The mammalian ER contains 23 members of the PDI superfamily. Their substrate specificity is largely unknown. TMX1 shows a preference for membrane-bound, cysteine-containing polypeptides.
Abstract
The endoplasmic reticulum (ER) is the site of maturation for secretory and membrane proteins in eukaryotic cells. The lumen of the mammalian ER contains >20 members of the protein disulfide isomerase (PDI) superfamily, which ensure formation of the correct set of intramolecular and intermolecular disulfide bonds as crucial, rate-limiting reactions of the protein folding process. Components of the PDI superfamily may also facilitate dislocation of misfolded polypeptides across the ER membrane for ER-associated degradation (ERAD). The reasons for the high redundancy of PDI family members and the substrate features required for preferential engagement of one or the other are poorly understood. Here we show that TMX1, one of the few transmembrane members of the family, forms functional complexes with the ER lectin calnexin and preferentially intervenes during maturation of cysteine-containing, membrane-associated proteins while ignoring the same cysteine-containing ectodomains if not anchored at the ER membrane. As such, TMX1 is the first example of a topology-specific client protein redox catalyst in living cells.
INTRODUCTION
The mammalian endoplasmic reticulum (ER) contains 23 members of the protein disulfide isomerase (PDI) family (Tannous et al., 2015). These are characterized by the presence of one or more thioredoxin (Trx)-like domains, which may contain an active site with a Cys-Xxx-Xxx-Cys (CXXC) consensus sequence. Enzymatically active PDIs catalyze formation, reduction, and isomerization of intramolecular or intermolecular covalent bonds between luminal cysteine residues of newly synthesized polypeptides entering the secretory pathway, conferring structural stability on the native proteome (Ellgaard and Ruddock, 2005; Appenzeller-Herzog and Ellgaard, 2008; Bulleid, 2012; Oka and Bulleid, 2013). The reason for the high number of PDIs in the ER and their individual roles in protein biogenesis is unclear (Bulleid, 2012). Individual members of the PDI family show some substrate preference. For example, ERp57 forms functional complexes with calnexin (CNX) and calreticulin (CRT; Oliver et al., 1997; Zapun et al., 1998; Frickel et al., 2002; Pollock et al., 2004; Jessop et al., 2007). CNX and CRT are ER lectins that bind newly synthesized polypeptides displaying monoglucosylated, N-linked oligosaccharides (Hammond et al., 1994; Hebert et al., 1995). ERp57 acts on their ligands, that is, viral glycoproteins (Molinari and Helenius, 1999; Solda et al., 2006) or endogenous glycoproteins sharing common structural domains (Jessop et al., 2007), thereby promoting the formation of native disulfide bonds. In contrast, P5 targets BiP-bound proteins (Jessop et al., 2009), and PDI, ERp72, and ERdj5 facilitate dislocation of misfolded proteins or toxin subunits from the ER lumen into the cytosol (Majoul et al., 1997; Tsai et al., 2001; Molinari et al., 2002; Forster et al., 2006).
The majority of the PDIs are soluble luminal proteins characterized by a KDEL-like retention signal and transcriptional up-regulation in response to activation of the unfolded protein response (Tasanen et al., 1992; Roy and Lee, 1999; Anelli et al., 2002; Cunnea et al., 2003; Lee et al., 2003; Chichiarelli et al., 2007; Galligan and Petersen, 2012). Five members of the family, TMX1–5, are anchored at the ER membrane. Although no information is available for TMX5, for TMX1–4 it has been reported that they are not induced by cellular stresses (Haugstetter et al., 2005; Matsuo et al., 2009; Kozlov et al., 2010b; Sugiura et al., 2010; Koritzinsky et al., 2013).
Here we focus on TMX1, which is highly expressed in kidney, liver, placenta, and lungs (Matsuo et al., 2001). The presence of a proline residue at position 2 of the TMX1 catalytic site (CPAC, residues 56–59) hints at a possible role of TMX1 as an ER reductase (Hatahet and Ruddock, 2009). Consistent with such a role, TMX1 facilitates cell intoxication by ricin and abrin, which requires a reductive step promoting dislocation of the catalytic subunit to the cell cytosol (Pasetto et al., 2012) and reduces insulin disulfides in vitro (Matsuo et al., 2001). Deletion of the TMX1 gene results in susceptibility to liver damage in mice (Matsuo et al., 2013). At the cellular level, TMX1 deletion has no phenotype, indicating the activation of compensatory mechanisms (unpublished results; Matsuo et al., 2013). In this study, we report on a mass spectrometry analysis with a trapping mutant version of TMX1 expressed in mammalian cultured cells that reveals the selective association of TMX1 with a series of endogenous membrane-bound client proteins. Systematic analysis with membrane-bound and soluble model polypeptides confirmed the exquisite preference of TMX1 for membrane-bound substrates and identified TMX1 as the first example of topology-specific PDI transiently associating with both folding-competent and folding-defective membrane-bound polypeptides.
RESULTS
TMX1 preferentially associates with membrane-bound substrates
During the reaction cycles leading to disulfide bond formation, disassembly, or rearrangement, active-site cysteines of PDIs transiently form short-lived mixed disulfides with surface-exposed cysteine residues of proteins expressed in the ER lumen (Huppa and Ploegh, 1998; Molinari and Helenius, 1999). The replacement of the resolving carboxy-terminal cysteine residue in the PDI catalytic site efficiently traps mixed disulfides in the reductive pathway (Figure 1A; Hatahet and Ruddock, 2009). Thus PDI trapping mutants have been used to identify endogenous substrates of select ER-resident oxidoreductases such as ERp57, PDI, P5, ERp18, ERp72, ERp46, and ERdj5 (Dick and Cresswell, 2002; Jessop et al., 2007, 2009; Schulman et al., 2010; Oka et al., 2013).
FIGURE 1:
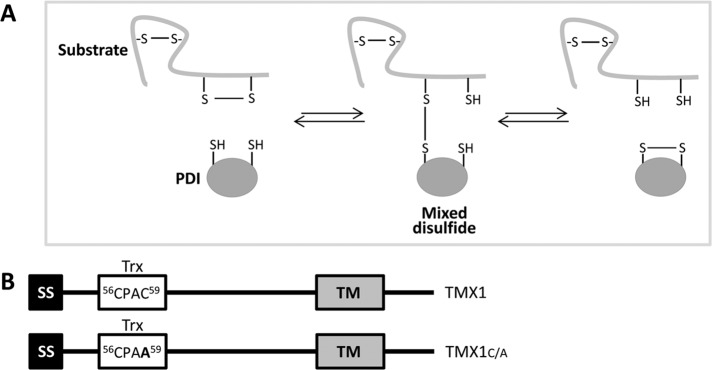
TMX1 trapping mutant to stabilize TMX1–substrate complexes. (A) Mechanism of reduction of disulfide bonds catalyzed by members of the PDI family. The formation of a mixed disulfide by the nucleophilic attack of the N-terminal active-site cysteine on a substrate disulfide bond. The resulting mixed disulfide can be resolved upon nucleophilic attack of the enzyme C-terminal active-site cysteine on the mixed disulfide. (B) TMX1 and TMX1C/A constructs. The replacement of the resolving cysteine (amino acid 59 of the TMX1 sequence) with an alanine residue substantially stabilizes mixed disulfides between TMX1 and its substrates. SS, signal sequence; Trx, thioredoxin-like domain; TM, transmembrane domain. The sequence of the active site of TMX1 and TMX1C/A and the position of the active site residues are shown.
To identify endogenous substrates of TMX1, we expressed the trapping mutant TMX1C/A (Figure 1B) in mouse embryonic fibroblasts (MEFs) and performed a mass spectrometry analysis of the cellular proteins coimmunoisolated with the ectopically expressed reductase. In contrast to the analyses performed for other PDIs, which revealed both soluble and membrane-bound proteins as endogenous substrates (Dick and Cresswell, 2002; Jessop et al., 2007, 2009; Schulman et al., 2010; Oka et al., 2013), TMX1C/A selectively trapped a series of cysteine-containing membrane proteins (Table 1). To determine whether the selective immunoisolation of membrane proteins was symptomatic of the intrinsic specificity of TMX1 for membrane-bound substrates, we used a series of model polypeptides characterized by the presence of a cysteine-containing ectodomain tethered or nontethered at the ER membrane (Figure 2A).
TABLE 1:
Endogenous substrates of TMX1 identified by mass spectrometry analysis.
| Gene name | Protein name | Entrynumber | Luminal cysteines | Predicted glycans | Protein topology |
|---|---|---|---|---|---|
| Itgb1 | Integrin β-1 | P09055 | 56 | 14 | Single-pass type I |
| Slc3a2 | 4F2 cell-surface antigen heavy chain | P10852 | 2 | 10 | Single-pass type II |
| Ece1 | Endothelin-converting enzyme 1 | Q4PZA2 | 12 | 13 | Single-pass type II |
| Lrrc59 | Leucine-rich repeat–containing protein 59 | Q922Q8 | 2 | 0 | Single-pass type II |
| Ncstn | Nicastrin | P57716 | 11 | 17 | Single-pass type I |
| Atp6ap1 | V-type proton ATPase subunit S1 | Q9R1Q9 | 2 | 8 | Single-pass type I |
| Tspan3 | Tetraspanin-3 | Q9QY33 | 6 | 4 | Multipass |
| Lrp10 | Low-density lipoprotein receptor–related protein 10 | Q7TQH7 | 30 | 4 | Single-pass type I |
| Pld4 | Phospholipase D4 | Q8BG07 | 5 | Several | Single-pass type II |
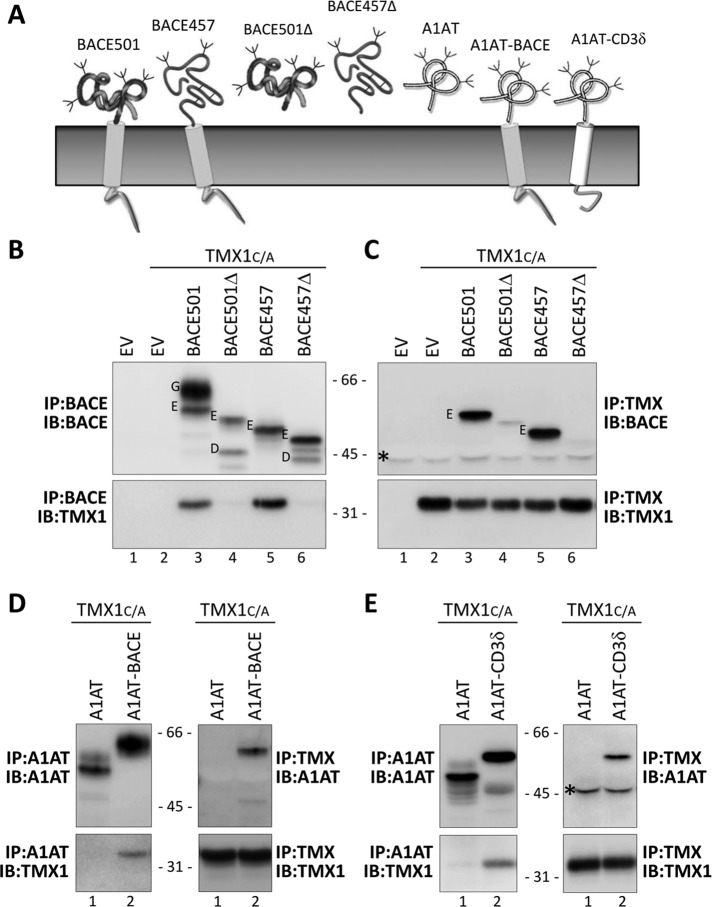
FIGURE 2:
TMX1 preferentially associates with membrane-bound substrates. (A) Model polypeptides used in this study. (B) MEFs transfected with empty vectors (EV, lane 1), β1-tagged TMX1C/A (lane 2), or TMX1C/A in combination with HA-tagged BACE501, BACE501Δ, BACE457, or BACE457Δ (lanes 3–6, respectively). The HA-tagged model substrates were immunoisolated from cell lysates. Top, WB with an anti-HA antibody to reveal the model substrates; bottom, WB with a TMX1-specific antibody to reveal TMX1C/A. (C) Same as B, but TMX1C/A was immunoisolated from the same cell lysates with an anti-β1 antibody to verify the presence of the model proteins (top) in the immunocomplexes. (D) Same as B and C, for the soluble HA-tagged A1AT and the membrane-anchored A1AT-BACE (lanes 1 and 2, respectively). (E) Same as D, for the soluble HA-tagged A1AT and the membrane-anchored A1AT-CD3δ (lanes 1 and 2, respectively). G, mature Golgi form of BACE proteins; E, immature ER form; D, deglycosylated form; asterisk, antibody heavy chain recognized by the secondary antibody in WB.
First, MEFs were mock transfected (EV [empty vector], Figure 2B, lane 1) or transfected with a plasmid for expression of β1-tagged TMX1C/A (lane 2) or with plasmids for expression of TMX1C/A and the folding-competent, membrane-bound BACE501 (lane 3), the folding-competent and soluble BACE501Δ (lane 4; Solda et al., 2007), the folding-defective, membrane-bound BACE457 (lane 5), or the folding-defective, soluble BACE457Δ (lane 6; Molinari et al., 2002). Cells were detergent solubilized, and the ectopically expressed, hemagglutinin (HA)-tagged BACE variants were immunoisolated from postnuclear supernatants. The immunocomplexes were separated in SDS–PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. The presence of the BACE variants was revealed by Western blot (WB) with anti-HA antibodies (Figure 2B, top). The association of TMX1C/A was assessed in the same PVDF membrane upon decoration of the membrane with anti-TMX1 antibodies (Figure 2B, bottom).
The membrane-bound BACE501 is separated into two forms (G is the Golgi, mature, endoglycosidase H [EndoH]–resistant form, and E is the ER, immature, EndoH-sensitive form of the protein; Figure 2B, lane 3; Solda et al., 2007). In the cell lysate, the soluble BACE501Δ is only present in the E form, as the G form is rapidly released in the extracellular medium (Solda et al., 2007). BACE457 and BACE457Δ are only present in their E form, since they are ER-retained, folding-defective polypeptides (Molinari et al., 2002). The experiment reveals that TMX1C/A associates with the membrane-bound versions of BACE (i.e., BACE501 and BACE457; Figure 2B, bottom, lanes 3 and 5). In contrast, TMX1C/A is not found in immunocomplexes containing the soluble variants of BACE (i.e., BACE501Δ and BACE457Δ; Figure 2B, bottom, lanes 4 and 6, respectively). The selectivity of TMX1 for membrane-bound polypeptides was confirmed upon immunoisolation from the cell lysates of the ectopically expressed, β1-tagged TMX1C/A (Figure 2C, bottom), which shows the abundant coprecipitation of BACE501 and BACE457 (top, lanes 3 and 5). Confirming the specificity of this assay of the G and the E forms of BACE501, only the latter, which is in the ER, is in the TMX1C/A-containing immunocomplexes (Figure 2C, lane 3). The association with both BACE501 and BACE457 shows that TMX1 does not discriminate between folding-competent and folding-defective polypeptides.
To further confirm the preference of TMX1 for association with membrane-bound proteins, we assessed association of the TMX1 trapping mutant with the soluble protein A1AT (Figure 2A; Perlmutter, 2011) and of two variants of A1AT that were artificially tethered at the ER membrane with two different membrane anchors (Figure 2A, A1AT-BACE and A1AT-CD3δ). Consistent with a preferential association of TMX1 with membrane-bound polypeptides, TMX1C/A associated with A1AT only when tethered at the membrane (Figure 2, D and E, lane 1 vs. lane 2). Of interest, and in contrast with the case of BACE proteins, the A1AT ectodomain contains a single cysteine residue. Hence, there are no intramolecular disulfides to be attacked by TMX1. However, there is evidence that the A1AT cysteine undergoes various reversible modifications, such as S-nitrosylation, S-glutathionylation, sulfenic acid formation, S-cysteinylation, and oxidation, which induce polymerization on the A1AT cysteine (Glaser et al., 1982; Tyagi and Simon, 1992; Miyamoto et al., 2000; Griffiths et al., 2002; Alam et al., 2011; Grek et al., 2012). Each of these modifications could be attacked by the substrate trap mutant of TMX1 to form a mixed disulfide.
TMX1 establishes mixed disulfides with newly synthesized membrane-bound BACE501
An active involvement of TMX1 in determining the fate of the associating proteins should involve formation of mixed disulfides as reaction intermediates (Figure 1A). To assess this directly, we performed a pulse-and-chase experiment. MEFs were transfected with two empty vectors (Figure 3, A– D, EV, lane 1), with expression vectors for BACE501, BACE501Δ, TMX1, or TMX1C/A and an empty vector (lanes 2–5), or with expression vectors for BACE501 and TMX1 (lane 6), BACE501 and TMX1C/A (lane 7), BACE501Δ and TMX1 (lane 8), or BACE501Δ and TMX1C/A (lane 9). Transfected cells were pulsed with [35S]methionine/cysteine for 13 min and chased for 10 min. At this time of chase, the newly synthesized BACE501 variants are still folding in the ER, as confirmed by the EndoH sensitivity of their oligosaccharides (Supplemental Figure S1A; Solda et al., 2007). BACE (Figure 3, A and B) or TMX1 variants (Figure 3, C and D) were immunoisolated from detergent extracts, and complexes were analyzed in nonreducing (Figure 3, A and C) and reducing SDS–PAGE (Figure 3, B and D).
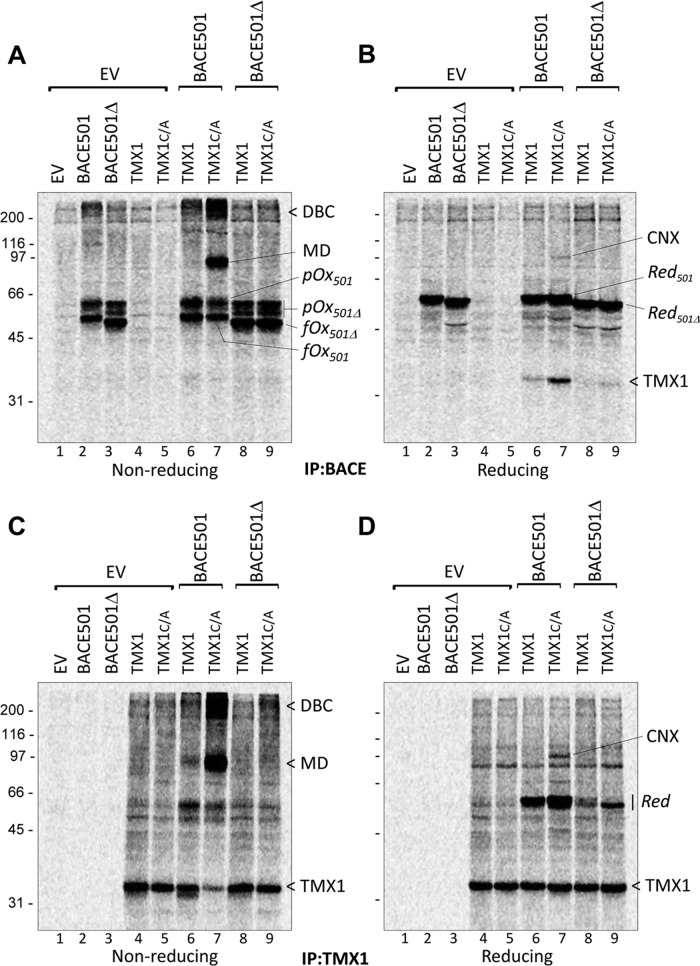
FIGURE 3:
TMX1 selectively establishes mixed disulfides with membrane-bound BACE501. (A) MEFs were transfected with empty vector (EV), BACE501, BACE501Δ, HA-tagged TMX1, or TMX1C/A (lanes 1–5), BACE501 in combination with HA-tagged TMX1 or TMX1C/A (lanes 6 and 7), or BACE501Δ in combination with HA-tagged TMX1 or TMX1C/A (lanes 8 and 9). The [35S]methionine/cysteine-radiolabeled model substrates were immunoisolated from cell lysates with anti-BACE antibodies. The immunocomplexes were separated under nonreducing conditions. (B) Same as A, but immunocomplexes were separated under reducing conditions. (C) Same as A, for complexes immunoisolated with anti-HA. (D) Same as C, analysis under reducing conditions. pOx501, partially oxidized BACE501; pOx501Δ, partially oxidized BACE501Δ; fOx501Δ, fully oxidized BACE501Δ; fOx501, fully oxidized BACE501; Red501, reduced BACE501; Red501Δ, reduced BACE501Δ; DBC, disulfide-bonded complexes; MD, mixed disulfides; CNX, calnexin.
In nonreducing gels, BACE501 and BACE501Δ expressed alone (Figure 3A, lanes 2 and 3) or in combination with TMX1 (lanes 6–9) are separated in a series of radiolabeled bands corresponding to various oxidation forms (Figure 3A and Supplemental Figure S1B, fully oxidized [fOx] and partially oxidized [pOx] for BACE501 and BACE501Δ, respectively). The bands with faster electrophoretic mobility correspond to BACE forms with lower hydrodynamic radius due to the presence of intramolecular disulfide bonds (fOx in Figure 3A and Supplemental Figure S1B) that relapse into the reduced (Red) BACE501 or BACE501Δ forms in the reducing gel (Figure 3, B and D, and Supplemental Figure S1B; Braakman et al., 1992).
In cells in which the membrane-bound BACE501 was cotransfected with the trapping mutant TMX1C/A, separation of the BACE immunoisolates under nonreducing conditions revealed an abundant labeled polypeptide band with an apparent molecular weight (MW) of ∼90 kDa (MD; Figure 3A, lane 7). Sample reduction dissociated the radiolabeled 90-kDa polypeptide into its components: BACE501, with an apparent MW of ∼60 kDa, and a radiolabeled polypeptide, with an apparent MW of ∼35 kDa (Figure 3B, lane 7). Both the 90-kDa polypeptide (Figure 3A, lane 7) and the 35-kDa polypeptide (TMX1; Figure 3B, lane 7) are much less abundant when BACE501 is cotransfected with the wild-type form of TMX1, which is unable to stabilize the mixed disulfide with the substrate. This led us to conclude that the 90-kDa polypeptide is a mixed disulfide (MD; Figure 3A, lane 7) containing BACE501 and TMX1C/A, which is dissociated under reducing conditions, releasing the radiolabeled TMX1 polypeptide of 35 kDa (Figure 3B, lane 7). Consistent with this hypothesis, mixed disulfides were significantly more abundant when the BACE501 was coexpressed with the trapping mutant than with the wild-type version of TMX1 (Figure 3C, lane 7 vs. lane 6). Moreover, both the MDs in the nonreducing gel (Figure 3A, lane 9) and the 35-kDa polypeptide in the reducing gel were virtually absent when the trapping mutant of TMX1 was cotransfected with the soluble BACE501Δ (Figure 3B, lane 9), which does not associate with TMX1 (Figure 2, B and C). Again, MDs were separated into their BACE501 and TMX1 constituents in the reducing gel (Figure 3D, lane 7). This confirms that the TMX1 trapping mutant stabilizes the otherwise short-lived MD intermediate of the BACE501 redox reaction. Cells expressing TMX1C/A and BACE501 also contained larger disulfide-bonded complexes of >200 kDa (Figure 3, A and C, lane 7).
Reduction of the functional complexes containing BACE501 and TMX1C/A revealed a radiolabeled polypeptide with an apparent MW of 97 kDa in Figure 3, B and D, lane 7. This polypeptide was identified by WB and specific immunoprecipitation as the lectin chaperone CNX. The increased presence of CNX in the BACE and in the TMX1C/A immunoisolates from cells overexpressing BACE501 (lane 7 in Figure 3, B and D, respectively) compared with cells that are not overexpressing BACE501 (lanes 5 and 9) led us to propose that CNX is a component of TMX1 functional complexes, which are assembled or stabilized in the presence of TMX1 substrates.
BACE501:TMX1:CNX complexes
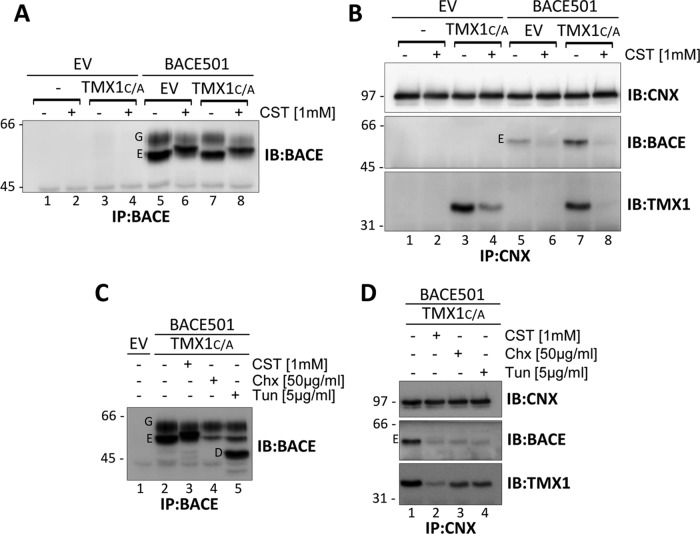
To assess whether CNX forms functional complexes with TMX1, we examined the consequences of cell exposure to castanospermine (CST), a glucose analogue that prevents association of newly synthesized proteins with CNX (Hammond et al., 1994). MEFs were mock transfected (EV; Figure 4, A and B, lanes 1 and 2), cotransfected with EV and a plasmid for expression of TMX1C/A (lanes 3 and 4), or cotransfected with EV and a plasmid for expression of BACE501 (lanes 5 and 6) or with plasmids for expression of TMX1C/A and BACE501 (lanes 7 and 8). Before solubilization and immunoisolation of ectopic BACE501 (Figure 4A) or of endogenous CNX with the associated polypeptides (Figure 4B), cells were incubated for 10 h in the absence (–) or presence (+) of CST to clear the CNX chaperone system of endogenous substrates (Figure 4, A and B, lanes 2 and 4) or of endogenous substrates and ectopically expressed BACE501 (lanes 6 and 8). In the absence of CST, CNX strongly interacted with TMX1C/A, which is a nonglycosylated protein (Figure 4B, bottom, lanes 3 and 7). This association was substantially reduced upon CST treatment (bottom, lanes 4 and 8; TMX1 is not glycosylated, thus excluding the possibility of its direct, glycan-lectin association with CNX). Thus inhibition of endogenous (Figure 4B, lane 4) and endogenous plus ectopic (lane 8) substrates access to the CNX chaperone system substantially reduces the fraction of TMX1 coprecipitated (i.e., participating in a functional complex) with CNX. Taken together, the data in Figures 3 and 4 show that CNX and TMX1 may form a functional complex, which is stabilized by client substrates. This conclusion is supported by the hampered association of TMX1 with CNX in cells with reduced protein synthesis (Figures 4, C, lane 4, and D, lane 3), or with defective N-glycosylation upon exposure to tunicamycin (Figure 4, C, lane 5, and D, lane 4).
FIGURE 4:
Client-mediated association between TMX1 and CNX. (A) MEFs were cotransfected with an empty vector (EV, lanes 1 and 2), an empty vector and HA-tagged TMX1C/A (3 and 4), an empty vector and BACE501 (5 and 6), or BACE501 and HA-tagged TMX1C/A (7 and 8). Cells were incubated for 10 h in the absence (–) or presence (+) of CST (1 mM). The expression level of BACE501 was checked upon WB of the immunoisolated ectopic protein. (B) Same as A, but endogenous CNX with associated proteins was immunoisolated from cell lysates. Immunocomplexes were analyzed under reducing conditions. Ectopically expressed BACE501 and TMX1C/A were revealed with an anti-BACE and an anti-HA antibody, respectively. (C) Same as A, in cells treated with CST, cycloheximide (Chx), or tunicamycin (Tun) for 3 h. (D) Same as B for cells treated with CST, Chx, or Tun. G, mature Golgi form of BACE501; E, immature ER form; D, deglycosylated form.
Characterization of TMX1C/A:BACE501 mixed disulfides by WB
To further confirm the selective involvement of TMX1 in mixed disulfides with membrane-bound clients, we expressed BACE501 alone (Figure 5A, lanes 1 and 2), with TMX1 (lanes 3 and 4), or with TMX1C/A (lanes 5 and 6). After immunoisolation of the HA-tagged bait, the immunocomplexes were separated in SDS–PAGE under nonreducing (NR; Figure 5A, lanes 1, 4, and 5) and reducing conditions (R; lanes 2, 3, and 6). We then transferred proteins to a PVDF membrane. BACE501 (Figures 5A, lanes 1–6) or TMX1 (lanes 7–12) were revealed with HA- or TMX1-specific antibodies.
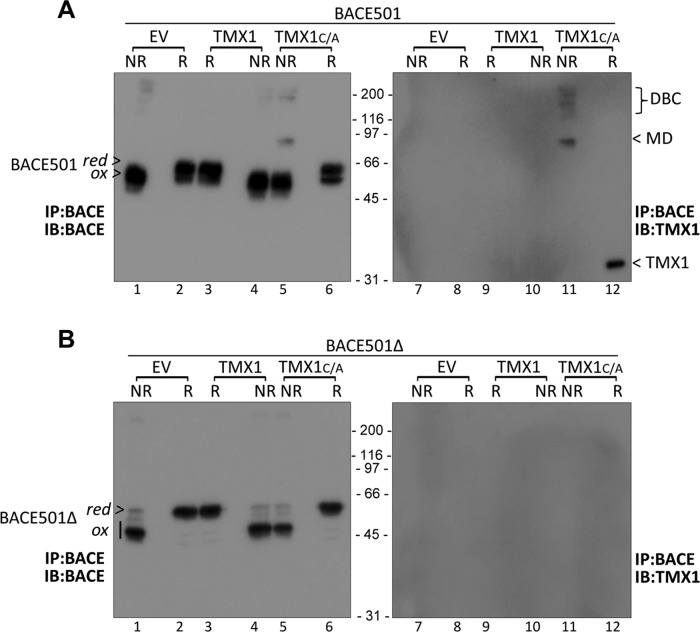
FIGURE 5:
Characterization of TMX1C/A-BACE501 mixed disulfides by WB. (A) MEFs were cotransfected with BACE501 and an empty vector (EV, lanes 1 and 2), TMX1 (3 and 4), or TMX1C/A (5 and 6). BACE501 was immunoisolated from cell lysates and immunocomplexes were analyzed under nonreducing (NR) or reducing (R) conditions. (B) Same as A, for BACE501Δ. red, reduced BACE; ox, oxidized BACE; DBC, disulfide-bonded complexes; MD, mixed disulfides.
Confirming the data obtained with radiolabeled proteins (Figure 3), the MD with the apparent MW of 90 kDa was only seen upon separation under nonreducing conditions of immunoisolates of cells cotransfected with BACE501 and TMX1C/A (Figure 5A, lane 5 for the BACE501 component of the MD and lane 11 for the TMX1 component). The undetectable level of immunoreactivity in cells expressing the wild-type form of TMX1 (Figure 5A, lanes 3/4 and 9/10) confirms that TMX1C/A stabilizes the otherwise short-lived reaction intermediate. Disassembly upon sample reduction revealed BACE501 and TMX1 as the constituents of the mixed disulfides (Figure 5A, lanes 6 and 12, respectively). The absence of mixed disulfides when the same experiment was performed with the soluble BACE501Δ variant further supported the selectivity of TMX1 for membrane-bound substrates (Figure 5B).
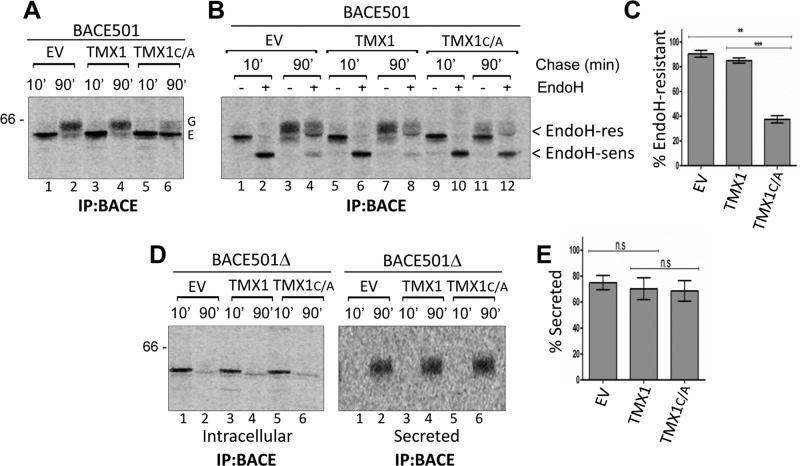
TMX1 and BACE501 maturation
Mixed disulfides are short-lived intermediates formed during the productive interaction between an oxidoreductase and its substrates (Huppa and Ploegh, 1998; Molinari and Helenius, 1999). Their stabilization upon mutation of the resolving cysteine residue in the oxidoreductase’s catalytic site is expected to delay substrate release from the ER. As for all glycoproteins, release of BACE501 from the ER can be monitored by the modification of protein-bound oligosaccharides that occurs during transit in the Golgi compartment, which reduces the electrophoretic mobility of the polypeptide chain and confers resistance to EndoH cleavage (Rothman et al., 1984). To determine whether the coexpression of the TMX1 trapping mutant delays the attainment of the BACE501’s EndoH-resistant status, MEFs expressing only BACE501, BACE501 and TMX1, or BACE501 and TMX1C/A were pulse labeled and chased for 10 (Figure 6A, lanes 1, 3, and 5) or 90 min (lanes 2, 4, and 6). After 10 min of chase, radiolabeled BACE501 expressed alone (Figure 6B, lanes 1 and 2) or coexpressed with TMX1 (lanes 5 and 6), or with TMX1C/A (lanes 9 and 10) is sensitive to EndoH cleavage. This is consistent with the ER localization of the newly synthesized polypeptide (Solda et al., 2007). The analysis after 90 min of chase showed that when expressed alone or with the wild- type form of TMX1, >85% of radiolabeled BACE501 displayed EndoH-resistant oligosaccharides (Figure 6, B, lanes 3/4 and 7/8, respectively, and C). This is consistent with the efficient export to the Golgi of this protein (Solda et al., 2007). The stabilization of the TMX1:BACE501 mixed disulfides upon coexpression of TMX1C/A dramatically reduced the fraction of EndoH-resistant BACE501 to <40% of the radiolabeled protein (Figure 6, B, lanes 11and 12, and C). In contrast, and consistent with the selectivity of TMX1 for membrane-bound polypeptides (Figures 2–4), the secretion of BACE501Δ (Figure 6, D, right, and E) was unaffected by coexpression of TMX1C/A.
FIGURE 6:
Coexpression of TMX1C/A selectively delays BACE501 maturation. (A) MEFs were cotransfected with BACE501 and an empty vector (EV, lanes 1 and 2), TMX1 (3 and 4), or TMX1C/A (5 and 6). Transfected cells were pulsed with [35S]methionine/cysteine for 13 min and chased for 10 or 90 min. Ectopically expressed BACE501 was immunoisolated from cell lysates with an anti-BACE antibody. (B) MEFs were cotransfected with BACE501 and an empty vector (lanes 1–4), TMX1 (5–8), or TMX1C/A (9–12). The maturation of immunoisolated radiolabeled BACE501 (i.e., the attainment of EndoH-resistant oligosaccharides upon arrival in the Golgi complex) was monitored after 10- and 90-min chases. (C) Quantification of the EndoH-resistant fraction of BACE501 after 90-min chase. Error bars show SDs of four independent experiments. (D) MEFs were cotransfected with BACE501Δ and an empty vector (EV, lanes 1 and 2), TMX1 (3 and 4), or TMX1C/A (5 and 6). Left, disappearance of immunoisolated BACE501Δ from the cell lysates (Intracellular), which correlates with its secretion in the extracellular media (right, Secreted). (E) Quantification of secreted BACE501Δ after 90-min chase. The error bars show SD of three independent experiments. Significance analyzed by paired t test; n.s., not significant; **p < 0.01; ***p < 0.001. G, mature Golgi form of BACE501; E, immature ER form.
It is of interest that only the coexpression of the TMX1 trapping mutant substantially reduced attainment of EndoH-resistant oligosaccharide as a measure of delayed BACE501 secretion (Figures 6 and 7, A, lanes 3 and 4, and B). ERdj5C/A did associate with BACE501 but only marginally delayed secretion (by 10–15%; Figure 7, A, lanes 5 and 6, and B). BACE501 coexpression with ERp57C/A (Figure 7, A, lanes 7 and 8, and B), ERp72C/A (Figure 7, A, lanes 9 and 10, and B), PDIC/A (Figure 7, A, lanes 11 and 12, and B) or P5C/A (Figure 7, A, lanes 13 and 14, and B; Ellgaard and Ruddock, 2005; Jessop et al., 2009; Rutkevich et al., 2010; Oka et al., 2013) had no consequences. Of note, and in contrast with TMX1 (Figure 6, D and E), ERdj5 also associated with BACE501Δ, thereby weakly reducing (by 20%) the secretion of this soluble model protein (Supplemental Figures S2, A, lanes 5/6 and 11/12, and B, and S3).
FIGURE 7:
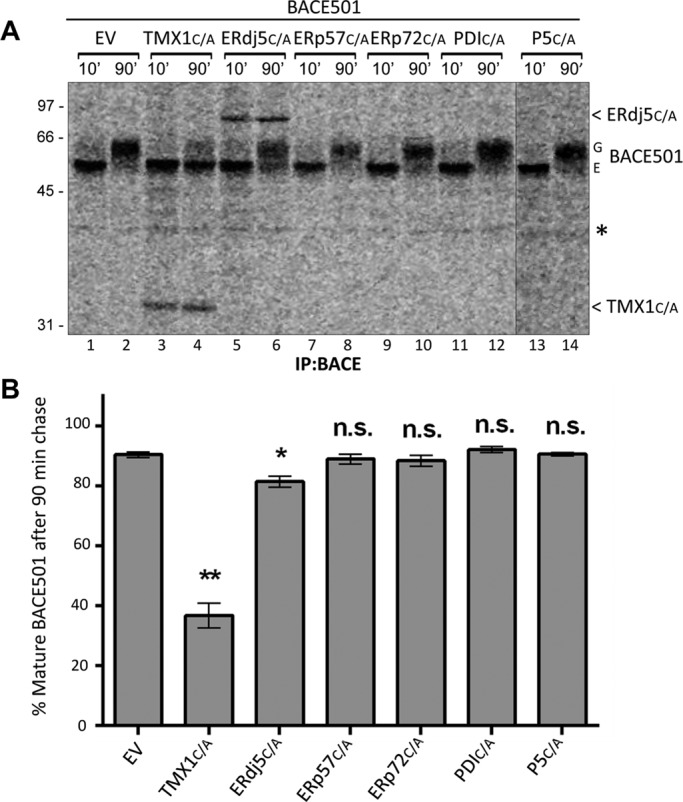
Trapping mutants of several PDI members and BACE501 maturation. (A) MEFs were cotransfected with BACE501 in combination with an empty vector (EV, lanes 1 and 2), TMX1C/A (3 and 4), ERdj5C/A (5 and 6), ERp57C/A (7 and 8), ERp72C/A (9 and 10), PDIC/A (11 and 12), or P5C/A (13 and 14). Ectopically expressed BACE501 was immunoisolated from cell lysates with an anti-BACE antibody. The maturation of radiolabeled BACE501 was monitored as in Figure 6B. (B) Quantification of the EndoH-resistant fraction of BACE501 after 90-min chase. Error bars show SDs of three independent experiments. Significance analyzed by paired t test; n.s., not significant; *p < 0.05; **p < 0.01. G, mature Golgi form of BACE501; E, immature ER form.
DISCUSSION
The mammalian ER contains 23 members of the PDI superfamily, marked by the presence of one or more thioredoxin-like domains (Ellgaard and Ruddock, 2005; Kozlov et al., 2010b; Galligan and Petersen, 2012; Tannous et al., 2015). Apart from this common feature, PDIs display different active-site compositions, enzymatic properties, domain arrangement, interacting partners, and subcompartmental and tissue distribution (Bulleid, 2012; Galligan and Petersen, 2012). This high degree of divergence encourages the study of the individual PDI proteins because it suggests that they might have peculiar substrate specificity and/or participate in distinct oxidative, reductive, or isomerase pathways. Although informative, studies with purified enzymes fail to recapitulate the complex environment of the ER, where molecular crowding, participation in supramolecular complexes, and variable redox conditions in different compartmental microdomains may substantially affect the action of individual PDIs. Studies performed in living cells, where substrate specificity has been determined by using trapping mutants that substantially retard resolution of the mixed disulfide formed as intermediate during the folding or unfolding process, reveal a certain degree of redundancy (i.e., different PDIs may engage the same substrate in mixed disulfides; Bulleid, 2012). The high redundancy of the PDI system is also highlighted by the hardly detectable phenotypes in cell lines derived from knockout mice where surrogate PDIs can functionally replace PDIs that have been deleted (e.g., ERp72 efficiently replaces ERp57 in assisting maturation of model glycoproteins (Solda et al., 2006), and other examples have been reported (Appenzeller-Herzog and Ellgaard, 2008; Kang et al., 2009; Zhang et al., 2009; Rutkevich et al., 2010).
The studies performed in living cells, however, also underscore the preference of PDIs for specific substrates or specific classes of substrates (e.g., ERp57 for glycoproteins entering the CNX/CRT cycle or ERp18 for proteins forming interchain disulfides; Oliver et al., 1997; Zapun et al., 1998; Molinari and Helenius, 1999; Frickel et al., 2002; Pollock et al., 2004; Solda et al., 2006; Jessop et al., 2007, 2009). Our finding that the membrane-bound member of the PDI family, TMX1, shows selectivity for transmembrane polypeptides and virtually ignores the same cysteine- containing ectodomains when not tethered at the ER membrane, regardless of their capacity to eventually attain the native structure, represents, to our knowledge, the first example of topology-determined substrate selection of a PDI family member. The proline residue at position 2 of the catalytic site, the capacity to reduce insulin disulfides in vitro, and the role in translocation of catalytic toxin subunits across the ER membrane (Matsuo et al., 2001; Hatahet and Ruddock, 2009; Pasetto et al., 2012) support a role of TMX1 as an ER reductase and imply possible involvement of TMX1 in ERAD, where reduction of intermolecular and intramolecular disulfide bonds is a crucial step for misfolded protein retrotranslocation across the ER membrane (Hebert et al., 2010).
Of note, it was shown that access of folding polypeptides to the CNX chaperone system leads to assembly/stabilization of functional complexes between CNX and the oxidoreductase ERp57 (Frickel et al., 2002; Ellgaard and Frickel, 2003; Jessop et al., 2007) or between CNX and the peptidyl-prolyl isomerase CypB (Kozlov et al., 2010a). Here we show that inhibition of endogenous or ectopic transmembrane protein association with CNX leads to disassembly/destabilization of functional complexes between CNX and TMX1 (Figure 4B, bottom, lane 3 vs. lane 4 for endogenous proteins and lane 7 vs. lane 8 for ectopic BACE501). On the other hand, sequestration of substrates in mixed disulfides with the trapping mutant version of TMX1 stabilizes the complexes between CNX and substrates (Figure 4B, middle, lane 5 vs. lane 7). The fact that TMX1 is not glycosylated supports the conclusion that the membrane-bound oxidoreductase TMX1 participates in functional complexes with the membrane-bound lectin CNX. Having PDI members with different topologies possibly determining their substrate selection is reminiscent of the lectin chaperone system, in which the membrane-bound CNX and the soluble CRT show different substrate specificity to broaden the range of newly synthesized gene products that can be assisted early after synthesis in the ER (Wada et al., 1995; Hebert et al., 1997; Danilczyk et al., 2000; Molinari et al., 2004). Of note, topology-determined preferences have also been reported for the supramolecular complexes that regulate delivery at and dislocation across the ER membrane of misfolded polypeptides to be degraded into the cytosol, where soluble misfolded polypeptides show strict requirement for members of the HRD1 complex (Bernasconi et al., 2010; Ninagawa et al., 2011). Thus accumulating evidence shows that substrate topology is a key factor in determining engagement of folding, quality control, and degradation pathways to ensure production of the cellular proteome in appropriate quantity and quality.
MATERIALS AND METHODS
Antibodies, expression plasmids, and inhibitors
Antibodies to TMX1 and HA were from Sigma-Aldrich (Buchs, Switzerland) and antibody to V5 from Invitrogen (Lucerne, Switzerland). The rabbit polyclonal antisera 855 and 809 (used to recognize membrane-bound and soluble BACE variants, respectively) were kind gifts of P. Paganetti (Neurocentro Svizzera italiana, Taverne, Switzerland). Genes encoding the HA-tagged TMX1 and TMX1C/A were subcloned in pCDNA3. The β1-tagged TMX1 and TMX1C/A variants were created by replacing the HA tag with an EFRH epitope, which is recognized by a monoclonal β1 antibody (Paganetti et al., 2005). Plasmids encoding the membrane-bound and soluble BACE are described in Molinari et al. (2002). V5-tagged ERdj5C/A-, ERp57C/A-, ERp72C/A-, PDIC/A-, and P5C/A-expressing vectors are described in Jessop et al. (2009) and Oka et al. (2013). Tunicamycin, cycloheximide, and CST (Sigma-Aldrich) were used at final concentrations of 5 μg/ml, 50 μg/ml, and 1 mM, respectively.
Cell lines and transient transfection
MEFs were cultured in DMEM supplemented with 10% fetal bovine serum. Cells grown on 3.5/6 cm culture dishes were transfected with 3 μg/6 μg of total plasmid DNA, using the jetPrime reagent (Polyplus transfection). Experiments were performed 17 h after transfection.
Cell lysis and Western blots
Cells were washed with phosphate-buffered saline (PBS) containing 20 mM N-ethylmaleimide (NEM) for 1 min and then lysed with 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS; Anatrace) in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–buffered saline, pH 6.8, supplemented with 20 mM NEM and protease inhibitors for 20 min on ice. Postnuclear supernatants (PNSs) were collected by centrifugation at 10,000 × g for 10 min. Samples were denatured and reduced in dithiothreitol (DTT)-containing sample buffer for 10 min at 65°C and separated by SDS–PAGE. Proteins were transferred to PVDF membranes with the Trans-Blot Turbo Transfer System (Bio-Rad, Cressier, Switzerland). Membranes were blocked with 10% (wt/vol) nonfat dry milk (Bio-Rad) and stained with the aforementioned primary antibodies and horseradish peroxidase–conjugated secondary antibodies. Membranes were developed using the Luminata Forte ECL detection system (Millipore, Schaffhausen, Switzerland), signals were detected with the ImageQuant LAS 4000 system in the standard acquisition mode (GE Healthcare Life Sciences, Glattbrugg, Switzerland), and bands were quantified using the Multi Gauge Analysis tool (Fujifilm). For each antigen, the linearity of the detected signal range was ensured with appropriate loading controls.
Metabolic labeling, immunoprecipitations, and EndoH treatment
Cells were pulse labeled with 0.1 mCi of [35S]-methionine/cysteine and chased in DMEM supplemented with 5 mM unlabeled methionine and cysteine. Cells were lysed as described, and PNS and extracellular medium were collected by centrifugation at 10,000 × g for 10 min and precleared with protein A beads (Sigma-Aldrich; 1:10 [wt/vol] swollen in PBS) for 1 h at 4°C. Immunoprecipitation was performed with protein A beads and specific antibody overnight at 4°C. After extensive washing of the immunoprecipitates with 0.5% CHAPS, beads were resuspended in sample buffer without (nonreducing conditions) or with DTT (reducing conditions) and denatured for 10 min at 65°C. Samples were subjected to SDS–PAGE. After exposure of the gels to autoradiography films (GE Healthcare, Fuji), films were scanned with the Typhoon FLA 9500 software, version 1.0. Bands were quantified using ImageQuant software (Molecular Dynamics, GE Healthcare). For EndoH (New England Biolabs, Allschwil, Switzerland) treatment, immunoisolated proteins were split into two aliquots and incubated in the presence or absence of 5 mU of EndoH for 2h at 37°C. Samples were then analyzed by reducing SDS–PAGE.
Mass spectrometry
Confluent MEFs transfected with an empty vector or transfected with HA-tagged TMX1C/A were rinsed with PBS and 20 mM NEM. Cells were lysed with 2% CHAPS (Anatrace) in HEPES-buffered saline, pH 6.8, supplemented with 20 mM NEM and protease inhibitors for 20 min on ice. Immunoisolates were washed three times with lysis buffer. Mass spectrometry analysis was performed at the Protein Analysis Facility, University of Lausanne, Lausanne, Switzerland.
Supplementary Material
Acknowledgments
We acknowledge P. Paganetti and M. Quadroni for discussions and the analysis shown in Table 1. M.M. is supported by Signora Alessandra, the Foundation for Research on Neurodegenerative Diseases, the Swiss National Science Foundation, and the Comel and Gelu Foundations. N.J.B. is supported by Wellcome Trust Grant 88053. L.W.R. is supported by Biocenter Oulu.
Abbreviations used:
- CNX
calnexin
- CRT
calreticulin
- DBC
disulfide-bonded complexes
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- MD
mixed disulfide
- PDI
protein disulfide isomerase
- Trx
thioredoxin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0321) on August 5, 2015.
REFERENCES
- Alam S, Li Z, Janciauskiene S, Mahadeva R. Oxidation of Z alpha1-antitrypsin by cigarette smoke induces polymerization: a novel mechanism of early-onset emphysema. Am J Respir Cell Mol Biol. 2011;45:261–269. doi: 10.1165/rcmb.2010-0328OC. [DOI] [PubMed] [Google Scholar]
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–844. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Solda T, Galli C, Pertel T, Luban J, Molinari M. Cyclosporine A-sensitive, cyclophilin B-dependent endoplasmic reticulum-associated degradation. PLoS One. 2010;5:e13008. doi: 10.1371/journal.pone.0013008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- Bulleid NJ. Disulfide bond formation in the mammalian endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2012;4:a013219. doi: 10.1101/cshperspect.a013219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichiarelli S, Ferraro A, Altieri F, Eufemi M, Coppari S, Grillo C, Arcangeli V, Turano C. The stress protein ERp57/GRP58 binds specific DNA sequences in HeLa cells. J Cell Physiol. 2007;210:343–351. doi: 10.1002/jcp.20824. [DOI] [PubMed] [Google Scholar]
- Cunnea PM, Miranda-Vizuete A, Bertoli G, Simmen T, Damdimopoulos AE, Hermann S, Leinonen S, Huikko MP, Gustafsson JA, Sitia R, Spyrou G. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J Biol Chem. 2003;278:1059–1066. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- Danilczyk UG, Cohen-Doyle MF, Williams DB. Functional relationship between calreticulin, calnexin, and the endoplasmic reticulum luminal domain of calnexin. J Biol Chem. 2000;275:13089–13097. doi: 10.1074/jbc.275.17.13089. [DOI] [PubMed] [Google Scholar]
- Dick TP, Cresswell P. Thiol oxidation and reduction in major histocompatibility complex class I-restricted antigen processing and presentation. Methods Enzymol. 2002;348:49–54. doi: 10.1016/s0076-6879(02)48625-9. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Frickel EM. Calnexin, calreticulin, and ERp57: teammates in glycoprotein folding. Cell Biochem Biophys. 2003;39:223–247. doi: 10.1385/CBB:39:3:223. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster ML, Sivick K, Park YN, Arvan P, Lencer WI, Tsai B. Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J Cell Biol. 2006;173:853–859. doi: 10.1083/jcb.200602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci USA. 2002;99:1954–1959. doi: 10.1073/pnas.042699099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics. 2012;6:6. doi: 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser CB, Karic L, Huffaker T, Chang R, Martin J. Studies on the disulfide region of alpha 1-protease inhibitor. Int J Peptide Protein Res. 1982;20:56–62. doi: 10.1111/j.1399-3011.1982.tb02652.x. [DOI] [PubMed] [Google Scholar]
- Grek CL, Townsend DM, Uys JD, Manevich Y, Coker WJ, 3rd, Pazoles CJ, Tew KD. S-glutathionylated serine proteinase inhibitors as plasma biomarkers in assessing response to redox-modulating drugs. Cancer Res. 2012;72:2383–2393. doi: 10.1158/0008-5472.CAN-11-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths SW, King J, Cooney CL. The reactivity and oxidation pathway of cysteine 232 in recombinant human alpha 1-antitrypsin. J Biol Chem. 2002;277:25486–25492. doi: 10.1074/jbc.M203089200. [DOI] [PubMed] [Google Scholar]
- Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- Haugstetter J, Blicher T, Ellgaard L. Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J Biol Chem. 2005;280:8371–8380. doi: 10.1074/jbc.M413924200. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M. ERAD substrates: which way out. Semin Cell Dev Biol. 2010;21:526–532. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The eS-Sence of -SH in the ER. Cell. 1998;92:145–148. doi: 10.1016/s0092-8674(00)80907-1. [DOI] [PubMed] [Google Scholar]
- Jessop CE, Chakravarthi S, Garbi N, Hammerling GJ, Lovell S, Bulleid NJ. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Park B, Oh C, Cho K, Ahn K. A role for protein disulfide isomerase in the early folding and assembly of MHC class I molecules. Antioxid Redox Signal. 2009;11:2553–2561. doi: 10.1089/ars.2009.2465. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M, Levitin F, van den Beucken T, Rumantir RA, Harding NJ, Chu KC, Boutros PC, Braakman I, Wouters BG. Two phases of disulfide bond formation have differing requirements for oxygen. J Cell Biol. 2013;203:615–627. doi: 10.1083/jcb.201307185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Bastos-Aristizabal S, Maattanen P, Rosenauer A, Zheng F, Killikelly A, Trempe JF, Thomas DY, Gehring K. Structural basis of cyclophilin B binding by the calnexin/calreticulin P-domain. J Biol Chem. 2010a;285:35551–35557. doi: 10.1074/jbc.M110.160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Maattanen P, Thomas DY, Gehring K. A structural overview of the PDI family of proteins. FEBS J. 2010b;277:3924–3936. doi: 10.1111/j.1742-4658.2010.07793.x. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoul I, Ferrari D, Soling HD. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 1997;401:104–108. doi: 10.1016/s0014-5793(96)01447-0. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Akiyama N, Nakamura H, Yodoi J, Noda M, Kizaka-Kondoh S. Identification of a novel thioredoxin-related transmembrane protein. J Biol Chem. 2001;276:10032–10038. doi: 10.1074/jbc.M011037200. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Irie K, Kiyonari H, Okuyama H, Nakamura H, Son A, Lopez-Ramos DA, Tian H, Oka S, Okawa K, Kizaka-Kondoh S, et al. The protective role of the transmembrane thioredoxin-related protein TMX in inflammatory liver injury. Antioxid Redox Signal. 2013;18:1263–1272. doi: 10.1089/ars.2011.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Masutani H, Son A, Kizaka-Kondoh S, Yodoi J. Physical and functional interaction of transmembrane thioredoxin-related protein with major histocompatibility complex class I heavy chain: redox-based protein quality control and its potential relevance to immune responses. Mol Biol Cell. 2009;20:4552–4562. doi: 10.1091/mbc.E09-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Akaike T, Maeda H. S-nitrosylated human alpha(1)-protease inhibitor. Biochim Biophys Acta. 2000;1477:90–97. doi: 10.1016/s0167-4838(99)00264-2. [DOI] [PubMed] [Google Scholar]
- Molinari M, Eriksson KK, Calanca V, Galli C, Cresswell P, Michalak M, Helenius A. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell. 2004;13:125–135. doi: 10.1016/s1097-2765(03)00494-5. [DOI] [PubMed] [Google Scholar]
- Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Helenius A. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature. 1999;402:90–93. doi: 10.1038/47062. [DOI] [PubMed] [Google Scholar]
- Ninagawa S, Okada T, Takeda S, Mori K. SEL1L is required for endoplasmic reticulum-associated degradation of misfolded luminal proteins but not transmembrane proteins in chicken DT40 cell line. Cell Struct Funct. 2011;36:187–195. doi: 10.1247/csf.11018. [DOI] [PubMed] [Google Scholar]
- Oka OB, Bulleid NJ. Forming disulfides in the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2425–2429. doi: 10.1016/j.bbamcr.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Oka OB, Pringle MA, Schopp IM, Braakman I, Bulleid NJ. ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol Cell. 2013;50:793–804. doi: 10.1016/j.molcel.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, van der Wal FJ, Bulleid NJ, High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. Beta-site specific intrabodies to decrease and prevent generation of Alzheimer’s Abeta peptide. J Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasetto M, Barison E, Castagna M, Della Cristina P, Anselmi C, Colombatti M. Reductive activation of type 2 ribosome-inactivating proteins is promoted by transmembrane thioredoxin-related protein. J Biol Chem. 2012;287:7367–7373. doi: 10.1074/jbc.M111.316828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter DH. Alpha-1-antitrypsin deficiency: importance of proteasomal and autophagic degradative pathways in disposal of liver disease-associated protein aggregates. Annu Rev Med. 2011;62:333–345. doi: 10.1146/annurev-med-042409-151920. [DOI] [PubMed] [Google Scholar]
- Pollock S, Kozlov G, Pelletier MF, Trempe JF, Jansen G, Sitnikov D, Bergeron JJ, Gehring K, Ekiel I, Thomas DY. Specific interaction of ERp57 and calnexin determined by NMR spectroscopy and an ER two-hybrid system. EMBO J. 2004;23:1020–1029. doi: 10.1038/sj.emboj.7600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Urbani LJ, Brands R. Transport of protein between cytoplasmic membranes of fused cells: correspondence to processes reconstituted in a cell-free system. J Cell Biol. 1984;99:248–259. doi: 10.1083/jcb.99.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Lee AS. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 1999;27:1437–1443. doi: 10.1093/nar/27.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkevich LA, Cohen-Doyle MF, Brockmeier U, Williams DB. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol Biol Cell. 2010;21:3093–3105. doi: 10.1091/mbc.E10-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman S, Wang B, Li W, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci USA. 2010;107:15027–15032. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda T, Galli C, Kaufman RJ, Molinari M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell. 2007;27:238–249. doi: 10.1016/j.molcel.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Solda T, Garbi N, Hammerling GJ, Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- Sugiura Y, Araki K, Iemura S, Natsume T, Hoseki J, Nagata K. Novel thioredoxin-related transmembrane protein TMX4 has reductase activity. J Biol Chem. 2010;285:7135–7142. doi: 10.1074/jbc.M109.082545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous A, Pisoni GB, Hebert DN, Molinari M. N-linked sugar-regulated protein folding and quality control in the ER. Semin Cell Dev Biol. 2015;41:79–89. doi: 10.1016/j.semcdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasanen K, Oikarinen J, Kivirikko KI, Pihlajaniemi T. Promoter of the gene for the multifunctional protein disulfide isomerase polypeptide. Functional significance of the six CCAAT boxes and other promoter elements. J Biol Chem. 1992;267:11513–11519. [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Tyagi SC, Simon SR. Role of disulfide exchange in alpha 1-protease inhibitor. Biochem. 1992;31:10584–10590. doi: 10.1021/bi00158a022. [DOI] [PubMed] [Google Scholar]
- Wada I, Imai S, Kai M, Sakane F, Kanoh H. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J Biol Chem. 1995;270:20298–20304. doi: 10.1074/jbc.270.35.20298. [DOI] [PubMed] [Google Scholar]
- Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kozlov G, Pocanschi CL, Brockmeier U, Ireland BS, Maattanen P, Howe C, Elliott T, Gehring K, Williams DB. ERp57 does not require interactions with calnexin and calreticulin to promote assembly of class I histocompatibility molecules, and it enhances peptide loading independently of its redox activity. J Biol Chem. 2009;284:10160–10173. doi: 10.1074/jbc.M808356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.