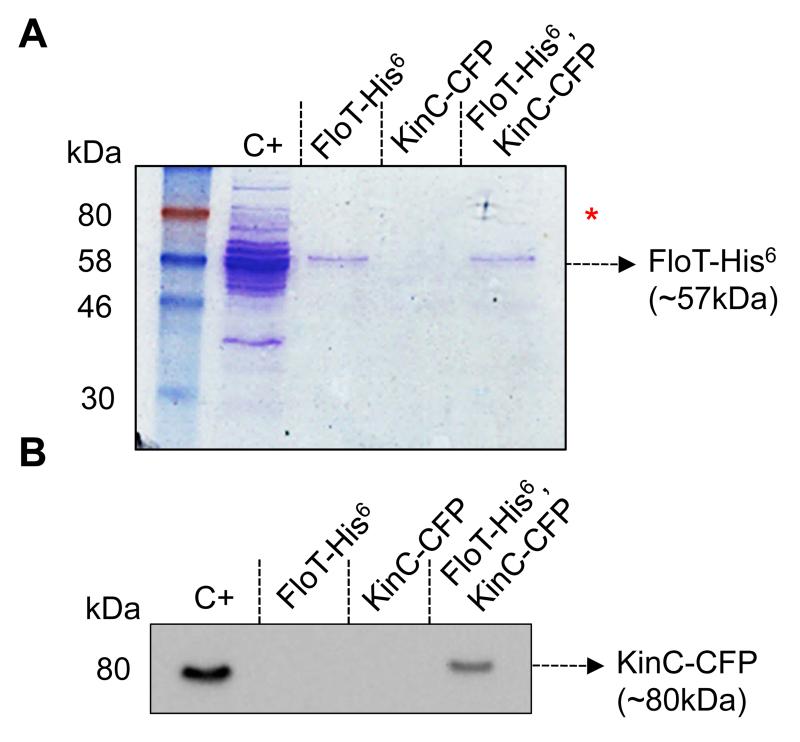
FloT-His6 KinC-CFP double-labeled strain and FloT-His6 and KinC-CFP single-labeled strains were grown to stationary phase in LB medium and their membrane fraction was purified and solubilized using 0.2% of DDM. This detergent treatment allows disaggregation of the membrane without affecting the oligomerization of protein complexes (Casey & Reithmeier, 1993). The samples were loaded onto a column of nickel-charged resin (Qiagen) that selectivity binds His6-tagged proteins and the proteins that are directly or indirectly bound to them. The pool of proteins bound to the resin was eluted from the column using an imidazole-containing buffer and run in a SDS-PAGE (Fig. 2a). The Coomassie stained gel showed a protein band attributable to FloT-His6 in the lane that corresponds to the FloT-His6 KinC-CFP double-labeled strain. The protein band migrated to approximately 57 kDa molecular weight (MW), which is the MW expected for FloT-His6. Moreover, the protein band was detected in the lane of the FloT-His6 single-labeled control strain. This indicates that the purification of FloT was successfully performed. Additionally, the lane of the FloT-His6 KinC-CFP double-labeled strain showed extra protein bands that were concentrated enough to be detected in the Coommasie-stained gel. These extra bands located at approximately 80 kDa, which is the putative MW of KinC-CFP (Fig. 1a red asterisk). Given that these protein bands were not detected in the lanes of the FloT-His6 and KinC-CFP single-labeled strains, it is likely that these proteins were coeluted with FloT-His6.
Figure 2. KinC physically interacts with FloT in B. subtilis membranes.
(A) Coomassie stained SDS-PAGE of the distinct pulled down protein samples. Positive control (C+) is the wild-type membrane fraction. The elution fraction from the FloT-His6 single-labeled strain was loaded into FloT-His6 lane. The elution fraction from the KinC-CFP single-labeled strain was loaded into KinC-CFP lane. Lane FloT-His6, KinC-CFP is the elution fraction from the FloT-His6, KinC-CFP double-labeled strain. The arrow indicates the presence of a band with the size predicted for FloT-His6. The red asterisk indicates the presence of extra protein bands in C+ and FloT-His6, KinC-CFP lanes with the size predicted for KinC-CFP. (B) Western blot assay using polyclonal antibodies against CFP to detect the presence of the KinC-CFP translational fusion in protein samples pulled down with the FloT-His6 translational fusion. The arrow indicates the presence of a band with the size predicted for KinC-CFP. Positive control (C+) is the wild-type membrane fraction. Lane KinC-CFP is the elution fraction from a nickel-charged column loaded with a sample of the membrane fraction from the KinC-CFP single-labeled strain. In the absence of FloT-His6, KinC-CFP does not bind to the column. Lane FloT-His6 is the elution fraction from the membrane fraction of a FloT-His6 single-labeled strain. No signal is detected in the absence of KinC-CFP. Lane FloT-His6, KinC-CFP is the elution fraction from the membrane fraction of a FloT-His6, KinC-CFP double-labeled strain. This lane shows a band with a molecular weight attributable to KinC-CFP, according to the C+ lane.
We performed immunoblotting to semi-quantitatively detect the presence of KinC-CFP in the samples, using polyclonal antibodies against the CFP epitope. We detected a signal attributable to KinC-CFP in the positive control sample that contained the purified membrane fraction of KinC-CFP labeled cells (Fig. 2b). In contrast, the elution fraction of KinC-CFP single-labeled strain showed no detectable signal, suggesting that KinC-CFP did not bind to the resin column in a non-specific manner (Fig. 2b). Moreover, the elution fraction of FloT-His6 single-labeled strain showed no detectable signal, which is indicative that the signal we detected by immunoblotting was attributable to KinC-CFP (Fig. 2b). Importantly, a fluorescence band attributable to KinC-CFP was detected in the eluted fraction of the column of the FloT-His6 KinC-CFP double-labeled strain (Fig. 2b), suggesting that KinC-CFP co-eluted with FloT-His6. This result indicates the existence of a physical interaction between FloT and KinC in B. subtilis cells.
FloT interacts with KinC in a selective fashion
Having determined that FloT interacted with KinC, we investigated whether the interaction was specific or whether, in contrast, FloT was able to interact with other different membrane-bound sensor kinases. We used a bacterial two-hybrid assay (BTH) assay, in which FloT was tagged to the T25 catalytic domain of an adenylate cyclase and a pool of different membrane-associated sensor kinases of B. subtilis were tagged to the T18 catalytic domain of the adenylate cyclase. We selected a pool of structurally different membrane-bound sensor kinases that represent the structural diversity of sensor kinases that exists in B. subtilis membranes (Fig. S1). Upon interaction of FloT with a sensor kinase, the two catalytic domains of the adenylate cyclase reconstitute the entire enzyme (Karimova et al., 1998). A fully active adenylate cyclase produces cAMP, which accumulates in the cytoplasm of the cell and triggers the expression of a cAMP-inducible lacZ reporter gene that is integrated in the chromosome of the cells (Karimova et al., 1998).
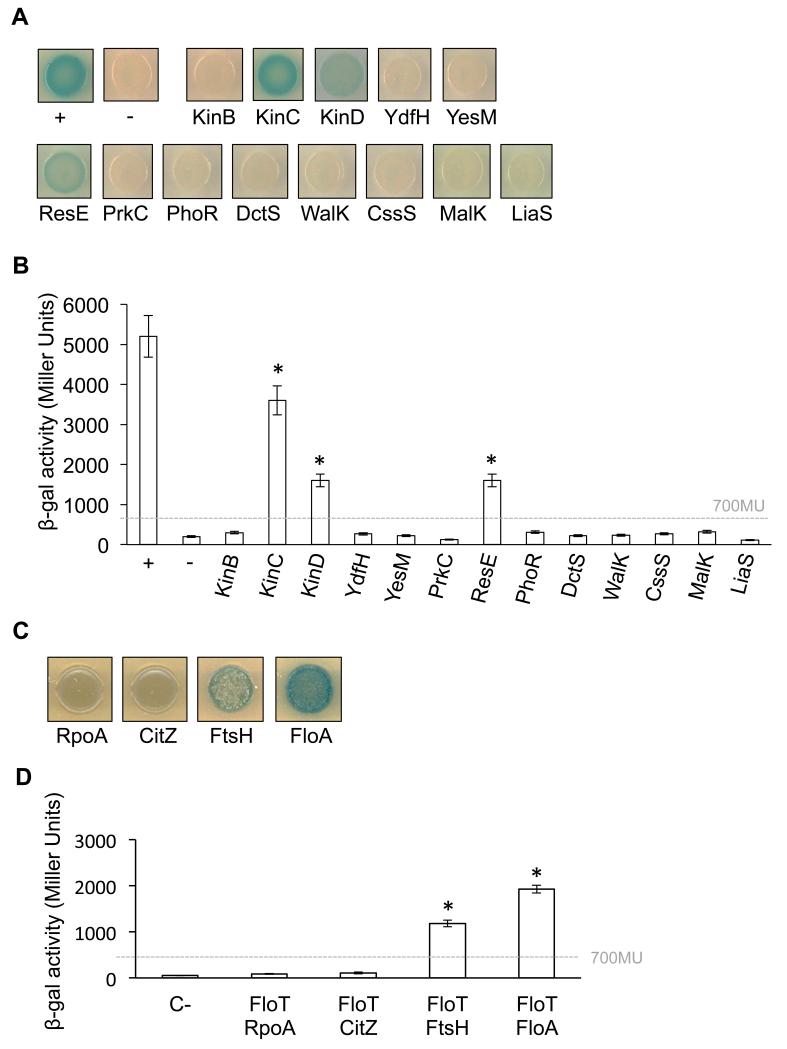
The BTH assay a strong positive interaction signal between FloT and KinC and with the sensor kinases ResE and KinD to a lesser extent (Fig. 3a). The BTH assay did not show an interaction signal between FloT and the rest of the selected membrane-bound sensor kinases (Fig. 3a), which indicates the existence of a specific interaction signal between flotillin and some sensor kinases. FloT–KinC, FloT–KinD and FloT–ResE interaction signals were detected in solid agar medium supplemented with X-gal (50 μM) (Fig. 3a). These results are consistent with previously published experiments that demonstrate that KinC and ResE are part of the protein cargo of the FMMs of B. subtilis (Lopez & Kolter, 2010a; Schneider et al., 2015). ResE is part of the ResD–ResE two-component regulatory system that activates the res regulon under oxygen-limiting conditions and triggers the expression of genes responsible for nitrate respiration (Baruah et al., 2004; Geng et al., 2007; Nakano et al., 1996; Nakano et al., 1999; Nakano & Zhu, 2001). KinC and KinD are two of the five sensor kinases (KinA–E) that phosphorylate Spo0A in B. subtilis (Jiang et al., 2000; LeDeaux et al., 1995). Because KinB did not show any interaction signal with FloT under the conditions tested, it seems that the interaction of FloT with the pool of membrane-bound kinases that activate the Spo0A is predominantly with KinC and to a lesser extent with KinD.
Figure 3. Interaction affinity of FloT to several membrane-bound sensor kinases of B. subtilis.
(a) BTH analysis to study the interactions between FloT and a pool of selected membrane-bound sensor kinases of B. subtilis. Interaction activates lacZ and this degrades X-gal; the product of the degradation is blue. The two cytoplasmic domains of a leucine-zipper represent a positive control (pKT25-zip + pUT18C-zip). The negative control is represented by the strains harboring empty plasmids (pKNT25 + pUT18). (b) Quantitative β-galactosidase activity assay of the BTH system. Degradation of the substrate ONPG by β-galactosidase generates a reaction product that can be monitored using a colorimetric assay. The activity of the enzyme is represented in Miller Units. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer (EuroMedex). Results represent a mean of three independent experiments. Statistical analysis was performed using Student’s t-test. An asterisk denotes p ≤ 0.05. The positive control is represented by a strain harboring two cytoplasmic domains of a leucine-zipper protein that are known to interact (pKT25-zip and pUT18C-zip). The negative control strain harbors empty plasmids (pKNT25 and pUT18). (b) BTH qualitative analysis to study the interactions between FloT and a selected proteins known to interact with FloT (FtsH and FloA) and known to not interact with FloT (RpoA and CitZ). (c) BTH quantitative analysis of the interactions between FloT and proteins known to interact with FloT (FtsH and FloA) and known to not interact with FloT (RpoA and CitZ). The β-galactosidase activity of the enzyme is represented in Miller Units. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) according to the instructions of the manufacturer (EuroMedex).
To quantitatively determine the level of interaction between FloT and the pool of sensor kinases, cultures of these strains were grown in LB medium and the cells extracts were used to determine β-galactosidase (β-gal) activity. Instructions of the manufacturer define a signal as positive if the β-gal activity is above the threshold of 700 Miller Units (Karimova et al., 1998). Consistently, the interactions that had negative results in solid agar plus X-gal were also negative according to the β-gal assay (< 400 Miller Units) (Fig. 2b). Likewise, the interaction of FloT with KinC exhibited a strong interaction signal in the β-gal assay (> 3500 Miller Units) (Fig. 2b), greater than the interaction signal of FloT with ResE and KinD (~1600 and 1100 Miller Units, respectively) (Fig. 3b). Based on these results, the interaction between FloT and KinC is the strongest interaction we detected using a heterologous system.
To validate the abovementioned results, we used a BTH assay to quantitatively determine several already reported interactions of proteins with FloT in B. subtilis (Fig. 3c). Specifically, the membrane-bound AAA protease FtsH is known to physically interact with FloT (Bach & Bramkamp, 2013; Schneider et al., 2015; Yepes et al., 2012). Likewise, Bach and Bramkamp recently reported that the second flotillin of B. subtilis, FloA, strongly interacts with FloT (Bach & Bramkamp, 2013). In contrast, the same report identified a pool of proteins that do not interact with FloT. Among those proteins are the RNA polymerase RpoA and the citrate synthase CitZ proteins. Both proteins are indeed cytoplasmic proteins and therefore, it is not surprising that did not show interaction with FloT. Accordingly, we performed BTH assays to test the interaction between FloT and the interacting proteins FtsH and FloT. We used a similar approach to test whether FloT interacts with non-interacting proteins such as RpoA and CitZ in our BTH assay. The BTH assay showed interaction signal between FloT-FtsH and FloT-FloA, which were confirmed using a β-gal quantification assay (Fig. 3c,d). Moreover, we did not detect any interaction signal between between FloT and the non-interacting proteins RpoA and CitZ (Fig. 3c,d). Thus, the interaction pattern of FloT in our BTH assay is in agreement with the data that has been already reported.
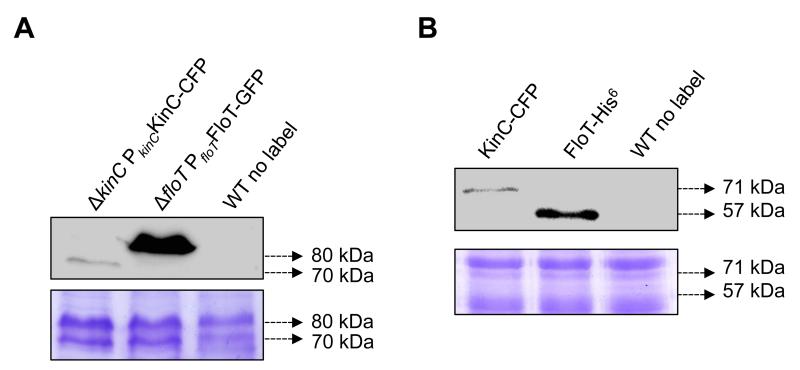
Having validated our BTH results to those published in the literature, we performed a number of experiments to determine the topology and relative expression levels of FloT and KinC in the BTH assay in comparison to the endogenous expression of the protein in B. subtilis cells. While it has already been published that the subcellular distribution pattern of FloT in E. coli is similar to the original pattern in B. subtilis cells (Schneider et al., 2015), we performed additional experiment to test whether there exist any alternation in the subcellular distribution pattern of KinC when expressed in E. coli. E. coli cells labeled with the translational fusion KinC-CFP were analyzed at the fluorescence microscope. The distribution of the fluorescence signal was detected in several discrete puncta across the cellular membrane, similar to the subcellular distribution pattern reported for KinC in B. subtilis (Fig. S2). Moreover, we performed immunoblot assays to determine if the relative protein expression levels of KinC and FloT in B. subtilis cells are comparable to those in the heterologous system. Wildtype, ΔkinC PkinCKinC-CFP and ΔfloT PfloTFloT-GFP strains were grown in MSgg medium, the proteins from the membrane fraction were purified and equal concentrations of the protein samples were resolved in a SDS-PAGE. Immunoblot detection of the fluorescence protein was performed using commercially available polyclonal antibodies. We detected a signal attributable to KinC-CFP in the ΔkinC PkinCKinC-CFP lane and a signal attributable to FloT-GFP in the ΔfloT PfloTFloT-GFP. No signal was detected in the wildtype lane that was used as negative control. We detected higher concentration of FloT than KinC in B. subtilis cells in a relation of approximately 10:1 (Fig. 4a). Next, we expressed KinC-CFP and FloT-His6 using the same heterologous system that we used to perform BTH assay. E. coli strains expressing KinC and FloT were grown in LB medium, the proteins from the membrane fraction were purified and equal concentrations of the protein samples were resolved in SDS-PAGE. Immunoblot detection of the proteins expressed unravel that FloT is expressed at higher concentration than KinC and that the relative expression level of these two proteins in E. coli is approximately 6:1 (Fig. 4b). In summary, both expression systems produced significantly more FloT than KinC and showed close relative expression levels.
Figure 4. KinC/FloT relative expression is comparable in B. subtilis and in the BTH assay.
(a) Immunoblot detection of relative levels of KinC-CFP (left lane) and FloT-GFP (center lane) in B. subtilis under the expression of their native promoters. Right lane is unlabeled wildtype strain (WT) that served as negative control. SDS-PAGE is shown as the loading control. (b) Immunoblot detection of relative levels of KinC-CFP (left lane) and FloT-His6 (center lane) in E. coli BTH strain under the expression of inducible promoters. Right lane is unlabeled wildtype strain (WT) that served as negative control. SDS-PAGE is shown as the loading control.
FloT facilitates the oligomerization of KinC
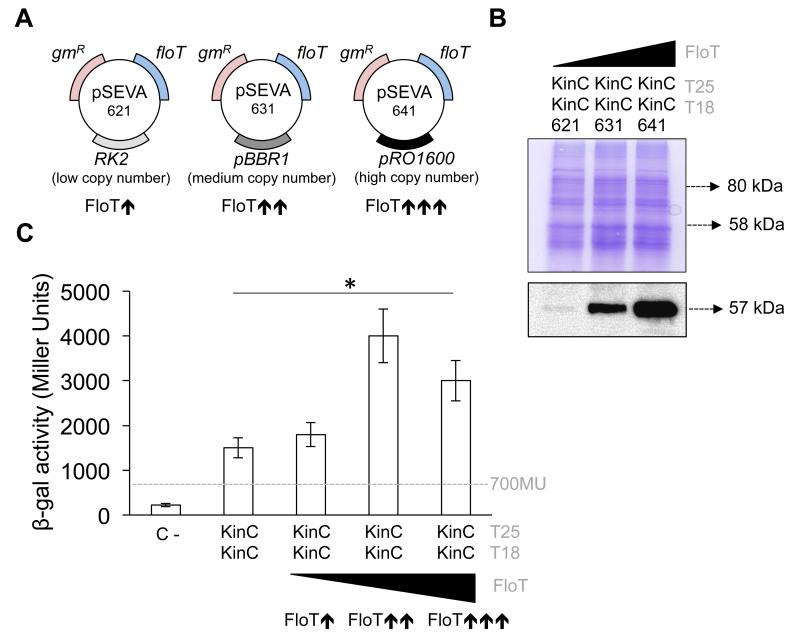
How does the scaffold activity of FloT influence the activity of KinC? The most direct hypothesis is that scaffold proteins promote the stability of protein complexes through tethering of interacting partners and increasing the likelihood of interaction (Good et al., 2011). To investigate the effect of FloT on the oligomerization of KinC, we used a BTH assay that quantitatively monitored the homo-dimerization of KinC under increasing concentrations of FloT (henceforth referred to as B3H assay). First, we semi-quantitatively determined the homo-oligomerization capacity of KinC using a BTH assay. We detected a positive interaction signal between KinC proteins (~1500 Miller Units; Fig. 3c), which points to the hypothesis that KinC is prone to self-interact and oligomerize in our heterologous system. Next, the BTH assay that tested the interaction efficiency of KinC–KinC was supplemented with a pSEVA modulable vector system (Silva-Rocha et al., 2013) (Fig. 5a). In pSEVA vectors, a FloT-His6 tagged variant was cloned under the expression of its own promoter in a suite of vectors that contained distinct replication origins and therefore replicated at different copy numbers (Fig. 5a). This generated several B3H strains that produced lower, medium and higher levels of FloT as a direct function of the copy number of floT gene (Fig. 5a). We performed western blot analyses to semi-quantitatively confirm the concentration of FloT in the B3H strains generated, using antibodies against the His6 epitope. The KinC–KinC strain carrying a lower copy plasmid showed lower concentration of FloT (Fig. 5b). The KinC–KinC strain that carried a medium-copy plasmid showed medium concentration of FloT (Fig. 5b), and the strain that carried a high-copy plasmid showed higher concentration of FloT (Fig. 5b).
Figure 5. The presence of FloT favors the self-interaction of KinC.
(a) Schematic representation of the three different pSEVA plasmids that were used to express FloT at different concentration levels. pSEVA 621, 631 and 641 maintain similar backbones and contain a gene that provides resistance to gentamicin (gmR) and the construct FloT-His6 The plasmids carry a different replication origin. pSEVA 621 carries the low-copy number replication origin RK2. pSEVA 631 carries the medium-copy number replication origin pBR1. pSEVA 641 carries the high-copy number replication origin pRO1600. The strains that carry each of these plasmids produce FloT at different concentration levels, in direct function of the number of floT genes that are expressed in the strain. (b) Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells when carrying lower- (pSEVA-621), medium- (pSEVA-631) and high-copy (pSEVA-641) plasmids expressing His6-tagged FloT. Strains produced lower ( ), medium (
), medium ( ) and higher (
) and higher ( ) concentration of FloT in the B3H assay, respectively. SDS-PAGE is shown as the loading control. (c) B3H assay to quantify the interaction of KinC under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
) concentration of FloT in the B3H assay, respectively. SDS-PAGE is shown as the loading control. (c) B3H assay to quantify the interaction of KinC under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
When we assayed the interaction affinity of KinC–KinC in the presence of distinct concentrations of FloT, the B3H assay showed a no improvement in the interaction efficiency of KinC with lower concentration of FloT. Self-interaction of KinC improved significantly with medium concentration of FloT and showed some decrease with higher concentration of FloT (Fig. 5c). These results are consistent with the typical limitation of the activity of scaffold proteins that has been described in other systems (Good et al., 2011; Levchenko et al., 2000). It is known that optimal concentration of the scaffold protein is necessary to efficiently tether interaction partners while higher concentrations of scaffolds titrate interacting partners into separate complexes, thus inhibiting their interaction (Good et al., 2011; Levchenko et al., 2000). This effect has been experimentally demonstrated in the scaffold protein Ste5 in yeast (Chapman & Asthagiri, 2009) and the JIP1 scaffold human cells (Dickens et al., 1997). Our results supports the hypothesis that FloT acts as typical scaffold protein to tether KinC interaction partners and promote its homo-oligomerization, similar to results reported in other scaffold proteins.
FloT does not facilitate oligomerization of non-interacting kinases
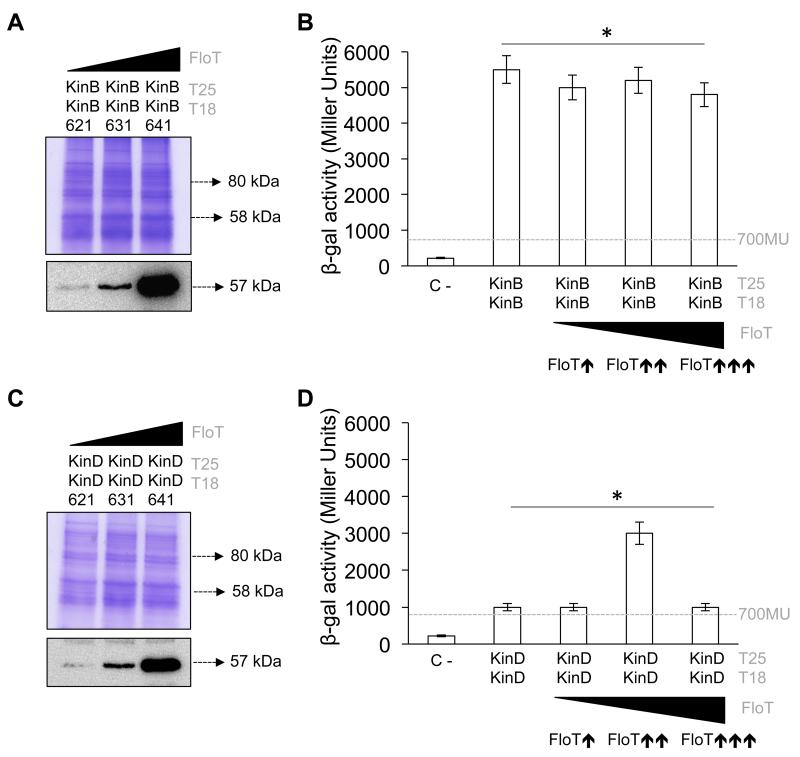
Our heterologous systems showed that FloT interacted slightly with KinD and did not interact with KinB (Fig. 3). To better understand the mechanism of action of FloT, we semi-quantitatively determined the homo-oligomerization capacity of these two kinases using a B3H assay. We hypothesized that KinB–KinB interaction efficiency should not be affected by the presence of FloT. In contrast, the interaction efficiency of KinD–KinD should show a similar pattern to the one observed with KinC–KinC. Indeed, our BTH assay showed interaction signals between KinB–KinB (~ 5500 Miller Units) and KinD–KinD (~ 1500 Miller Units) (Figs. 6a–d), suggesting that KinB and KinD are able to self-interact in our heterologous system.
Figure 6. FloT disfavors self-interaction of KinB but favors self-interaction of KinD.
(a) B3H assay showing interaction of KinC under different concentrations of FloT. Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells carrying lower- (pSEVA-621), medium- (pSEVA-631) and high-copy (pSEVA-641) plasmids expressing His6-tagged FloT. Strains produced lower ( ), medium (
), medium ( ) and higher (
) and higher ( ) concentration of FloT in the BTH assay, respectively. SDS-PAGE is shown as the loading control. (b) B3H assay to quantify the interaction of KinB under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05). (c) B3H assay showing interaction of KinD under different concentrations of FloT. Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells carrying lower- (pSEVA-621), medium- (pSEVA-631) and high-copy (pSEVA-641) plasmids expressing His6-tagged FloT. Strains produced lower (
) concentration of FloT in the BTH assay, respectively. SDS-PAGE is shown as the loading control. (b) B3H assay to quantify the interaction of KinB under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05). (c) B3H assay showing interaction of KinD under different concentrations of FloT. Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells carrying lower- (pSEVA-621), medium- (pSEVA-631) and high-copy (pSEVA-641) plasmids expressing His6-tagged FloT. Strains produced lower ( ), medium (
), medium ( ) and higher (
) and higher ( ) concentrations of FloT in the B3H assay, respectively. SDS-PAGE is shown as the loading control. (d) B3H assay to quantify the interaction of KinD under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
) concentrations of FloT in the B3H assay, respectively. SDS-PAGE is shown as the loading control. (d) B3H assay to quantify the interaction of KinD under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
To investigate whether FloT affects the homo-oligomerization of KinB and KinD, we generated a B3H assay to test the interaction efficiency of these kinases in the presence of increasing concentrations of FloT. We generated a number of B3H strains that, according to our immunoblot detection assay, produced lower, medium and higher levels of FloT (Figs. 6a,c). There was no improvement in the interaction efficiency of the KinB–KinB interaction in B3H strains at any concentration of FloT tested. This suggests that oligomerization of KinB occurs in a very efficient manner in the absence of FloT and the presence of FloT does not improve the interaction efficiency of this kinase. In contrast, when we tested the KinD–KinD interaction in the B3H strains, we observed no improvement in interaction efficiency at the lower concentration of FloT but significant improvements at the medium concentration of FloT. Similar to the results for KinC, a higher concentration of FloT resulted in significant decrease in the interaction efficiency of the KinD–KinD interaction (Figs. 6c,d), according to the typical limitation of the activity of scaffold proteins and their capacity to titrate reaction components at higher concentrations (Good et al., 2011; Levchenko et al., 2000).
FloT prevents unspecific interactions of KinC
One of the most important roles of scaffold proteins is to physically interact with specific proteins by guiding the assembly of proteins into productive complexes to prevent unproductive interactions, such as non-specific aggregation and the formation of dead-end intermediates (Daley, 2008). Our B3H assay provided us with an interesting approach to explore whether FloT favored interaction specificity and prevented non-specific aggregation with other proteins. We tested interaction efficiency of KinC with KinB and KinD in the presence and absence of FloT. If interactions were detected, we assumed they were the consequence of non-specific aggregation. Given that, with a few exceptions in which hetero-associations between histidine kinases have been reported (Goodman et al., 2009), sensor kinases generally show a preference for homo-oligomerization (Laub & Goulian, 2007).
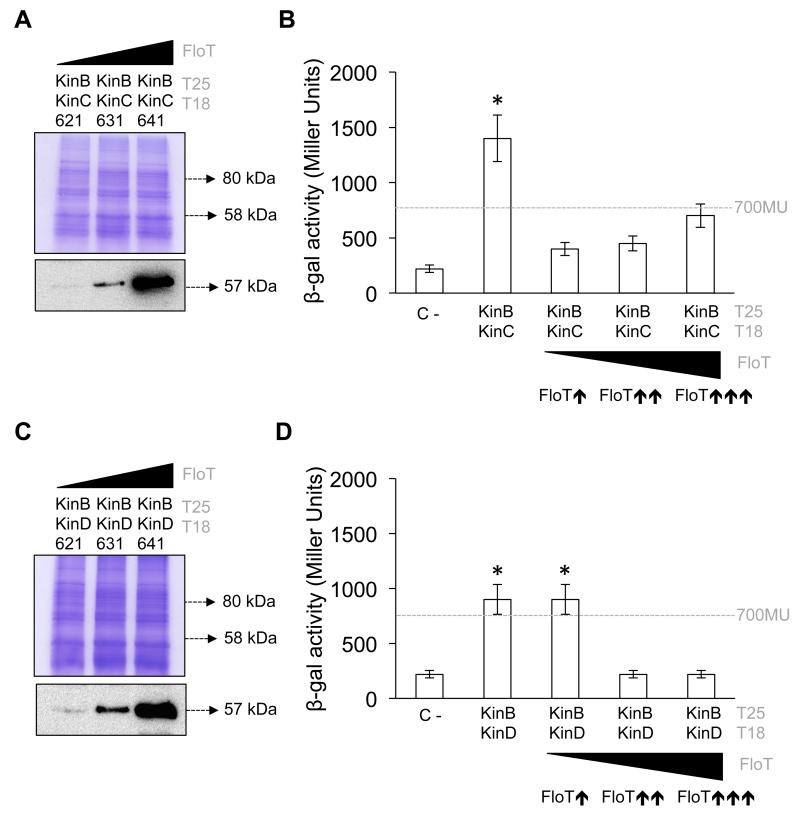
Using a BTH assay, we detected a significant interaction signal between KinC and KinB and also between KinD and KinB (Figs. 7a,b). We did not detect any interaction signal between KinC and KinD. The interactions were assayed in both solid agar medium supplemented with X-gal (50 μM) (Fig. 7 i panels) and β-gal activity from LB liquid cultures (~1100 and 900 Miller Units for KinC–KinB and KinD–KinB interactions, respectively) (Fig. 7 ii panels). The KinC–KinD interaction showed a negative detection signal of approximately 400 Miller Units, similar to the value observed in the negative control (Fig. 7c). Therefore, it is possible that KinB has a non-specific preference for aggregation with KinC and KinD in this heterologous system.
Figure 7. KinB aggregates with KinC and KinD in a non-specific manner.
(a) (i) BTH analysis to study the interactions between KinC and KinB. Interaction activates lacZ, which degrades the substrate X-gal; the degradation product is blue. (ii) Quantitative β-galactosidase activity assay of the BTH system. The activity of the enzyme is represented in Miller Units. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05). (b) (i) BTH analysis to study the interactions between KinB and KinD. (ii) Quantitative β-galactosidase activity assay of the BTH system. The activity of the enzyme is represented in Miller Units. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05). (C) (i) BTH analysis to study the interactions between KinC and KinD. (ii) Quantitative β-galactosidase activity assay of the BTH system. The activity of the enzyme is represented in Miller Units. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
We next investigated whether FloT played any role in preventing the formation of non-specific aggregates between KinB and KinC. We hypothesized that the selective binding of FloT to KinC may protect KinC from non-specific aggregation and unproductive titration. To explore this hypothesis, we generated a B3H assay to test the interaction efficiency of KinC–KinB under increasing concentrations of FloT. Our immunoblot detection assays showed that the strains produced increasing levels of FloT, using antibodies against the His6 epitope (Fig. 8a). When we assayed the interaction affinity of KinC and KinB in the B3H assay, we observed a significant decrease in the interaction signal in the presence of medium and higher concentrations of FloT (Fig. 8b), suggesting that the presence of increasing concentrations of FloT inhibited non-specific aggregation of KinC. Likewise, we used a B3H assay to test the role of FloT in preventing non-specific aggregation of KinB and KinD (Figs. 8c,d). The interaction signal was also significantly decreased in presence of low concentration of FloT, similar to that observed with the KinB–KinC interaction. Furthermore, we semi-quantitatively determined the efficiency of oligomerization between KinC and KinD using a B3H assay. As these proteins showed no interaction affinity, the B3H assay demonstrated that the presence of increasing concentrations of FloT did not affect this negative interaction (Fig. S3). Overall, our biochemical approach showed that FloT prevented the formation of non-productive intermediates of its interaction partners and also prevented the titration of these proteins in the formation of non-specific aggregates.
Figure 8. The presence of FloT prevents unspecific aggregation of KinB with KinC and KinD.
(a) B3H assay showing aggregation of KinB with KinC under different concentrations of FloT. Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells carrying lower- (pSEVA-621), medium- (pSEVA-631) and high-copy (pSEVA-641) plasmids expressing His6-tagged FloT. Strains produced lower ( ), medium (
), medium ( ) and higher (
) and higher ( ) concentration of FloT in the B3H assay, respectively. SDS-PAGE is shown as the loading control. (b) B3H assay to quantify KinB–KinC aggregation under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05). (c) B3H assay showing aggregation of KinB with KinD under different concentrations of FloT. Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells when carrying lower, medium and high-copy plasmids expressing His6-tagged FloT. Strains produced lower (
) concentration of FloT in the B3H assay, respectively. SDS-PAGE is shown as the loading control. (b) B3H assay to quantify KinB–KinC aggregation under different concentrations of FloT. Dashed line indicates the threshold limit that defines a positive (≥ 700 Miller Units) or a negative interaction signal (≤ 700 Miller Units) according to the instructions of the manufacturer. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05). (c) B3H assay showing aggregation of KinB with KinD under different concentrations of FloT. Immunoblot analysis showing the distinct concentrations of FloT that were produced in B3H cells when carrying lower, medium and high-copy plasmids expressing His6-tagged FloT. Strains produced lower ( ), medium (
), medium ( ) and higher (
) and higher ( ) concentrations of FloT in the BTH assay, respectively. SDS-PAGE is shown as the loading control. (d) B3H assay to quantify KinB–KinD aggregation under different concentrations of FloT. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
) concentrations of FloT in the BTH assay, respectively. SDS-PAGE is shown as the loading control. (d) B3H assay to quantify KinB–KinD aggregation under different concentrations of FloT. Results represent a mean of three independent experiments (* Student’s t-test, p ≤ 0.05).
The interaction pattern of KinC is altered in a flotillin-deficient B. subtilis
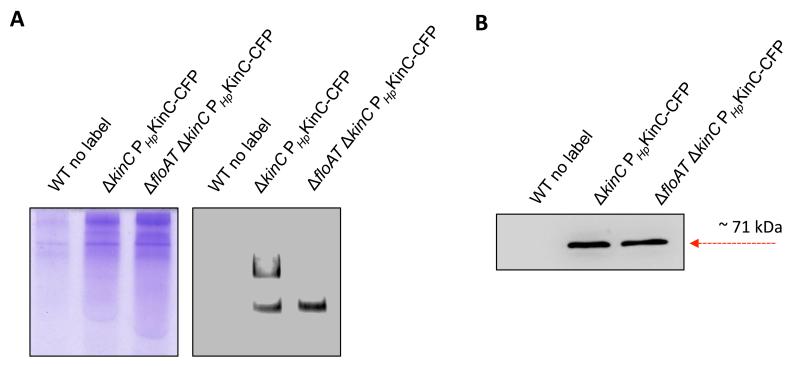
We were interested in exploring the influence of flotillins on the oligomerization of KinC in B. subtilis cells. The B3H assay showed that the presence of FloT favors more efficient homo-oligomerization of KinC. Thus, we hypothesized that B. subtilis cells lacking flotillins would show a different pattern of oligomerization of KinC compared with wild-type (WT) cells. We explored this hypothesis by comparing the different oligomeric states of KinC in wild-type cells and flotillin-deficient B. subtilis cells. To perform this experiment, we used a B. subtilis strain lacking the two flotillins, FloA and FloT, since it is known that flotillins play redundant role and B. subtilis could compensate the absence of one flotillin with the overproduction of the other flotillin (Lopez & Kolter, 2010b). Using this approach, a CFP-tagged version of KinC was expressed in B. subtilis WT and flotillin-deficient mutants. The membrane fraction was collected and solubilized using 1% DDM. The samples were resolved using native PAGE (Wittig & Schagger, 2005; Wittig & Schagger, 2008). This technique of protein separation is capable of resolving proteins of a broad range of MW, from individual proteins to protein complexes and supercomplexes. Proteins are resolved based on their charge to mass ratios and maintain the protein conformation and biological activity while achieving higher sensitivity of protein detection in later steps using immunoblot analysis. We used native PAGE to resolve membrane extracts of wild-type cells. Immunoblot detection showed two important detection signals that were attributable to the different oligomeric states of KinC (Fig. 9a,b). Importantly, membrane extracts from the ΔfloAT mutant were resolved in native PAGE and immunoblot detection performed to detect KinC. The extracts from the ΔfloAT mutant showed one single detection signal of KinC attributable to the lower MW signal that occurred in wildtype samples, suggesting that the higher MW oligomeric state of KinC is compromised in the absence of flotillin in B. subtilis cells.
Figure 9. The oligomerization pattern of KinC is different in B. subtilis cells lacking flotillin.
(a) Native PAGE to resolve the membrane fraction of B. subtilis cells. : Left lane is unlabeled wildtype strain (WT) that served as negative control. The coomassie-stained gel is shown as loading control (left panel). DDM-solubilized membrane samples from different genetic backgrounds were run on an 8% polyacrylamide gel in a tris-glycine buffered system. Higher-state KinC oligomers do not form in the absence of flotillins. The ΔkinC PHpKinC-CFP lane resolves the membrane fraction of a complemented strain labeled with the KinC-CFP translational fusion. The ΔfloAT ΔkinC PHpKinC-CFP lane resolves the membrane fraction of a clean-off flotillin-double mutant complemented strain that is labeled with the KinC-CFP translational fusion. Right panel shows an immunoblot detection assay of KinC-CFP using antibodies against CFP, to detect the oligomeric states of KinC in the different genetic backgrounds. (b) Immunoblot detection of KinC-CFP of different genetic backgrounds after SDS PAGE.
While protein resolution in native PAGE allows a more sensitive detection of the proteins of interest, the resolution of proteins depends on the intrinsic charge of the protein, which prevents a precise estimation of the MW of the resolved protein complexes. Because of this, the samples were also resolved using blue native PAGE (BN-PAGE). BN-PAGE allows the separation of the membrane-bound protein complexes in their native state (Wittig et al., 2006). This approach for protein resolution is less sensitive than native PAGE but it confers the advantage that proteins are resolved based on a charge shift, which allows an estimation of the MW of the resolved oligomeric proteins. Our BN-PAGE assays used a polyacrylamide gradient of 4–20% to allow the resolution of membrane-bound protein complexes with a molecular weight up to 1000 kDa (Fig. S5). BN-PAGE coupled with immunoblotting, using antibodies against the CFP tag, was used to identify a number of membrane-associated protein complexes that contained KinC. Using this approach, we also detected two important detection signals in the membrane extracts of wild-type cells that were attributable to the different oligomeric states of KinC (Fig. S5a). These two detection signals had molecular weights (MW) of approximately 80 kDa and 160 kDa (Fig. S5b). Given that the MW of KinC-CFP is approximately 80 kDa, we hypothesized that the two detection bands represented the monomeric and the dimeric state of KinC in B. subtilis. This is consistent with mounting evidence demonstrating that sensor kinases require dimerization to become active (Capra & Laub, 2012; Krell et al., 2010). We did not detect any additional detection signal beyond a tentative dimeric state of KinC, which suggests that the dimeric form of KinC was the most abundant oligomeric state of KinC in the membrane of B. subtilis under the conditions tested. In contrast, membrane extracts of the flotillin-deficient mutants showed one single signal attributable to the monomeric state of KinC-CFP (Fig. S5b). Furthermore, this approach does not allow detection KinC:KinC-CFP heterodimer due to the lower concentration of KinC in B. subtilis, which prevents the detection of KinC-CFP that is bound to KinC using this approach. The abrogation of detection signal for KinC oligomerization in flotillin-deficient cells is consistent with our hypothesis that flotillin plays an important role in facilitating the dimerization and thus, the activation of KinC in B. subtilis.
Discussion
Scaffold proteins play an important role in controlling and regulating the assembly of interacting protein components in both eukaryotic and prokaryotic life (Bashor et al., 2008; Bhattacharyya et al., 2006; Chapman & Asthagiri, 2009; Dueber et al., 2009; Good et al., 2011; Levchenko et al., 2000; Milano et al., 2002; Park et al., 2003). However, a question that remains to be answered is how scaffold proteins tether specific protein components and how this benefits the interaction of these components. In this study, we investigated the scaffold activity of the membrane-bound flotillin protein FloT of the bacterium Bacillus subtilis. The genetic tractability of bacterial systems allowed us to precisely dissect the molecular mechanism underlying the interaction of scaffold proteins with their interaction partners. Flotillin is a scaffold protein typically found in the lipid rafts of eukaryotic cells (Bickel et al., 1997; Dermine et al., 2001; Lang et al., 1998) but also in the FMMs of bacteria, which are structurally and functionally similar to eukaryotic lipid rafts (Bramkamp & Lopez, 2015). Our relatively simple biochemical approaches demonstrated that the flotillin protein FloT of B. subtilis behaves in a similar way to other scaffold proteins described in eukaryotic cells, by specifically increasing the interaction efficiency of interaction partners at lower concentrations, or titrating and preventing the interaction of the same interaction partners at higher concentrations (Chapman & Asthagiri, 2009; Dickens et al., 1997; Good et al., 2011; Levchenko et al., 2000). This raises the possibility that flotillin proteins of eukaryotic lipid rafts behave like typical scaffold proteins, similar to the bacterial flotillins that we describe in this study.
One of the interaction partners of FloT in B. subtilis is the membrane-bound sensor kinase KinC. In this study, we used a pull-down assay coupled to a BTH assay to show that FloT physically binds to and interacts with KinC. This interaction is consistent with previous publications showing that KinC is one of the proteins harbored in the FMMs of B. subtilis along with FloT (Lopez & Kolter, 2010b). KinC could be interaction partner of the second flotillin FloA that is harbored in the FMMs of B. subtilis. However, we performed BTH analyses to test this particular interaction and results were inconclusive, suggesting that the study FloA-KinC interaction may require a different biochemical approach. Interestingly, our BTH assay showed a newly discovered interaction between FloT and the sensor kinase KinD (Banse et al., 2011). It is not surprising that FloT interacts with and mediates the activity of other sensor kinases in B. subtilis membranes, given that the activity of many different cellular processes, such as biofilm formation, motility, competence, protease secretion, antibiotic resistance and sporulation is affected in B. subtilis cells lacking flotillin (Bach & Bramkamp, 2013; Bach & Bramkamp, 2015; Dempwolff et al., 2012a; Dempwolff et al., 2012b; Donovan & Bramkamp, 2009; Huang et al., 1997; Huang et al., 1998; Huang et al., 1999; Lee et al., 2012; Lopez & Kolter, 2010b; Mann et al., 2013; Mielich-Süss et al., 2013; Yepes et al., 2012) (Fig. S3).
Furthermore, the B3H assay that we designed to test the scaffold activity of FloT shows an increase in the efficiency of KinC–KinC and KinD–KinD interactions in the presence of lower and medium concentrations of FloT. Dimerization is commonly required for bacterial sensor kinases to become active (Capra & Laub, 2012; Krell et al., 2010). Thus, it is possible that the scaffold activity of FloT facilitates the dimerization of KinC and KinD. However, while the activity of scaffold proteins assists in the interaction of protein partners, scaffold proteins are not absolutely required for the activity of proteins. Thus, the oligomerization efficiency of protein partners may vary with different experimental conditions (Devi et al., 2015). We have shown that FloT benefited the interaction of KinC under two experimental conditions: in a heterologous system; and also directly in B. subtilis membranes. BN-PAGE coupled to immunoblotting suggested that KinC preferentially formed dimers similar to the majority of sensor kinases (Capra & Laub, 2012; Krell et al., 2010) and dimerization was abrogated in flotillin-defective cells. It has recently been suggested that KinC is capable of forming tetramers using a soluble variant of KinC that lacks the N-terminal membrane-anchoring region (Devi et al., 2015). However, using a membrane-associated KinC we found that KinC preferred dimerization under our test conditions. We cannot rule out the possibility that tetramerization occurs in a discrete fraction of KinC that we were not able to detect in our assay, but the majority of the membrane-associated KinC oligomerized in a dimeric form in the conditions we tested.
The co-localization of KinC with FloT on the FMMs may increase dimerization of KinC. Additionally, localization of KinC within the FMMs may protect KinC from non-specific aggregation with other membrane proteins. In this study, we used a BTH assay to show that the KinC and KinD were able to interact with KinB, a membrane sensor kinase that is not associated with FMMs. These types of interactions are unlikely to occur in a specific fashion but rather as consequence of unspecific aggregation, given that sensor kinases show a strong preference for homo-oligomerization (Laub & Goulian, 2007), with a few exceptions in which hetero-oligomerization has been reported (Goodman et al., 2009). Interestingly, the presence of FloT prevented non-specific aggregation of KinC and KinD, probably by interfering with non-specific binding. However, these results should be cautiously interpreted, as this protein–protein interaction approach used a heterologous system with limitations that prevent us from predicting whether this type of interaction occurs in B. subtilis cells. Our results raise the intriguing possibility that flotillins may play a dual role in bacterial cells, not only promoting the interaction of specific interacting protein partners but also preventing non-specific interactions that may titrate protein partners and reduce their efficiency in the activation of their respective signal transduction cascades (Fig. 8).
The scaffold activity of FloT over the physical assembly of KinC adds to other recent examples in which scaffold proteins participate in coordinating the assembly of two-component signal transduction pathways in bacteria. For instance, the universal stress protein UspC acts as a scaffold protein of the KdpDE two-component signal cascade in Escherichia coli, which responds to potassium uptake under potassium limiting growth conditions (Heermann et al., 2009). Likewise, the scaffold activity of ApsX in Staphylococcus aureus facilitates the assembly of the ApsSR (antimicrobial peptide sensor) two-component signal transduction cascade that responds to cell wall stress (Li et al., 2007). Additionally, the protein CheW scaffolds the assembly of the basic structural unit for bacterial chemotaxis along with the chemotaxis kinase CheA and chemoreceptors (Li & Hazelbauer, 2011; Underbakke et al., 2011). Overall, scaffold proteins seem to play an important role in bacterial signal transduction.
Supplementary Material
Acknowledgments
This work was funded by the European Research Council ERC-StG 335568-BacRafts and BFU2014-55601-P (MINECO, Spain). Benjamin Mielich-Süss was supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg.
References
- Babuke T, Tikkanen R. Dissecting the molecular function of reggie/flotillin proteins. Eur J Cell Biol. 2007;86:525–532. doi: 10.1016/j.ejcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Bach JN, Bramkamp M. Flotillins functionally organize the bacterial membrane. Mol Microbiol. 2013;88:1205–1217. doi: 10.1111/mmi.12252. [DOI] [PubMed] [Google Scholar]
- Bach JN, Bramkamp M. Dissecting the molecular properties of prokaryotic flotillins. PLoS One. 2015;10:e0116750. doi: 10.1371/journal.pone.0116750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banse AV, Hobbs EC, Losick R. Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein med in Bacillus subtilis. J Bacteriol. 2011;193:3949–3955. doi: 10.1128/JB.05199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah A, Lindsey B, Zhu Y, Nakano MM. Mutational analysis of the signal-sensing domain of ResE histidine kinase from Bacillus subtilis. J Bacteriol. 2004;186:1694–1704. doi: 10.1128/JB.186.6.1694-1704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- Bauer M, Pelkmans L. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 2006;580:5559–5564. doi: 10.1016/j.febslet.2006.08.077. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- Bodrikov V, Solis GP, Stuermer CA. Prion protein promotes growth cone development through reggie/flotillin-dependent N-cadherin trafficking. J Neurosci. 2011;31:18013–18025. doi: 10.1523/JNEUROSCI.4729-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramkamp M, Lopez D. Exploring the Existence of Lipid Rafts in Bacteria. Microbiol Mol Biol Rev. 2015;79:81–100. doi: 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, Reithmeier RA. Detergent interaction with band 3, a model polytopic membrane protein. Biochemistry. 1993;32:1172–1179. doi: 10.1021/bi00055a023. [DOI] [PubMed] [Google Scholar]
- Chapman SA, Asthagiri AR. Quantitative effect of scaffold abundance on signal propagation. Mol Syst Biol. 2009;5:313. doi: 10.1038/msb.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TY, Liu PH, Ruan CT, Chiu L, Kung FL. The intracellular domain of amyloid precursor protein interacts with flotillin-1, a lipid raft protein. Biochem Biophys Res Commun. 2006;342:266–272. doi: 10.1016/j.bbrc.2006.01.156. [DOI] [PubMed] [Google Scholar]
- Daley DO. The assembly of membrane proteins into complexes. Curr Opin Struct Biol. 2008;18:420–424. doi: 10.1016/j.sbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- DeLoache WC, Dueber JE. Compartmentalizing metabolic pathways in organelles. Nat Biotechnol. 2013;31:320–321. doi: 10.1038/nbt.2549. [DOI] [PubMed] [Google Scholar]
- Dempwolff F, Moller HM, Graumann PL. Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins. J Bacteriol. 2012a;194:4652–4661. doi: 10.1128/JB.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempwolff F, Wischhusen HM, Specht M, Graumann PL. The deletion of bacterial dynamin and flotillin genes results in pleiotrophic effects on cell division, cell growth and in cell shape maintenance. BMC Microbiol. 2012b;12:298. doi: 10.1186/1471-2180-12-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J Biol Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- Devi SN, Vishnoi M, Kiehler B, Haggett L, Fujita M. In vivo functional characterization of the transmembrane histidine kinase KinC in Bacillus subtilis. Microbiology. 2015 doi: 10.1099/mic.0.000054. [DOI] [PubMed] [Google Scholar]
- Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- Diekmann Y, Pereira-Leal JB. Evolution of intracellular compartmentalization. Biochem J. 2013;449:319–331. doi: 10.1042/BJ20120957. [DOI] [PubMed] [Google Scholar]
- Donovan C, Bramkamp M. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology. 2009;155:1786–1799. doi: 10.1099/mic.0.025312-0. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Erwin KN, Nakano S, Zuber P. Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx. J Bacteriol. 2005;187:4042–4049. doi: 10.1128/JB.187.12.4042-4049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Zuber P, Nakano MM. Regulation of respiratory genes by ResD-ResE signal transduction system in Bacillus subtilis. Methods Enzymol. 2007;422:448–464. doi: 10.1016/S0076-6879(06)22023-8. [DOI] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori C, Asai M, Onishi H, Sasagawa N, Hashimoto Y, Saido TC, Maruyama K, Mizutani S, Ishiura S. BACE1 interacts with lipid raft proteins. J Neurosci Res. 2006;84:912–917. doi: 10.1002/jnr.20981. [DOI] [PubMed] [Google Scholar]
- Heermann R, Weber A, Mayer B, Ott M, Hauser E, Gabriel G, Pirch T, Jung K. The universal stress protein UspC scaffolds the KdpD/KdpE signaling cascade of Escherichia coli under salt stress. J Mol Biol. 2009;386:134–148. doi: 10.1016/j.jmb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Huang X, Decatur A, Sorokin A, Helmann JD. The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J Bacteriol. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Fredrick KL, Helmann JD. Promoter recognition by Bacillus subtilis sigmaW: autoregulation and partial overlap with the sigmaX regulon. J Bacteriol. 1998;180:3765–3770. doi: 10.1128/jb.180.15.3765-3770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Gaballa A, Cao M, Helmann JD. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G, Yepes A, Forstner KU, Wermser C, Stengel ST, Modamio J, Ohlsen K, Foster KR, Lopez D. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158:1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- Lang DM, Lommel S, Jung M, Ankerhold R, Petrausch B, Laessing U, Wiechers MF, Plattner H, Stuermer CA. Identification of reggie-1 and reggie-2 as plasmamembrane-associated proteins which cocluster with activated GPI-anchored cell adhesion molecules in non-caveolar micropatches in neurons. J Neurobiol. 1998;37:502–523. doi: 10.1002/(sici)1097-4695(199812)37:4<502::aid-neu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Langhorst MF, Reuter A, Stuermer CA. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- LeDeaux JR, Yu N, Grossman AD. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–863. doi: 10.1128/jb.177.3.861-863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Kingston AW, Helmann JD. Glutamate Dehydrogenase Affects Resistance to Cell Wall Antibiotics in Bacillus subtilis. J Bacteriol. 2012;194:993–1001. doi: 10.1128/JB.06547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci U S A. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Gontang EA, Kolter R. Potassium sensing histidine kinase in Bacillus subtilis. Methods Enzymol. 2010;471:229–251. doi: 10.1016/S0076-6879(10)71013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010a;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010b;24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JM, Carabetta VJ, Cristea IM, Dubnau D. Complex formation and processing of the minor transformation pilins of Bacillus subtilis. Mol Microbiol. 2013;90:1201–1215. doi: 10.1111/mmi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin R, Rojo JA, Fabelo N, Fernandez CE, Diaz M. Lipid raft disarrangement as a result of neuropathological progresses: a novel strategy for early diagnosis? Neuroscience. 2013;245:26–39. doi: 10.1016/j.neuroscience.2013.04.025. [DOI] [PubMed] [Google Scholar]
- McLoon AL, Kolodkin-Gal I, Rubinstein SM, Kolter R, Losick R. Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J Bacteriol. 2011;193:679–685. doi: 10.1128/JB.01186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel V, Bakovic M. Lipid rafts in health and disease. Biol Cell. 2007;99:129–140. doi: 10.1042/BC20060051. [DOI] [PubMed] [Google Scholar]
- Mielich-Süss B, Schneider J, Lopez D. Overproduction of flotillin influences cell differentiation and shape in Bacillus subtilis. MBio. 2013;4:e00719–00713. doi: 10.1128/mBio.00719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratories; Cold Spring Harbor: 1972. Cold Spring Harbor N.Y. edn. [Google Scholar]
- Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Zuber P, Glaser P, Danchin A, Hulett FM. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zhu Y, Haga K, Yoshikawa H, Sonenshein AL, Zuber P. A mutation in the 3-phosphoglycerate kinase gene allows anaerobic growth of Bacillus subtilis in the absence of ResE kinase. J Bacteriol. 1999;181:7087–7097. doi: 10.1128/jb.181.22.7087-7097.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zhu Y. Involvement of ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J Bacteriol. 2001;183:1938–1944. doi: 10.1128/JB.183.6.1938-1944.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Kuster-Schock E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GP, Nichols BJ. The roles of flotillin microdomains--endocytosis and beyond. J Cell Sci. 2011;124:3933–3940. doi: 10.1242/jcs.092015. [DOI] [PubMed] [Google Scholar]
- Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- Reusch RN, Hiske TW, Sadoff HL. Poly-beta-hydroxybutyrate membrane structure and its relationship to genetic transformability in Escherichia coli. J Bacteriol. 1986;168:553–562. doi: 10.1128/jb.168.2.553-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Rajendran L, Honsho M, Gralle M, Donnert G, Wouters F, Hell SW, Simons M. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Klein T, Mielich-Suss B, Koch G, Franke C, Kuipers OP, Kovacs AT, Sauer M, Lopez D. Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium. PLoS Genet. 2015;11:e1005140. doi: 10.1371/journal.pgen.1005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Chai Y. A Combination of Glycerol and Manganese Promotes Biofilm Formation in Bacillus subtilis via Histidine Kinase KinD Signaling. J Bacteriol. 2013;195:2747–2754. doi: 10.1128/JB.00028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Rocha R, Martinez-Garcia E, Calles B. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013;41:D666–675. doi: 10.1093/nar/gks1119. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Stuermer CA, Plattner H. The ‘lipid raft’ microdomain proteins reggie-1 and reggie-2 (flotillins) are scaffolds for protein interaction and signalling. Biochem Soc Symp. 2005:109–118. doi: 10.1042/bss0720109. [DOI] [PubMed] [Google Scholar]
- Stuermer CA. Reggie/flotillin and the targeted delivery of cargo. J Neurochem. 2011;116:708–713. doi: 10.1111/j.1471-4159.2010.07007.x. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Driscoll M, Kyrpides NC. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- Underbakke ES, Zhu Y, Kiessling LL. Protein footprinting in a complex milieu: identifying the interaction surfaces of the chemotaxis adaptor protein CheW. J Mol Biol. 2011;409:483–495. doi: 10.1016/j.jmb.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Features and applications of blue-native and clear-native electrophoresis. Proteomics. 2008;8:3974–3990. doi: 10.1002/pmic.200800017. [DOI] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepes A, Schneider J, Mielich B, Koch G, Garcia-Betancur JC, Ramamurthi KS, Vlamakis H, Lopez D. The biofilm formation defect of a Bacillus subtilis flotillin-defective mutant involves the protease FtsH. Mol Microbiol. 2012;86:457–471. doi: 10.1111/j.1365-2958.2012.08205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepes A, Koch G, Waldvogel A, Garcia-Betancur JC, Lopez D. Reconstruction of mreB expression in Staphylococcus aureus via a collection of new integrative plasmids. Appl Environ Microbiol. 2014;80:3868–3878. doi: 10.1128/AEM.00759-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Li Z, Tsudome M, Ito S, Takami H, Horikoshi K. An alkali-inducible flotillin-like protein from Bacillus halodurans C-125. Protein J. 2005;24:125–131. doi: 10.1007/s10930-004-1519-3. [DOI] [PubMed] [Google Scholar]
- Zhao F, Zhang J, Liu YS, Li L, He YL. Research advances on flotillins. Virol J. 2011;8:479. doi: 10.1186/1743-422X-8-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.