Abstract
Analysis of rumen microbial community structure based on small-subunit rRNA marker genes in metagenomic DNA samples provides important insights into the dominant taxa present in the rumen and allows assessment of community differences between individuals or in response to treatments applied to ruminants. However, natural animal-to-animal variation in rumen microbial community composition can limit the power of a study considerably, especially when only subtle differences are expected between treatment groups. Thus, trials with large numbers of animals may be necessary to overcome this variation. Because ruminants pass large amounts of rumen material to their oral cavities when they chew their cud, oral samples may contain good representations of the rumen microbiota and be useful in lieu of rumen samples to study rumen microbial communities. We compared bacterial, archaeal, and eukaryotic community structures in DNAs extracted from buccal swabs to those in DNAs from samples collected directly from the rumen by use of a stomach tube for sheep on four different diets. After bioinformatic depletion of potential oral taxa from libraries of samples collected via buccal swabs, bacterial communities showed significant clustering by diet (R = 0.37; analysis of similarity [ANOSIM]) rather than by sampling method (R = 0.07). Archaeal, ciliate protozoal, and anaerobic fungal communities also showed significant clustering by diet rather than by sampling method, even without adjustment for potentially orally associated microorganisms. These findings indicate that buccal swabs may in future allow quick and noninvasive sampling for analysis of rumen microbial communities in large numbers of ruminants.
INTRODUCTION
Intensive farming of ruminant livestock for the production of human food and everyday commodities has wide implications for the environment (1). Apart from the well-described impacts of animals and their effluents on soils and waterways through nitrate leaching (2, 3), ruminants also represent a major anthropogenic source of the potent greenhouse gas methane through the microbial processes occurring in the rumen during fermentation of ingested feed (4). There is also great interest in the role of rumen microorganisms in feed conversion to animal products and in the association of differences in feed provided or animal genetics with the rumen microbial community. With the rapid development of highly resolving high-throughput sequencing technologies, there has been increased interest and opportunity to better understand the structure of bacterial, archaeal, and eukaryotic microbial communities in the rumen and to resolve the contributions of individual taxa to methane emission or animal productivity. Over the past years, this knowledge has provided valuable leads to new methane mitigation strategies, such as the use of low-greenhouse-gas feeds, feed supplements, vaccines, and inhibitors, and selective breeding for animals that show a naturally low-methane-emission phenotype (for a recent review, see reference 5). For example, it has been hypothesized that the microbial communities in low- and high-methane-emitting animals are ultimately controlled by the host's genome. This hypothesis is supported by the finding that low-methane animals appear to have a smaller rumen (6) and a higher turnover rate (6, 7), which would theoretically result in higher hydrogen concentrations in the rumen (8), thus explaining the selection for a bacterial community that produces less hydrogen (9). In a recent study involving 118 animals, two different possible low-methane communities were identified in sheep (9), meaning that there could be multiple underlying animal genotypes. Similar studies of dairy cows and steers with differential residual feed intake (RFI) suggest a correlation between RFI and certain rumen bacterial (10–12) and archaeal (13) taxa. However, due to the logistical and financial challenges of maintaining large dairy or beef research herds, the numbers of animals used in these studies were limited. To identify the host genetic loci that could be responsible for the low-methane or low-RFI trait, microbial community structures need to be correlated with animal genotyping data. Correlation of phenotypes with genotypes in animals of mixed genetic backgrounds typically involves the use of thousands of animals (14). Unlike some typical phenotypes, rumen microbial community structures are not static, and variation within a group of animals on a defined diet could be observed for various reasons, e.g., differences in animal condition, history, and sampling time after feeding, all of which will add variability to what otherwise might be very similar community types. Thus, a large number of samples will be needed to achieve ample statistical power. The practicality of the rumen sampling method and the affordability of large-scale parallel sequencing will therefore be major factors for ensuring the feasibility of a research project. In earlier studies, sampling of rumen contents from small numbers of animals through a fistula or at slaughter was considered the gold standard for obtaining a representative sample of the rumen microbial community, but detailed analysis of the microbial community by use of microscopic or low-throughput molecular fingerprinting methods was highly laborious (15). Overcoming this limitation by using bar-coded next-generation sequencing technologies (16) allowed quick and inexpensive sequence analysis of microbial communities in rumen samples from animal trials with larger numbers of animals per treatment group. This in turn stimulated research into alternative sampling techniques that retain a living, intact animal. Stomach tubing has become a routine method for sample collection, and its suitability for sampling a variety of different ruminants has been demonstrated (17–20). This method, however, can be technically challenging and time-consuming, and DNA extraction from samples collected in this way requires a lengthy process involving freeze-drying, grinding, and subsampling of rumen samples (17). These sampling and processing procedures currently represent a major bottleneck that hampers rapid and high throughput of rumen samples. It would therefore be desirable to identify more efficient work flows for rumen sampling and microbial community structure analysis.
Because ruminants regurgitate large amounts of rumen material to their oral cavities when they eat, oral swab samples may contain good representations of the rumen microbiota and could be used in lieu of rumen samples to study rumen microbial communities. In the present study, we collected buccal swabs from 24 animals feeding on four different diets by using three different swabbing methods and a simplified sample preparation protocol and compared the apparent bacterial, archaeal, and eukaryotic microbial communities and diet-related differences with those in the same animals sampled by use of a stomach tube.
MATERIALS AND METHODS
Collection of rumen samples and representative samples from the buccal cavity.
Samples were collected from a total of 24 sheep. The use of animals, including welfare, feeding, experimental procedures, and the collection of rumen samples used for this study, was approved by the AgResearch Grasslands Animal Ethics Committee (application 13015) and complied with the institutional Code of Ethical Conduct for the Use of Animals in Research, Testing and Teaching, as prescribed in the New Zealand Animal Welfare Act of 1999 and its amendments. The sheep belonged to four different dietary treatment groups: 100% lucerne (alfalfa) silage (100LS; n = 6), 100% maize silage (100MS; n = 6), 25% maize grain-75% lucerne silage (25MG; n = 6), and 65% maize grain-35% lucerne silage (65MG; n = 6). Four different sampling methods were used within approximately 30 min on any individual animal, as described below, and all animals on the same diet were sampled within approximately 2 h in the morning of the same day, resulting in a total of 96 samples.
Sampling methods. (i) Stomach tubing (rumen).
Samples were collected from the rumen of the animals via a polystyrene tube that was inserted through the mouth into the rumen. Approximately 100 g of rumen content was collected per animal. The sample was immediately placed on ice and transferred to −20°C. The sample was subsequently freeze-dried and homogenized, and 30 mg of rumen sample was weighed into a bead-beating vial containing 0.7 g of sterile zirconia-silica beads (0.1-mm diameter; Dnature, Auckland, New Zealand) for DNA extraction.
(ii) Buccal swabbing using an Omnigene kit (buccal OM).
A sterile cotton roll (10 by 38 mm; Dental Store, Auckland, New Zealand) held with a sterile forceps (25 cm long) was inserted into the mouth of the animal and swabbed several times across the inner side of the left cheek. The cotton roll was then placed in an Omnigene test tube containing a stabilizing solution (DNA Genotek Inc., Ottawa, Canada), and the tube was sealed according to the manufacturer's instructions. Samples were stored at room temperature for 3 weeks and then transferred to and stored at −20°C. The Omnigene tubes, each containing one swab, were first heated in a water bath at 50°C for 1 h according to the manufacturer's protocol. In the meantime, a sterile 15-ml tube was prepared by cutting off the bottom tip (approximately 5 mm) and placing it without the lid inside a sterile 50-ml tube. The cotton roll was transferred from the collection tube into the bottomless 15-ml tube. To extract saliva from the cotton roll, the sample was centrifuged at 1,000 × g for 3 min, and the liquid that accumulated in the 50-ml tube was transferred to a sterile 1.5-ml reaction tube. Saliva samples were stored at −20°C until further analysis. A total of 300 μl of saliva sample was transferred into a bead-beating vial containing zirconia-silica beads as described above for DNA extraction.
(iii) Buccal swabbing using a Performagene kit (buccal PG).
The collection sponge included in a Performagene sample collection kit (14.5 cm long) was inserted into the mouth of the animal and swabbed several times across the inner side of the left cheek. The collection sponge was then placed in a Performagene test tube containing a stabilizing solution (DNA Genotek Inc.), and the tube was sealed according to the manufacturer's instructions. Samples were stored at room temperature for 3 weeks and then transferred to and stored at −20°C. The Performagene tubes were heated in a water bath at 50°C for 1 h according to the manufacturer's protocol, and then the collection sponge was squeezed against the inner wall of the collection tube to release most of the liquid. The liquid sample was transferred to a sterile 1.5-ml Eppendorf reaction tube and stored at −20°C until further analysis. A total of 300 μl of saliva sample was transferred into a bead-beating vial containing zirconia-silica beads as described above for DNA extraction.
(iv) Buccal swabbing using direct DNA extraction from a cotton roll (buccal SD).
A sterile cotton roll (10 by 38 mm; Dental Store) held with a sterile forceps (25 cm long) was inserted into the mouth of the animal and swabbed several times across the inner side of the left cheek. The cotton roll was placed in a sterile 15-ml tube. Tubes were placed on ice and then transferred to −20°C. The cotton roll was removed from the tube with a sterile forceps, and approximately 1/3 of the swab was cut off, using a sterile pair of scissors, and immediately placed into a bead-beating vial containing zirconia-silica beads as described above for DNA extraction.
Extraction of nucleic acids.
DNA extraction was carried out as described previously (21), and DNA was quantified using a NanoDrop ND-1000 UV-visible (UV-Vis) spectrophotometer (NanoDrop Technologies, Wilmington, DE). Larger quantities of DNA were extracted from samples collected via stomach tubing than from samples collected via buccal swabbing. Prior to PCR amplification, the former DNA samples were diluted to approximately 45 ng/μl with sterile water. DNAs in samples collected via the buccal OM, buccal PG, and buccal SD methods were generally present at lower concentrations (approximately 30 ng/μl). These samples were used undiluted.
PCR amplification and high-throughput sequencing of microbial marker genes.
For 454 Titanium pyrosequencing, bacterial and archaeal 16S rRNA genes and ciliate protozoal 18S rRNA genes were amplified from 96 DNA samples by using the primers listed in Table 1 (9, 22). Bar coding, quantification, pooling of amplification products at equimolar concentrations, and pooling of microbial groups at a ratio of 1:1:1 (bacteria:archaea:protozoa) were performed as described previously (9). The sample was sent to Eurofins Genomics (Ebersberg, Germany) for 454 amplicon pyrosequencing using Titanium chemistry.
TABLE 1.
Oligonucleotides used for amplification of bacterial and archaeal 16S rRNA, ciliate protozoal 18S rRNA, and anaerobic fungal ITS1 genesa
| Primer name | Target group | Sequence (5′-3′) | Adapterb |
|---|---|---|---|
| Illumina-Ba9F | Bacteria | GAGTTTGATCMTGGCTCAG | A |
| Illumina-Ba515Rmod1 | Bacteria | CCGCGGCKGCTGGCAC | B |
| Illumina-Ar915aF | Archaea | AGGAATTGGCGGGGGAGCAC | B |
| Illumina-Ar1386R | Archaea | GCGGTGTGTGCAAGGAGC | A |
| Illumina-RP841F | Rumen ciliates | GACTAGGGATTGGARTGG | B |
| Illumina-Reg1302R | Rumen ciliates | AATTGCAAAGATCTATCCC | A |
| Illumina-MN100F | Anaerobic fungi | TCCTACCCTTTGTGAATTTG | B |
| Illumina-MNGM2 | Anaerobic fungi | CTGCGTTCTTCATCGTTGCG | A |
According to the work of Kittelmann et al. (22).
Primers had Illumina adapter A (5′-TCG TCG GCA GCG TCA GAT GTG AT AAG AGA CAG-3′) or B (5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G-3′) attached to the 5′ end.
For Illumina MiSeq PE300 sequencing, PCR amplification of bacterial and archaeal 16S rRNA genes, ciliate 18S rRNA genes, and anaerobic fungal internal transcribed spacer 1 (ITS1) genes was carried out as described above (9, 22). However, no bar codes were used during initial amplification, and the forward and reverse primers contained Illumina adapters at the 5′ and 3′ ends (Table 1). Each DNA sample was amplified in triplicate in a 96-well plate, and one well per plate served as a negative control for the master mix. Triplicate amplification products were pooled, and the correct sizes of PCR products and the absence of amplicons in the negative controls were verified by agarose gel electrophoresis. Amplicons were quantified using a Quant-iT dsDNA BR assay kit (Invitrogen, Carlsbad, CA) and a fluorometer (BioTek Instruments, Winooski, VT), and the four amplicons from the four microbial groups per DNA sample were pooled at a ratio of 1:1:1:0.2 (bacteria:archaea:protozoa:fungi). The amplicon pool for each sample was individually column purified using a MinElute96 UF PCR purification kit (Qiagen), and DNA was eluted in 20 μl sterile water. Purified amplicons were quantified using a Quant-iT dsDNA BR assay kit (Invitrogen) and normalized to contain 1 ng DNA in a total volume of 25 μl, adjusted with sterile water. The samples were sent to New Zealand Genomics Limited (Palmerston North, New Zealand) for sequencing using Illumina MiSeq PE300 chemistry.
Analysis of sequence data.
Sequence data obtained by 454 Titanium technology were processed and analyzed following the procedure described by Caporaso et al. (23). Sequence reads were assigned to corresponding samples by examining the 12-bp error-correcting Golay bar codes and default parameters in QIIME v1.8 (23), apart from protozoal sequences, for which no ambiguous bases were allowed (−a 0). Samples represented by fewer than 1,000 sequence reads for bacteria or fewer than 100 sequence reads for archaea and ciliate protozoa were excluded from further analysis. Quality-filtered sequence reads were clustered into operational taxonomic units (OTUs) at 97% (bacteria) or 99% (archaea) sequence similarity, using uclust as described previously (22, 24, 25). Ciliate protozoal sequences were clustered using the prefix-suffix method (22). Representative OTUs were subjected to BLAST searches against a newly assembled BLAST database composed of the following databases: a modified Greengenes gg_13_5 database, which was depleted of all sequences with taxonomy assignments beginning with “k__Archaea” (26); RIM-DB (25); and databases for intestinal ciliate protozoa (27) and anaerobic fungi (28). Relative abundance tables were generated at the species level (bacteria and archaea) or genus level (ciliate protozoa). Taxa that did not contribute ≥1% of the total community in at least one sample were excluded from further analyses.
Sequence data obtained by Illumina MiSeq PE300 technology were processed as follows. Raw sequences were trimmed to the longest contiguous segment with a quality score above a cutoff of 0.01, using the software DynamicTrim from the SolexaQA package (http://solexaqa.sourceforge.net/). If a sequence was trimmed to a length of 0 bases, a single base was left in the file in its place to maintain the order of reads. The program fastq-join was used with default settings to assemble contigs from paired-end sequence reads (29). Read 1 (containing Illumina adapter A) (Table 1) and read 2 (containing Illumina adapter B) (Table 1) were analyzed individually as well as after assembly. For individual analysis of reads 1 and 2, only reads with a minimum length of 200 bp were included. Samples represented by fewer than 10,000 sequence reads for bacteria or fewer than 100 sequence reads for archaea, ciliate protozoa, and anaerobic fungi were excluded from further analyses. All reads were subjected to BLAST searches against the combined database described above, and relative abundance tables were generated at the species level (bacteria and archaea) or genus level (ciliate protozoa and anaerobic fungi). Taxa that did not contribute ≥1% of the total community in at least one sample were excluded from further analyses.
Statistical analysis.
Good's coverage estimator was used to evaluate the completeness of taxon sampling at the analyzed level for each microbial group. This parameter returns the probability that a randomly selected taxon from a sample has already been sequenced (30, 31). Good's coverage (30) was calculated in Excel (version 2010; Microsoft Corp., Redmond, WA) and is presented as the average percentage ± standard deviation (SD) for each microbial group.
Principal coordinate analysis was performed using the Bray-Curtis dissimilarity metric in QIIME (23). The Bray-Curtis dissimilarity metric is a value between 0 and 1, with 0 indicating that the two samples have the same composition and 1 indicating that the two samples do not share any taxa at the level analyzed (32). The coefficients of the principal coordinates were imported into SigmaPlot v13.0 (Systat Software Inc., San Jose, CA) to visualize the clustering of samples within and between treatment groups (by diet and sampling method). Analysis of similarity (ANOSIM) was used in QIIME to test for significant differences between two or more treatment groups. The ANOSIM statistic R is based on the difference in mean ranks between and within treatment groups. R is a value of −1 to +1, with 0 indicating completely random grouping. Log-transformed relative abundance tables were scaled by row, and heat maps were generated by Pearson correlation and average linkage clustering, using the gplots package in R (33). Relative abundance data were subjected to Kruskal-Wallis one-way analysis of variance (ANOVA), and Bonferroni post hoc tests were carried out with R software (33), using the agricolae package (34), to identify taxa that had significantly different relative abundances between treatment groups.
Phylogenetic analysis.
A total of 126 Streptococcus 16S rRNA gene sequences were exported from ARB (SSURef_119_SILVA_14_07_14_opt.arb) (35, 36), using the ssuref:bacteria filter and Escherichia coli positions 28 to 1388, and an RAxML tree with 1,000 bootstrap replications was calculated. Several sequences belonging to the genus Fusobacterium were used as an outgroup. Representative sequences of the 28 most dominant OTUs (≥0.5% in at least one sample) that were assigned to the genus Streptococcus and were ≥260 bp long were filtered from our Illumina data set, aligned against the SILVA database by using SINA aligner (SSURef_119_SILVA_14_07_14_opt.arb) (35, 37), and imported into ARB (36). These sequences were added to the tree by use of the ARB parsimony insertion tool with the ssuref:bacteria filter and E. coli positions 28 to 288. The tree was exported in Newick format and prepared for publication using iTOL software (38).
Nucleotide sequence accession number.
Titanium 454 and Illumina MiSeq sequencing data obtained in this study were deposited in the NCBI BioProject database under accession number PRJNA282696.
RESULTS
Our aim was to determine whether buccal swabs collected from sheep can validly describe the microbial communities in their rumens. To do this, we compared bacterial, archaeal, and eukaryotic community structures in DNAs extracted from buccal swabs to those in DNAs from samples collected directly from the rumen by use of a stomach tube from sheep on four different diets. Because there are also bacteria that are part of the normal microbiota of the oral cavity, we expected the true rumen bacteria to make up only a part of the sequence data from buccal swab samples. Achieving sufficient coverage after bioinformatic depletion of sequences stemming from oral bacteria was therefore an important prerequisite. Using Illumina MiSeq PE300 sequencing technology allowed us to achieve an approximately 10 times larger number of sequence reads per sample and per analyzed microbial marker gene, and thus greater taxon coverage, than 454 Titanium pyrosequencing (see Text S1 and Tables S1 and S2 in the supplemental material). Moreover, the results obtained from Illumina MiSeq read 1 sequence data were highly similar to those obtained using 454 technology (see Text S2 and Fig. S1). Therefore, we subsequently used Illumina MiSeq read 1 sequence data to evaluate the microbial community structure in samples obtained using buccal swabs in comparison to that in parallel samples obtained via the common stomach tube method. We used samples from animals receiving one of a graduated range of four diets to allow differentiation between clustering of microbial communities due to the sampling methods used or due to the anticipated diet-driven differences.
Comparison of apparent bacterial community structures in samples collected via buccal swabs and stomach tubing.
In total, 113 bacterial taxa each contributed ≥1% of the total bacterial community in at least 1 of the 96 samples. Among these, all of the taxa that occurred in at least two samples collected via stomach tubing were also found in at least one sample collected by each of the buccal swabbing methods for each of the four studied diets.
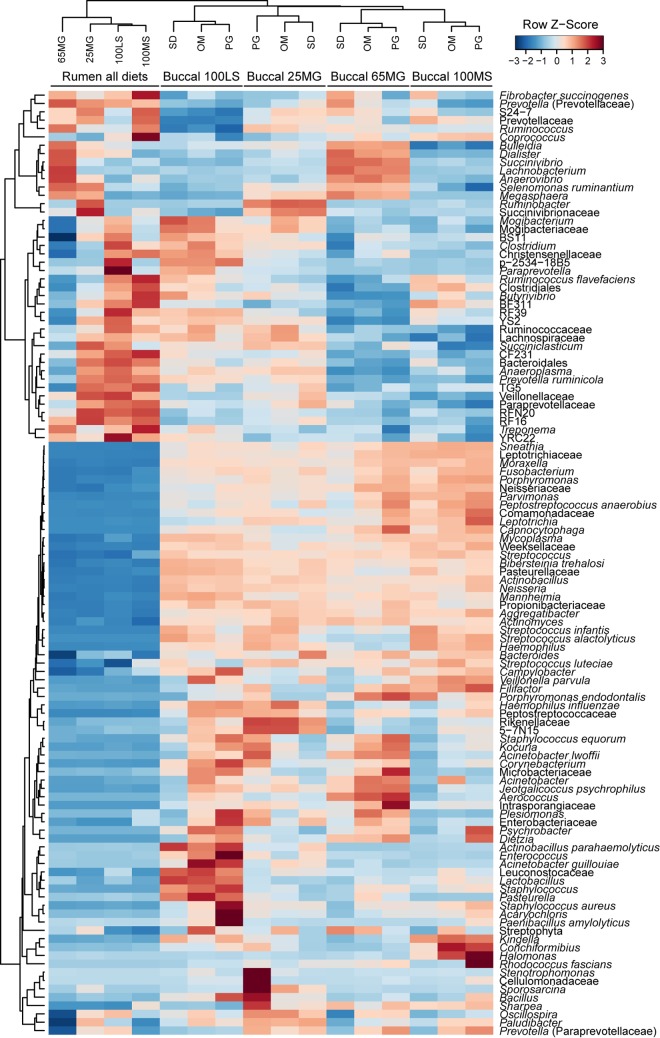
Bacterial communities in the samples collected by stomach tubing clustered separately from those in samples collected via buccal swabs (Fig. 1; see Fig. S2 in the supplemental material). Samples collected using the three different buccal swab methods clustered together, while four distinct clusters were formed according to the diets of the animals (Fig. 1). Clustering of bacterial taxa resulted in a large cluster of potentially orally associated or minor rumen bacteria (containing 71 taxa) and a smaller cluster of potentially rumen-associated bacteria (containing 42 taxa) (Fig. 1). Analysis of similarity revealed that while there was a significant difference between the diets (P ≤ 0.001; R = 0.17) (see Fig. S2), there was also a significant difference between sampling methods (P ≤ 0.001; R = 0.28) (see Fig. S2). This appeared to be caused by the high relative abundances of potential oral bacteria in the samples collected using buccal swabs. For this reason, the data set was explored in more detail to determine taxa that belonged predominantly to oral bacteria.
FIG 1.
Heat map of average bacterial taxon abundances in samples collected via buccal swabs (OM, PG, and SD) and stomach tubing (rumen) from sheep feeding on four different diets (100LS, 25MG, 65MG, and 100MS). Strong red colors indicate high standardized relative abundance values (row Z-scores), while dark blue colors indicate low standardized relative abundance values. Samples and taxa were clustered using Pearson correlation and hierarchical clustering with the average linkage method (n = 6 samples per diet and sampling method combination).
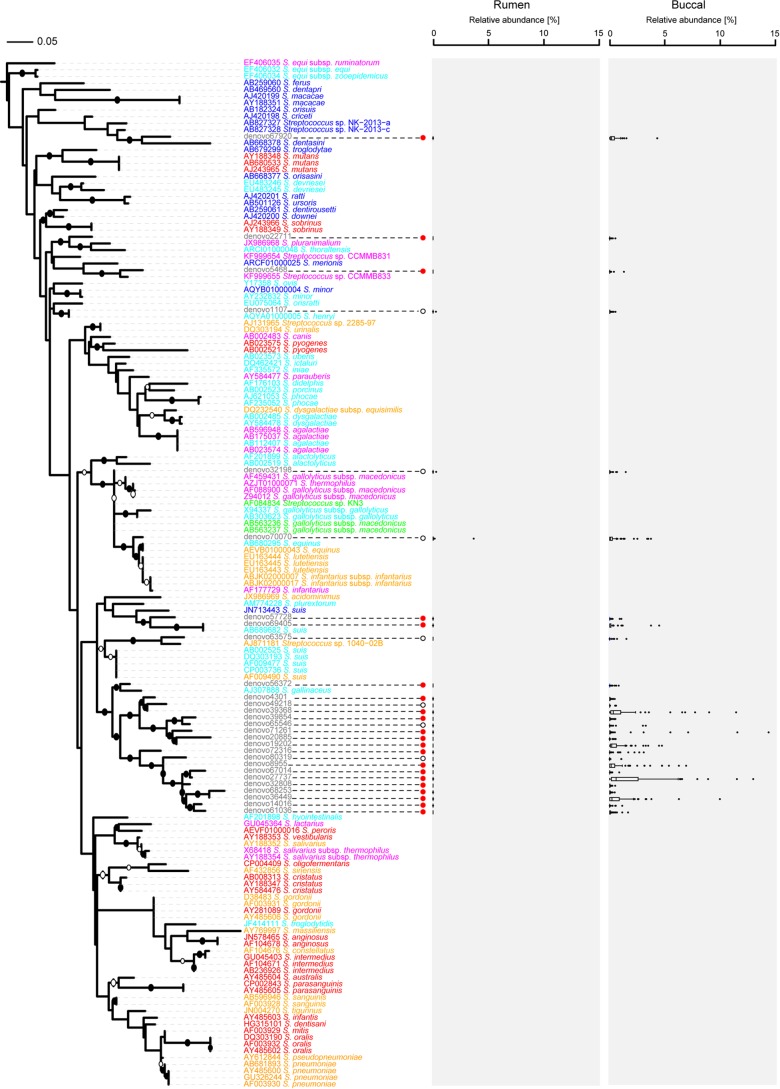
The data set was screened for potential oral bacteria based on the assumption that bacterial taxa with a maximum relative abundance in buccal swab samples that was ≥1% greater than that in any sample collected via stomach tubing (arbitrary cutoff) were likely to be “true” oral bacteria (mathematical filtering approach). We also used a second screening method based on each taxon's environmental distribution as reported in the literature (manual filtering approach) and compared the two different approaches. Using a manual approach would in future allow conclusions to be made purely based on microbial community structure data derived from buccal swabs, without the need to collect samples via stomach tubing, but it relies on knowledge from other studies. Based on the mathematical and manual filtering approaches, 73 and 64 of a total of 113 species-level taxa, respectively, were deemed potential oral bacteria and removed from the data set (see Table S3 in the supplemental material). The mathematical approach identified 69 of the 71 taxa of potentially orally associated or minor rumen bacteria identified using hierarchical clustering (Fig. 1). The two exceptions were Paludibacter and Oscillospira, which were not detected using the mathematical approach. For the mathematical and manual filtering approaches, the excluded taxa, on average, made up 63.7% and 43.7% of the buccal samples, respectively, but only 17.9% and 0.7% of the samples collected via stomach tubing. Following normalization of the remaining 40 or 49 “true” rumen species-level taxa to account for a total of 100% in each sample, analysis of β-diversity obtained after applying the mathematical approach resulted in a distinct clustering of the samples by diet (P ≤ 0.001; R = 0.44; ANOSIM) rather than by sampling method (P = 0.002; R = 0.06). After application of the manual approach, there was a weak clustering by diet (P ≤ 0.001; R = 0.29), but the sampling method still had a significant impact on the apparent community structure (P ≤ 0.001; R = 0.12). The mathematical approach eliminated the taxon “Streptococcus with unknown species affiliation,” because its maximum relative abundance in any sample collected via buccal swabbing was as high as 21.7%, while its maximum relative abundance in any sample collected via stomach tubing was only 0.5%. However, this taxon was initially not excluded by the manual filtering approach, as certain species of the genus Streptococcus appear to occur in the rumen (39). To determine whether the group of sequences assigned to Streptococcus with an unknown species affiliation could validly be eliminated from the data set, we created a phylogenetic tree of the genus Streptococcus, including the sequences obtained in this study (Fig. 2). We found that 16 of the 21 most abundant Streptococcus OTUs with unknown species affiliations from our study clustered among the OTUs that were assigned to Streptococcus luteciae (e.g., OTUs denovo65546 and denovo61036) and separately from strains isolated from the rumen, such as Streptococcus equinus (Fig. 2; see Table S4). The exception was OTU denovo32198, which clustered most closely with sequences from rumen isolates. This potentially true rumen OTU, however, tended to occur at higher relative abundances in samples collected via buccal swabs (average, 0.06%) than in those collected via stomach tubing (average, 0.01%) (P = 0.10; Student's t test). The remaining four Streptococcus OTUs with unknown species affiliations clustered closely with previously isolated oral strains, such as Streptococcus dentasini, Streptococcus merionis, or Streptococcus minor (Fig. 2; see Table S4). This result suggested that this taxonomic group predominantly contained non-rumen-derived sequences and should be excluded from the analysis. The manual approach was thus repeated without the taxon Streptococcus with unknown species affiliation, with the result that samples showed much improved clustering by diet (P ≤ 0.001; R = 0.37) (Fig. 3B and D; see Fig. S3) rather than by sampling method (P ≤ 0.001; R = 0.07) (see Fig. S3A and C).
FIG 2.
Phylogenetic tree of 126 Streptococcus reference sequences, constructed using RAxML, with 1,000 bootstrap replications. Representative sequences from 28 OTUs (gray font) assigned to the genus Streptococcus were added using the ARB parsimony insertion tool with the ssuref:bacteria filter and E. coli positions 28 to 288. Bootstraps with values of >70% and >90% were inferred and are marked as open and closed circles, respectively, on the branches. Several sequences belonging to the genus Fusobacterium were used as an outgroup. Species names are colored according to the habitats from which the species were isolated: red, human oral species; orange, human-associated species; blue, animal oral species; light blue, animal-associated species; magenta, dairy product species; and green, rumen species. Filled red circles indicate significant differences in relative abundances between samples collected using the two different methods, while open circles indicate no significant difference. Box plots show the median relative abundances and lower (25%) and upper (75%) percentiles for samples collected via stomach tubing (rumen) or buccal swabs (buccal). Outliers are shown as individual dots. The scale bar indicates 0.05 nucleotide substitution per nucleotide position.
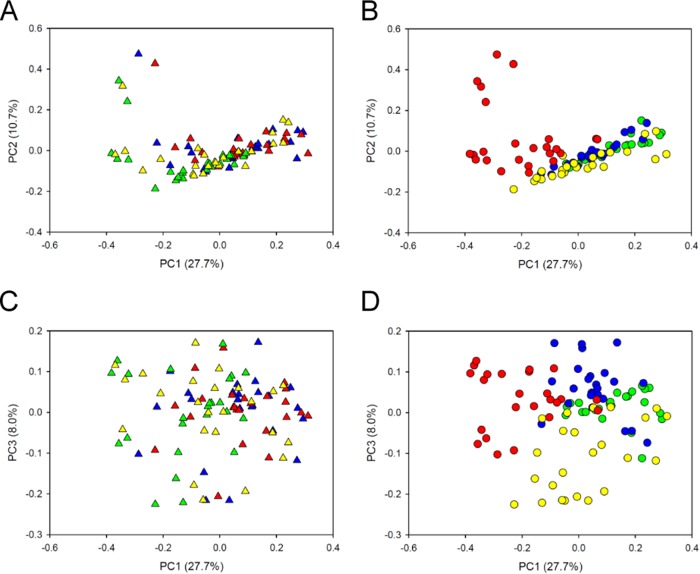
FIG 3.
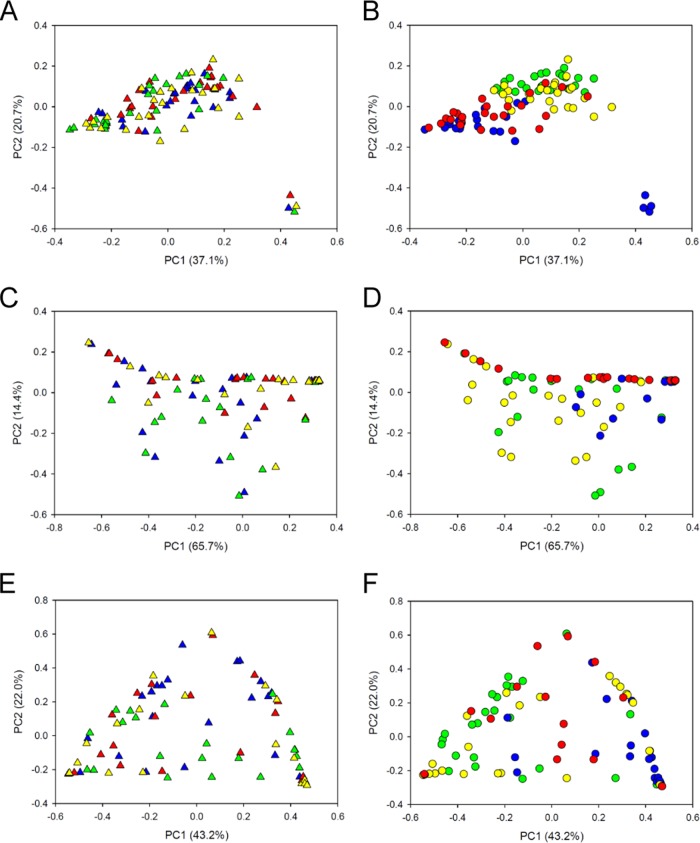
Principal coordinate analysis of bacterial communities in 96 samples collected via four different sampling methods from sheep feeding on four different diets after exclusion of potential oral taxa (by the manual filtering approach and excluding Streptococcus isolates with an unknown species affiliation). Sequence analysis was performed using Illumina MiSeq PE300 chemistry, and read 1 data were used. Principal coordinate 1 (PC1) is plotted against PC2 (A and B) or PC3 (C and D). Each point represents one sample. Different sampling methods are indicated by different colored triangles in panels A and C: red triangles, buccal PG; green triangles, rumen; blue triangles, buccal OM; and yellow triangles, buccal SD. Different diets are indicated by different colored circles in panels B and D: red circles, 65MG; green circles, 100LS; blue circles, 100MS; and yellow circles, 25MG. The left and right panels show the same plots, with the points colored in different ways.
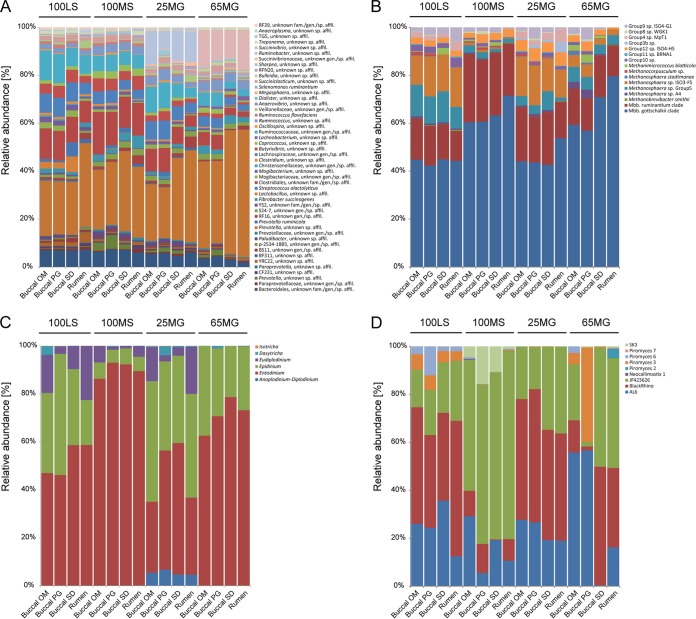
After elimination of the orally associated taxa, the bacterial phyla represented in the 96 samples were, in order of average relative abundance, Bacteroidetes (47.5%), Firmicutes (41.3%), Proteobacteria (6.9%), Fibrobacteres (2.2%), Spirochaetes (0.8%), Tenericutes (0.7%), Cyanobacteria (0.4%), and Synergistetes (0.2%). Analysis of variance and Bonferroni post hoc tests revealed the taxa that were significantly different in relative abundance between diets at the phylum (see Table S5 in the supplemental material) and species (see Table S6) levels. As yet unclassified members of the orders Bacteroidales and Clostridiales and the genus Butyrivibrio, for example, were significantly more abundant in sheep on diets consisting of 100% lucerne or maize silage (100LS and 100MS) (Fig. 4A; see Table S6), while as yet unclassified members of the genera Prevotella and Succinivibrio were significantly more abundant when animals were fed a diet with a high inclusion level of maize grain (65MG) (Fig. 4A; see Table S6). Members of the genus Ruminobacter were prominent in animals feeding on the 25MG diet (Fig. 4A; see Table S6).
FIG 4.
Average relative abundances of bacterial taxa at the species level (A), archaeal taxa at the species level (B), ciliate protozoal taxa at the genus level (C), and anaerobic fungal taxa at the genus level (D) in rumen samples collected via four different sampling methods (rumen, buccal OM, buccal PG, and buccal SD) from animals feeding on four different diets (100LS, 100MS, 25MG, and 65MG). For bacteria and archaea, each bar represents the average for 6 animals per method-diet combination. For ciliate protozoa and anaerobic fungi, each bar represents the average for at least 5 animals per method-diet combination, except for the anaerobic fungi analyzed in the following method-diet combinations: buccal SD-100MS (average for 4 animals), buccal SD-65MG (average for 2 animals), and buccal PG-65MG (average for 2 animals). Taxa are indicated by colored segments in the bars, and those contributing less than 1% to the microbial community in all samples are not included.
Fewer taxa were observed to vary significantly between the sampling methods used (see Table S7 in the supplemental material).
Comparison of apparent archaeal community structures in samples collected via buccal swabs and stomach tubing.
Archaeal communities were composed of a total of 16 taxa that each contributed ≥1% to the total archaeal community in at least 1 of the 96 samples. All taxa that occurred in at least two samples collected via stomach tubing were also found in at least one sample collected by each of the buccal swabbing methods for each of the four diets.
Archaeal communities did not cluster by sampling method (P = 0.12; R = 0.02) (Fig. 5A). Instead, the four different sampling methods (three buccal swabbing methods and the stomach tube method) provided very similar results and indicated clear differences between the four diets (P ≤ 0.001; R = 0.39) (Fig. 5B). The archaeal community was composed of members of the orders Methanobacteriales (average relative abundance, 89.5%), Methanomassiliicoccales (9.2%), Methanosarcinales (0.8%), and Methanomicrobiales (0.4%). The different diets resulted in significantly different community profiles, with animals feeding on 100LS or 25MG harboring significantly higher relative abundances of members of the Methanomassiliicoccales than animals feeding on 100MS or 65MG (Fig. 4B; see Table S6 in the supplemental material). Methanosphaera sp. group 5 was most abundant in sheep fed 100LS, and Methanosphaera ISO3-F5 showed high relative abundances in sheep on both the 100LS and 25MG diets (Fig. 4B; see Table S6). In contrast, members of the Methanobrevibacter gottschalkii clade showed significantly higher relative abundances in animals feeding on 100MS and 65MG (Fig. 4B; see Table S6). Few methanogen species appeared to be affected by the sampling method used (see Table S7).
FIG 5.
Principal coordinate analysis of archaeal communities in 96 samples (A and B), ciliate protozoal communities in 94 samples (C and D), and anaerobic fungal communities in 84 samples (E and F), collected via four different sampling methods (triangles; panels A, C, and E) from sheep feeding on four different diets (circles; panels B, D, and F). Sequence analysis was performed using Illumina MiSeq PE300 chemistry, and read 1 data were used. Each point represents one sample. Red triangles, buccal PG; green triangles, rumen; blue triangles, buccal OM; yellow triangles, buccal SD; red circles, 65MG; green circles, 100LS; blue circles, 100MS; yellow circles, 25MG.
Comparison of apparent ciliate community structures in samples collected via buccal swabs and stomach tubing.
Ciliate communities were composed of only 6 taxa, each contributing ≥1% to the total community in at least 1 of the 96 samples. Almost all taxa that occurred in at least two samples collected via stomach tubing were also found in at least one sample collected by each of the buccal swabbing methods for each of the four diets. The exception was the Anoplodinium-Diplodinium clade, which was not detected in samples collected using the buccal OM and buccal PG methods from sheep feeding on 100LS. However, their average relative abundance in the corresponding stomach tube samples was also low (0.01% ± 0.03%).
Ciliate communities appeared to cluster by diet (P ≤ 0.001; R = 0.22) rather than by sampling method (P = 0.48; R = −0.003) (Fig. 5C and D). Across all diets, Entodinium represented the dominant genus in sheep samples collected via stomach tubing and buccal swabs (average relative abundance, 64.1%) (Fig. 4C). Other genera identified in these samples were Epidinium (27.4%), Eudiplodinium (6.5%), Anoplodinium-Diplodinium (1.4%), Dasytricha (0.6%), and Isotricha (0.04%) (Fig. 4C). Samples from animals that had been feeding on a diet supplemented with 65% grain (65MG) contained significantly higher relative abundances of members of the genus Entodinium than those from sheep feeding on 100LS or 25MG (Fig. 4C; see Table S6). Samples collected from animals feeding on 100MS showed the highest relative abundances of Entodinium species. Sampling method, in contrast, had little impact on the apparent protozoal community structure (see Table S7 in the supplemental material).
Comparison of apparent anaerobic fungal community structures in samples collected via buccal swabs and stomach tubing.
In total, 9 anaerobic fungal taxa contributed ≥1% to the total anaerobic fungal community in at least 1 of the 96 samples. Piromyces 2, Piromyces 7, and SK3 were inconsistently retrieved from buccal swab samples when their relative abundances were also low in samples collected via stomach tubing. However, all other taxa that occurred in at least two samples collected via stomach tubing were also found in at least one sample collected by each of the buccal swabbing methods for each of the four diets.
Anaerobic fungal communities showed clustering by diet (P ≤ 0.001; R = 0.23), while the sampling method did not have a significant impact on community structure (P = 0.10; R = 0.03) (Fig. 5E and F). The anaerobic fungal taxa represented in the samples belonged to the JF423626 (average relative abundance, 36.3%), BlackRhino (33.4%), AL6 (23.6%), Piromyces 6 (2.8%), Piromyces 7 (1.9%), SK3 (1.6%), Piromyces 2 (0.3%), Piromyces 3 (0.04%), and Neocallimastix 1 (0.03%) groups (Fig. 4D). Anaerobic fungi assigned to the AL6, BlackRhino, and JF423626 groups were ubiquitously present in sheep on all administered diets and were the only taxa detected in sheep feeding on 25MG. In addition to these taxa, samples from animals feeding on 100LS contained members of the genus Piromyces, whereas those from animals feeding on 100MS contained members of the SK3 group (Fig. 4D; see Table S6 in the supplemental material). Animals that were administered the 65MG diet showed no clear diet-related pattern (Fig. 4D). This could be due in part to the difficulty of obtaining sufficient PCR products for sequencing from certain samples or to true animal-to-animal variation in this diet group. The sampling method significantly affected only two anaerobic fungal taxa, Piromyces 2 and 7, which showed slightly lower and higher relative abundances, respectively, in samples collected via stomach tubing (see Table S7).
DISCUSSION
The ease of the sampling method and the cost-effectiveness of high-throughput microbial community structure analysis are two important considerations for streamlining laboratory work flows to increase the rate of sample throughput and the number of samples analyzed at an affordable cost. The present study compared Illumina MiSeq PE300 sequencing to 454 Titanium pyrosequencing and then used the Illumina sequencing data set, which generated >10 times more sequence data, for evaluation of buccal swabs as a sampling method alternative to stomach tubing to assess bacterial, archaeal, and eukaryotic community structures in the rumen.
Feasibility of different buccal swabbing and sample processing methods.
Obtaining swab samples from the buccal cavity of ruminants is a promising sampling method, as it is noninvasive and quicker than the routine stomach tube method. In this study, three different buccal swab methods were compared with the routinely used stomach tube method. Each method was ranked based on its advantages and disadvantages in regard to several parameters of interest, such as ease of handling, time and effort, costs, and DNA yield (Table 2). Two of the methods used commercially available sampling kits (buccal OM and buccal PG), while the third (buccal SD) used simpler materials. While swabbing of the cheeks was done with sterile cotton buds held with a forceps for two of the three methods (buccal OM and buccal SD), the kit used for the buccal PG method included a sterile collection swab. It was noted, however, that the collection swab was shorter than the forceps and less robust. Reaching into the mouth of the animal to obtain sufficient material without contaminating or damaging the swab was therefore challenging. Overall, the buccal swab methods had the advantage that no freeze-drying, grinding, and weighing of samples were required prior to DNA extraction. However, collection of saliva from cotton swabs as done for the buccal OM method was highly laborious. An important advantage of using the Omnigene (buccal OM method) or Performagene (buccal PG method) kit was that there was no need for the sample to be transferred to −20°C immediately after collection. According to the manufacturer's specifications, once sealed, the sample can be stored at room temperature for up to 12 months after sampling. A combination of the buccal OM and buccal SD methods, where a sterile cotton roll would be swabbed across the inner side of the cheeks and then placed in an Omnigene test tube prior to direct extraction of nucleic acids from the cotton swab, may provide maximum benefit with minimum effort. This combined method would allow researchers, veterinarians, or farmers, even those in remote locations, to collect buccal swab samples and send them for diagnosis of rumen microbial community type/host phenotype for thousands of animals without the need for freezing and then storing and transporting frozen samples, and at the same time, it would guarantee fast and effective sample processing in the laboratory.
TABLE 2.
Comparison of the five different sampling methods based on ease of sampling and storage, cost of materials, preparation time, and DNA yield
| Parameter | Score for sampling methoda |
|||
|---|---|---|---|---|
| Rumen | Buccal OM | Buccal PG | Buccal SD | |
| Ease of sample collection | + | +++ | + | +++ |
| Ease of storage | + | +++ | +++ | + |
| Cost of materials (kits) | +++ | + | + | +++ |
| Preparation for DNA extraction | + | + | +++ | +++ |
| DNA yield | +++ | ++ | + | + |
| Total score (score/total possible) | 9/15 | 10/15 | 9/15 | 11/15 |
+++, very good; ++, good; +, to be improved or inefficient.
Samples collected by buccal swabbing produced lower yields of DNA and were less reliable in yielding amplification products, especially for anaerobic fungal ITS1 genes. Since repetition of DNA extraction and PCR amplification is impractical for analyzing thousands of samples, the potential loss of individual samples due to low DNA yields needs to be considered when setting up experiments. Despite the loss of individual samples due to poor DNA yields, we obtained ample sequence data with Illumina MiSeq technology from a sufficient number of animals to assess whether buccal swabs would provide an effective alternative to stomach tubing. Based on microbial community structure analysis, we evaluated whether diet effects could be detected readily by using the different sampling methods.
Representation of rumen microbiota in buccal swab samples.
Overall, buccal swab samples recovered the vast majority of bacterial, archaeal, ciliate protozoal, and anaerobic fungal taxa that were detected in samples collected via stomach tubing. Exceptions were ciliate protozoal members of the Anoplodinium-Diplodinium clade, which could not be recovered consistently from buccal swab samples from sheep on 100LS, and anaerobic fungal members of the clades Piromyces 2, Piromyces 7, and SK3, which could not be recovered consistently from buccal swab samples from sheep on three or four of the diets. These clades, however, were also detected at very low relative abundances in stomach tube samples (average abundance of Anoplodinium-Diplodinium in stomach tube samples from sheep on 100LS, 0.01% ± 0.03% [standard deviation]; average abundance of Piromyces 2 in stomach tube samples from sheep across all four diets, 0.99% ± 4.74%; average abundance of Piromyces 7 in stomach tube samples from sheep across all four diets, 0.12% ± 0.48%; and average abundance of SK3 in stomach tube samples from sheep across all four diets, 0.38% ± 1.24%). It is likely that these taxa were not detected consistently in buccal swab samples due to their very low relative abundances in the rumen on certain diets, rather than being specifically retained in the rumen, as some (e.g., Anoplodinium-Diplodinium and SK3) were readily detectable in buccal swab samples from sheep on the different diets in cases where their average relative abundances in stomach tube samples were at least 1%.
Applicability of buccal swabs to the study of rumen microbial community composition.
In general, buccal swab samples contained rumen bacterial taxa with a diversity similar to that in the corresponding samples collected by stomach tubing. Even some bacterial taxa known to be strongly associated with the solids fraction, such as Butyrivibrio spp. (40), Fibrobacter spp. (41), and Ruminococcus spp. (41), could be detected using buccal swabs. However, data obtained for samples collected via buccal swabs did not immediately provide a valid representation of rumen-inhabiting bacterial communities. This was because a large fraction of the taxa detected in buccal samples belonged to groups known to be typical oral bacteria, such as, e.g., Actinobacillus, Bibersteinia, Fusobacterium, Haemophilus, Mannheimia, Moraxella, and Neisseria. It is assumed that as more time passes between regurgitation and sampling, the proportion of oral bacteria will become larger. This factor may add to animal-to-animal variation of the microbiome. In our study, all 24 animals were randomly sampled and could reasonably be expected to be at different time points during the rumination cycle. Despite this, or perhaps because of it, we were able to detect even subtle diet-related differences between treatment groups.
We used two different approaches, a mathematical and a manual filtering approach, to remove orally associated taxa from the data set. While the mathematical approach bore the risk of falsely eliminating true rumen taxa, the highly conservative manual approach may have falsely retained true oral taxa in the data set. Despite these potential drawbacks, both the mathematical and manual filtering approaches to eliminate potential oral taxa from the data set, combined with detailed phylogenetic analysis of the questionable taxon Streptococcus with unknown species affiliation, resulted in a clear clustering by diet, independently of the sampling method used. In future, further work is needed to facilitate the identification of typical oral taxa in DNAs obtained from buccal swabs. The mathematical approach relies on taking rumen samples in parallel with buccal swabs, which defeats the purpose of buccal swabbing. Future application of buccal swabbing for rumen bacterial community analysis will rely on a literature-informed bioinformatic approach. Better knowledge of the oral microbial community structure, e.g., by comprehensive analysis of the oral microbiota of young ruminants or adult animals several hours after feeding or even of oral bacteria from nonruminating herbivores, such as horses, would allow the establishment of a reference database of oral bacteria. This database could serve as a tool for depletion of oral taxa from a buccal data set. Similarly, common environmental contaminants can be traced back to their sources and selectively filtered out of a data set by using a source tracker tool (42), or microbial metagenomic and transcriptomic data sets from host-associated environments can be searched against the host genome to eliminate host-derived sequences (43, 44). Alternatively, a curated reference database of 16S rRNA genes from rumen bacteria could be developed and used to eliminate all sequences from the data set that are not classified in the rumen-specific database, with due regard to checking that new, true rumen bacterial groups are not being eliminated. At this stage, such databases are not available, and until they are, buccal swabbing must be considered a promising technique for the assessment of rumen bacterial community structure. In very large studies of animals on a common diet, a small part of the animal population could be assessed using both direct rumen sampling and buccal swabbing, and this information could then be used to develop criteria for bioinformatically filtering out potential oral bacteria.
In contrast to the case for the bacterial community, the study of archaeal, ciliate protozoal, and anaerobic fungal communities in the rumen by using buccal swabs offers a useful alternative to stomach tubing, without the requirement to compare against a database of specific oral or rumen-associated microorganisms. With the information gathered in our study from buccal swab samples from sheep, diet effects were clearly detectable for all three groups of microorganisms. It should be noted, however, that specific orally associated methanogens, and perhaps even eukaryotic microorganisms, may be present, as occurs in humans (45–47), and this warrants further investigation in ruminants. More studies along the lines of ours will be needed to confirm that oral and rumen methanogen communities are always very similar before the method can be used with full confidence.
Diet-driven differences in rumen microbial community structure as assessed using buccal swabs.
Microbial community structures in the rumens of sheep feeding on four selected natural diets (pasture with different inclusion levels of maize grain or maize silage) were found to be consistently different for samples collected via either stomach tubing or buccal swabs. Differences in microbial communities in response to the four diets were mostly gradual, as expected from the gradient in diets, and consisted mainly of differences in relative abundances of the same reoccurring taxa (Fig. 1; see Table S6 in the supplemental material). This finding is in agreement with a previous study on the characterization of the rumen microbiota in cattle during transition from forage to a high-concentrate diet (48). Members of the highly diverse genus Prevotella, for example, contributed considerably to the bacterial community in sheep on all diets, but they showed the highest relative abundances by far in sheep on the high-grain diet (65MG). Similarly, members of the archaeal Methanobrevibacter gottschalkii clade were ubiquitously present in sheep on all diets but made up larger proportions of the community when the animals were fed on 100MS or 65MG, while members of the order Methanomassiliicoccales and Methanosphaera sp. ISO3F5 followed the reverse trend, with the lowest relative abundances in sheep on 100MS and 65MG. Methanogens (among them species of the order Methanomassiliicoccales) and ciliate protozoa have been reported to live in close proximity in the rumen (49, 50), and the advantages of a symbiotic relationship for both the archaeal and ciliate protozoal partners are well recognized (51). Our study confirms that high-grain diets are generally characterized by lower ciliate protozoal diversity. In contrast to the 100LS, 100MS, and 25MG diets, protozoal communities were composed of only two major genera, namely, Entodinium and Epidinium, in samples from animals feeding on 65MG. Comparable trends from the bovine rumen have led to the hypothesis that a decreased relative abundance of Methanomassiliicoccales in animals on high-grain-containing diets could be due to the loss of the symbiotic protozoal partner (50). However, the similar responses of Methanomassiliicoccales and some protozoa to dietary changes could also be due to physiological or metabolic changes in the rumen environment that adversely affect both Methanomassiliicoccales and certain protozoal genera independently of each other (50). In congruence with the observations by Petri et al. (48), but in contrast to early reports by Hungate et al. (52), we found significantly lower relative abundances of members of the phylum Firmicutes (which contains mostly Gram-positive bacteria) in animals feeding on medium (25MG)- and high (65MG)-grain diets, while Bacteroidetes and Proteobacteria were present at significantly higher relative abundances.
While the majority of microorganisms gradually changed with diet, some bacterial taxa, such as the amylolytic genera Ruminobacter and Succinivibrio, appeared to thrive only under highly specific conditions and were almost absent from sheep fed the other diets in our study. Ruminobacter spp. were most abundant in sheep on the lucerne silage diet with 25% inclusion of maize grain. Elevated levels of Ruminobacter spp. in sheep on this diet are not surprising, as an increasing inclusion of concentrate has been found to result in higher butyrate concentrations (53), which in turn have a stimulating effect on Ruminobacter populations (54). Intriguingly, this genus seemed to be replaced almost entirely and abruptly by Succinivibrio spp. in sheep fed the diet with a 65% inclusion of maize grain. Our finding of a significantly higher relative abundance of Succinivibrio spp. in samples from animals that were fed the 65MG diet corroborates the results from previous studies (48). Members of this genus were also detected at higher relative abundances in the rumen epimural bacterial community in beef cattle feeding on a high-grain diet (55) and have been reported to be the predominant isolates from the rumen when the diet of the animal is high in starch (56). Our findings suggest that the rumen microbial community contains a wide range of microorganisms with different abilities to adapt to dietary change. Alongside a core of highly versatile taxa that fluctuate depending on the conditions, the rapid expansion of highly specialized, more sensitive microorganisms upon niche development appears to be supported by our data, even if these specialists are initially present only in small numbers.
Conclusions.
Our study compared microbial community compositions in buccal swabs and rumen content samples obtained by stomach tubing from sheep feeding on four different diets. Our results suggest that buccal swabs can provide accurate information on the composition of the animal's rumen microbiome, composed of bacteria, archaea, ciliate protozoa, and anaerobic fungi. To further refine the analysis of sequence data derived from buccal swabs, in particular for studying rumen bacterial communities, a detailed exploration of the oral microbiome or of the differences between the rumen microbiome and its apparent composition as assessed using buccal swabs should be undertaken using ruminant animals under different dietary conditions. Alternatively, a broad database of rumen-specific taxa could be generated for calibration of data obtained from buccal swabs. In future, this method may be applied broadly to investigate the interplay between relevant animal phenotypes (e.g., methane emission or productivity), animal genotypes, and the rumen microbiota.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the joint science program of the New Zealand Agricultural Greenhouse Gas Research Centre and the Pastoral Greenhouse Gas Research Consortium. Funding for the animal trial from which samples were collected was provided by the Sustainable Land Management and Climate Change research program. S.K. was awarded a sequencing grant from New Zealand Genomics Limited, under the Illumina Miseq “Pilot the Possibilities” program.
We thank Edgar Sandoval for help with stomach tube sampling and German Molano and Savannah Devente (all from AgResearch) for assistance with the collection of buccal swabs. We are grateful to Christina Moon and two independent reviewers for constructive criticism of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02385-15.
REFERENCES
- 1.Steinfeld H, Gerber P, Wassenaar T, Castel V, Rosales M, de Haan C. 2006. Livestock's long shadow—environmental issues and options. Food and Agriculture Organisation of the United Nations, Rome, Italy. [Google Scholar]
- 2.Ryden JC, Ball PR, Garwood EA. 1984. Nitrate leaching from grassland. Nature 311:50–53. doi: 10.1038/311050a0. [DOI] [Google Scholar]
- 3.Di HJ, Cameron KC. 2000. Nitrate leaching in temperate agroecosystems: sources, factors and mitigating strategies. Nutr Cycling Agroecosyst 64:237–256. [Google Scholar]
- 4.Ripple WJ, Smith P, Haberl H, Montzka SA, McAlpine C, Boucher DH. 2014. Ruminants, climate change and climate policy. Nat Clim Change 4:2–5. [Google Scholar]
- 5.McAllister T, Meale SJ, Valle E, Guan LL, Zhou M, Kelly WJ, Henderson G, Attwood GT, Janssen PH. 2015. Use of genomics and transcriptomics to identify strategies to lower ruminal methanogenesis. J Anim Sci 93:1431–1449. doi: 10.2527/jas.2014-8329. [DOI] [PubMed] [Google Scholar]
- 6.Goopy JP, Donaldson A, Hegarty R, Vercoe PE, Haynes F, Barnett M, Oddy VH. 2014. Low-methane yield sheep have smaller rumens and shorter rumen retention time. Br J Nutr 111:578–585. doi: 10.1017/S0007114513002936. [DOI] [PubMed] [Google Scholar]
- 7.Pinares-Patiño CS, Ulyatt MJ, Lassey KR, Barry TN, Holmes CW. 2003. Rumen function and digestion parameters associated with differences between sheep in methane emissions when fed chaffed lucerne hay. J Agric Sci 140:205–214. doi: 10.1017/S0021859603003046. [DOI] [Google Scholar]
- 8.Janssen PH. 2010. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim Feed Sci Technol 160:1–22. doi: 10.1016/j.anifeedsci.2010.07.002. [DOI] [Google Scholar]
- 9.Kittelmann S, Pinares-Patiño CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC, Janssen PH. 2014. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. doi: 10.1371/journal.pone.0103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Sanabria E, Guan LL, Goonewardene LA, Li M, Mujibi DF, Stothard P, Moore SS, Leon-Quintero MC. 2010. Correlation of particular bacterial PCR-denaturing gradient gel electrophoresis patterns with bovine ruminal fermentation parameters and feed efficiency traits. Appl Environ Microbiol 76:6338–6350. doi: 10.1128/AEM.01052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Sanabria E, Goonewardene LA, Wang Z, Durunna ON, Moore SS, Guan LL. 2012. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl Environ Microbiol 78:1203–1214. doi: 10.1128/AEM.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jewell KA, McCormick C, Odt CL, Weimer PJ, Suen G. 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710. doi: 10.1128/AEM.00720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou M, Hernandez-Sanabria E, Guan LL. 2009. Assessment of microbial ecology of ruminal methanogens in cattle with different feed efficiency. Appl Environ Microbiol 75:6524–6533. doi: 10.1128/AEM.02815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kemper KE, Goddard ME. 2012. Understanding and predicting complex traits: knowledge from cattle. Hum Mol Genet 21:R45–R51. doi: 10.1093/hmg/dds332. [DOI] [PubMed] [Google Scholar]
- 15.Makkar HPS, McSweeney CS. 2005. Methods in gut microbial ecology for ruminants. Springer, Dordrecht, Netherlands. [Google Scholar]
- 16.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson G, Cox F, Kittelmann S, Miri VH, Zethof M, Noel SJ, Waghorn GC, Janssen PH. 2013. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One 8:e74787. doi: 10.1371/journal.pone.0074787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodge-Ivey SL, Browne-Silva J, Horvath MB. 2009. Technical note: bacterial diversity and fermentation end products in rumen fluid samples collected via oral lavage or rumen cannula. J Anim Sci 87:2333–2337. doi: 10.2527/jas.2008-1472. [DOI] [PubMed] [Google Scholar]
- 19.Ramos-Morales E, Arco-Pérez A, Martín-García AI, Yáñez-Ruiz DR, Frutos P, Hervás G. 2014. Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim Feed Sci Technol 198:57–66. doi: 10.1016/j.anifeedsci.2014.09.016. [DOI] [Google Scholar]
- 20.Shen J, Chai Z, Song L, Liu J, Wu Y. 2012. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J Dairy Sci 95:5978–5984. doi: 10.3168/jds.2012-5499. [DOI] [PubMed] [Google Scholar]
- 21.Rius AG, Kittelmann S, Macdonald KA, Waghorn GC, Janssen PH, Sikkema E. 2012. Nitrogen metabolism and rumen microbial enumeration in lactating cows with divergent residual feed intake fed high-digestibility pasture. J Dairy Sci 95:5024–5034. doi: 10.3168/jds.2012-5392. [DOI] [PubMed] [Google Scholar]
- 22.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzales-Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widman J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 25.Seedorf H, Kittelmann S, Henderson G, Janssen PH. 2014. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J 2:e494. doi: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kittelmann S, Devente SR, Kirk MR, Seedorf H, Dehority BA, Janssen PH. 2015. Phylogeny of intestinal ciliates, including Charonina ventriculi, and comparison of microscopy and 18S rRNA gene pyrosequencing for rumen ciliate community structure analysis. Appl Environ Microbiol 81:2433–2444. doi: 10.1128/AEM.03697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koetschan C, Kittelmann S, Lu J, Al-Halbouni D, Jarvis GN, Müller T, Wolf M, Janssen PH. 2014. Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS One 9:e91928. doi: 10.1371/journal.pone.0091928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aronesty E. 2011. Command-line tools for processing biological sequencing data. http://code.google.com/p/ea-utils.
- 30.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. [Google Scholar]
- 31.Esty WW. 1986. The efficiency of Good's nonparametric coverage estimator. Ann Stat 14:1257–1260. doi: 10.1214/aos/1176350066. [DOI] [Google Scholar]
- 32.Bray JR, Curtis JT. 1957. An ordination of upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 33.R Core Team. 2014. R: a language and environment for statistical computing. R Core Team, R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 34.de Mendiburu F. 2014. agricolae: statistical procedures for agricultural research. R package, version 1.2-1 http://CRAN.R-project.org/package=agricolae.
- 35.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Gloeckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig W, Strunk O, Westram R, Richter L, Meier H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 39.Hungate RE. 1966. The rumen and its microbes. Academic Press Inc, New York, NY. [Google Scholar]
- 40.Koike S, Yoshitani S, Kobayashi Y, Tanaka K. 2003. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol Lett 229:23–30. doi: 10.1016/S0378-1097(03)00760-2. [DOI] [PubMed] [Google Scholar]
- 41.Koike S, Kobayashi Y. 2009. Fibrolytic rumen bacteria: their ecology and functions. Asian Aust J Anim Sci 22:131–138. doi: 10.5713/ajas.2009.r.01. [DOI] [Google Scholar]
- 42.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmieder R, Edwards R. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S, Fan C, Deutsch S, Gagic D, Seedorf H, Kelly WJ, Atua R, Sang C, Soni P, Li D, Pinares-Patiño CS, McEwan JC, Janssen PH, Chen F, Visel A, Wang Z, Attwood GT, Rubin EM. 2014. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belay N, Johnson R, Rajagopal BS, Conway de Macario E, Daniels L. 1988. Methanogenic bacteria from human dental plaque. Appl Environ Microbiol 54:600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horz H-P, Conrads G. 2010. The discussion goes on: what is the role of Euryarchaeota in humans? Archaea 2010:967271. doi: 10.1155/2010/967271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. 2004. Methanogenic archaea and human periodontal disease. Proc Natl Acad Sci U S A 101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petri RM, Schweiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS One 8:e83424. doi: 10.1371/journal.pone.0083424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irbis C, Ushida K. 2004. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J Gen Appl Microbiol 50:203–212. doi: 10.2323/jgam.50.203. [DOI] [PubMed] [Google Scholar]
- 50.Tymensen LD, Beauchemin KA, McAllister TA. 2012. Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology 158:1808–1817. doi: 10.1099/mic.0.057984-0. [DOI] [PubMed] [Google Scholar]
- 51.Hillman K, Lloyd D, Williams AG. 1988. Interactions between the methanogen, Methanosarcina barkeri, and rumen holotrich ciliate protozoa. Lett Appl Microbiol 7:49–53. doi: 10.1111/j.1472-765X.1988.tb01250.x. [DOI] [Google Scholar]
- 52.Hungate RE, Dougherty RW, Bryant MP, Cello RM. 1952. Microbiological and physiological changes associated with acute indigestion in sheep. Cornell Vet 42:423–449. [PubMed] [Google Scholar]
- 53.Moss AR, Givens DI, Garnsworthy PC. 1995. The effect of supplementing grass silage with barley on digestibility, in sacco degradability, rumen fermentation and methane production in sheep at two levels of intake. Anim Feed Sci Technol 55:9–33. doi: 10.1016/0377-8401(95)00799-S. [DOI] [Google Scholar]
- 54.Li RW, Wu S, Baldwin RL VI, Li W, Li C. 2012. Perturbation dynamics of the rumen microbiota in response to exogenous butyrate. PLoS One 7:e29392. doi: 10.1371/journal.pone.0029392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol 79:3744–3755. doi: 10.1128/AEM.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bryant MP. 1970. Normal flora-rumen bacteria. Am J Clin Nutr 23:1440–1450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.