Nuclear lamins counterbalance dynein forces on nuclear pore complexes through BICD2 and ensure even nuclear pore complex distribution and proper centrosome separation at prophase.
Abstract
Lamins, the type V nuclear intermediate filament proteins, are reported to function in both interphase and mitosis. For example, lamin deletion in various cell types can lead to an uneven distribution of the nuclear pore complexes (NPCs) in the interphase nuclear envelope, whereas deletion of B-type lamins results in spindle orientation defects in mitotic neural progenitor cells. How lamins regulate these functions is unknown. Using mouse cells deleted of different combinations or all lamins, we show that lamins are required to prevent the aggregation of NPCs in the nuclear envelope near centrosomes in late G2 and prophase. This asymmetric NPC distribution in the absence of lamins is caused by dynein forces acting on NPCs via the dynein adaptor BICD2. We further show that asymmetric NPC distribution upon lamin depletion disrupts the distribution of BICD2 and p150 dynactin on the nuclear envelope at prophase, which results in inefficient dynein-driven centrosome separation during prophase. Therefore lamins regulate microtubule-based motor forces in vivo to ensure proper NPC distribution in interphase and centrosome separation in the mitotic prophase.
INTRODUCTION
The nuclear lamins have been implicated in regulating many cellular functions in both interphase and mitosis (Dechat et al., 2008; Burke and Stewart, 2012; Butin-Israeli et al., 2012; Zheng, 2010). Mammals express a family of three lamin proteins. For example, the three mouse lamin genes, Lmnb1, Lmnb2, and Lmna, express lamin-B1, -B2, and -A/C, respectively. Each cell type often expresses more than one lamin, and in interphase, these proteins assemble into a dense network under the nuclear envelope. This lamin network interacts with a large number of other nuclear peripheral proteins to form the nuclear lamina, which is important for a number of nuclear functions. For example, lamin depletion causes defects in DNA replication (Meier et al., 1991; Spann et al., 1997; Shimi et al., 2008), gene transcription (Reddy et al., 2008; Zullo et al., 2012), and the distribution of some nuclear peripheral proteins, such as the nuclear pore complexes (NPCs; Sullivan et al., 1999; Kubben et al., 2011; Chen et al., 2013; Jung et al., 2013; Guo et al., 2014) and emerin (Vaughan et al., 2001; Chen et al., 2012; Guo et al., 2014). Of importance, lamin-mediated even distribution of nucleoporins (Nups) functions in somatic stem cells to support epidermal growth factor (EGF) signaling, which is essential for the development of the germline stem cells and the reproductive organ in Drosophila (Chen et al., 2013). The mechanism by which lamins regulate these interphase nuclear functions, however, is not well understood.
Of interest, lamin-B3, the major lamin found in Xenopus eggs, maintains the morphology of mitotic spindles assembled in Xenopus egg extracts (Tsai et al., 2006). Because lamin-B3 directly binds to the dynein regulator Nudel, this B-type lamin appears to aid the minus end–directed dynein motor to counterbalance the forces generated by the plus end–directed kinesin Eg5 on spindle microtubules (Ma et al., 2009; Goodman et al., 2010). The connection between lamin and microtubule-based motors in mitosis raises the question of whether these intermediate filament proteins also balance motor forces exerted on the interphase nucleus and, if so, whether this could explain some of lamin’s roles in the nucleus.
Microtubule cytoskeleton can exert forces on the interphase nucleus. For example, dynein interacts with NPCs during late G2 and prophase. The dynein adaptor bicaudal D2 (BICD2) binds to the Nup358 to recruit dynein to NPCs during late G2 and prophase, whereas another protein, CENP-F, brings dynein to NPCs in prophase by bridging the dynein regulator NudE or NudE-like (Nudel) to Nup133 (Splinter et al., 2010; Bolhy et al., 2011). The binding of dynein to NPCs suggests that as dynein moves toward the minus ends of the astral microtubules anchored at the centrosome, it exerts force on NPCs. If this force were not counterbalanced, all NPCs would be pulled toward the centrosomes during late G2 and prophase. The even distribution of NPCs indicates that some nuclear proteins and/or plus end–directed motors must counterbalance the dynein force.
Among the nuclear peripheral proteins, the structural components of the nuclear lamina, the lamins, could function to anchor NPCs because lamins have been shown to directly bind to nucleoporins (Smythe et al., 2000; Al-Haboubi et al., 2011; Lussi et al., 2011). By tethering NPCs to the nuclear lamina, lamins may counterbalance the dynein forces to ensure the even distribution of NPCs throughout the nuclear envelope. Using different mouse cells, we demonstrate that lamins ensure proper distribution of NPCs during late G2 and prophase by counteracting the dynein forces exerted on NPCs via BICD2. This in turn contributes to efficient centrosome separation in prophase. We will discuss the implications of these findings in the context of how lamins may use a conceptually similar means to balance motor forces exerted on the interphase nucleus and in the mitotic spindle.
RESULTS AND DISCUSSION
Each lamin, when expressed at a sufficient level, prevents asymmetric NPC distribution
To study the role of lamins, we previously created both mouse embryonic stem cells (ESCs) and mice deleted of individual or different combinations of Lmnb1, Lmnb2, or Lmna genes (Kim et al., 2011, 2013; Kim and Zheng, 2013). Using these ESCs, mouse embryonic fibroblasts (MEFs) derived from mouse embryos, and ESC-derived fibroblast-like cells (EDFCs), we have shown that the total lamin concentration, and not a specific lamin type, is required to maintain the even distribution of NPCs around the nuclear envelope in MEFs and EDFCs but not in ESCs (Guo et al., 2014). Further immunofluorescence analyses of EDFCs derived from lamin triple-knockout (TKO) ESCs using the antibody against Nup98 revealed that the uneven distribution of NPCs could be separated into two types—clustered and asymmetric distributions. In the clustered distribution, a number of NPCs cluster together to form small aggregates that distribute throughout the nuclear envelope, whereas in the asymmetric distribution, all NPCs aggregate toward one side of the nuclear envelope (Figure 1, A–C). Compared to wild-type EDFCs, which exhibited nearly 100% even distribution of NPCs, ∼40% of the lamin TKO EDFCs exhibited asymmetric NPC distribution, whereas ∼50% had clustered NPCs (Figure 1D).
FIGURE 1:
Asymmetric NPC distribution upon depletion of all lamins. (A) The localization of examined Nups in NPCs. (B) Immunofluorescence of Nup98 in EDFCs. (C) Representative images of nuclei exhibiting even, clustered, or asymmetric NPC distribution. (D) Quantification of NPC distribution in EDFCs. (E) Nup93, TPR, and mAb414 immunostaining pattern and Nup133 distribution in EDFCs. 3xGFP-Nup133 was imaged by 1-min exposure time. (F) Immunofluorescence of NPCs and lamins in TKO EDFCs that overexpress lamin-A, -B1, or -B2. Arrows, cells that overexpress each lamin exhibit rescued NPC distribution. (G) Quantification of NPC distribution in TKO EDFCs that overexpress each lamin. (H) Western blotting analysis of lamin-A/C (LA and LC) in wild-type (WT) and LBDKO tMEFs treated with control (siCtrl) or two different lamin-A/C (siLA/C 1 and siLA/C 2) siRNAs. Tubulin (Tub), loading control. (I) Immunofluorescence of NPCs in tMEFs. (J) Quantification of NPC distribution in tMEFs. Scale bars, 5 μm. For all quantifications, three independent experiments. Error bars, SD. More than 100 cells were counted in each group. n.s., not significant; ***p < 0.001 by Student’s t test.
Because lamin-B loss in the Drosophila cyst stem cells and cyst cells resulted in the aggregation of some Nups without affecting the distribution of others (Chen et al., 2013), we used antibodies that recognize nucleoporin Nup93 or TPR, and the monoclonal antibody mAb414, which recognizes four FG-Nups—Nup358, Nup214, Nup153, and Nup62—to probe NPC distribution (see Figure 1A for the localization of all the Nups probed). All of these Nups exhibited either clustered (unpublished data) or asymmetric distribution in the TKO EDFCs (Figure 1E). We also transfected a plasmid expressing Nup133 tagged by three tandem green fluorescent proteins (3xGFPs) in wild-type or lamin TKO EDFCs. Although the signal of 3xGFP-Nup133 is very weak, by increasing the exposure time, we found that the 3xGFP-Nup133–marked NPCs also exhibited clustered (unpublished data) or asymmetric distributions (Figure 1E). As expected, overexpression of lamin-B1, -B2, or -A ameliorated both NPC clustering and asymmetric distributions (Figure 1, F and G).
After differentiation from ESCs, the EDFCs proliferate very slowly. To explore further whether the asymmetric NPC distribution phenotype also occurs in cells that proliferate rapidly, we derived transformed MEFs (tMEFs) by stably expressing the SV40 large T antigen in primary MEFs. Proliferation analyses showed that the wild-type and lamin-B1/lamin-B2 double-knockout (LBDKO) tMEFs exhibited similar morphology and high growth rates (Supplemental Figure S1). Depletion of lamin-A/C by two different small interfering RNAs (siRNAs) did not affect the proliferation of the wild-type or LBDKO tMEFs (Supplemental Figure S1B). We therefore analyzed the NPC distribution in the tMEFs treated by control or lamin-A/C siRNA. We found that reduction of lamin-A/C by two different siRNAs in the LBDKO tMEFs (Figure 1H) resulted in either clustered or asymmetric distribution of NPCs similar to those observed in the lamin TKO EDFCs (Figure 1, I and J). These studies demonstrate that lamins are required for the even distribution of NPCs, regardless of the growth rate of the cells.
Lamins prevent asymmetric NPC distribution near centrosomes during late G2 and prophase
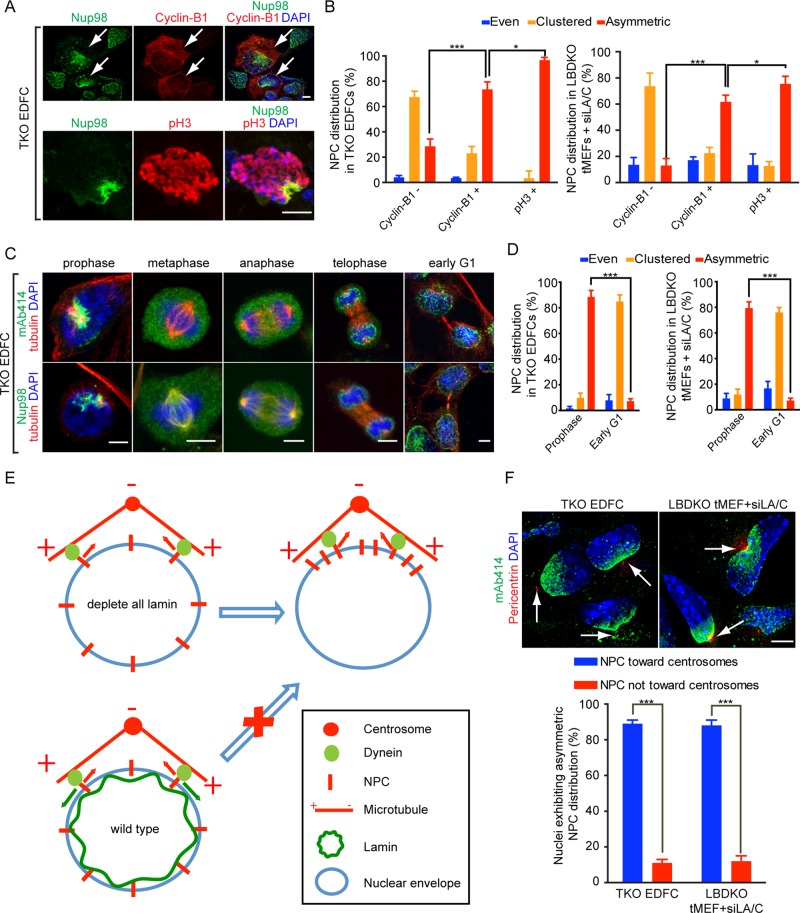
Previous studies showed that dynein is loaded onto NPCs during late G2 and prophase by dynein adaptor proteins (Splinter et al., 2010; Bolhy et al., 2011). Therefore dynein could exert forces on NPCs to pull them toward the minus ends of microtubules during late G2 and prophase. We reasoned that by analyzing the cell cycle stages when the NPC clustering or asymmetric distribution occurs, we could gain further insights into the cause of the NPC distribution defects upon lamin deletion. We performed immunofluorescence of cyclin-B1 and phosphorylated histone H3 (pH3) to label late G2 and prophase cells (Figure 2A, top, arrows) or only prophase cells (Figure 2A, bottom) in lamin TKO EDFCs, respectively. We found that ∼70% of cyclin-B1–positive and ∼95% of pH3-positive cells exhibited asymmetric NPC distribution in EDFCs, whereas the cyclin-B1– negative cells exhibited mostly the NPC clustering defect (Figure 2, A and B). Similar analyses in lamin-A/C siRNA–treated LBDKO tMEFs revealed that NPC asymmetric distribution occurred in late G2 and prophase (Figure 2B). In both EDFCs and tMEFs depleted of all lamins, Nup98 and other Nups recognized by mAb414 were distributed throughout the cytoplasm upon nuclear envelope breakdown (NEBD) during mitosis. In telophase, these Nups underwent grossly even reassembly into NPCs surrounding the decondensing chromosomes, and in early G1 cells, clustered NPCs appeared around the nuclear envelope (Figure 2, C and D). Thus the asymmetry of NPC distribution in prophase is resolved through NPC reassembly during cell division in the lamin-depleted EDFCs and tMEFs.
FIGURE 2:
NPCs cluster toward centrosomes at late G2 and prophase upon complete lamin loss. (A) Immunofluorescence of Nup98 in cyclin-B1−, cyclin-B1+ (top, arrows) and pH3+ (bottom) TKO EDFCs. (B) Quantification of NPC distribution in TKO EDFCs (left) and LBDKO tMEFs treated with lamin-A/C siRNAs (right). Cyclin-B1−, cyclin-B1+, and pH3+ cells were separately quantified. (C) Immunofluorescence of mAb414, Nup98, and tubulin in TKO EDFCs in M phase and early G1 phase. (D) Quantification of NPC distribution in TKO EDFCs (left) and LBDKO tMEFs treated with lamin-A/C siRNAs (right) in prophase and early G1 phase. (E) How dynein force could pull NPCs toward centrosomes. Lamin could resist dynein force and prevent asymmetric NPC clustering. (F) Immunofluorescence of mAb414 and pericentrin in TKO EDFCs and LBDKO tMEFs treated with lamin-A/C siRNAs (top). Arrows, centrosomes. Bottom, quantification of nuclei with NPC aggregation toward centrosomes. Scale bars, 5 μm. For all quantifications, three independent experiments. Error bars, SD. n.s., not significant; *p < 0.05 and ***p < 0.001 by Student’s t test.
We found, as expected, that microtubule arrays were anchored to centrosomes in EDFCs or tMEFs near the interphase nucleus in the presence or absence of lamins (Supplemental Figure S2). These microtubule arrays could exert forces on NPCs via the NPC-associated dynein during late G2 and prophase. If the lamin meshwork functions to counteract the dynein forces exerted on NPCs, lamin depletion would allow all NPCs to be pulled toward the centrosomes to assume the asymmetric distribution during late G2 and prophase (Figure 2E). To analyze this, we performed double immunostaining of the centrosomes and NPCs using the pericentrin antibody and mAb414, respectively. We found that NPCs asymmetrically aggregated toward the centrosomes (Figure 2F, arrows) in ∼90% of lamin-depleted EDFCs and tMEFs (Figure 2F). Because centrosomes serve as the major microtubule-organizing centers where microtubule minus ends reside, this result strongly suggests that NPCs are pulled toward the minus ends of microtubules in the lamin-null cells. Thus lamins prevent asymmetric aggregation of NPCs near the centrosomes during late G2 and prophase.
Microtubules mediate asymmetric NPC distribution in the absence of lamins
The foregoing findings suggest that dynein could pull NPCs toward centrosomes during late G2 and prophase in the absence of lamins. We therefore first tested whether depolymerizing microtubules by nocodazole could prevent the asymmetric aggregation of NPCs at the centrosome. The lamin TKO EDFCs or the LBDKO tMEFs depleted of lamin-A/C by RNA interference (RNAi) were incubated with 1 μM nocodazole for 1, 4, 8, and 16 h. Depolymerizing microtubules caused a gradual reduction of asymmetric NPC distribution in these lamin-depleted cells (Figure 3, A and B). We also quantified the effect of microtubule depolymerization on NPC clustering in the lamin-depleted EDFCs or tMEFs. We found that nocodazole treatment caused a gradual increase of cells exhibiting clustered NPCs in both cell types (Figure 3, A and B).
FIGURE 3:
Microtubule depolymerization partially rescues asymmetric NPC distribution in lamin-depleted cells. (A) Immunofluorescence of Nup98 and tubulin in lamin-depleted EDFCs and tMEFs treated with DMSO or nocodazole. (B) Quantification of NPC distribution upon nocodazole treatment at indicated time points in lamin-depleted EDFCs and tMEFs. (C) Costaining of mAb414 and cyclin-B1 in lamin-depleted EDFCs and tMEFs treated with DMSO or nocodazole. (D) Quantification of NPC distribution in nocodazole-treated, cyclin-B1+, lamin-depleted EDFCs and tMEFs. (E) Immunofluorescence images of Nup98 in lamin-depleted EDFCs and tMEFs treated with DMSO or blebbistatin. (F) Quantification of NPC distribution upon blebbistatin treatment in lamin-depleted EDFCs and tMEFs. Scale bars, 5 μm. For all quantifications, error bars, SD. Three independent experiments. More than 100 cells were counted at each time point for each experiment. n.s., not significant; *p < 0.05, **p < 0.01, and ***p < 0.001 by Student’s t test.
We also labeled cells with cyclin-B1 and mAb414 and quantified the percentages of symmetric, asymmetric, and clustered NPCs in lamin-depleted late G2 and prophase cells that were treated with dimethyl sulfoxide (DMSO) or nocodazole. Asymmetric NPC distribution caused by lamin depletion was rescued in late G2 and prophase cells by nocodazole treatment (Figure 3, C and D). Disrupting the actomyosin activity using the myosin II inhibitor blebbistatin did not affect NPC clustering or NPC asymmetric distribution in these lamin-depleted cells (Figure 3, E and F). Therefore, of the two NPC distribution defects caused by lamin depletion, the asymmetric NPC distribution toward centrosomes during late G2 and prophase is due to the microtubule-based activity. It is unclear how small NPC clusters form throughout the nucleus in the absence of lamins, but the actomyosin system does not appear to play a role.
Lamins counteract the BICD2-based dynein force to prevent asymmetric NPC positioning
Dynein is known to be loaded onto NPCs during late G2 and prophase by either BICD2 or CENP-F (Splinter et al., 2010; Bolhy et al., 2011). The microtubule-based asymmetric NPC distribution toward the centrosomes in the absence of lamins suggests that the NPC-localized dynein could move toward the minus ends of microtubules, exerting forces on NPCs. Because NPCs are known to directly interact with lamins, the lamin meshwork could resist the dynein forces to prevent the movement of NPCs (Figure 2E). To dissect whether dynein mediates the asymmetric distribution of NPCs and whether the dynein adaptor BICD2 or CENP-F or both are responsible for this dynein force, we depleted each of these proteins by two different siRNAs in lamin-depleted EDFCs or tMEFs. Approximately 40–80% reduction of dynein heavy chain (DHC), BICD2, and CENP-F were achieved (Figure 4, A and B). Reduction of DHC and BICD2 significantly rescued NPC asymmetry in lamin-depleted cells, but CENP-F reduction had no effect (Figure 4, C and D). Similar to nocodazole treatment (Figure 3), the reduction of NPC asymmetry upon DHC or BICD2 depletion resulted in a corresponding increase of clustered NPC distribution (Figure 4, C and D).
FIGURE 4:
Dynein drives asymmetric NPC distribution through BICD2 pathway. (A, B) Western blotting analysis of DHC, BICD2, and CENP-F in TKO EDFCs (A) and LBDKO tMEFs (B) upon indicated RNAi treatment. Tubulin, loading control. (C, D) Quantification of NPC distribution in TKO EDFCs and LBDKO tMEFs upon RNAi knockdown of DHC (C), BICD2 (D), and CENP-F (D). Three experiments. Error bars, SD. More than 100 nuclei were counted for each experiment. n.s., not significant; **p < 0.01, and ***p < 0.001 by Student’s t test. (E) Immunofluorescence of p150 and BICD2 in pH3+ LBDKO tMEFs. (F) Colocalization of p150 or BICD2 with NPCs in LBDKO tMEFs treated with lamin-A/C siRNAs. (G) Dual-color immunofluorescence of p150 and Nup98 in pH3+ LBDKO tMEFs upon indicated RNAi treatment. Scale bar, 5 μm.
Because BICD2 is known to recruit dynein to NPCs (Splinter et al., 2010; Bolhy et al., 2011; Hu et al., 2013), RNAi of BICD2 could perturb dynein loading onto NPCs, thereby preventing the dynein force from pulling NPCs toward the centrosomes. To test this possibility, we first attempted to detect the localization of dynein or its regulator dynactin and BICD2 on NPCs by immunofluorescence. We found that 1 h of nocodazole treatment was sufficient to reveal the nuclear envelope localization of p150, a subunit of dynactin and BICD2 in pH3-positive tMEFs (Figure 4E), which is consistent with previous studies (Splinter et al., 2010; Bolhy et al., 2011; Jodoin et al., 2013). RNAi of lamin-A/C in LBDKO tMEFs resulted in asymmetric distribution of p150 and BICD2, which colocalized with the asymmetrically aggregated NPCs (Figure 4F). In both control tMEFs and tMEFs depleted of all lamins, RNAi of BICD2, but not of CENP-F, greatly reduced p150 localization to the nuclear envelope, which also ameliorated NPC asymmetric distribution in lamin-depleted tMEFs (Figure 4G). We also analyzed the localization of Sun1 and Sun2 in TKO EDFCs because they are part of the link of nucleoskeleton and cytoskeleton (LINC) complex, which can also interact with NPCs and dynein at the nuclear envelope. Whereas Sun2 did not exhibit asymmetric distribution in cells with NPC asymmetric localization (Supplemental Figure S3, A and B), nuclear Sun1 signal was greatly diminished in the absence of lamins (Supplemental Figure S3C). These studies demonstrate that BICD2, but not CENP-F or the LINC complex, serves as an adaptor between dynein and NPCs, which allows dynein forces to pull NPCs toward the centrosome during late G2 and prophase in lamin-depleted cells.
Lamins facilitate centrosome separation by distributing the NPC-associated dynein forces
Multiple pathways are involved in centrosome separation during prophase and prometaphase in tissue culture cells. For example, the docking of dynein onto NPCs via BICD2 during prophase facilitates the separation of duplicated centrosomes by pulling on the astral microtubules nucleated from each centrosome (Figure 5A; Tanenbaum et al., 2008; Raaijmakers et al., 2012; van Heesbeen et al., 2013). On the other hand, the plus end–directed kinesin Eg5 is known to act on the antiparallel microtubules between the two microtubule asters to drive apart the duplicated centrosomes (Figure 5A; Tanenbaum et al., 2008; Raaijmakers et al., 2012; van Heesbeen et al., 2013). The asymmetric distribution of p150 and BICD2 in lamin-depleted cells (Figure 4F) suggests that dynein forces are limited to a small area of the astral microtubules, which could lead to inefficient separation of the duplicated centrosomes during prophase (Figure 5A). To test this, we analyzed prophase centrosome separation by immunostaining using pericentrin and pH3 antibodies in EDFCs and tMEFs. Depletion of lamins in these two cell types resulted in significantly shorter intercentrosome distance in prophase cells (Figure 5, B and C). To confirm this result in live cells, we performed dual-color live imaging of LBDKO tMEFs stably expressing GFP-EB3 and mCherry-Histone H2B (Figure 5D). GFP-EB3 concentrates on centrosomes during late G2 and prophase, allowing us to localize centrosomes in tMEFs. A fraction of GFP-EB3 that diffusely localizes in the cytoplasm in interphase and prophase diffuses into the nuclear space upon NEBD, which allows accurate determination of the time of NEBD. We measured centrosome distance in prophase right before NEBD based on GFP-EB3 and mCherry-Histone H2B in tMEFs (Figure 5D, bracket) and found that lamin depletion reduced centrosome distance in prophase within 1.5 min before NEBD (Figure 5E). However, lamin depletion did not alter the duration of prophase or prometaphase plus metaphase (Figure 5F). Thus the prophase centrosome separation defect in lamin-depleted tMEFs is not caused by shortened prophase in these cells.
FIGURE 5:
Asymmetric NPC distribution in the absence of lamin disrupts centrosome separation. (A) Illustration of microtubule motors that drive prophase centrosome separation. Lamin depletion disrupts dynein distribution and impairs dynein-driven centrosome separation. (B, C) Immunofluorescence of pericentrin in pH3+ EDFCs (B) and tMEFs (C) allows measurement of centrosome distance (brackets) at prophase (left). Right, quantifications of centrosome distance. (D) Time-lapse images of GFP-EB3 and mCherry-H2B in LBDKO tMEFs. Condensation of chromosomes (marked by mCherry-H2B) indicates the start of prophase. Entry of EB3 into nucleus shows the occurrence of NEBD. The bracket shows the centrosome distance in the frame before NEBD. (E) Quantification of centrosome distance in LBDKO tMEFs in the first frame before NEBD. (F) Quantification of mitotic timing at prophase or prometaphase plus metaphase in LBDKO tMEFs. (G) Quantification of centrosome distance in LBDKO tMEFs depleted of lamin-A/C, DHC, or both. (H) Quantification of prophase centrosome distance in LBDKO tMEFs treated with control or lamin-A/C RNAi. The centrosome distance in lamin-A/C RNAi–treated cells were separately quantified as those exhibiting symmetric (sym) or asymmetric (asym) NPC distribution. Scale bars, 5 μm. For all quantifications, >50 cells in three independent experiments. Error bars, SD. n.s., not significant; ***p < 0.001 by Student’s t test.
Codepletion of dynein and lamins in tMEFs did not cause additional decrease of centrosome distance compared with the depletion of lamins alone (Figure 5G), which is consistent with the idea that lamin is required for the proper distribution of dynein and therefore proper dynein forces on microtubules for centrosome separation. Depletion of lamins did not change the protein expression levels of Eg5, DHC, and the dynein adaptors BICD2 and CENP-F (Supplemental Figure S4A). Lamin depletion also did not alter the localization of Eg5 in prophase cells (Supplemental Figure S4B). Inhibition of Eg5 by a chemical inhibitor, S-trityl-l-cysteine (STLC; Brier et al., 2004) resulted in an additional shortening of centrosome distance in LBDKO tMEFs treated by either control or lamin-A/C RNAi (Supplemental Figure S3C). Thus lamin and Eg5 forces are additive in driving centrosome separation in prophase (Figure 5A), which is consistent with previous reports (van Heesbeen et al., 2013; Tanenbaum et al., 2008; Raaijmakers et al., 2012).
If lamins facilitated prophase centrosome separation by ensuring even distribution of dynein on NPCs throughout the nuclear envelope, we would expect a strict correlation between asymmetric NPC distribution and the shortening of centrosome distance. We measured prophase centrosome distance in the lamin-depleted tMEFs with or without NPC asymmetry and found that lamin-depleted cells lacking NPC asymmetry had similar centrosome distance as the control cells, whereas the lamin-depleted cells exhibiting asymmetric NPC distribution had significantly shortened centrosome distance (Figure 5H). These findings show that lamins facilitate prophase centrosome separation by regulating dynein distribution throughout the nuclear envelope by the NPC-bound BICD2.
The asymmetric distribution of NPCs has been observed in lamin-depleted Caenorhabditis elegans (Liu et al., 2000) and cells from mouse brain explants (Jung et al., 2013), but neither the cause nor the consequence of this distribution defect is clear. By analyzing the effect of cell cycle, microtubules, and dynein on the distribution of NPCs, we demonstrate that lamins anchor NPCs on the nuclear envelope, which allows NPCs to resist dynein forces during late G2 and prophase of the cell cycle, thereby preventing the asymmetric aggregation of NPCs. The even distribution of NPCs regulated by lamins also ensures that BICD2 can recruit dynein to NPCs throughout the nuclear envelope during prophase. This in turn allows dynein to efficiently capture astral microtubules to separate the duplicated centrosomes. Of interest, Drosophila cyst stem cells and cyst cells lacking lamin-B exhibit aggregation of some nucleoporins, but NPCs appear to be properly distributed (Chen et al., 2013). It is possible that a small amount of A-type lamin in these cells is sufficient to ensure the even distribution of NPCs. Alternatively, dynein in this cell type may not be loaded onto NPCs. It will be important to dissect further how different cells in different organisms use lamins to regulate NPC distribution and function.
The direct binding of the dynein regulator Nudel to lamin-B appears to help dynein to counteract the forces generated by Eg5 to ensure spindle pole focusing in spindles assembled in Xenopus egg extracts (Ma et al., 2009; Goodman et al., 2010), where centrosomes do not play a major role at the spindle poles. Of interest, in cells in which centrosomes are important for spindle pole organization, lamin and dynein regulate spindle orientation but not spindle pole focusing (Yingling et al., 2008; Kim et al., 2011). Although the role of dynein in spindle orientation is well understood, how lamins regulate spindle orientation is unknown. Because the cortical capture of microtubules nucleated from centrosomes is important for spindle orientation, it is tempting to speculate that the defects in centrosome separation in the lamin-null cells could result in defects in spindle orientation. We have not been able to test this idea due to the highly random spindle orientation in the tMEFs with or without lamins and the extremely low proliferation rate of differentiating EDFCs. However, the findings we report here should provide an opportunity to dissect further whether lamins regulate spindle orientation through dynein and centrosomes by employing cell types that are more amenable for studying spindle positioning.
MATERIALS AND METHODS
Derivation and culture of EDFCs and tMEFs
The derivation and culture of EDFCs and primary MEFs were previously described (Guo et al., 2014). To generate tMEFs, 1.25 × 105 primary MEFs at passage two or three were seeded in a well of a six-well plate and cultured for 24 h. A 2-μg amount of pBsSVD2005 plasmid (plasmid #21826; Addgene, Cambridge, MA) expressing SV40 large T antigen was transfected into MEFs using Lipofectamine 2000 (11668-027; Invitrogen, Carlsbad, CA). At 24 h after transfection, cells were split into two 10-cm tissue culture dishes and cultured for ∼10 d until all nontransformed cells had died and colonies of tMEFs formed. Cells were pooled and frozen as passage one tMEFs. tMEFs were cultured using MEF medium: knockout DMEM (10829-018) supplemented with 15% fetal bovine serum (26140-079), 100 mM β-mercaptoethanol (21985-023), 2 mM l-glutamine (GlutaMAX, 35050-061), 0.1 mM nonessential amino acids (11140-050) and 50 U/ml penicillin/streptomycin (15140-122; all from Invitrogen). tMEFs were split every 48 h at 1:10 dilution.
RNA interference
Silencer Select siRNAs (4390771; Invitrogen) were used for all RNAi experiments. Lamin-A/C siRNAs, 5′-GGCUUGUGGAGAUCGAUAAtt-3′ and 5′-CCACCGAAGUUCACCCUAAtt-3′; DHC siRNAs, 5′-GGAACGAAUGAAUACCCUUtt-3′ and 5′-GAAAGAUCCGCAACACGAAtt-3′; BICD2 siRNAs, 5′-CGACAGACCUCGUUGGAUAtt-3′ and 5′-GCAUCAACGAUUCCUUCUAtt-3′; and CENP-F siRNAs, 5′-GCUUGACUUUAAACGAAGAtt-3′ and 5′-GCCUGAGAAUUUUCGAAUUtt-3′. Control siRNA was purchased from Invitrogen (4390847).
Transfection of siRNAs was performed using RNAiMAX transfection reagents (13778030; Invitrogen) following the manufacturer’s instructions. For siRNA treatment of EDFCs, 5 × 104/well ESCs were seeded on gelatinized coverglasses in 24-well plates. ESCs were induced to differentiate into EDFCs in MEF medium containing 0.4 μM retinoic acid. siRNA (10 nM) was transfected at 24 h after induction of differentiation. Cells were analyzed 48–72 h after siRNA transfection.
For RNAi in tMEFs, 1.25 × 105/well tMEFs were seeded in gelatinized six-well plates, followed by the first transfection. At 48 h after the first transfection, 2.5 × 104/well tMEFs were split on gelatin-coated coverglass in 24-well plates, followed by the second transfection. At 48 h after the second transfection, cells were analyzed. For single RNAi in tMEFs, 10 nM siRNA was transfected. For double-RNAi treatments, 5 nM each of the indicated siRNA was used.
Nocodazole, STLC, and blebbistatin treatments
Nocodazole, 1 μM (Sigma-Aldrich, St. Louis, MO), was added into the culture medium of tMEFs or EDFCs and incubated for 1, 4, 8, or 16 h to depolymerize microtubules. Different concentrations (0.125, 0.22, 0.5, 1, or 2 μM) of STLC (Sigma-Aldrich) were added into culture medium to inhibit Eg5 activity for 8 h. Blebbistatin, 5 μM (Sigma-Aldrich), was used to treat cells for 8 h.
Immunofluorescence microscopy and quantifications
Primary antibodies used for immunofluorescence microscopy are rat monoclonal anti-Nup98 (1:200; ab50610; Abcam, Cambridge, UK), mouse monoclonal antibody 414 (mAb414, 1:1000; ab24609; Abcam), rabbit polyclonal anti-TPR (1:200; ab84516; Abcam), mouse monoclonal anti-Nup93 (1:200; sc-81343; Santa Cruz Biotechnology, Dallas, TX), rabbit polyclonal anti-Sun1 (1:200; ab74758; Abcam), rabbit polyclonal anti-Sun2 (1:200; ab87036; Abcam), rabbit polyclonal anti–cyclin-B1 (1:200; sc-752; Santa Cruz Biotechnology), mouse monoclonal anti–p-histone H3 (1:1000; 9706S; Cell Signaling, Danvers, MA), rabbit polyclonal anti-pH3 (1:2000; 09797; Millipore), mouse monoclonal anti–α-tubulin (1:500; DM1 α; Sigma-Aldrich), rabbit polyclonal anti-pericentrin (M8, 1:1000; Doxsey et al., 1994), mouse monoclonal anti-p150 (1:1000; 610473; BD, Franklin Lakes, NJ), rabbit polyclonal anti-BICD2 antibodies (1:200; 81488; Novus, Littleton, CO), and rabbit polyclonal anti-Eg5 (1:200; sc-66873; Santa Cruz Biotechnology). Secondary antibodies used were Alexa 488 goat anti-mouse, Alexa 488 goat anti-rat, Alexa 488 goat anti-rabbit, Alexa 568 goat anti-mouse, Alexa 568 goat anti-rabbit, and Alexa 633 goat anti-mouse antibodies (Invitrogen).
EDFCs and tMEFs were grown on gelatin-coated coverslips before immunofluorescence. For immunostaining of cyclin-B1, microtubules, or BICD2, cells were fixed by −20°C methanol for 5 min. Otherwise, cells were fixed by 4% paraformaldehyde (15711; EMS, Hatfield, PA) for 10 min, followed by permeabilization with 0.1% Triton-100 (T8532; Sigma-Aldrich) in phosphate-buffered saline (PBS) for 10 min. After fixation, samples were blocked by 4% bovine serum albumin (BSA; A4503; Sigma-Aldrich) in PBS for 1 h. The cells were incubated with primary antibodies at 4°C overnight. Then cells were washed with 4% BSA in PBS, followed by incubation with secondary antibodies for 1 h at room temperature. Cells were washed twice with PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI), washed twice with PBS, and mounted with Immu-Mount (99-094-12; Thermo Scientific, Waltham, MA).
Confocal images were acquired using a laser scanning confocal microscope (Leica SP5) with a 63×/1.4 objective and an electron-multiplying charge-coupled device (CCD) camera. Images were processed using ImageJ software (National Institutes of Health, Bethesda, MD). A Nikon Eclipse E800 epifluorescence microscope with a PL APO 63×/1.4 oil objective was used for quantification.
To measure the distance between the two centrosomes, a prophase cell was identified using DAPI and pH3 staining. The positions of the two centrosomes were manually defined at the center of pericentrin staining. MetaMorph’s caliber tools were used to measure the distance between the two centrosomes. For all quantifications, Excel (Microsoft) was used to calculate percentages, mean, and SD. All plots were drawn using Prism software.
Live imaging of GFP-EB3 and mCherry-H2B tMEF
To generate LBDKO tMEFs stably expressing GFP-EB3 and mCherry-H2B, LBDKO tMEFs were first transduced with GFP-EB3 lentivirus (LentiBrite biosensor; Millipore, Billerica, MA). Single GFP+ LBDKO cells were isolated by fluorescence-activated cell sorting (FACS; FACSAriaIII; BD) to generate GFP-EB3 single-cell colonies. GFP-EB3 LBDKO tMEFs were next transfected with plasmids expressing mCherry-H2B (55055; Addgene). GFP-EB3/mCherry-H2B double-positive tMEFs were again isolated by FACS (FACSAriaIII).
GFP-EB3/mCherry-H2B LBDKO tMEFs were transfected with control and lamin-A/C siRNAs using the same RNAi protocol used for tMEFs. After 72 h of RNAi, GFP-EB3/mCherry-H2B LBDKO tMEFs were imaged using a TE2000 inverted microscope equipped with a 20×/0.45 objective and a CCD camera. A LiveCell controller (Pathology Devices, Westminster, MD) was used to maintain the temperature, CO2 level, and humidity during imaging. Cells were imaged at 1.5-min intervals for 24 h using MetaMorph’s multidimensional acquisition. GFP (150-ms exposure time) and mCherry (75-ms exposure time) images were taken sequentially for each frame. MetaMorph’s caliber tools were used to measure the distance between the two EB3-GFP spots one frame before NEBD occurred. Mitotic timing was measured by counting the number of frames in which cells were in prophase or prometaphase plus metaphase (from NEBD to chromosome separation).
Western blotting analyses
RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholic acid, 10 mg/ml DNase I) was used to generate whole-cell lysates at 5000–10,000 cells/μl, which was further diluted in SDS sample buffer. Cell lysates were analyzed by PAGE using 10% or 4–15% gradient polyacrylamide gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk and probed with one of the following primary antibodies: mouse monoclonal anti–lamin-A/C (1:5000; 39287; Active Motif), rabbit polyclonal anti–CENP-F (1:2000, ab5; Abcam), rabbit polyclonal anti-DHC (1:2000, made in house; Wang and Zheng, 2011; Wang et al., 2013), rabbit polyclonal anti-BICD2 (1:2000, 81488; Novus), mouse monoclonal anti–α- tubulin (1:5000; DM1 α; Sigma-Aldrich), rabbit polyclonal anti-Nudel (1:2000, made in house; Ma et al., 2009), and rabbit polyclonal anti-Eg5 (1:1000; sc-365593; Santa Cruz Biotechnology). Horseradish peroxidase–conjugated anti-mouse (1:5000) or anti-rabbit (1:5000) antibodies (31432 and 31462; Thermo Scientific) and West Pico Chemiluminescent Substrate (34078; Thermo Scientific) were used for detection.
Expression of 3xGFP-Nup133 and individual lamins in TKO EDFCs
The 3×GFP-Nup133 plasmid was a kind gift from Valerie Doye, Institut Jacques Monod, France (Bolhy et al., 2011). To express 3×GFP-Nup133 in EDFCs, the plasmid was used to transfect TKO EDFCs at 3 d after the induction of differentiation on a glass-bottom dish using Lipofectamine 2000 (Invitrogen). At 48–72 h after transfection, cells were imaged using an inverted Nikon TE200 microscope with a PL APO 63×/1.4 oil objective. Expression of individual lamins in TKO EDFCs was described previously (Guo et al., 2014).
Supplementary Material
Acknowledgments
We thank Ona Martin for technical assistance, Mahmud Siddiqi for help in microscopy analysis, and all members in the Zheng lab for critical suggestions and discussions. This study was supported by National Institutes of Health Grants R01GM056312 and R01GM06023 to Y.Z.
Glossary
Abbreviations used:
- BICD2
bicaudal D2
- EDFC
ESC-derived fibroblast-like cell
- ESC
embryonic stem cell
- LBDKO
lamin-B1/lamin-B2 double knockout
- MEF
mouse embryonic fibroblast
- NEBD
nuclear envelope breakdown
- NPC
nuclear pore complex
- TKO
lamin triple knockout.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0482) on August 5, 2015.
REFERENCES
- Al-Haboubi T, Shumaker DK, Köser J, Wehnert M, Fahrenkrog B. Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus. 2011;2:500–509. doi: 10.4161/nucl.2.5.17913. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier S, Lemaire D, Debonis S, Forest E, Kozielski F. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry. 2004;43:13072–13082. doi: 10.1021/bi049264e. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2012;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Chi Y-H, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang K-T. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen X, Zheng Y. The nuclear lamina regulates germline stem cell niche organization via modulation of EGFR signaling. Cell Stem Cell. 2013;13:73–86. doi: 10.1016/j.stem.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Goodman B, Channels W, Qiu M, Iglesias P, Yang G, Zheng Y. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. J Biol Chem. 2010;285:35238–35244. doi: 10.1074/jbc.M110.140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell. 2014;25:1287–1297. doi: 10.1091/mbc.E13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DJ-K, Baffet AD, Nayak T, Akhmanova A, Doye V, Vallee RB. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell. 2013;154:1300–1313. doi: 10.1016/j.cell.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodoin JN, Sitaram P, Albrecht TR, May SB, Shboul M, Lee E, Reversade B, Wagner EJ, Lee LA. Nuclear-localized Asunder regulates cytoplasmic dynein localization via its role in the integrator complex. Mol Biol Cell. 2013;24:2954–2965. doi: 10.1091/mbc.E13-05-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H-J, Nobumori C, Goulbourne CN, Tu Y, Lee JM, Tatar A, Wu D, Yoshinaga Y, de Jong PJ, Coffinier C, et al. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc Natl Acad Sci USA. 2013;110:E1923–E1932. doi: 10.1073/pnas.1303916110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan C-M, Gaiano N, Ko MSH, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng Y. Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem Biophys Res Commun. 2013;440:8–13. doi: 10.1016/j.bbrc.2013.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res. 2013;23:1420–1423. doi: 10.1038/cr.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N, Voncken JW, Konings G, van Weeghel M, van den Hoogenhof MM, Gijbels M, van Erk A, Schoonderwoerd K, van den Bosch B, Dahlmans V, et al. Post-natal myogenic and adipogenic developmental: defects and metabolic impairment upon loss of A-type lamins. Nucleus. 2011;2:195–207. doi: 10.4161/nucl.2.3.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi YC, Hügi I, Laurell E, Kutay U, Fahrenkrog B. The nucleoporin Nup88 is interacting with nuclear lamin A. Mol Biol Cell. 2011;22:1080–1090. doi: 10.1091/mbc.E10-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Tsai M-Y, Wang S, Lu B, Chen R, Iii JRY, Zhu X, Zheng Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat Cell Biol. 2009;11:247–256. doi: 10.1038/ncb1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopus eggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S-I, Pack C-G, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 2000;19:3918–3931. doi: 10.1093/emboj/19.15.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-Y, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- van Heesbeen RGHP, Raaijmakers JA, Tanenbaum ME, Medema RH. Nuclear envelope-associated dynein cooperates with Eg5 to drive prophase centrosome separation. Commun Integr Biol. 2013;6:e23841. doi: 10.4161/cib.23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A, Alvarez-Reyes M, Bridger JM, Broers JL, Ramaekers FC, Wehnert M, Morris GE, Whitfield WGF, Hutchison CJ. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J Cell Sci. 2001;114:2577–2590. doi: 10.1242/jcs.114.14.2577. [DOI] [PubMed] [Google Scholar]
- Wang S, Ketcham SA, Schön A, Goodman B, Wang Y, Yates J, Freire E, Schroer TA, Zheng Y. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell. 2013;24:3522–3533. doi: 10.1091/mbc.E13-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y. Identification of a novel dynein binding domain in nudel essential for spindle pole organization in Xenopus egg extract. J Biol Chem. 2011;286:587–593. doi: 10.1074/jbc.M110.181578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol. 2010;11:529–535. doi: 10.1038/nrm2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.