Abstract
The plasmid pGKE75-catA138T, which comprises pUC18 and the catA138T gene encoding thermostable chloramphenicol acetyltransferase with an A138T amino acid replacement (CATA138T), serves as an Escherichia coli-Geobacillus kaustophilus shuttle plasmid that confers moderate chloramphenicol resistance on G. kaustophilus HTA426. The present study examined the thermoadaptation-directed mutagenesis of pGKE75-catA138T in an error-prone thermophile, generating the mutant plasmid pGKE75αβ-catA138T responsible for substantial chloramphenicol resistance at 65°C. pGKE75αβ-catA138T contained no mutation in the catA138T gene but had two mutations in the pUC replicon, even though the replicon has no apparent role in G. kaustophilus. Biochemical characterization suggested that the efficient chloramphenicol resistance conferred by pGKE75αβ-catA138T is attributable to increases in intracellular CATA138T and acetyl-coenzyme A following a decrease in incomplete forms of pGKE75αβ-catA138T. The decrease in incomplete plasmids may be due to optimization of plasmid replication by RNA species transcribed from the mutant pUC replicon, which were actually produced in G. kaustophilus. It is noteworthy that G. kaustophilus was transformed with pGKE75αβ-catA138T using chloramphenicol selection at 60°C. In addition, a pUC18 derivative with the two mutations propagated in E. coli at a high copy number independently of the culture temperature and high plasmid stability. Since these properties have not been observed in known plasmids, the outcomes extend the genetic toolboxes for G. kaustophilus and E. coli.
INTRODUCTION
ColE1-type plasmids, such as pBR322 and pUC, replicate in Escherichia coli autonomously with substantial copy numbers and are extensively utilized in microbial genetic engineering. As shown by pBR322 (Fig. 1), the replicon of ColE1-type plasmids generally contains genes for a precursor of RNA primer (RNA II), a replication-regulatory RNA (RNA I), and a replication-regulatory protein (Rom). The precursor RNA II is transcribed from 555 bp upstream of the replication origin and adopts a stem-loop structure that forms a persistent hybrid at the origin. This structure is subsequently cleaved by RNase H to serve as a primer for plasmid replication (1). RNA I consists of 108 nucleotides transcribed from 445 bp upstream of the origin to near the initiation site for RNA II synthesis (2–4). Because RNA I synthesis proceeds in the direction opposite to that of RNA II synthesis, RNA I can hybridize to RNA II and trigger conformational changes in RNA II. This event, which prevents RNA II from hybridization at the replication origin, inhibits plasmid replication and decreases the plasmid copy number. Plasmid replication is also negatively regulated by the Rom protein (2, 5–8), which facilitates the initial interaction between RNA I and RNA II. Thus, the copy number of ColE1-type plasmids is under the tripartite control of RNA I, RNA II, and the Rom protein.
FIG 1.
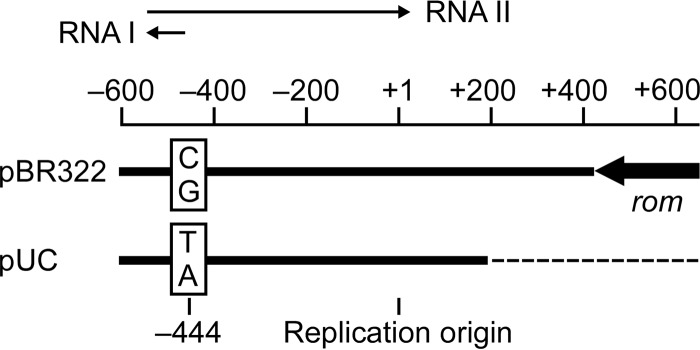
pBR322 and pUC replicons. The replication origin is indicated at position +1. RNA I and RNA II are indicated, with their transcription directions. Compared with the pBR322 replicon, the pUC replicon lacks a rom (also known as rop) gene and has a point mutation (C·G→T·A transition) at position −444.
Genetic alterations in or near the replicon are known to affect the copy number of ColE1-type plasmids (9–12). Although the pUC plasmids are pBR322 derivatives, their replicons lack a rom gene and have a point mutation in the RNA II gene (Fig. 1). Consequently, pUC plasmids are present in higher copy numbers than pBR322 (9). Both of the alterations are essential for pUC plasmids to achieve high copy numbers; pBR322 derivatives that have either the rom deletion or the point mutation show copy numbers comparable to that of pBR322 (9). It is known that the copy numbers of pUC plasmids change depending on the culture temperature of the host E. coli cells. This appears to arise from a temperature-dependent secondary structure within RNA II (9).
Geobacillus kaustophilus HTA426 is an aerobic, Gram-positive, Bacillus-related thermophile that was isolated from the deep-sea sediments of the Mariana Trench (13, 14). We have studied this strain as a model for Geobacillus spp., because basal genetic tools (15–18) and a whole-genome sequence (19) are available for the strain. In a previous study (20), we constructed G. kaustophilus MK480 from the HTA426 strain by inactivating DNA repair genes and demonstrated that MK480, an error-prone thermophile that exhibits frequent mutations in vivo, can generate mutant genes encoding more thermostable variants than the parent enzymes during periods of cell growth at high temperatures. This approach, thermoadaptation-directed evolution, was further employed on the plasmid pGKE75 carrying the cat gene (pGKE75-cat) (Fig. 2A) (21). The cat gene encodes the chloramphenicol acetyltransferase (CAT) from Staphylococcus aureus, which confers chloramphenicol (Cm) resistance on host bacteria (22). pGKE75 is an E. coli-G. kaustophilus shuttle plasmid that includes the pUC18 plasmid (Fig. 2B). The thermoadaptation-directed evolution was performed by successive propagation of pGKE75-cat in MK480 cells under growth inhibition by Cm pressure, resulting in the generation of a mutant plasmid, pGKE75-catA138T, which encodes a CAT variant (CATA138T) having enhanced thermostability due to an A138T amino acid replacement (21).
FIG 2.
Structures of pGKE75-cat (A), pUC18 (B), and pUC replicons of their derivatives (C). The replication origin is indicated at position +1. Mutation sites are indicated by open circles. pGKE75-cat and pGKE75-catA138T shared the usual pUC replicon with pUC18. pGKE75αβ-catA138T and pUC18αβ carry both a G·C→T·A transversion at +175 and a G·C→A·T transition at −252. pGKE75α-catA138T and pUC18α carry a G·C→T·A transversion at +175. pGKE75β-catA138T and pUC18β carry a G·C→A·T transition at −252. pUC18γ carries a T·A→C·G transition at −444 and thus has a pBR322 replicon lacking the rom gene. Pgk704, a promoter functional in G. kaustophilus (18); bla, ampicillin resistance gene functional in E. coli; TK101, kanamycin resistance gene functional at high temperatures (40); pUC, pUC replicon functional in E. coli; pBST1, pBST1 replicon functional in Geobacillus spp. (38); and oriT, conjugative-transfer origin from pRK2013 (44).
Antibiotic resistance genes are commonly used as selectable markers during complex genetic modifications of microbes. However, a single kanamycin resistance gene has been the only antibiotic resistance marker used in G. kaustophilus HTA426 (15–18, 20). Although cat and catA138T genes may serve as selectable markers, neither pGKE75-cat nor pGKE75-catA138T conferred Cm resistance on G. kaustophilus at temperatures higher than 65°C (21). Therefore, this study was designed to generate catA138T mutants that function at higher temperatures via the further thermoadaptation-directed evolution of pGKE75-catA138T. Here, we report a new plasmid that confers Cm resistance at 65°C, which was generated via unexpected mutations in the pUC replicon of pGKE75-catA138T.
MATERIALS AND METHODS
Bacterial strains and media.
G. kaustophilus strains MK242 and MK480 were constructed previously (20). Strain MK480 was used as an error-prone thermophile for thermoadaptation-directed evolution, and MK242 was used for genetic characterization because of its genetic stability. These strains were essentially grown at 60°C in Luria-Bertani (LB) medium, but G. kaustophilus(pGKE75 derivative) was grown in LB medium supplemented with 5 mg liter−1 of kanamycin (LK5). E. coli DH5α (TaKaRa Bio, Otsu, Japan) was used for DNA manipulation. E. coli DH5α(pUC18) or E. coli DH5α(pGKE75-cat derivative) and E. coli DH5α(pUB307) were grown at 37°C in LB medium supplemented with ampicillin (50 mg liter−1) and kanamycin (25 mg liter−1), respectively.
Plasmids and primers.
Table 1 lists the plasmids used, and Fig. 2 shows their structures. Primers are summarized in Table 2. pGKE75-cat and pGKE75-catA138T were constructed previously (21). pUC18αβ was constructed from pUC18 (TaKaRa Bio) by site-directed mutagenesis as follows. The pUC18 replicon was amplified using primers colEF1 and colER2. PCR was performed using PrimeStar HS DNA polymerase (TaKaRa Bio) in accordance with the manufacturer's protocol. Using the two strands of the resulting fragment (458 bp) as mutagenesis primers, pUC18 was replicated in vitro in a mixture (25 μl) that contained the fragment (0.2 μg), pUC18 (0.1 μg), deoxynucleoside triphosphates (dNTPs) (0.4 mM each), 1× buffer, and PrimeStar HS DNA polymerase (1.2 units). The reaction was performed in 17 cycles of 98°C for 10 s, 55°C for 10 s, and 72°C for 8.5 min. The template pUC18 was digested with DpnI. Subsequently, the resulting DNA was propagated in E. coli to obtain pUC18αβ. This approach was also used to construct the plasmids pUC18α, pUC18β, pUC18γ, pGKE75α-catA138T, and pGKE75β-catA138T. pUC18α, pUC18β, and pUC18γ were generated from pUC18 using mutagenesis primers colEF1 and colER1, colEF2 and colER2, and colEF3 and colER3, respectively. pGKE75α-catA138T and pGKE75β-catA138T were generated from pGKE75-catA138T using primers colEF1 and colER1, and colEF2 and colER2, respectively.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pGKE75-cat | E. coli-Geobacillus shuttle plasmid; pUC pBST1 TK101 bla cat oriT | 21 |
| pGKE75-catA138T | pGKE75-cat derivative carrying catA138T instead of cat | 21 |
| pGKE75αβ-catA138T | pGKE75-catA138T derivative carrying two mutations in the pUC replicon | |
| pGKE75α-catA138T | pGKE75-catA138T derivative carrying one mutation in the pUC replicon | |
| pGKE75β-catA138T | pGKE75-catA138T derivative carrying one mutation in the pUC replicon | |
| pUC18 | E. coli plasmid; pUC replicon; bla lacZα | |
| pUC18αβ | pUC18 derivative carrying two mutations in the pUC replicon | |
| pUC18α | pUC18 derivative carrying one mutation in the pUC replicon | |
| pUC18β | pUC18 derivative carrying one mutation in the pUC replicon | |
| pUC18γ | pUC18 derivative carrying pBR322 replicon without rom | |
| pUB307 | Derivative of IncP-1 plasmid RP1; tra oriT kan tet | 43 |
Plasmid structures are shown in Fig. 2. pUC and pBST1 denote replicons functional in E. coli and Geobacillus spp., respectively. TK101 is a kanamycin resistance gene functional in G. kaustophilus (40). bla, cat, kan, and tet are ampicillin, chloramphenicol, kanamycin, and tetracycline resistance genes, respectively, for E. coli. oriT is the conjugative-transfer origin from pRK2013 (44). pUB307 carries tra genes responsible for conjugative DNA transfer.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| blaF | TGCTGAAGATCAGTTGGGTG |
| blaR | TTGTTGCCGGGAAGCTAGAG |
| catF1 | GGACGACGATGACAAAATGCAATTTAATAAAATTG |
| catF2 | GCGCATGCTGGACTACAAGGACGACGATGAC |
| catR | GCCGGATCCTTATAAAAGCCAGTCATTAG |
| colEF1 | CGCTCCAAGCTGGGTTGTGTGCACGAACCC |
| colER1 | GGGTTCGTGCACACAACCCAGCTTGGAGCG |
| colEF2 | GTATTGGGCGCTCTTCAGCTTCCTCGCTCACTG |
| colER2 | CAGTGAGCGAGGAAGCTGAAGAGCGCCCAATAC |
| colEF3 | CGGCTACACTAGAAGGACAGTATTTGGTAT |
| colER3 | ATACCAAATACTGTCCTTCTAGTGTAGCCG |
| ECrpoAF | GCCTTTAGAGCGTGGCTTTG |
| ECrpoAR | TTTCGATGACCAGCTTGTCC |
| GKrpoAF | CAACCTTAGGGAACTCCTTG |
| GKrpoAR | TTCGGCCCAATGCTTCCATC |
| repF1 | GCTTGCAAACAAAAAAACCACCGCTACCAG |
| repR1 | GTTTTTCCATAGGCTCCGCCCCCCTGACGAG |
| repR2 | GAGAGGCGGTTTGCGTATTGGGCGCTCTTC |
Plasmid introduction into G. kaustophilus.
pGKE75-cat derivatives were introduced into G. kaustophilus using conjugative DNA transfer from E. coli DH5α (16). G. kaustophilus was used as the DNA recipient. E. coli(pGKE75-cat derivative) and E. coli(pUB307) were used as the DNA donor and conjugation helper, respectively. These strains were cultured until the optical density at 600 nm (OD600) reached 0.3. The cultures were then mixed and spotted on LB plates, followed by incubation at 37°C to achieve conjugation. In this process, E. coli(pGKE75-cat derivative) transfers the plasmid to G. kaustophilus with the mediation of E. coli(pUB307). Subsequently, the cell mixture was recovered from LB plates and incubated at 60°C on LK5 plates to isolate G. kaustophilus transformants.
Thermoadaptation-directed evolution of pGKE75-catA138T.
LK5 plates containing Cm (10 mg liter−1) were used as the solid medium. G. kaustophilus MK480(pGKE75-catA138T) was cultured on solid medium at 60°C for 24 h. The resulting cells were collected and incubated for 1 h at 30°C in the presence of 20 mM hydrogen peroxide to induce mutations (cell survival rate, 1%). The surviving cells (104 cells) were regrown on solid medium at 60°C. This process was repeated three more times. Subsequently, the cells were cultivated on solid medium at 60°C, followed by cell collection. This process was performed successively at 65°C and 70°C. The plasmid mixtures were then extracted from the resultant cells and reintroduced into G. kaustophilus MK242. The transformants were incubated on solid medium at 65°C to obtain clones obviously resistant to Cm. pGKE75-catA138T derivatives were isolated from these clones and subjected to DNA sequence analysis.
Cm resistance assay.
G. kaustophilus MK242(pGKE75-cat derivative) was precultured in liquid LK5. Equal-volume aliquots of culture were incubated at various temperatures for 24 h on LK5 plates with or without Cm (5 mg liter−1). The resulting colonies were counted to calculate the Cm resistance efficiency, which is the ratio of Cm-resistant colonies (grown on LK5 with Cm) to the total number of colonies (grown on LK5 without Cm). The efficiency was determined from four independent experiments and is presented as the mean and standard deviation (SD).
CAT activity assay.
G. kaustophilus MK242(pGKE75-cat derivative) was cultured at 65°C for 18 h on LK5 plates. Cells were collected and suspended in buffer (50 mM potassium phosphate, pH 7.0). The suspension was homogenized by sonication and then centrifuged to remove cell debris. The supernatant (cell extract) was used for further analysis. The protein concentration in cell extracts was analyzed using the Bradford method. CAT activity was assayed at 60°C according to an established method (21). One unit was defined as the amount of enzyme that produces 1 μmol of coenzyme A (CoA) per min.
Acetyl-CoA assay.
The concentration of acetyl-CoA in cell extracts (see above) was determined using the PicoProbe Acetyl-CoA Fluorometric Assay kit (BioVision Inc., CA, USA). The data were normalized by the protein concentration of cell extracts.
Assay of plasmid copy number.
G. kaustophilus MK242(pGKE75-cat derivative) was cultured on LK5 plates at 65°C for 18 h. E. coli DH5α(pUC18 derivative) was cultured in liquid LB at 20 to 42°C until the OD600 reached 1.0. Total DNA was extracted from cells using the method of Wu and Welker (23). Plasmid copy numbers of pGKE75-cat and pUC18 derivatives in total DNA were analyzed using quantitative-competitive-PCR (QC-PCR) (24). The plasmid concentration was determined using primers blaF and blaR, which amplify a portion of the bla gene (positions in the open reading frame, 99 to 612; 514 bp). Artificial short fragments that consisted of positions 99 to 251 and 459 to 612 of the bla gene were used as competitor DNA. The chromosome concentration was determined using primers to amplify a portion of the rpoA gene. Primers GKrpoAF and GKrpoAR were used to amplify G. kaustophilus rpoA (positions 101 to 620; 520 bp). E. coli rpoA (positions 87 to 613; 527 bp) was amplified using ECrpoAF and ECrpoA. Competitor DNA consisted of positions 101 to 240 and 440 to 620 of G. kaustophilus rpoA and positions 87 to 249 and 445 to 613 of E. coli rpoA. QC-PCR was performed using Quick Taq HS DyeMix (Toyobo, Osaka, Japan) in the presence of competitor DNA (1.0 pM to 54 nM) in 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min. The products were separated by agarose gel electrophoresis and visualized using ethidium bromide. DNA bands were quantified using the ImageJ program (http://rsb.info.nih.gov/ij). These data were used to calculate the ratio of bla to rpoA copies in total DNA, which was defined as the plasmid copy number. The data are presented as mean and SD (n = 3 or 4).
Transcriptional analysis of the mutant pUC replicon.
G. kaustophilus MK242(pGKE75αβ-catA138T) was cultured on LK5 plates at 65°C for 18 h. Total RNA was prepared from the resulting cells using an RNeasy minikit with RNAprotect Bacteria Reagent (Qiagen, Venlo, Netherlands). The RNA obtained from this procedure was treated with gDNA Eraser (TaKaRa Bio) to eliminate genomic DNA and then used for two-step reverse transcription (RT)-PCR to detect transcripts from the mutant pUC replicon. Reverse transcription was performed using the PrimeScript RT reagent kit (TaKaRa Bio) with primer repF1 or repR2. PCR was performed using Quick Taq HS DyeMix in 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min. Primers repF1 and repR1 were used to detect transcripts between −559 and +1, and repF1 and repR2 were used for those between −559 and +205.
Plasmid stability assay of pUC18 derivatives.
E. coli(pUC18 derivative) was precultured in liquid LB medium supplemented with ampicillin. An aliquot of culture (103 cells) was then cultured at 20 to 42°C in liquid LB without antibiotics until the culture reached stationary phase. This culture was repeated successively two more times. The resulting cells were grown at 37°C on LB plates, and then, 100 colonies were screened for ampicillin resistance to calculate the plasmid retention rate. The data are presented as means and SD (n = 3).
Plasmid transformation of G. kaustophilus using Cm selection.
pGKE75-cat derivatives were introduced into G. kaustophilus by conjugative DNA transfer (see above). The E. coli donor and conjugation helper were cultured in 10 ml of LB medium; G. kaustophilus MK242 was cultured in 100 ml. These cultures were mixed and incubated on LB plates at 37°C for 18 h, followed by incubation at 60°C for 1 h. The resulting cells were collected in LB medium, and equal-volume aliquots (G. kaustophilus cells, 107 to 108) were incubated at 60°C for 24 h on LB plates supplemented with Cm (10 mg liter−1) and LK5 plates. Colonies were counted to calculate the selection efficiency, which is the ratio of the number of colonies grown using Cm selection (grown on LB plates with Cm) to the number of colonies grown using kanamycin selection (grown on LK5 plates). The data are presented as the means and SD (n = 5).
RESULTS
Generation of pGKE75αβ-catA138T.
We first examined thermoadaptation-directed evolution of pGKE75-catA138T by simple successive cultures of G. kaustophilus MK480(pGKE75-catA138T) under Cm selection pressure. While the strain could intrinsically grow at 65°C with tiny colonies only when spread at high cell density, this approach provided no colonies that were more Cm resistant than the parent cells (population rate, <0.01%). The result led us to treat cells with hydrogen peroxide between successive cultures for more frequent mutations. This approach produced many colonies that showed substantial Cm resistance at 70°C following 7 successive cultures (population rate, approximately 1%). pGKE75-catA138T was extracted from these colonies in a mixture and reintroduced into G. kaustophilus MK242 to eliminate false positives (21, 25, 26). Using this process, we identified four clones that showed obvious Cm resistance at 65°C in dozens of clones. The pGKE75-catA138T derivatives were isolated from these clones, and their sequences in the Pgk704-catA138T region were determined. Since none of the four sequences contained mutations, we further analyzed the entire sequence of the plasmid that conferred the most efficient Cm resistance on G. kaustophilus. The mutant plasmid, designated pGKE75αβ-catA138T, contained two mutations in the pUC replicon: a C·G→A·T transversion and a C·G→T·A transition (Fig. 2). The C·G→T·A transition was presumably due to a spontaneous mutation in MK480, because this type of mutation is intrinsically frequent in G. kaustophilus (20). Meanwhile, the C·G→A·T transversion was attributable to hydrogen peroxide, because it facilitates generation of the aberrant base 7,8-dihydro-8-oxoguanine (27), which causes C·G→A·T transversions (28, 29).
Cm resistance conferred by pGKE75-cat derivatives.
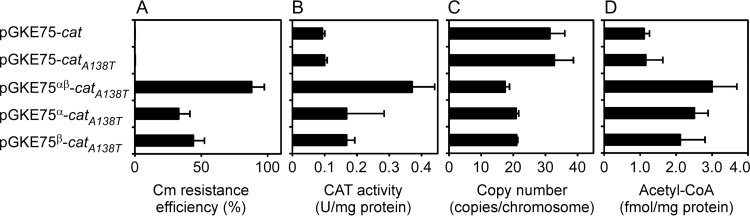
G. kaustophilus MK242(pGKE75-cat) showed efficient Cm resistance at 50°C, and MK242(pGKE75-catA138T) showed it at 55°C; however, MK242(pGKE75αβ-catA138T) showed substantial Cm resistance at 60 to 65°C (Table 3). The pGKE75-catA138T derivatives with either of the two mutations, pGKE75α-catA138T and pGKE75β-catA138T, conferred moderate Cm resistance at 65°C (Fig. 3A). The observation suggests that both mutations in the mutant pUC replicon participate in the process by which pGKE75αβ-catA138T confers efficient Cm resistance.
TABLE 3.
Cm resistance efficiencies conferred by pGKE75-cat derivatives
| Plasmid | Cm resistance efficiency (%) at culture temp (°C)a: |
||||
|---|---|---|---|---|---|
| 50 | 55 | 60 | 65 | 70 | |
| pGKE75-cat | 66 ± 25 | 37 ± 8 | 27 ± 3 | <1 | <1 |
| pGKE75-catA138T | 56 ± 18 | 77 ± 4 | 46 ± 6 | <1 | <1 |
| pGKE75αβ-catA138T | 21 ± 4 | 20 ± 6 | 71 ± 6 | 88 ± 12 | 2 ± 1 |
G. kaustophilus MK242(pGKE75-cat derivative) was precultured in liquid LK5 and incubated at the indicated temperature on LK5 plates with and without Cm (5 mg liter−1). Cm resistance efficiency was defined as the ratio of Cm-resistant colonies to the total number of colonies. The data are presented as means ± SD (n = 4).
FIG 3.
Effects of pGKE75-cat derivatives on G. kaustophilus. G. kaustophilus MK242(pGKE75-cat derivative) was grown at 65°C on LK5 plates and analyzed for Cm resistance efficiency (A), intracellular CAT activity (B), plasmid copy number (C), and acetyl-CoA concentration (D). (A) Cells were incubated at 65°C on LK5 plates with and without Cm to determine the Cm resistance efficiency. The data are presented as means and SD (n = 4). (B) Cell extract was prepared and analyzed for CAT specific activity. The data are presented as means and SD (n = 5). (C) Total DNA was extracted from cells and used to analyze the plasmid copy number, which was determined using the ratio of bla (in pGKE75-cat derivatives) to rpoA (in the G. kaustophilus chromosome). The data are presented as means and SD (n = 3). (D) Cell extract was prepared and analyzed for acetyl-CoA concentration. The data were normalized by the protein concentration and are presented as means and SD (n = 4).
CAT activity in G. kaustophilus.
To examine the reasons for efficient Cm resistance conferred by pGKE75αβ-catA138T, we analyzed the intracellular CAT activity in G. kaustophilus MK242(pGKE75-cat derivative) (Fig. 3B). MK242(pGKE75αβ-catA138T) had higher CAT activity than MK242(pGKE75-cat or pGKE75-catA138T). pGKE75α-catA138T and pGKE75β-catA138T caused activity between that of pGKE75αβ-catA138T and those of pGKE75-cat and pGKE75-catA138T. Because the specific activities of the CAT and CATA138T proteins were comparable (21), it is likely that these data reflect the intracellular concentration of active CAT protein. Thus, there was a correlation between Cm resistance efficiency and the CAT concentration in MK242(pGKE75-cat derivative).
Plasmid copy numbers of pGKE75-cat derivatives in G. kaustophilus.
Plasmid copy numbers often affect gene expression from plasmids. In general, a higher number of copies leads to increased gene expression (30). Consequently, we analyzed the copy numbers of pGKE75-cat derivatives in G. kaustophilus MK242 using QC-PCR (Fig. 3C). The copy number of pGKE75αβ-catA138T was 17 ± 2 copies per chromosome, which was lower than those of pGKE75-cat and pGKE75-catA138T, contrary to our expectation. pGKE75α-catA138T and pGKE75β-catA138T showed copy numbers between that of pGKE75αβ-catA138T and those of pGKE75-cat and pGKE75-catA138T. Thus, there was a negative correlation between the Cm resistance efficiencies conferred by pGKE75-cat derivatives and their copy numbers in G. kaustophilus.
Acetyl-CoA concentration in G. kaustophilus.
Because CAT inactivates Cm using acetyl-CoA as the substrate, we analyzed the acetyl-CoA concentration in G. kaustophilus MK242(pGKE75-cat derivative) (Fig. 3D). MK242(pGKE75αβ-catA138T) had more abundant acetyl-CoA than MK242(pGKE75-cat or pGKE75-catA138T). MK242(pGKE75α-catA138T) and MK242(pGKE75β-catA138T) showed acetyl-CoA concentrations higher than those of MK242(pGKE75-cat) and MK242(pGKE75-catA138T) but less than that of MK242(pGKE75αβ-catA138T). These data indicate a positive correlation between Cm resistance efficiencies and acetyl-CoA concentrations in MK242(pGKE75-cat derivative).
Transcription from the mutant pUC replicon.
RNA transcribed from the mutant pUC replicon in G. kaustophilus MK242(pGKE75αβ-catA138T) was analyzed using RT-PCR (see Fig. S1 in the supplemental material). The analysis detected a transcript from −559 to +1 in the mutant pUC replicon (RNA−559/+1) and its complementary transcript (RNA+1/−559). RT-PCR also detected a longer transcript from −559 to +205 (RNA−559/+205), which could contain the two mutations, although not its complement. The observation suggests that the mutant pUC replicon is actually transcribed in G. kaustophilus, even though it arises from incidental promoter sequences.
G. kaustophilus transformation using Cm selection.
To examine whether G. kaustophilus can be transformed with pGKE75αβ-catA138T using Cm selection, the plasmid was introduced into G. kaustophilus MK242 by conjugative DNA transfer. A cell mixture containing the donor, recipient, and conjugation helper was incubated at 37°C for conjugation and subsequently preincubated at 60°C. An aliquot of cells was then incubated at 60°C on LB plates supplemented with Cm, providing 53 transformants (conjugation efficiency, 10−5 to 10−6 recipient−1). An equal aliquot provided 122 transformants on LK5 plates. Five repeated analyses indicated that transformants obtained using Cm selection accounted for 36% ± 13% of those obtained using kanamycin selection. Preincubation was essential for transformant growth, probably because of sufficient production of CATA138T prior to Cm exposure. Almost all the transformants (>95%) obtained using Cm selection were resistant to kanamycin, confirming that the number of false positives was negligible. pGKE75-cat and pGKE75-catA138T provided few transformants using Cm selection. Thus, pGKE75αβ-catA138T is a unique plasmid for G. kaustophilus; its introduction can be selected using Cm resistance at 60°C.
Plasmid copy numbers of pUC18 derivatives in E. coli.
The plasmid pUC18αβ, which contains the mutant pUC replicon of pGKE75αβ-catA138T, was constructed from pUC18 and analyzed for plasmid copy numbers (Fig. 4). QC-PCR analysis indicated that pUC18 propagates with 310 ± 12 copies at 37°C. The copy number increased with increasing culture temperature, in agreement with previous observations (9). However, pUC18αβ exhibited a substantial but temperature-independent copy number (120 to 210 copies at 25 to 42°C). pUC18α and pUC18β showed temperature-dependent and independent profiles, respectively, but both had lower copy numbers than pUC18αβ. Because a pBR322 derivative lacking a rom gene is also known to exhibit a temperature-independent copy number, we analyzed the copy number of pUC18γ, which had the replicon of pBR322 lacking the rom gene. Although pUC18γ exhibited a temperature-independent profile, like pUC18αβ, its copy number was much lower at all temperatures examined. Thus, pUC18αβ had a unique copy number profile different from those of known pUC plasmids.
FIG 4.

Plasmid copy number of pUC18 (top; solid bars), pUC18αβ (top; open bars), pUC18α (bottom; solid bars), pUC18β (bottom; open bars), and pUC18γ (bottom; gray bars) in E. coli. E. coli(pUC18 derivative) was cultured at the indicated temperatures. Total DNA was extracted from the cells and used to analyze the plasmid copy number, which is the ratio of bla (in pUC18 derivatives) to rpoA (in the E. coli chromosome). The data are presented as means and SD (n = 3).
Plasmid stability of pUC18 derivatives in E. coli.
The plasmid stability of pUC18 derivatives was assessed on the basis of plasmid retention rates following 3 successive cultures of E. coli(pUC18 derivative). pUC18 and pUC18α were notably unstable at 42°C and 20 to 30°C, respectively (Table 4). However, pUC18αβ, pUC18β, and pUC18γ showed excellent stability at 20 to 42°C. pUC18αβ, pUC18β, and pUC18γ were completely retained even following 7 successive cultures at 37°C, whereas pUC18 and pUC18α were retained at rates of 79% ± 7% and 55% ± 8%, respectively.
TABLE 4.
Plasmid retention rates of pUC18 derivatives following successive cultures
| Plasmid | Plasmid retention rate (%) at culture temp (°C)a: |
||||
|---|---|---|---|---|---|
| 20 | 25 | 30 | 37 | 42 | |
| pUC18 | 98 ± 0 | 97 ± 3 | 98 ± 2 | 96 ± 5 | 3 ± 5 |
| pUC18αβ | >99 | >99 | >99 | >99 | >99 |
| pUC18α | <1 | <1 | <1 | 94 ± 4 | 92 ± 11 |
| pUC18β | >99 | 97 ± 5 | 97 ± 5 | >99 | >99 |
| pUC18γ | >99 | >99 | >99 | >99 | >99 |
E. coli(pUC18 derivative) was successively cultured in liquid LB three times. The resulting cells were screened for ampicillin resistance to calculate the plasmid retention rate. The data are presented as means ± SD (n = 3).
DISCUSSION
A new plasmid responsible for Cm resistance at high temperatures, pGKE75αβ-catA138T, was generated from pGKE75-catA138T through a directed evolution process. The mutations in pGKE75αβ-catA138T were found in the pUC replicon and caused efficient Cm resistance at high temperatures, although the pUC replicon has no role, in theory, in G. kaustophilus. An immediate reason for this is that the mutation increased the intracellular concentration of active CATA138T protein. An increased concentration of intracellular acetyl-CoA could also contribute to the Cm resistance because acetyl-CoA is the substrate for CAT reaction. This idea is supported by the fact that some E. coli strains exhibit Cm sensitivity despite sufficient cat expression because of a low intracellular concentration of acetyl-CoA (31, 32). The increased concentrations of CATA138T and acetyl-CoA can be attributed to the decreased copy number of pGKE75αβ-catA138T. It is known that plasmid maintenance places a metabolic burden on bacteria and/or inhibits cell growth (30, 33–35), presumably because of the considerable bioenergy required for replication. In addition, acetyl-CoA is often used during the synthesis of metabolites that store redundant bioenergy in bacteria, such as polyhydroxybutyrate (36) and fatty acids. Therefore, it is possible that the decrease in the plasmid copy number leads to redundant bioenergy and facilitates the biosynthesis of both CATA138T and acetyl-CoA. It is unlikely that the decreased copy number resulted in much slower CATA138T production, thereby increasing soluble and active CATA138T, because, when analyzed by Western blotting, CATA138T seemed to be more abundantly produced from pGKE75αβ than pGKE75 as soluble forms while being equally produced as insoluble forms (see Fig. S2 in the supplemental material).
We note that the actual copy numbers of the intact pGKE75-cat derivatives could be much lower than the values determined in this study. We previously determined the copy number of pUCG18T, which is a parent plasmid of pGKE75-cat (16, 21), and another plasmid, pSTE33T, using the method of Wu and Welker (23). This method, using agarose gel electrophoresis, clearly detected the pSTE33T band in the agarose gel and determined its copy number as 16 copies. Meanwhile, the copy number of pSTE33T was determined to be 7 ± 3 (n = 4) using QC-PCR, confirming that QC-PCR can determine the plasmid copy number as well as the Wu and Welker method. However, the Wu and Welker method could not detect pUCG18T, indicating a lower abundance of the plasmid (17), although QC-PCR determined the copy number to be 13 ± 9 (n = 4). QC-PCR also determined that there were >17 copies of each pGKE75-cat derivative, whereas none of the pGKE75-cat derivatives were detected by agarose gel electrophoresis. These observations imply the presence of incomplete forms of pUCG18T and pGKE75-cat derivatives, suggesting that the decrease in pGKE75αβ-catA138T copies, which was indicated by QC-PCR analysis, may reflect the decrease in incomplete forms. Taken together, efficient Cm resistance conferred by pGKE75αβ-catA138T can be explained by an increase in intracellular concentrations of CATA138T and acetyl-CoA following a decrease in the number of incomplete plasmids.
We speculate that the incomplete forms are single-stranded DNA rather than randomly truncated DNA, because they were undetectable by agarose gel electrophoresis and PCR analysis exclusively detected the intact cat gene, but not partial fragments, in G. kaustophilus MK242(pGKE75-cat derivative). It is unclear why the mutations in the pUC replicon suppress production of single-stranded plasmids. However, the pBST1 replicon in pGKE75-cat derivatives shows sequence similarity to the replicon of a plasmid from Bacillus megaterium, pBM300, which is thought to replicate via a theta-type mechanism, as do ColE1-type plasmids (37–39). In theta-type mechanisms, plasmids replicate continuously on the leading strand but discontinuously on the lagging strand. Therefore, single-stranded DNA may accumulate when the plasmid is replicated efficiently on the leading strand but not on the lagging strand. This hypothesis allows us to speculate that RNA species from the mutant pUC replicon, which were actually produced in G. kaustophilus(pGKE75αβ-catA138T), may form thermostable secondary structures and hybridize with the leading strand at the pUC replication origin, as they do in E. coli, under high-temperature conditions and that this event may inhibit DNA elongation on the leading strand to reduce incomplete plasmids.
A kanamycin resistance gene is currently in use as an efficient selectable marker for Geobacillus spp. at temperatures above 65°C (17, 38, 40, 41). Although cat can be used as a selectable marker (23, 42), neither cat nor catA138T conferred Cm resistance on G. kaustophilus at temperatures above 65°C (21). Therefore, it is noteworthy that pGKE75αβ-catA138T conferred Cm resistance at 65°C. Moreover, G. kaustophilus was efficiently transformed with pGKE75αβ-catA138T using Cm selection at 60°C. The selection was performed by incubation for only 1 day without producing false positives. These observations suggest this new plasmid is useful for genetic modifications of G. kaustophilus and perhaps other Geobacillus spp.
In addition to pGKE75αβ-catA138T, this study provided a unique plasmid for E. coli, pUC18αβ, which shows high plasmid stability and a temperature-independent high copy number. pUC18αβ has two mutations compared with pUC18. Based on the properties of pUC18α and pUC18β, which have either of the two mutations, both mutations obviously participate in the process that results in the high plasmid stability and high copy number of pUC18αβ. Mutant RNA species from the mutant pUC replicon are probably responsible for the unique profile of pUC18αβ. One mutation in the RNA II sequence can affect secondary structures within RNA II, thereby affecting plasmid replication. Although the other mutation is outside the RNA II sequence, the mutation can affect the secondary structure of extended RNAs that are partially transcribed beyond the origin during plasmid replication (1). The temperature-independent plasmid copy number of pUC18αβ may arise from thermostable secondary structures of mutant RNA species, in contrast to temperature-sensitive RNAs from pUC18.
In summary, this study generated two plasmids: pGKE75αβ-catA138T and pUC18αβ. Because of their unique properties, these plasmids extend the genetic toolboxes for G. kaustophilus and E. coli. The results also suggest that thermoadaptation-directed evolution using an error-prone thermophile, G. kaustophilus MK480, can generate not only mutant genes encoding thermostable enzyme variants, but also plasmids with unique properties, via unexpected mutations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jun Ishii of Kobe University for advice on Western blotting.
This work was supported by the following organizations: Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, Japan; the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry, Japan; JSPS KAKENHI (grant number 25450105); and the Institute for Fermentation, Osaka, Japan.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01574-15.
REFERENCES
- 1.Itoh T, Tomizawa J. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomizawa J. 1986. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell 47:89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- 3.Masukata H, Tomizawa J. 1986. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell 44:125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- 4.Tomizawa J, Itoh T, Selzer G, Som T. 1981. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A 78:1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eguchi Y, Tomizawa J. 1990. Complex formed by complementary RNA stem-loops and its stabilization by a protein: function of ColE1 Rom protein. Cell 60:199–209. doi: 10.1016/0092-8674(90)90736-X. [DOI] [PubMed] [Google Scholar]
- 6.Tomizawa J. 1990. Control of ColE1 plasmid replication. Interaction of Rom protein with an unstable complex formed by RNA I and RNA II. J Mol Biol 212:695–708. [DOI] [PubMed] [Google Scholar]
- 7.Cesareni G, Cornelissen M, Lacatena RM, Castagnoli L. 1984. Control of pMB1 replication: inhibition of primer formation by Rop requires RNA1. EMBO J 3:1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomizawa J, Som T. 1984. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell 38:871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- 9.Lin-Chao S, Chen W, Wong T. 1992. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol Microbiol 6:3385–3393. doi: 10.1111/j.1365-2958.1992.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 10.Wong EM, Muesing MA, Polisky B. 1982. Temperature-sensitive copy number mutants of ColE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci U S A 79:3570–3574. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupski JR, Projan SJ, Ozaki LS, Godson GN. 1986. A temperature-dependent pBR322 copy number mutant resulting from a Tn5 position effect. Proc Natl Acad Sci U S A 83:7381–7385. doi: 10.1073/pnas.83.19.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelfand DH, Shepard HM, O'Farrell PH, Polisky B. 1978. Isolation and characterization of a ColE1-derived plasmid copy-number mutant. Proc Natl Acad Sci U S A 75:5869–5873. doi: 10.1073/pnas.75.12.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takami H, Inoue A, Fuji F, Horikoshi K. 1997. Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett 152:279–285. doi: 10.1111/j.1574-6968.1997.tb10440.x. [DOI] [PubMed] [Google Scholar]
- 14.Takami H, Nishi S, Lu J, Shinamura S, Takaki Y. 2004. Genomic characterization of thermophilic Geobacillus species isolated from the deepest sea mud of the Mariana Trench. Extremophiles 8:351–356. doi: 10.1007/s00792-004-0394-3. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki H, Murakami A, Yoshida K. 2012. Counterselection system for Geobacillus kaustophilus HTA426 through disruption of pyrF and pyrR. Appl Environ Microbiol 78:7376–7383. doi: 10.1128/AEM.01669-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki H, Wada K, Furukawa M, Doi K, Ohshima T. 2013. A ternary conjugation system for the construction of DNA libraries for Geobacillus kaustophilus HTA426. Biosci Biotechnol Biochem 77:2316–2318. doi: 10.1271/bbb.130492. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H, Yoshida K. 2012. Genetic transformation of Geobacillus kaustophilus HTA426 by conjugative transfer of host-mimicking plasmids. J Microbiol Biotechnol 22:1279–1287. doi: 10.4014/jmb.1203.03023. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Yoshida K, Ohshima T. 2013. Polysaccharide-degrading thermophiles generated by heterologous gene expression in Geobacillus kaustophilus HTA426. Appl Environ Microbiol 79:5151–5158. doi: 10.1128/AEM.01506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takami H, Takaki Y, Chee G-J, Nishi S, Shimamura S, Suzuki H, Matsui S, Uchiyama I. 2004. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res 32:6292–6303. doi: 10.1093/nar/gkh970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H, Kobayashi J, Wada K, Furukawa M, Doi K. 2015. Thermoadaptation-directed enzyme evolution in an error-prone thermophile derived from Geobacillus kaustophilus HTA426. Appl Environ Microbiol 81:149–158. doi: 10.1128/AEM.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi J, Furukawa M, Ohshiro T, Suzuki H. 2015. Thermoadaptation-directed evolution of chloramphenicol acetyltransferase in an error-prone thermophile using improved procedures. Appl Microbiol Biotechnol 99:5563–5572. doi: 10.1007/s00253-015-6522-4. [DOI] [PubMed] [Google Scholar]
- 22.Shaw WV, Brodsky RF. 1968. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol 95:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Welker NE. 1989. Protoplast transformation of Bacillus stearothermophilus NUB36 by plasmid DNA. J Gen Microbiol 135:1315–1324. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu E, Kato H, Nakagawa Y, Kodama T, Futo S, Minegishi Y, Watanabe T, Akiyama H, Teshima R, Furui S, Hino A, Kitta K. 2008. Development of a screening method for genetically modified soybean by plasmid-based quantitative competitive polymerase chain reaction. J Agric Food Chem 56:5521–5527. doi: 10.1021/jf073348n. [DOI] [PubMed] [Google Scholar]
- 25.Mankin AS, Garrett RA. 1991. Chloramphenicol resistance mutations in the single 23S rRNA gene of the archaeon Halobacterium halobium. J Bacteriol 173:3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMurry LM, George AM, Levy SB. 1994. Active efflux of chloramphenicol in susceptible Escherichia coli strains and in multiple-antibiotic-resistant (Mar) mutants. Antimicrob Agents Chemother 38:542–546. doi: 10.1128/AAC.38.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibutani S, Takeshita M, Grollman AP. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 29.Tajiri T, Maki H, Sekiguchi M. 1995. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat Res 336:257–267. doi: 10.1016/0921-8777(94)00062-B. [DOI] [PubMed] [Google Scholar]
- 30.Seo J, Balley JE. 1985. Effects of recombinant plasmid content on growth properties and cloned gene product formation in Escherichia coli. Biotechnol Bioeng 27:1668–1674. doi: 10.1002/bit.260271207. [DOI] [PubMed] [Google Scholar]
- 31.Potrykus J, Wegrzyn G. 2001. Chloramphenicol-sensitive Escherichia coli strain expressing the chloramphenicol acetyltransferase (cat) gene. Antimicrob Agents Chemother 45:3610–3612. doi: 10.1128/AAC.45.12.3610-3612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potrykus J, Wegrzyn G. 2003. The acrAB locus is involved in modulating intracellular acetyl coenzyme A levels in a strain of Escherichia coli CM2555 expressing the chloramphenicol acetyltransferase (cat) gene. Arch Microbiol 180:362–366. doi: 10.1007/s00203-003-0592-x. [DOI] [PubMed] [Google Scholar]
- 33.Ow DS, Nissom PM, Philip R, Oh SK, Yap MG. 2006. Global transcriptional analysis of metabolic burden due to plasmid maintenance in Escherichia coli DH5α during batch fermentation. Enzyme Microb Technol 39:391–398. doi: 10.1016/j.enzmictec.2005.11.048. [DOI] [Google Scholar]
- 34.Helling RB, Kinney T, Adams J. 1981. The maintenance of plasmid-containing organisms in populations of Escherichia coli. J Gen Microbiol 123:129–141. [DOI] [PubMed] [Google Scholar]
- 35.Jones KL, Kim S, Keasling JD. 2000. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab Eng 2:328–338. doi: 10.1006/mben.2000.0161. [DOI] [PubMed] [Google Scholar]
- 36.Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunnimalaiyaan M, Vary PS. 2005. Molecular characterization of plasmid pBM300 from Bacillus megaterium QM B1551. Appl Environ Microbiol 71:3068–3076. doi: 10.1128/AEM.71.6.3068-3076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor MP, Esteban CD, Leak DJ. 2008. Development of a versatile shuttle vector for gene expression in Geobacillus spp. Plasmid 60:45–52. doi: 10.1016/j.plasmid.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson DM, Kunnimalaiyaan M, Müller K, Vary PS. 1998. Characterization of a theta plasmid replicon with homology to all four large plasmids of Bacillus megaterium QM B1551. Plasmid 40:175–189. doi: 10.1006/plas.1998.1359. [DOI] [PubMed] [Google Scholar]
- 40.Liao H, McKenzie T, Hageman R. 1986. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc Natl Acad Sci U S A 83:576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narumi I, Nakayama N, Nakamoto S, Kimura T, Yanagisawa T, Kihara H. 1993. Construction of a new shuttle vector pSTE33 and its stabilities in Bacillus stearothermophilus, Bacillus subtilis, and Escherichia coli. Biotechnol Lett 15:815–820. [Google Scholar]
- 42.Imanaka T, Fujii M, Aramori I, Aiba S. 1982. Transformation of Bacillus stearothermophilus with plasmid DNA and characterization of shuttle vector plasmids between Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol 149:824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett PM, Grinsted J, Richmond MH. 1977. Transposition of TnA does not generate deletions. Mol Gen Genet 154:205–211. doi: 10.1007/BF00330839. [DOI] [PubMed] [Google Scholar]
- 44.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.