Background: LHCSR protein in algae and mosses is essential for NPQ.
Results: Expression and characterization of Physcomitrella patens LHCSR1 protein upon heterologous expression in N. benthamiana and N. tabacum was obtained.
Conclusion: LHCSR1 is the first member of LHC protein family lacking Chlorophyll b. It is active in NPQ.
Significance: LHCSR1 isolation is crucial for the elucidation of the NPQ mechanism.
Keywords: light-harvesting complex (antenna complex), photobiology, photosynthesis, photosynthetic pigment, recombinant protein expression, LHCSR, NPQ, Physcomitrella, tobacco
Abstract
Oxygenic photosynthetic organisms evolved mechanisms for thermal dissipation of energy absorbed in excess to prevent formation of reactive oxygen species. The major and fastest component, called non-photochemical quenching, occurs within the photosystem II antenna system by the action of two essential light-harvesting complex (LHC)-like proteins, photosystem II subunit S (PSBS) in plants and light-harvesting complex stress-related (LHCSR) in green algae and diatoms. In the evolutionary intermediate Physcomitrella patens, a moss, both gene products are active. These proteins, which are present in low amounts, are difficult to purify, preventing structural and functional analysis. Here, we report on the overexpression of the LHCSR1 protein from P. patens in the heterologous systems Nicotiana benthamiana and Nicotiana tabacum using transient and stable nuclear transformation. We show that the protein accumulated in both heterologous systems is in its mature form, localizes in the chloroplast thylakoid membranes, and is correctly folded with chlorophyll a and xanthophylls but without chlorophyll b, an essential chromophore for plants and algal LHC proteins. Finally, we show that recombinant LHCSR1 is active in quenching in vivo, implying that the recombinant protein obtained is a good material for future structural and functional studies.
Introduction
Photosynthesis depends on light; however, excess light can be harmful for the photosynthetic apparatus when its intensity exceeds the activity of the photosynthetic electron transport chain. Indeed, light in excess with respect to the photochemical quenching activity increases the lifetime of chlorophyll singlet excited states (1Chl*),2 thus increasing the probability of triplet chlorophyll (3Chl*) formation. Long living triplets react with O2 to yield singlet oxygen (1O2), leading to photodamage and photoinhibition (1–3). Plants developed multiple photoprotection mechanisms against photooxidation. A major mechanism for response to changes in light availability is non-photochemical quenching (NPQ), which is activated upon exposure to excess light and leads to the dissipation of excess energy absorbed as heat (4, 5). NPQ includes several components whose relative importance for photoprotection depends on species and physiological state. The energy quenching (qE) component is reversibly activated within seconds upon an increase in light intensity and has the highest potential for quenching. Slowly relaxing components are included under the definition of “inhibitory quenching” (qI) and are activated within several minutes of excess irradiance and relax within 1–2 h in the dark (5–7). qI includes qZ, the result of binding of zeaxanthin to light-harvesting complex (LHC) antenna proteins, which decreases their fluorescence yield. Zeaxanthin is produced in excess light from pre-existing violaxanthin and is photoprotective (8). Finally, the actual quenching from photoinhibition (9), which is relaxed upon repair of photodamaged photosystem II (PSII) (10). An intermediate kinetic component, qM, which relaxes in 8–20 min, has been attributed to the chloroplast avoidance relocation in the cell, implying that this fluorescence decay is not dependent on quenching (11). Among quenching mechanisms, the most studied and yet obscure is qE whose mechanistic details are still a matter of intense debate some 50 years after its first report (12). Forward genetic analysis showed that qE requires one of two lhc-like gene products, PSBS and LHCSR, respectively, in plants and algae. These are essential for sensing pH of the thylakoid lumen (13, 14) and transduction into quenching events. Although PSBS is not a typical pigment-binding protein (15, 16), making it unlikely as a site for quenching reactions that have been reported to occur in interacting LHC proteins (17–19), LHCSR does bind Chl and xanthophylls (20). In Physcomitrella patens, LHCSR is responsible for most of the quenching activity (21), which is strongly enhanced by zeaxanthin binding (22), making LHCSR the ideal system for structure-function studies aimed at elucidating the properties of this molecular switch that regulates photon use efficiency in unicellular algae and mosses resuming the two functions of pH detection and quenching of the chlorophyll excited states (14, 20). However, the level of LHCSR is low in thylakoids, and its physicochemical properties are similar to the highly abundant LHCs, hampering purification so far. Heterologous expression systems have been exploited to obtain a recombinant protein for refolding in vitro with pigments in a second stage (20, 23), but this method is expensive and did not allow successful crystallization.3 With the aim of scaling up production of this gene product, we attempted heterologous expression in two Nicotiana species, which proved to be a useful system, yielding LHCSR as a pigment protein whose properties are consistent with previous reports. Also, we show that LHCSR is active in quenching in vivo, suggesting that the recombinant product obtained closely features the protein responsible for NPQ in mosses, therefore providing a solid basis for future structural and functional studies.
Experimental Procedures
Construction of Plant Expression Vectors
cDNA obtained from P. patens protonema grown in control conditions was used as starting material. Coding sequence for LHCSR1 (XM_001776900.1) with and without the His tag was amplified by PCR and used as a template to construct the pENTRTM/D-TOPO-based vectors. The following primers were used: forward primer, 5′-CACCCTCGCTCTGCAACTTTCCTTT-3′ for both the constructs; reverse primer for LHCSR1 without His tag, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGACTGCGAATCAATCAGAA-3′; and reverse primer for LHCSR1 His tag, 5′-TCAGTGGTGGTGATGATGATGGCTGCCGCGCGGCACCAGCAGGCCCAATCTCTTGAACAA-3′. The pENTR.LHCSR1 and pENTR.LHCSR1 His tag vectors obtained were used for LR recombination reactions with the GATEWAY destination vector pK7WG2 (24), generating the final vectors pK7WG2.LHCSR1 and pK7WG2.LHCSR1 His tag. The two vectors were transformed in Agrobacterium tumefaciens EHA105 strain.
Nicotiana tabacum Leaf Transformation
Tobacco (N. tabacum cv. Petit Havana SR1) leaf disks were transformed with pK7WG2-based constructs as described (25). Kanamycin-resistant transgenic lines were selected and tested by immunoblot analysis using homemade polyclonal α-PpLHCSR antibody.
Agroinfiltration of Nicotiana benthamiana Leaves
Bacterial suspension in infiltration buffer (10 mm MES, 10 mm MgCl2, 100 μm acetosyringone, pH 5.6) at a final A600 of 0.8 was used for syringe infiltration of 5–6-week-old N. benthamiana plants. Five leaves were infiltrated for each plant and for each construct. The recombinant protein time course accumulation of P. patens LHCSR1 (PpLHCSR1) protein was estimated by immunoblotting total protein extract starting from a disk of infected leaves collected for 7 days after agroinfiltration and ground in loading buffer (62.5 mm Tris, pH 6.8, 10% glycerol, 2% SDS, and 3% β-mercaptoethanol).
Thylakoid Extraction and Pigment-binding Complex Purification
Thylakoids were purified from N. tabacum and N. benthamiana leaves following a protocol for seed plants (26).
For separation of pigment-binding complex, 500 μg Chl of thylakoid membranes were washed with 5 mm EDTA and resuspended at a final concentration of 1 mg/mlChl in 10 mm Hepes, pH 7.5. Samples were then solubilized at a final concentration of 0.5 mg/ml Chl, adding 1.6% n-dodecyl α-d-maltopyranoside (α-DM) and 10 mm Hepes, pH 7.5 and vortexing for 1 min. After 10 min of incubation on ice, thylakoid membranes were centrifuged at 15,000 × g for 10 min to eliminate unsolubilized material. Fractionation occurred upon ultracentrifugation on a 0.1–1 m sucrose gradient containing 0.03% α-DM and 10 mm Hepes, pH 7.5 (22 h at 280,000 × g at 4 °C).
Deriphat-PAGE
Non-denaturing Deriphat-PAGE was performed according to (27) with minor modifications. The stacking and resolving gels contained, respectively, 3.5 (w/v) and 7% (w/v) acrylamide (38:1 acrylamide/bisacrylamide). Thylakoids concentrated at 1 mg/ml Chl were solubilized with 0.8% α-DM. 500 μg of Chl were loaded in each wide well for comparing LHCSR1-expressing (oeLHCSR1) plants with the corresponding wild type (WT).
Nickel Affinity Chromatography
30 mg of Chl of thylakoid membranes from oeLHCSR1 with histidine tail-expressing (oeLHCSR1 His tag) plants were washed with 5 mm EDTA and resuspended at a final concentration of 1 mg/ml in 10 mm Hepes, pH 7.5. Samples were then solubilized with 1% α-DM and centrifuged at 15,000 × g for 10 min to eliminate unsolubilized material. The supernatant was loaded onto a nickel affinity column equilibrated with the same buffer, and the resin was washed with 2–3 column volumes of washing buffer (10 mm Hepes, pH 7.5, 0.15 m NaCl, 10 mm imidazole, 0.03% α-DM). Further step elutions were performed by increasing the imidazole concentration (25, 50, and 250 mm).
Pigment Composition
The pigment composition of the complexes was analyzed by fitting the spectrum of the 80% acetone-extracted pigments with the spectra of the individual pigments in acetone and by HPLC as described previously (28, 29).
Absorption Spectroscopy
Samples collected from the sucrose gradients were analyzed by absorption spectroscopy using an SLM-Aminco DW-2000 (Aminco) spectrophotometer at room temperature. Spectra were also collected for native Chl-binding complexes after extraction from the acrylamide gel by grinding the slices in a buffer containing 10 mm Hepes, pH 7.5, 0.03% α-DM.
SDS-PAGE and Immunoblotting Analysis
SDS-PAGE analyses were performed as described (30). An acrylamide/bisacrylamide ratio of 75:1 and a total concentration of acrylamide of 4.5 and 15% were used, respectively, for stacking and running gels. Urea (6 m) was also added into the running gel. Following SDS-PAGE, polypeptides were transferred onto an Immobilon polyvinylidene difluoride (PVDF) membrane (Millipore) using a Mini Trans-Blot cell (Bio-Rad) and detected by homemade polyclonal antibodies.
NPQ Measurements
In vivo Chl fluorescence of N. tabacum leaf disks of WT and oeLHSCR1 plants was measured at room temperature on a FluorCam imaging fluorometer (Photon Systems Instruments) with saturating light at 4000 μmol m−2 s−1 and actinic light at 1200 μmol m−2 s−1. Before measurements, plants were dark-adapted for 40 min at room temperature. Parameters Fv/Fm and NPQ were calculated as (Fm − Fo)/Fm and (Fm − Fm′)/Fm′ (31). Data are presented as means ± S.D. of at least three independent experiments.
Results
Expression of P. patens LHCSR1 in N. benthamiana and N. tabacum Plants
The full-length ORFs of LHCSR1 with or without thrombin-cleavable histidine tail (His tag) at the C terminus of the protein were introduced by LR recombination into the binary GATEWAY vector pK7WG2 under the control of the constitutive cauliflower mosaic virus 35S promoter. The resulting vectors, named pK7WG2.LHCSR1 and pK7WG2.LHCSR1 His tag, were used for stable (25) and transient (32, 33) A. tumefaciens-mediated transformation of plants.
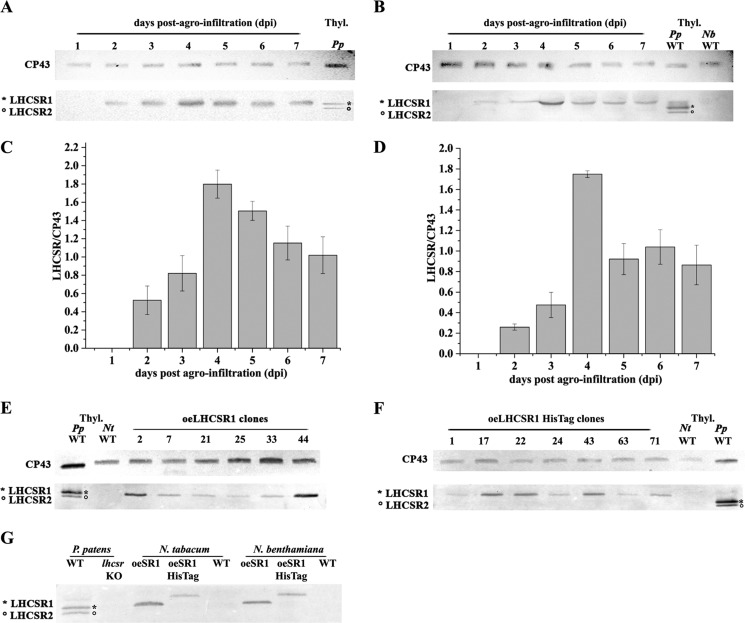
N. benthamiana was used as host for transient expression (34). Five leaves of 5–6 week-old plants were agroinfiltrated. Upon infection, a time course analysis was performed by collecting leaf disks for 7 days postagroinfiltration (dpi) to identify the peak of recombinant protein accumulation. Total proteins were analyzed by SDS-PAGE and immunoblotting with an α-PpLHCSR1 polyclonal antibody. α-CP43, a subunit of PSII core, was used as a loading control. Fig. 1 shows the time course analysis of PpLHCSR1 (Fig. 1A) and PpLHCSR1 His tag (Fig. 1B) expression in N. benthamiana. α-PpLHCSR1 antibody revealed a band at ∼23-kDa apparent molecular mass in N. benthamiana leaf disks, whereas two bands were detected in the WT P. patens sample, which contains both LHCSR1 and LHCSR2 (21). The LHCSR1 signal had the same apparent molecular weight in both N. benthamiana and P. patens, implying that the maturation of the precursor recombinant protein in N. benthamiana occurred as in the homologous moss system. In the case of oeLHCSR1 His tag N. benthamiana plants, the molecular weight of LHCSR1 is slightly higher with respect to P. patens LHCSR1. This difference is due to the presence of the thrombin-cleavable histidine tail affecting the mobility of the protein in SDS-PAGE. No reaction was observed in the untransformed WT N. benthamiana sample. In Fig. 1, C and D, the accumulation of PpLHCSR1 with and without His tag is shown. In both constructs, the protein was detectable at 2 dpi and reached a maximum at 4 dpi.
FIGURE 1.
Expression analysis of N. benthamiana and N. tabacum transformation using two different PpLHCSR1 constructs and localization of PpLHCSR1 in thylakoid membranes of tobacco. A–D, time course PpLHCSR1 accumulation trial in N. benthamiana upon transient expression. Leaf disks collected daily postagroinfiltration were ground in loading buffer, fractionated by SDS-PAGE, and transferred to a PVDF membrane. Immunoblot analyses using a homemade α-PpLHCSR antibody of oeLHCSR1 (A) or oeLHCSR1 His tag (B) are reported. The time course of LHCSR1 for oeLHCSR1 (C) or oeLHCSR1 His tag (D) is shown. Data are presented as means ± S.D. (error bars) of three independent experiments. E and F, immunoblot analysis of oeLHCSR1 (E) or oeLHCSR1 His tag (B) plants of stably transformed N. tabacum. Antibody α-PpLHCSR was used. An equal volume for each leaf disk was loaded. Immunodetection using α-CP43 antibody is shown as a control of equal loading. P. patens and N. tabacum WT thylakoids (1 μg of Chl) were loaded as positive and negative controls, respectively. Thyl., thylakoids; Nb, N. benthamiana; Nt, N. tabacum. G, immunoblot analysis of thylakoid membranes purified from WT, oeLHCSR1 (oeSR1), and oeLHCSR1 His tag (oeSR1 HisTag).
Alternatively, we used N. tabacum cv. Petit Havana SR1 for stable transformation into the nuclear genome. Transgenic plants were generated by Agrobacterium-mediated transformation of leaf disks and in vitro regeneration of plants under kanamycin selection. About 80 regenerated plants were analyzed for each construct. Fig. 1E shows the accumulation of PpLHCSR1 protein by immunoblotting analysis in oeLHCSR1 plants of N. tabacum. Similar to results with N. benthamiana, the reaction in the WT moss had the same apparent molecular weight as the recombinant PpLHCSR1. PpLHCSR1 protein from oeLHCSR1 His tag plants showed slightly slower migration than LHCSR1 from oeLHCSR1 plants due to additional sequence encoding the tag and thrombin target site (Fig. 1F). The protein accumulation levels varied significantly between independently transformed lines, likely reflecting the positional effects of random insertion (35). The protein level was used to select T1 transgenic lines (elite lines) for a breeding program to reach homozygosis of the inserted transgene(s), which was initiated by self-crossing the best performing T1 oeLHCSR1 and oeLHCSR1 His tag transgenic plants. In the following paragraphs, the results obtained with T1 plants are reported.
Subcellular Localization of LHCSR1 in Thylakoid Membranes and Aggregation State Analysis
The intracellular localization of recombinant PpLHCSR1 was analyzed in N. benthamiana and N. tabacum plants. In mosses, LHCSR1 is encoded by the nuclear genome, translated in the cytoplasm, and targeted toward the chloroplast (36). The transit peptide is then removed, and the apoprotein co-insertionally assembles in the thylakoid membranes with Chl and carotenoids (37). To study the intracellular fate of the recombinant protein in Nicotiana sp., thylakoid membranes were purified from N. benthamiana-infected leaves 4 dpi and N. tabacum transgenic plants. Fig. 1G shows the results of immunoblotting analysis on thylakoid membranes using α-PpLHCSR antibody. LHCSR1 and LHCSR1 His tag recombinant proteins were detected in thylakoids of both N. benthamiana and N. tabacum as well as in thylakoids from WT P. patens but not in WT thylakoids of N. benthamiana and N. tabacum or in the lhcsr knock-out (KO) of P. patens, implying that Nicotiana sp. machinery is competent for the targeting of PpLHCSR1 into the thylakoid compartment.
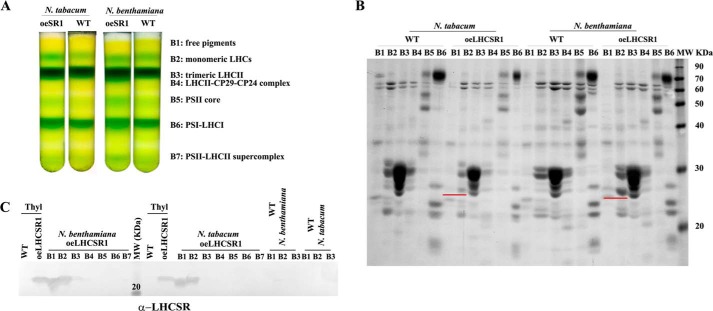
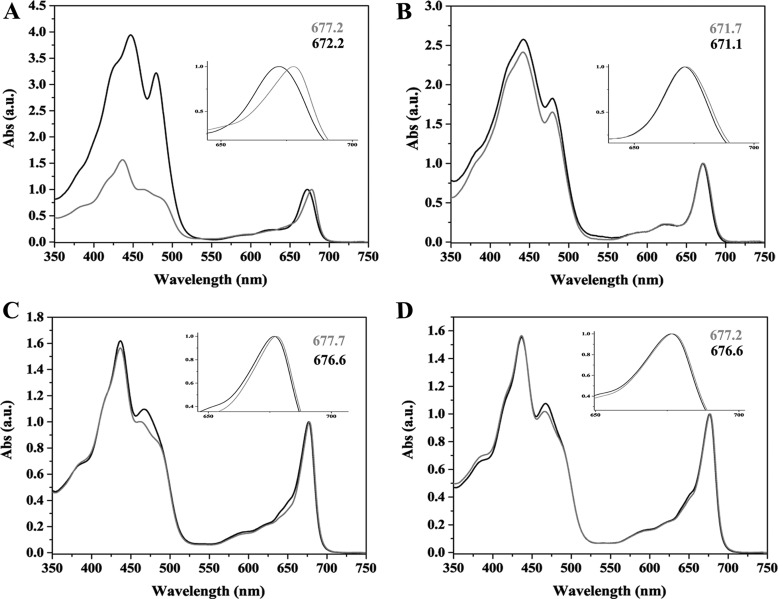
Next, we tested the aggregation state of recombinant tobacco-produced PpLHCSR1 as compared with the native P. patens LHCSR1 (22). Thylakoids were solubilized with the mild detergent α-DM and loaded onto a sucrose gradient, which was centrifuged at 280,000 × g for ∼22 h. The separation pattern, consisting of seven green bands, was similar in WT and oeLHCSR1 N. benthamiana or N. tabacum plants. Bands were collected from the gradient and analyzed by absorption spectroscopy and SDS-PAGE to identify their pigment protein content and polypeptide composition. Fig. 2A shows the result of the sucrose gradient: in the upper part of the gradient, a yellow top band contained carotenoid-enriched free pigments (band 1 (B1)), monomeric LHCs (B2), trimeric LHCII (B3), and LHCII-CP29-CP24 complex (B4). In the central part of the gradient, PSII core complex was found (B5), whereas PSI-LHCI (B6) and PSII-LHCII supercomplexes (B7) migrated in the lower part of the gradient. The separation pattern was very similar in WT versus oeLHCSR1. Absorption spectra of B1 and B2 of oeLHCSR1 showed a red-shifted Qy absorption peak when compared with the corresponding fractions from WT (Fig. 3, A–D); because a red-shifted spectrum is a typical characteristic of LHCSR1 protein (20, 22), this suggests LHCSR1 migrates in the region of B1 with monomeric LHCs. This characteristic was more evident in the case of N. benthamiana (Fig. 3A) where the Qy peak of B1 was shifted from 672.2 nm in WT to 677.7 nm in oeLHCSR1 but was less evident in N. tabacum plants (Fig. 3B). Absorption spectra of B2 also showed a small red shift of the Qy peak from 676.6 nm in WT to 677.7 nm in oeLHCSR1 of N. benthamiana but was almost absent in the case of N. tabacum (Fig. 3, C and D). No differences were found between the absorption spectra of WT and oeLHCSR1 of other bands (data not shown).
FIGURE 2.
Fractionation of WT and oeLHCSR1 solubilized thylakoids of N. benthamiana and N. tabacum. A, sucrose gradient of thylakoids from WT and oeLHCSR1 of N. benthamiana and N. tabacum plants solubilized with 0.8% α-DM. B, Coomassie Blue-stained SDS-PAGE analysis of green bands collected from the sucrose gradient. An equal volume (50 μl) of each band was loaded. A polypeptide corresponding to the molecular weight of LHCSR1, underlined in red, was clearly visible in B1 and B2 (corresponding to free pigments and monomeric LHCs, respectively). C, immunoblot analysis with α-PpLHCSR antibody. Thylakoids (Thyl) from oeLHCSR1 and WT were loaded as positive and negative controls, respectively. B1, B2, and B3 from WT thylakoids were loaded as additional negative controls.
FIGURE 3.
Absorption (Abs) spectra of fractions obtained from sucrose gradient. Spectra of B1 fraction from WT (black line) versus oeLHCSR1 (gray line) of N. benthamiana (A) and N. tabacum (B) are shown. Absorption spectra of B2 fraction from WT (black line) and oeLHCSR1 (gray line) of N. benthamiana (C) and N. tabacum (D) are shown. Peak wavelengths are indicated. a.u., arbitrary units.
Further assessment of protein distribution was performed by SDS-PAGE. For each band, an equal volume was loaded onto the gel, and the protein profile was analyzed after Coomassie Blue staining (Fig. 2B). A clear signal of ∼23 kDa was found in monomeric LHCs (B2) of N. tabacum oeLHCSR1 as well as in free pigments (B1) and monomeric LHCs (B2) of N. benthamiana oeLHCSR1 but was absent in the samples from WT. Immunoblotting analysis confirmed that the Coomassie-stained 23-kDa band was indeed LHCSR1 (Fig. 2C).
Expression and Purification of PpLHCSR1 His Tag by Affinity Chromatography
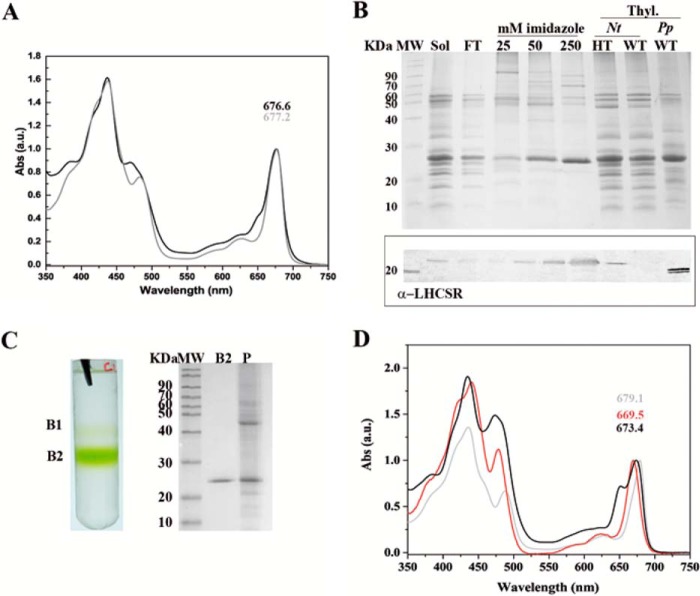
When Nicotiana sp. plants expressing the PpLHCSR1 His tag version were analyzed, a protein accumulation level far lower than that of the PpLHCSR1 version was observed on a Chl basis (see below). Nevertheless, we attempted the purification of the recombinant protein by nickel ion affinity chromatography. To this aim, thylakoid membranes purified from full grown tobacco leaves (about 30 mg of Chl) were solubilized with α-DM and loaded onto nickel-chelating Sepharose followed by step elution with 25, 50, and 250 mm imidazole, respectively. The absorption spectrum of solubilized thylakoids showed two peaks at around 470 and 650 nm, which are typical of Chl b, whereas that of the fraction eluted with 250 mm imidazole did not, implying a depletion in Chl b (Fig. 4A). SDS-PAGE analysis showed a major Coomassie-stained band at ∼25-kDa molecular mass that was recognized by antibodies directed against His tag (data not shown) and α-PpLHCSR (Fig. 4B), suggesting an enrichment in LHCSR1 with traces of high molecular weight contaminants. To eliminate these contaminants and the possible free pigments, the fraction eluted with 250 mm imidazole was concentrated to 300 μl and loaded onto a sucrose gradient, which was centrifuged in a SW60 rotor overnight at 360,000 × g, yielding the pattern shown in Fig. 4C consisting of a faint yellowish upper band (B1) and a lower green band (B2); a green pellet (P) was also observed. SDS-PAGE analysis of the fractions showed a single band (∼25 kDa) in B2 and multiple bands in P including a prominent ∼25-kDa component. The absorption spectrum of the green B2 obtained from the sucrose gradient was characterized by major peaks at 435 and 679.1 nm, respectively, in the Soret and Qy range and minor peaks at 488 and 628 nm, whereas deep troughs were detected at 470 and 650 nm, the typical absorption wavelengths for Chl b in LHC proteins (38), implying that the recombinant protein bound mainly Chl a and carotenoids and is depleted of Chl b.
FIGURE 4.
PpLHCSR1 His tag isolation and biochemical characterization. A, absorption (Abs) spectra of fraction eluted with 250 mm imidazole (gray line) after solubilization of oeLHCSR1 His tag thylakoids with α-DM. The absorption spectrum of solubilized thylakoids (black line) is also reported. a.u., arbitrary units. B, Coomassie Blue-stained SDS-PAGE gel analysis of fractions eluted with different imidazole concentrations. Samples containing 2 μg of Chl were loaded in each slot. Thylakoids (3 μg of Chl) of WT and oeLHCSR1 His tag (HT) of N. tabacum and WT of P. patens were also loaded as controls. Immunoblot analysis using antibody α-PpLHCSR is shown in the panel below the gel. Sol, solubilized thylakoids; FT, flow-through; Thyl., thylakoids; Nt, N. tabacum. C, sucrose gradient fractionation obtained by loading the fractions eluted from the Ni2+ column with 250 mm imidazole (on the left). Coomassie stain of SDS-PAGE of B2 and P fractions is shown on the right. D, absorption spectra of B1 (red line) and B2 (gray line) fractions from the sucrose gradient. LHCII (black line) is reported for comparison.
Recombinant LHCSR1 Expression Level
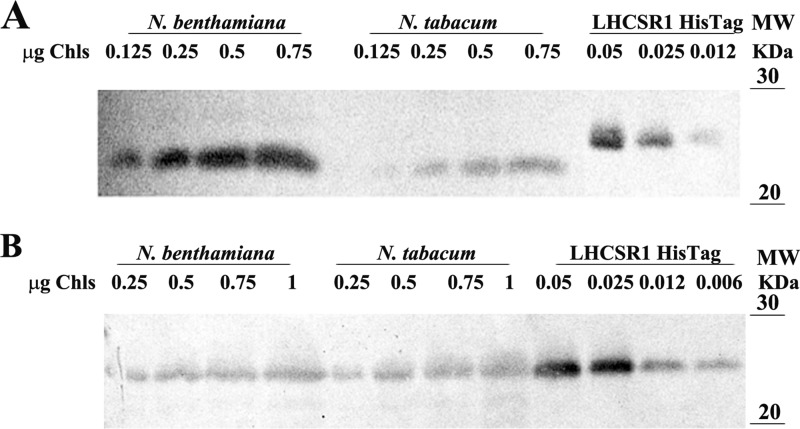
The abundance of the two recombinant proteins in the thylakoid fraction of both expression systems was determined by immunotitration (Fig. 5, A and B). For this aim, we used PpLHCSR1 His tag purified from N. tabacum as a reference (Fig. 4C). An SDS-polyacrylamide gel was loaded with serial dilutions of PpLHCSR1 His tag (0.012, 0.025, and 0.05 μg of Chl) and different amounts of oeLHCSR1 thylakoids (0.12, 0.25, 0.5, and 0.75 μg of Chl) from both N. benthamiana and N. tabacum. The intensity of the immunoreaction was not dependent on whether the determination was performed with recombinant LHCSR1 alone or mixed with WT thylakoids. The reactivity of α-LHCSR antibody was evaluated by densitometric analysis of immunoreactions. We calculated that in 1 mg of Chl of thylakoids PpLHCSR1 protein was present in amounts corresponding to 0.13 ± 0.025 and 0.036 ± 0.002 mg of Chl in N. benthamiana and N. tabacum, respectively (Fig. 5A). Similarly, we determined the abundance of recombinant LHCSR1 His tag in thylakoids obtained both from agro-infiltrated N. benthamiana and stable transformed N. tabacum plants. In this case, the resulting yields were 0.021 ± 0.001 and 0.011 ± 0.003 mg of Chl in N. benthamiana and N. tabacum, respectively (Fig. 5B).
FIGURE 5.
Abundance of the recombinant PpLHCSR1 protein in the different expression systems. A, LHCSR1 immunotitration of oeLHCSR1 thylakoids of N. benthamiana and N. tabacum. 0.125, 0.25, 0.5, and 0.75 μg of Chl were loaded for each sample. In the case of LHCSR1 His tag purified protein, 0.012, 0.025, and 0.05 μg of Chl were loaded. B, LHCSR1 immunotitration of oeLHCSR1 His tag thylakoids of N. benthamiana and N. tabacum. Filters were probed with α-PpLHCSR antibody.
Purification of Untagged PpLHCSR1 Using Deriphat-PAGE
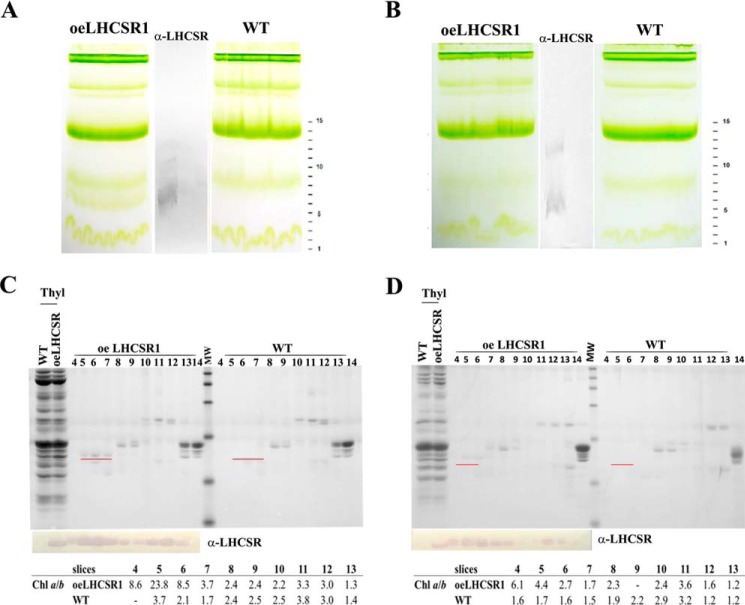
The untagged PpLHCSR1 version accumulated higher levels of LHCSR1 protein with respect to its tagged version in both N. tabacum and N. benthamiana (Fig. 5, A and B). To attempt purification, non-denaturing Deriphat-PAGE was applied to fractionate low molecular mass proteins with high resolution. Fig. 6 shows such a green gel of pigment-binding complexes from thylakoids of WT and oeLHCSR1 of N. benthamiana (Fig. 6A) and N. tabacum (Fig. 6B) after solubilization with 0.8% α-DM. In N. benthamiana, a clearly visible additional green band migrating below the monomeric LHC band was present in oeLHCSR1 plants but was absent in the WT sample (Fig. 6A). In the case of transgenic N. tabacum, a clear distinguishable band corresponding to recombinant LHCSR1 was less visible, but the monomeric LHC green band extended toward lower molecular mass when compared with the WT sample (Fig. 6B). The middle section of the gel including both samples was blotted onto a PVDF membrane, and the migration of the PpLHCSR1 protein was detected with an α-PpLHCSR antibody. A clear reaction in the oeLHCSR1 plants but not in the WT was detectable at the apparent molecular weight corresponding to the low molecular weight green band, suggesting that non-denaturing Deriphat-PAGE was effective in separating PpLHCSR1 from monomeric LHCs. Another clear reaction was also present in between monomers and trimers in the sample from oeLHCSR1 plants, suggesting the presence of a dimeric form of the protein. The remaining part of the gel was used for detailed fractionation. To this aim, the 20–100-kDa molecular mass range including monomeric LHCs and trimers was cut into thin slices that were eluted in a buffer solution containing 0.03% α-DM and 10 mm Hepes, pH 7.5. The eluate from each slice was submitted to HPLC pigment analysis, SDS-PAGE/immunoblotting (Fig. 6, C and D), and absorption spectroscopy (Fig. 7).
FIGURE 6.
PpLHCSR1 localization among pigment-binding complexes of N. benthamiana and N. tabacum. A and B, non-denaturing gel electrophoresis of pigment-binding complexes from thylakoids of oeLHCSR1 and WT of N. benthamiana (A) and N. tabacum (B) after solubilization with 0.8% α-DM. 500 μg of Chl were loaded for each sample. C and D, Coomassie blue staining and immunoblotting analysis of the fractions eluted from the non-denaturing gel slices shown in A and B and separated in a second dimension under denaturing conditions. The numbering of fractions reported on the top of each filter corresponds to that of gel slices cut from the non-denaturing gel. A polypeptide corresponding to the molecular weight of LHCSR1, underlined in red, was clearly visible in slices 5, 6, and 7 of oeLHCSR1 from both plant systems. Chlorophyll a/b ratios are indicated underneath each filter.
FIGURE 7.
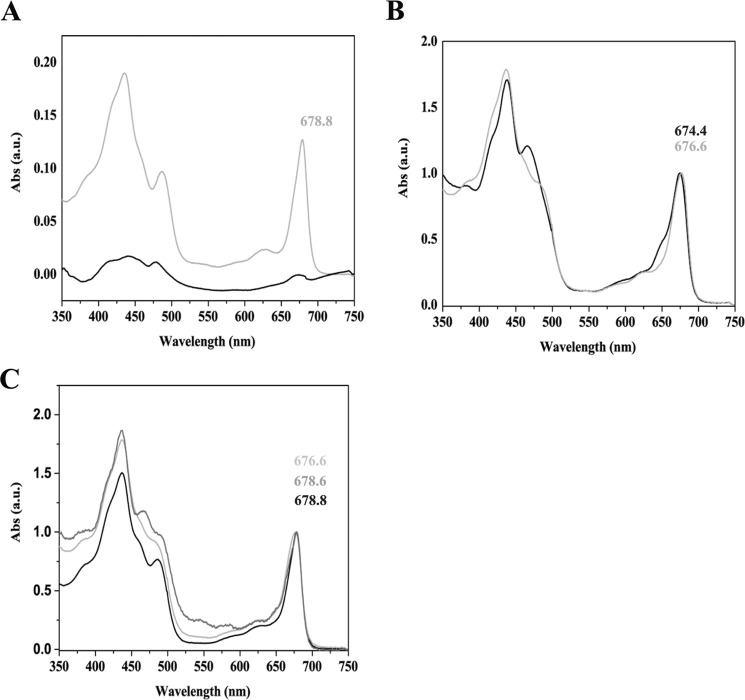
Spectroscopic analysis of recombinant PpLHCSR1 protein purified from Deriphat-PAGE. A, absorption (Abs) spectrum of slice 6 eluted from the gel. The sample containing PpLHCSR1 protein from oeLHCSR1 thylakoids (gray line) is compared with the corresponding fraction purified from N. benthamiana WT solubilized thylakoids (black line). B, absorption spectrum of slice 6 eluted from the gel of N. tabacum thylakoids. The sample containing PpLHCSR1 protein from oeLHCSR1 thylakoids (gray line) compared with the corresponding fraction purified from WT (black line). C, comparison among absorption spectra of fraction 6 containing LHCSR1 protein from N. benthamiana (black line) and N. tabacum (light gray line) and native LHCSR from P. patens (dark gray line) isolated in Pinnola et al. (22). a.u., arbitrary units.
Coomassie Blue staining showed that slice numbers 5, 6, and 7 from N. benthamiana and N. tabacum (Fig. 6, C and D, respectively), corresponding to the green band observed in Deriphat-PAGE, were strongly enriched in a polypeptide of an apparent molecular mass of ∼23 kDa that was absent in the corresponding sections from the gel loaded with WT thylakoids and was reactive to α-PpLHCSR antibody.
In addition, based on immunoblot analysis after Deriphat-PAGE, an oligomeric band was observed, suggesting the presence of LHCSR1 in a dimeric form in Nicotiana thylakoid membranes as reported previously for PSBS in plants (39). Considering all the fractions containing PpLHCSR1, the estimated yield (in Chl) of the recombinant protein was 0.13% and 0.03% in N. benthamiana and N. tabacum, respectively. HPLC analysis on eluted slices showed that fractions enriched in PpLHCSR1 protein were characterized by a very high Chl a/b ratio with values between 8 and 24 (Fig. 6, C and D), suggesting that PpLHCSR1 could bind Chl b in a substoichiometric amount only as reported previously for Chlamydomonas reinhardtii LHCSR3 proteins (20) and native LHCSR1 from P. patens (22).
Absorption spectra of LHCSR-enriched and LHCb-enriched fractions are shown in Fig. 7, A–C. In the case of N. benthamiana, the spectrum of the fraction eluted from the green gel and containing PpLHCSR1 protein was characterized by a Qy absorption peak red-shifted to 678.8 nm, whereas the eluate from the corresponding area of the gel in the WT lane only exhibited a weak and unresolved spectrum (Fig. 7A). Also in N. tabacum, the eluted fraction containing PpLHCSR1 protein was characterized by a Qy absorption peak red-shifted to 676.6 versus 674.4 nm of the corresponding area of the gel in the WT lane (Fig. 7B), closely featuring the native spectrum of LHCSR of P. patens (22) (Fig. 7C).
Isolation of LHCSR1 Using Membrane Fractionation and Sucrose Gradient Ultracentrifugation
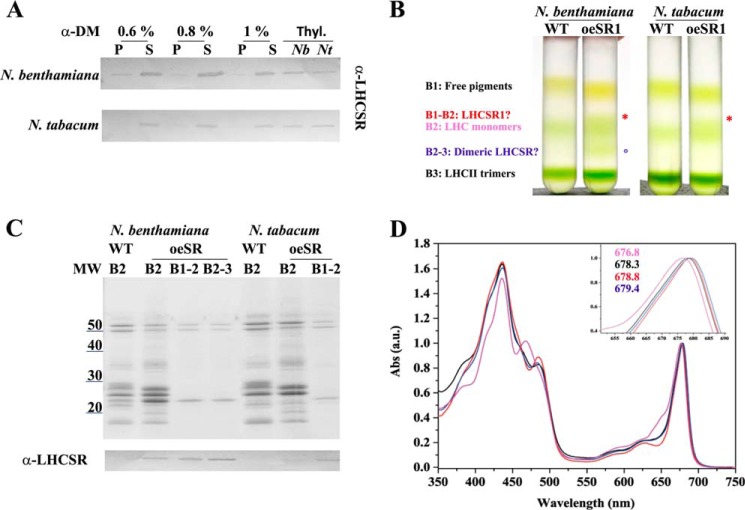
As an alternative approach to the isolation of recombinant PpLHCSR1, we fractionated stacked thylakoid membranes with α-DM (0.6, 0.8, and 1%) to obtain a supernatant (S) fraction enriched in stroma membrane proteins and a P fraction containing grana partitions in both systems. The pellet yield was between 40 and 70% Chl depending on detergent concentration. The Chl a/b was also in agreement with the Chl yield, showing a Chl a/b decrease in the P fraction by increasing the detergent concentration, whereas the Chl a/b of the S fraction was the specular image of Chl a/b of P (Table 1). Immunoblotting of the P versus S fractions showed that grana obtained with the highest concentration of detergent were depleted of LHCSR1, reflecting the finding that LHCSR1 is localized in stroma-exposed membranes in P. patens.4 At 0.8% α-DM, almost all LHCSR1 protein was in the S fraction, which yielded 52 and 41% Chl in N. benthamiana and N. tabacum, respectively (Fig. 8A). The S fractions from WT and oeLHCSR1 of both plant systems were fractionated by sucrose gradient ultracentrifugation (0.05–0.8 m sucrose). Results (Fig. 8B) showed that three bands were obtained from WT: the upper, yellowish band containing free pigments (B1), the second band with monomeric LHCs (B2), and the third band (B3) with trimeric LHCII. In the case of N. benthamiana (Fig. 8B, left), the sample from oeLHCSR1 plants contained two additional bands, one just above B2 (called B1–2) and one halfway between B2 and B3 (called B2–3). In contrast, in the case of N. tabacum, only one addition band (B1–2), just above B2, was found (Fig. 8B, right). Bands were carefully harvested with a syringe and analyzed by SDS-PAGE and absorption spectroscopy (Fig. 8, C and D). Coomassie staining showed that both the additional bands (B1–2 and B2–3) in N. benthamiana and the B1–2 band in N. tabacum contained an ∼23-kDa Coomassie-stained band with low levels of contaminants (Fig. 8C, upper panel) reactive to the α-PpLHCSR antibody (Fig. 8C, lower panel). Absorption spectra were very similar for B1–2 of both N. tabacum and N. benthamiana with the red-shifted peak at 678.3 and 678.8 nm, respectively (Fig. 8D). A similar spectrum was found for the lower LHCSR1 band (B2–3) of N. benthamiana that peaked at 679.4 nm, suggesting it contained a homodimeric form of LHCSR1. The lower level of B1–2 and the absence of B2–3 in N. tabacum could be related to the lower level of expression in this material. Anyway, considering the high biomass yield of N. tabacum plants despite the low amount of LHCSR1 protein, we also tried to improve the purification level of the PpLHCSR1 untagged version using N. tabacum plants. For this purpose, we applied a washing step of thylakoids with 1 m NaBr to remove CF1-ATPase (40) before the solubilization step. After the first sucrose gradient, the band containing LHCSR1 in monomeric form was loaded onto a second sucrose gradient to improve the purification and exclude the free pigment contamination (Fig. 9A). Coomassie staining showed that the PpLHCSR1 untagged version did not contain other contamination and migrated just below PpLHCSR1 His tag in SDS-PAGE (Fig. 9B). Absorption spectra of the two versions of the protein were very similar, suggesting that the histidine tag did not influence Car or Chl binding to the protein (Fig. 9C). Pigment binding was analized by HPLC, and the spectrum of the acetonic extracts were fitted with the spectra of purified pigments (28, 29) (Table 2). Both versions of the protein were characterized by a high Chl a/b ratio, and the pigment protein bound mainly Chl a and trace amounts of Chl b, lutein, and violaxanthin.
TABLE 1.
Chlorophyll a/b ratio of supernatant and pellet fractions obtained by solubilizing thylakoids of N. tabacum and N. benthamiana with different α-DM concentrations
Data are given as means ± S.D. of at least three independent experiments. Thyl., thylakoids.
| Thyl. | Pellet α-DM concentration |
Supernatant α-DM concentration |
|||||
|---|---|---|---|---|---|---|---|
| 0.6% | 0.8% | 1% | 0.6% | 0.8% | 1% | ||
| N. benthamiana | 2.60 ± 0.05 | 2.21 ± 0.04 | 2.44 ± 0.30 | 2.14 ± 0.08 | 4.76 ± 0.04 | 4.96 ± 0.10 | 4.43 ± 0.20 |
| N. tabacum | 2.54 ± 0.06 | 2.48 ± 0.02 | 2.29 ± 0.02 | 2.11 ± 0.03 | 4.64 ± 0.05 | 4.73 ± 0.10 | 4.22 ± 0.30 |
FIGURE 8.
Untagged PpLHCSR1 isolation and biochemical characterization. A, immunoblot of fractions obtained by solubilizing thylakoids by different concentration of α-DM probed with α-PpLHCSR antibody. An equal amount of Chl (0.25 μg) was loaded for each fraction. Thylakoids of oeLHCSR1 of N. benthamiana and N. tabacum are shown as reference. Thyl, thylakoids; Nb, N. benthamiana; Nt, N. tabacum. B, sucrose gradient of supernatant obtained by solubilizing thylakoids from WT and oeLHCSR1 (oeSR1) of both plant systems with 0.8% α-DM. C, polypeptide composition of fractions harvested from the sucrose gradient of oeLHCSR1 plants. 0.5 μg of Chl was loaded for B2, and 0.3 μg of Chl was loaded for other green bands. In the lower panel, the immunoblot with α-PpLHCSR antibody is shown. D, absorption (Abs) spectra of monomeric LHCs (pink line) and B1–2 (black line) from the sucrose gradient of N. tabacum and from B1–2 (red line) and B2–3 (blue line) from the sucrose gradient of N. benthamiana. a.u., arbitrary units.
FIGURE 9.
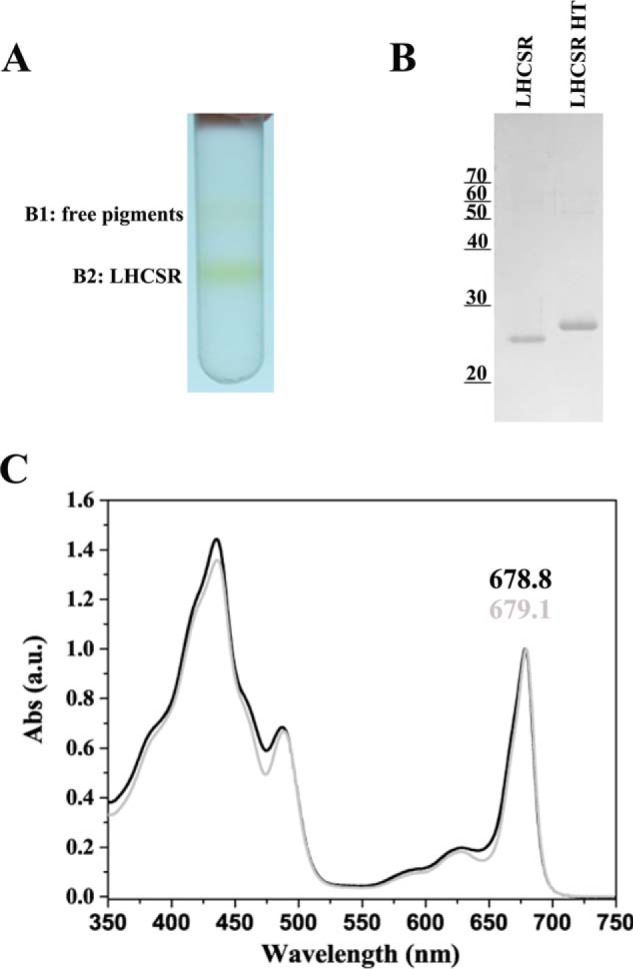
Purification of LHCSR1 without His tag. A, sucrose gradient of PpLHCSR1-enriched fraction. Enrichment was obtained by pooling the LHCSR-containing fractions from the first sucrose gradient (not shown). This second gradient allowed for separation of free pigments. B, Coomassie Blue-stained SDS-PAGE gel analysis of fraction B2. LHCSR His tag (LHCSR HT) was also loaded for reference. Loading was 0.3 μg of Chl/lane. C, absorption (Abs) spectra of LHCSR1 with and without His tag (gray and black lines, respectively). a.u., arbitrary units.
TABLE 2.
Pigment content and Chl a/b and Chl/Car ratios of purified LHCSR1 with and without His tag
The individual pigment value was normalized to 8, the proposed number of Chls per polypeptide. Neo, neoxanthin; Viola, violaxanthin; Ante, antheraxanthin; Lut, lutein.
| Chl a/Chl b | Chl/Car | Chl a + b normalization | Pigment content |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chl a | Chl b | Neo | Viola | Ante | Lut | No. Car | ||||
| LHCSR1 His tag | 37.94 | 2.02 | 8.00 | 7.79 | 0.21 | 0.00 | 1.83 | 0.09 | 2.04 | 3.96 |
| LHCSR1 | 12.71 | 2.32 | 8.00 | 7.42 | 0.58 | 0.00 | 1.37 | 0.02 | 2.04 | 3.43 |
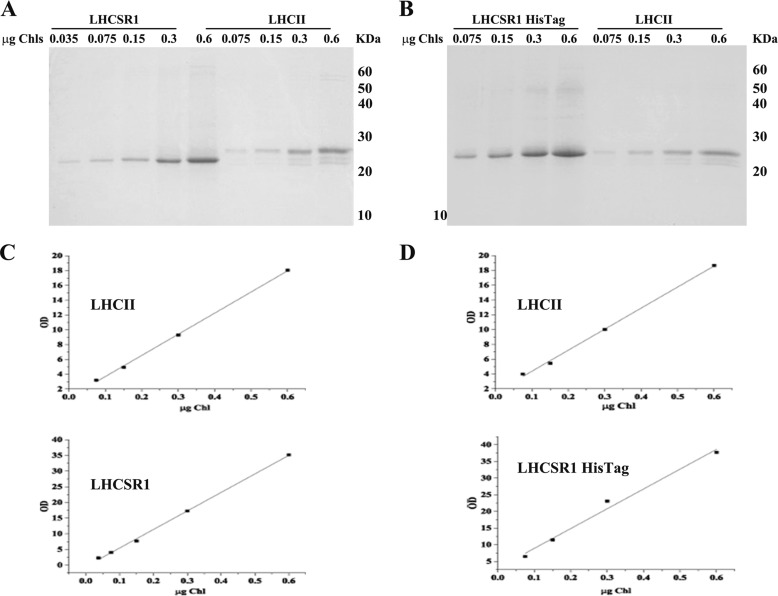
To estimate the number of Chls bound to LHCSR, we exploited the knowledge from the LHCII crystal structure showing 14 Chls per polypeptide (41) as a reference for stoichiometric analysis of the pigment to protein ratio. For this purpose, various dilutions (0.6, 0.3, 0.15, and 0.075 μg of Chl) of trimeric LHCII harvested from the sucrose gradient and isolated LHCSR1 protein were separated by SDS-PAGE and stained with Coomassie Blue (Fig. 10). We first verified the signal linearity for LHCII and LHCSR by densitometric analysis and found that it was very high in all samples analyzed (R2 ≥ 0.99) (Fig. 10, C and D). Because Coomassie dye binding depends on the basicity of a protein (the number of lysine, histidine, and arginine residues) (42), we normalized the densitometric signal to the basic amino acid content. We found that 6.29 ± 0.41 and 6.67 ± 0.28 Chls were bound to LHCSR1 with and without His tag, respectively.
FIGURE 10.
Determination of Chl to polypeptide ratio in LHCSR1. A and B, Coomassie-stained gels of various dilutions (0.6, 0.3, 0.15, 0.075, and 0.035 μg of Chl) of LHCSR, LHCSR His tag, and LHCII (0.6, 0.3, 0.15, and 0.075 μg of Chl). C, plot of Coomassie stain versus chlorophyll amount for LHCII (upper panel) and LHCSR (lower panel) showing linearity of the Coomassie binding (R2 ≥ 0.99). D, plot of Coomassie stain versus chlorophyll amount for LHCII (upper panel) and LHCSR His tag (lower panel) (R2 ≥ 0.99). The figure shows the result of a typical experiment.
Correlation between LHCSR1 Accumulation and NPQ Kinetics
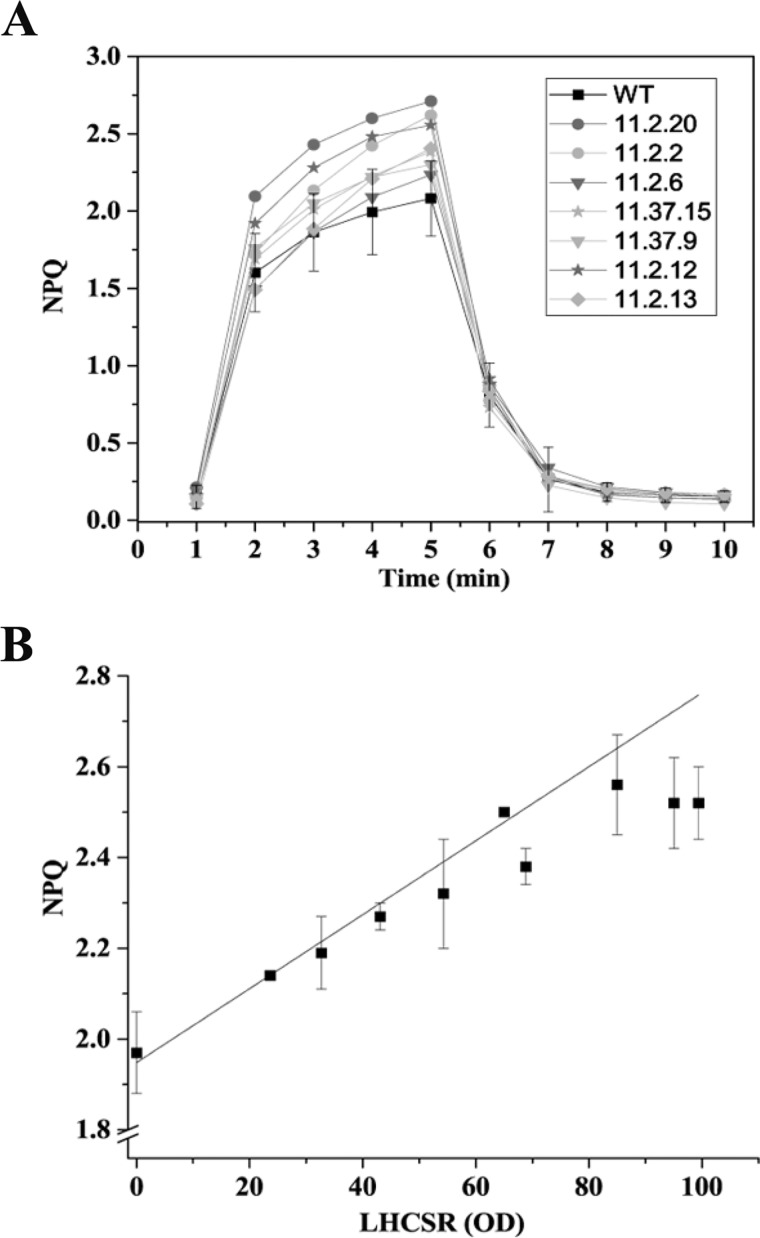
We further verified whether PpLHSCR1 protein was active in catalyzing NPQ in the heterologous tobacco expression system. Leaves from N. tabacum plants expressing different levels of LHCSR1 were analyzed for their NPQ kinetics as compared with WT plants lacking LHCSR1. WT leaf disks scored an NPQ activity of 1.97 upon 5 min of actinic light. oeLHCSR1 plants exhibited a somewhat higher NPQ than WT up to a value of 2.5 (Fig. 11A). To verify whether a correlation could be found between the LHCSR1 expression level and NPQ, we plotted LHCSR1 amount versus maximum NPQ at 5 min in plants expressing PpLHCSR1 to different levels (Fig. 11B). A positive correlation with an R2 value ≥0.96 was found between the NPQ and PpLHCSR1 level. The same procedure could not be performed for N. benthamiana because of the high variability of results on infected leaves subjected to the mechanical stress of the agroinfiltration procedure.
FIGURE 11.
Relationship between NPQ activity and accumulation of PpLHCSR1 protein. A, NPQ kinetic measurements on N. tabacum leaf disks. N. tabacum samples expressing different levels of PpLHCSR1 were compared with WT. B, correlation between NPQ activity at the last light point (maximum NPQ) of N. tabacum plants and LHCSR accumulation (R2 ≥ 0.96). Data are presented as a means ± S.D. (error bars) (n ≥ 3).
Discussion
Heterologous expression systems for functional membrane proteins have been previously reported based on bacteria, cyanobacteria, and yeasts (43, 44). However, the case of pigment-binding proteins of the LHC family is more complex because of the need for multiple chromophores essential for the folding of the holoprotein. Some chlorophyll-binding proteins, such as CP43, are able to fold only in the presence of Chl a (45), whereas LHCs also require Chl b and xanthophylls (46–49). This restricts the possible expression platforms to green algae and plants when aiming for the expression of active holocomplexes (50). Alternatively, the production of holocomplex can be divided into two steps, the first consisting of expression of apoprotein in bacteria followed by the second consisting of refolding in vitro with purified pigments (48, 51).
Here, the first approach, adopted to obtain folded and active recombinant protein, was the transient expression of PpLHCSR1 in N. benthamiana as a fast track option for suitability of the expression system for different DNA constructs before moving to higher throughput expression systems (33). In a second stage, a stable expression system in N. tabacum was developed. Indeed, N. tabacum has been reported to be a major tool for molecular farming and offers several practical advantages over other systems. Both systems contain chromophores needed for LHCSR1 protein folding according to the biochemical analysis of holoproteins (20, 22) and yielded very similar pigment-protein complexes. The highest protein level was obtained with the transient expression of PpLHCSR1 in N. benthamiana at 4 dpi. This yielded 0.13 ± 0.025 mg of Chl of LHCSR1/mg of Chl versus 0.036 ± 0.002 mg of Chl of thylakoids obtained from stably transformed N. tabacum (Fig. 5A). However, this gap is likely to be decreased when stably transformed plants, presently at the T2 generation, reach homozygosis upon self-crossing. In the case of PpLHCSR1 His tag, the amount of the protein was even lower, namely 0.021 ± 0.001 and 0.011 ± 0.003 mg of Chl/mg of Chl of thylakoids in N. benthamiana and N. tabacum, respectively (Fig. 5B).
The heterologous protein was correctly addressed to thylakoid membranes and processed to an apparent molecular weight indistinguishable from that observed in SDS-PAGE of moss thylakoids, strongly suggesting a correct targeting and processing of the preprotein encoded by the constructs. In addition, the fractionation of thylakoid membrane domains showed that PpLHCSR1 is located in stroma-exposed membranes including grana margins, which are solubilized by the α-DM treatment (53). This is in agreement with the localization of LHCSR1 in P. patens,4 implying that the protein elements (although unknown) that determine the partition of this LHC protein family member in the stroma membranes are acting in both Physcomitrella and Nicotiana. It should be noted that in fact most LHC proteins are located in grana membranes in plants unless they are tightly bound to PSI (54). Rather, LHCSR1 appears not to be part of a permanently assembled supercomplex because it is found as a low molecular weight pigment protein in the sucrose gradient upon solubilization with the mild detergent α-DM (Figs. 2 and 8).
The aggregation state of recombinant PpLHCSR1 expressed in Nicotiana heterologous systems was mostly monomeric as judged from mobility upon ultracentrifugation and in non-denaturing green gels (Figs. 2 and 6). Nevertheless, higher molecular weight forms of PpLHCSR1 were detected with lower mobility in green gels from both N. benthamiana and N. tabacum (Fig. 6, A and B). Upon loading the sample in a lower density sucrose gradient (0.05–0.8 m sucrose) and using a longer ultracentrifugation time, an LHCSR1 band was observed from both expression systems running at higher apparent molecular weight than monomeric LHCs (Fig. 8B). It can be asked whether this corresponded to homo- or hetero-oligomer. First, we observed that the oligomeric form migrated slower than the monomeric LHC band but faster than LHCII trimers in Deriphat-PAGE and conversely faster than monomeric LHCs but slower than PSI in the sucrose gradient. This is consistent with a dimer, although a trimeric aggregation state cannot entirely be excluded because the monomeric LHCSR1 is somehow smaller than monomeric LHCs (Figs. 2A; 6, A and B; and 8B). In the case of N. benthamiana, absorption spectra of the two bands from the gradient (Fig. 8D) are very similar if not identical, allowing for the effect of minor contaminations. Because all other LHCs bind Chl b and are blue-shifted in the absorption peak, heterodimerization would likely lead to a change in the spectra. Together with the presence of only very minor contaminations in SDS-polyacrylamide gels (Fig. 8C) this suggests that the most likely state of LHCSR1 is a homodimer. LHC proteins can be found as trimers (LHCII), monomers (CP29, CP26, and CP24), and dimers (LHCAs) (55–57). For both the LHC-like proteins involved in NPQ triggering, namely PSBS and LHCSR3 from Chlamydomonas, there have been reports of dimeric organization, although in most conditions, the most abundant form was a monomer (20, 22, 39). In this respect, the recombinant LHCSR1 purified from Nicotiana closely features the properties of the homologous systems.
The different purification methods that we used in this work were consistent in yielding a pigment protein highly enriched in Chl a with a red-shifted peak in the Qy range (679.1 nm), consistent with previous reports on recombinant C. reinhardtii LHCSR3 refolded in vitro (20, 23) or LHCSR1 purified from P. patens (22). The most pure preparation obtained, although in small amounts, had an exceedingly high Chl a/b ratio (Table 2). Based on a stoichiometry of seven Chls per polypeptide (20), only one LHCSR molecule in 12 actually binds a Chl b chromophore. The most likely interpretation is that LHCSR1 binds Chl a only and that the traces of Chl b, although undetectable in SDS-PAGE, come from contaminations. This result is in contrast with previous reports on C. reinhardtii LHCSR3 obtained from refolding in vitro with pigments that has a Chl a/b ratio of 7 (20, 23), suggesting that conditions during biogenesis of LHCSR in vivo are more stringent than refolding in vitro as for pigment selectivity in agreement with previous reports (48, 58). This is the first member of the LHC family so far reported that binds Chl a as the only porphyrin and is consistent with the accumulation of LHCSR3 in the Chl b-less Cbs3 strain of C. reinhardtii (20). The alternative hypothesis that Chl b is bound to LHCSR1 in vivo but is lost during purification cannot be excluded in principle. In this case, different purification procedures would yield an LHCSR1 preparation with different Chl b content. However, LHCSR1 purified by single step Deriphat-PAGE (Fig. 6) only binds Chl b in traces like LHCSR1 His tag whose purification involves repeated washing steps with detergent solutions (Fig. 4). Chl b is needed for folding of most LHCs (46) and can replace Chl a in most sites (58) yielding stable complexes. X-ray analysis of LHCII revealed that Chl b is involved in a network of H-bonds involving the carbonyl group at position C7 that stabilizes the structure of the pigment protein (41). Among LHC proteins, the Chl b-rich LHCII complex exhibits essentially a single 3.6-ns fluorescence lifetime, whereas decay kinetic from the Chl a-rich CP29 reveals at least three lifetimes with the amplitude of the shortest lifetime increasing upon zeaxanthin binding to site L2. (59). We suggest that the absence of Chl b might be important in easing the switching of LHCSR1 from unquenched to quenched conformation upon lumen acidification (20) and zeaxanthin binding (22). Besides Chl a, lutein and violaxanthin were found in very similar amounts, assuming that antheraxanthin (in traces) is bound to the same site(s) (likely site L2) as violaxanthin. With a Chl/Car ratio of 2.32 determined for LHCSR1 without His tag, the minimal hypothesis is two xanthophylls and four Chls. However, we obtained a 6.7 ± 0.29 Chls per LHCSR polypeptide ratio by Coomassie binding; this is half the chromophore complement of any other LHC protein so far reported upon crystallization (41, 60, 61). A consensus figure of 4 ± 1 xanthophylls and 8 ± 1 Chls is a likely possibility because up to four xanthophyll binding sites have been reported (41). In either case, the number of Chls of LHCSR1 is well below that of LHCII and CP29, consistent with the low level of sequence conservation between LHCII and LHCSR1 (24% identity and 37% similarity). It should be noted that PSBS, a member of the LHC family involved in energy dissipation, does not exhibit Chl binding sites (15), and its sequence identity with LHCII is 20.8%.
Structure-function studies require that the protein is functional. Indeed, the expression of recombinant PpLHCSR1 increased the high light-induced quenching activity in N. tabacum from 2 to 2.7 (Fig. 11A). It should be noted that WT N. tabacum is active in NPQ from resident PSBS. We therefore expected a background NPQ activity and a further increase in NPQ amplitude in oeLHCSR1 plants if the protein was active. The correlation of NPQ activity elicited by 5 min of high light treatment with the accumulation level of LHCSR1 was very good (R2 ≥ 0.96), intercepting the zero LHCSR1 protein line at NPQ = 1.97, i.e. the level of activity found in WT leaves. In the case of N. benthamiana, we could not directly prove the quenching activity of transiently expressed LHCSR1 because of the high variability of NPQ activity likely induced by the infiltration procedure. Indeed, a strong down-regulation of PSII quantum yield has been reported upon A. tumefaciens infection (62). However, the identical spectra of the protein preparations from N. tabacum and N. benthamiana and the same localization of LHCSR1 in stroma-exposed membranes strongly suggest that both expression systems yield the same preparation.
In this work, we purified PpLHCSR1 by (i) Ni2+ ion affinity chromatography (Fig. 4), (ii) non-denaturing Deriphat-PAGE (Fig. 6), or (iii) membrane fractionation followed by sucrose gradient ultracentrifugation (Figs. 8 and 9). We did not succeed in purification by ion exchange chromatography and preparative isoelectric focusing because of heavy co-fractionation with contaminants (not shown). The best purity was obtained by affinity chromatography (Fig. 4). Nevertheless, the low level of accumulation of the PpLHCSR1 His tag in thylakoids makes the final yield low (1:1000) on a Chl basis. Better yields corresponding to 5:1000 and 9:1000 were obtained using untagged versions of the constructs based on Chl molar ratios for N. tabacum and N. benthamiana, respectively.
We conclude that heterologous expression of PpLHCSR1 in Nicotiana yielded an active protein that has properties similar if not identical to those of LHCSR1 purified from the P. patens homologous system and is suitable for the study of the molecular basis of NPQ in algae and mosses with the further advantage that a single gene product rather than a mixture of two (LHCSR1 + LHCSR2) was obtained. Further studies are needed to understand the reasons for the lower specific NPQ activity in Nicotiana versus Physcomitrella. Likely the localization of the protein is in the stroma membranes of thylakoids, which are rich in LHCII in mosses but not in plants (52), and thus allows for quenching of a small fraction only of highly fluorescent LHCII in plants.
Author Contributions
A. P. performed the stable transformation of N. tabacum. L. G. and A. A. performed the transient expression of N. benthamiana. M. M., L. A., E. G., and M. P. provided technology and supervised the transformation and plant regeneration procedures. A. P., L. G., and S. Capaldi purified and characterized the protein. S. Cazzaniga performed HPLC pigment analysis and helped with data analysis. A. P. wrote the manuscript. R. B. designed the study, supervised the work, and revised the manuscript.
Acknowledgment
We thank Matteo Ballottari for helpful discussion.
This work was supported in part through the European Union Seventh Framework Program for Research Project 316427, Environmental Acclimation of Photosynthesis and the Italian Ministry of Agriculture, Food and Forestry Project HYDROBIO. The authors declare that they have no conflicts of interest with the contents of this article.
W. Chang, Z. Liu, M. Ballottari, and R. Bassi, unpublished results.
A. Pinnola, S. Cazzaniga, A. Alboresi, R. Nevo, S. Levin-Zaidman, Z. Reich, and R. Bassi, submitted manuscript.
- Chl
- chlorophyll
- Car
- carotenoid
- LHC
- light-harvesting complex
- LHCSR
- light-harvesting complex stress-related
- NPQ
- non-photochemical quenching
- PS
- photosystem
- PSBS
- photosystem II subunit S
- α-DM
- n-dodecyl α-d-maltopyranoside
- oeLHCSR1
- LHCSR1-overexpressing
- oeLHCSR1 His tag
- LHCSR1 with histidine tail-overexpressing
- dpi
- days postagroinfiltration
- PpLHCSR1
- P. patens LHCSR1
- S
- supernatant
- P
- pellet
- B
- band.
References
- 1. Barber J., Andersson B. (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem. Sci. 17, 61–66 [DOI] [PubMed] [Google Scholar]
- 2. Miller G., Shulaev V., Mittler R. (2008) Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi S., Murata N. (2008) How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 13, 178–182 [DOI] [PubMed] [Google Scholar]
- 4. Genty B., Briantais J. M., Baker N. R. (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990, 87–92 [Google Scholar]
- 5. Niyogi K. K. (2000) Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3, 455–460 [DOI] [PubMed] [Google Scholar]
- 6. Külheim C., Agren J., Jansson S. (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297, 91–93 [DOI] [PubMed] [Google Scholar]
- 7. Nilkens M., Kress E., Lambrev P., Miloslavina Y., Müller M., Holzwarth A. R., Jahns P. (2010) Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim. Biophys. Acta 1797, 466–475 [DOI] [PubMed] [Google Scholar]
- 8. Dall'Osto L., Caffarri S., Bassi R. (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17, 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Powles S. B., Björkman O. (1982) Photoinhibition of photosynthesis: effect on chlorophyll fluorescence at 77K in intact leaves and in chloroplast membranes of Nerium oleander. Planta 156, 97–107 [DOI] [PubMed] [Google Scholar]
- 10. Bruce D., Samson G., Carpenter C. (1997) The origins of nonphotochemical quenching of chlorophyll fluorescence in photosynthesis. Direct quenching by P680+ in photosystem II enriched membranes at low pH. Biochemistry 36, 749–755 [DOI] [PubMed] [Google Scholar]
- 11. Cazzaniga S., Dall'Osto L., Kong S.-G., Wada M., Bassi R. (2013) Interaction between avoidance of photon absorption, excess energy dissipation and zeaxanthin synthesis against photooxidative stress in Arabidopsis. Plant J. 76, 568–579 [DOI] [PubMed] [Google Scholar]
- 12. Duysens L. N. M., Sweers H. E. (1963) in Studies on Microalgae and Photosynthetic Bacteria (Japanese Society of Plant Physiologists, ed), pp. 352–372, University of Tokyo Press, Tokyo, Japan [Google Scholar]
- 13. Li X. P., Björkman O., Shih C., Grossman A. R., Rosenquist M., Jansson S., Niyogi K. K. (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395 [DOI] [PubMed] [Google Scholar]
- 14. Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 15. Dominici P., Caffarri S., Armenante F., Ceoldo S., Crimi M., Bassi R. (2002) Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J. Biol. Chem. 277, 22750–22758 [DOI] [PubMed] [Google Scholar]
- 16. Bonente G., Howes B. D., Caffarri S., Smulevich G., Bassi R. (2008) Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J. Biol. Chem. 283, 8434–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahn T. K., Avenson T. J., Ballottari M., Cheng Y.-C., Niyogi K. K., Bassi R., Fleming G. R. (2008) Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320, 794–797 [DOI] [PubMed] [Google Scholar]
- 18. Avenson T. J., Ahn T. K., Zigmantas D., Niyogi K. K., Li Z., Ballottari M., Bassi R., Fleming G. R. (2008) Zeaxanthin radical cation formation in minor light-harvesting complexes of higher plant antenna. J. Biol. Chem. 283, 3550–3558 [DOI] [PubMed] [Google Scholar]
- 19. Dall'Osto L., Holt N. E., Kaligotla S., Fuciman M., Cazzaniga S., Carbonera D., Frank H. A., Alric J., Bassi R. (2012) Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J. Biol. Chem. 287, 41820–41834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonente G., Ballottari M., Truong T. B., Morosinotto T., Ahn T. K., Fleming G. R., Niyogi K. K., Bassi R. (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9, e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alboresi A., Gerotto C., Giacometti G. M., Bassi R., Morosinotto T. (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl. Acad. Sci. U.S.A. 107, 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pinnola A., Dall'Osto L., Gerotto C., Morosinotto T., Bassi R., Alboresi A. (2013) Zeaxanthin binds to light-harvesting complex stress-related protein to enhance nonphotochemical quenching in Physcomitrella patens. Plant Cell 25, 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liguori N., Roy L. M., Opacic M., Durand G., Croce R. (2013) Regulation of light harvesting in the green alga Chlamydomonas reinhardtii: the C-terminus of LHCSR is the knob of a dimmer switch. J. Am. Chem. Soc. 135, 18339–18342 [DOI] [PubMed] [Google Scholar]
- 24. Karimi M., Inzé D., Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 [DOI] [PubMed] [Google Scholar]
- 25. Horsch R. B., Klee H. J., Stachel S., Winans S. C., Nester E. W., Rogers S. G., Fraley R. T. (1986) Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proc. Natl. Acad. Sci. U.S.A. 83, 2571–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bassi R., Simpson D. J. (1987) Chlorophyll-protein complexes of barley photosystem I. Eur. J. Biochem. 163, 221–230 [DOI] [PubMed] [Google Scholar]
- 27. Peter G. F., Takeuchi T., Philip Thornber J. (1991) Solubilization and two-dimensional electrophoretic procedures for studying the organization and composition of photosynthetic membrane polypeptides. Methods 3, 115–124 [Google Scholar]
- 28. Gilmore A. M., Yamamoto H. Y. (1991) Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96, 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Croce R., Canino G., Ros F., Bassi R. (2002) Chromophore organization in the higher-plant photosystem II antenna protein CP26. Biochemistry 41, 7334–7343 [DOI] [PubMed] [Google Scholar]
- 30. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31. Demmig-Adams B., Gilmore A. M., Adams W. W. 3rd (1996) Carotenoids 3: in vivo function of carotenoids in higher plants. FASEB J. 10, 403–412 [DOI] [PubMed] [Google Scholar]
- 32. Fischer R., Emans N. (2000) Molecular farming of pharmaceutical proteins. Transgenic Res. 9, 279–299 [DOI] [PubMed] [Google Scholar]
- 33. Kapila J., De Rycke R., Van Montagu M., Angenon G. (1997) An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122, 101–108 [Google Scholar]
- 34. Goodin M. M., Zaitlin D., Naidu R. A., Lommel S. A. (2008) Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant. Microbe Interact. 21, 1015–1026 [DOI] [PubMed] [Google Scholar]
- 35. Krysan P. J., Young J. C., Jester P. J., Monson S., Copenhaver G., Preuss D., Sussman M. R. (2002) Characterization of T-DNA insertion sites in Arabidopsis thaliana and the implications for saturation mutagenesis. OMICS 6, 163–174 [DOI] [PubMed] [Google Scholar]
- 36. Stengel A., Soll J., Bölter B. (2007) Protein import into chloroplasts: new aspects of a well-known topic. Biol. Chem. 388, 765–772 [DOI] [PubMed] [Google Scholar]
- 37. Hoober J., Eggink L. (1999) Assembly of light-harvesting complex II and biogenesis of thylakoid membranes in chloroplasts. Photosynth. Res. 61, 197–215 [Google Scholar]
- 38. Dainese P., Bassi R. (1991) Subunit stoichiometry of the chloroplast photosystem II antenna system and aggregation state of the component chlorophyll a/b binding proteins. J. Biol. Chem. 266, 8136–8142 [PubMed] [Google Scholar]
- 39. Bergantino E., Segalla A., Brunetta A., Teardo E., Rigoni F., Giacometti G. M., Szabò I. (2003) Light- and pH-dependent structural changes in the PsbS subunit of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 100, 15265–15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strotmann H., Kleefeld S., Lohse D. (1987) Control of ATP hydrolysis in chloroplasts. FEBS Lett. 221, 265–269 [Google Scholar]
- 41. Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 42. Tal M., Silberstein A., Nusser E. (1985) Why does Coomassie Brilliant Blue R interact differently with different proteins? A partial answer. J. Biol. Chem. 260, 9976–9980 [PubMed] [Google Scholar]
- 43. Bernaudat F., Frelet-Barrand A., Pochon N., Dementin S., Hivin P., Boutigny S., Rioux J.-B., Salvi D., Seigneurin-Berny D., Richaud P., Joyard J., Pignol D., Sabaty M., Desnos T., Pebay-Peyroula E., Darrouzet E., Vernet T., Rolland N. (2011) Heterologous expression of membrane proteins: choosing the appropriate host. PLoS One 6, e29191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosgaard L., de Porcellinis A. J., Jacobsen J. H., Frigaard N.-U., Sakuragi Y. (2012) Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J. Biotechnol. 162, 134–147 [DOI] [PubMed] [Google Scholar]
- 45. Ji Liu, Xie S.-S., Yue Luo, Zhu G.-F., Du L.-F. (2014) Soluble expression of Spinach psbC gene in Escherichia coli and in vitro reconstitution of CP43 coupled with chlorophyll a only. Plant Physiol. Biochem. 79, 19–24 [DOI] [PubMed] [Google Scholar]
- 46. Plumley F. G., Schmidt G. W. (1987) Reconstitution of chlorophyll a/b light-harvesting complexes: xanthophyll-dependent assembly and energy transfer. Proc. Natl. Acad. Sci. U.S.A. 84, 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Havaux M., Dall'osto L., Bassi R. (2007) Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145, 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giuffra E., Cugini D., Croce R., Bassi R. (1996) Reconstitution and pigment-binding properties of recombinant CP29. Eur. J. Biochem. 238, 112–120 [DOI] [PubMed] [Google Scholar]
- 49. Croce R., Weiss S., Bassi R. (1999) Carotenoid-binding sites of the major light-harvesting complex II of higher plants. J. Biol. Chem. 274, 29613–29623 [DOI] [PubMed] [Google Scholar]
- 50. Flachmann R., Kühlbrandt W. (1996) Crystallization and identification of an assembly defect of recombinant antenna complexes produced in transgenic tobacco plants. Proc. Natl. Acad. Sci. U.S.A. 93, 14966–14971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paulsen H., Finkenzeller B., Kühlein N. (1993) Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. Eur. J. Biochem. 215, 809–816 [DOI] [PubMed] [Google Scholar]
- 52. Pinnola A. (2014) Physcomitrella patens at the Crossroad between Algal and Plant Photosynthesis: a Tool for Studying the Regulation of Light Harvesting. Ph.D. thesis, University of Verona [Google Scholar]
- 53. Morosinotto T., Segalla A., Giacometti G. M., Bassi R. (2010) Purification of structurally intact grana from plants thylakoids membranes. J. Bioenerg. Biomembr. 42, 37–45 [DOI] [PubMed] [Google Scholar]
- 54. Simpson D. J. (1983) Freeze-fracture studies on barley plastid membranes. VI. Location of the P700-chlorophyll a-protein 1. Eur. J. Cell Biol. 31, 305–314 [DOI] [PubMed] [Google Scholar]
- 55. Dekker J. P., Boekema E. J. (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39 [DOI] [PubMed] [Google Scholar]
- 56. Wientjes E., Oostergetel G. T., Jansson S., Boekema E. J., Croce R. (2009) The role of Lhca complexes in the supramolecular organization of higher plant photosystem I. J. Biol. Chem. 284, 7803–7810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ballottari M., Girardon J., Dall'osto L., Bassi R. (2012) Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta 1817, 143–157 [DOI] [PubMed] [Google Scholar]
- 58. Croce R., Müller M. G., Bassi R., Holzwarth A. R. (2003) Chlorophyll b to chlorophyll a energy transfer kinetics in the CP29 antenna complex: a comparative femtosecond absorption study between native and reconstituted proteins. Biophys. J. 84, 2508–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moya I., Silvestri M., Vallon O., Cinque G., Bassi R. (2001) Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry 40, 12552–12561 [DOI] [PubMed] [Google Scholar]
- 60. Pan X., Li M., Wan T., Wang L., Jia C., Hou Z., Zhao X., Zhang J., Chang W. (2011) Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat. Struct. Mol. Biol. 18, 309–315 [DOI] [PubMed] [Google Scholar]
- 61. Ben-Shem A., Frolow F., Nelson N. (2003) Crystal structure of plant photosystem I. Nature 426, 630–635 [DOI] [PubMed] [Google Scholar]
- 62. Berger S., Sinha A. K., Roitsch T. (2007) Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 58, 4019–4026 [DOI] [PubMed] [Google Scholar]