Abstract
Deleterious mutations contribute to polymorphism even when selection effectively prevents their fixation. The efficacy of selection in removing deleterious mitochondrial mutations from populations depends on the effective population size (Ne) of the mitochondrial DNA and the degree to which a lack of recombination magnifies the effects of linked selection. Using complete mitochondrial genomes from Drosophila melanogaster and nuclear data available from the same samples, we reexamine the hypothesis that nonrecombining animal mitochondrial DNA harbor an excess of deleterious polymorphisms relative to the nuclear genome. We find no evidence of recombination in the mitochondrial genome, and the much-reduced level of mitochondrial synonymous polymorphism relative to nuclear genes is consistent with a reduction in Ne. Nevertheless, we find that the neutrality index, a measure of the excess of nonsynonymous polymorphism relative to the neutral expectation, is only weakly significantly different between mitochondrial and nuclear loci. This difference is likely the result of the larger proportion of beneficial mutations in X-linked relative to autosomal loci, and we find little to no difference between mitochondrial and autosomal neutrality indices. Reanalysis of published data from Homo sapiens reveals a similar lack of a difference between the two genomes, although previous studies have suggested a strong difference in both species. Thus, despite a smaller Ne, mitochondrial loci of both flies and humans appear to experience similar efficacies of purifying selection as do loci in the recombining nuclear genome.
Keywords: cytoplasmic sweep, mtDNA, neutrality index, tests of selection
The effective size of a population (Ne) impacts how effectively selection removes deleterious mutations and fixes advantageous mutations. The unique genetics of the mitochondrial genome (mitochondrial DNA; mtDNA) are thought to reduce its Ne relative to the nuclear genome, via haploid, uniparental inheritance, the mitochondrial bottleneck in the maternal germline, and a lack of recombination that decreases Ne via selection on linked sites (Hill and Robertson 1966; Maynard Smith and Haigh 1974; Gillespie 2000; Meiklejohn et al. 2007; White et al. 2008; Charlesworth 2012). In addition, cytoplasmic transmission can link the mtDNA to selfish cytoplasmic elements (e.g., Wolbachia in insects) that may sweep through populations, further decreasing mitochondrial Ne and possibly increasing mitochondrial substitution rates via the fixation of slightly deleterious mutations (Shoemaker et al. 2004). For these reasons it has been widely hypothesized that selection is less effective in mitochondrial genomes than in their nuclear counterparts and that mitochondrial genomes may accumulate greater numbers of deleterious substitutions (Lynch 1996, 1997). Analyses of sequence data in Drosophila and mammals have largely supported the conclusion that mtDNA harbors significant levels of slightly deleterious polymorphism (Ballard and Kreitman 1994; Rand and Kann 1996; Nachman 1998; Rand and Kann 1998; Weinreich and Rand 2000).
Ne is not the only evolutionary parameter that distinguishes mitochondrial and nuclear genomes. The distinct functional landscape of the mitochondrial genome likely affects the distribution of selective effects (s) of mutations that arise in this genome. Animal mitochondrial genomes typically encode regulatory information for replication and transcription nested within a hypervariable region (also known as the D-loop, control, or A+T-rich region), 22 transfer RNAs (tRNAs), two ribosomal components, and 13 protein-coding genes—all core components of oxidative phosphorylation (OXPHOS). Outside of the hypervariable region, there is little noncoding DNA in animal mtDNAs. In Drosophilids, 99% of the genome outside of the hypervariable region encodes DNA and RNA genes with highly conserved sequences that function in mitochondrial protein synthesis and aerobic respiration (Clary and Wolstenholme 1985; Wolstenholme and Clary 1985; Ballard 2000; Montooth et al. 2009), which suggests that the distribution of selective effects in the mtDNA may be shifted toward larger negative effects on fitness.
The mutational landscape of the mtDNA also differs from the nuclear genome. In most animal taxa, the mitochondrial mutation rate greatly exceeds that of the nuclear genome (Lynch et al. 2008), and the mitochondrial mutational process is also highly biased (Montooth and Rand 2008). For example, nearly all mitochondrial mutations in D. melanogaster change a G:C base pair to an A:T (Haag-Liautard et al. 2008). When combined with the strong A+T-bias in this mitochondrial genome, where 95% of third codon positions are an A or a T (Montooth et al. 2009), this indicates that the most commonly occurring mutations in protein-coding loci of the Drosophila mtDNA will change an amino acid. Relative to the nuclear genome, animal mitochondrial genomes thus experience a greater mutational pressure that can also be biased in some taxa toward nonsynonymous mutations; these are likely to have deleterious effects in a molecule that encodes such highly conserved functions.
Some of the strongest population genetic patterns in support of distinct selective pressures acting on mitochondrial and nuclear genomes come from analyses of the neutrality index (NI) (Rand and Kann 1996; Nachman 1998; Rand and Kann 1998; Weinreich and Rand 2000). NI is a summary statistic of the deviation from the neutral expectation in the McDonald-Kreitman (MK) test (McDonald and Kreitman 1991; Rand and Kann 1996) and is calculated from counts of synonymous and nonsynonymous polymorphic and divergent sites within and between related species. Weakly deleterious nonsynonymous mutations that segregate in the population, but that will not contribute to divergence, lead to a value of NI greater than 1. When the efficacy of selection is decreased, the expectation is that the number of segregating weakly deleterious polymorphisms will increase; this is the pattern that has been observed in mtDNA. Meta-analyses of MK tables and their associated NI values for mitochondrial and nuclear loci in animals have concluded that NI is predominantly greater than the neutral expectation of 1 for the mtDNA (Rand and Kann 1996; Nachman 1998; Rand and Kann 1998; Betancourt et al. 2012) and exceeds the average NI of the nuclear genome (Weinreich and Rand 2000). Although the relative sparseness of the data was recognized early on (Nachman 1998), and the conclusions were largely limited to how selection shapes animal mtDNA, these patterns are often taken as evidence that selection is largely ineffective in the mtDNA because of its reduced Ne and that mitochondrial genomes are expected to harbor more deleterious polymorphisms than do their nuclear counterparts.
Here we revisit this pattern using new, complete mitochondrial genomes from D. melanogaster that we compare with published nuclear data from the same samples (Langley et al. 2012) and with available human data from both mitochondrial and nuclear genomes (Bustamante et al. 2005; Just et al. 2008; Rubino et al. 2012). We find little evidence that the effects of purifying selection differ on average between mitochondrial and nuclear genomes within flies or within humans, despite evidence that there is much reduced Ne due to a lack of recombination and linkage with the cytoplasm. We discuss reasons why NI is, on average, similar between mitochondrial and nuclear loci, despite the distinct population genetic properties of these two genomes.
Materials and Methods
D. melanogaster mtDNA assembly, annotation, and estimates of sequence diversity
Raw sequence read files from 38 genetic lines of D. melanogaster from Raleigh, North Carolina (Mackay et al. 2012), sequenced by the 50 Genomes subproject of the Drosophila Population Genomics Project (Langley et al. 2012) were downloaded from the National Center for Biotechnology Information Sequence Read Archive. We used the Burrows-Wheeler Aligner, and specifically the fast and accurate short read alignment with Burrows-Wheeler Transform (Li and Durbin 2009), to map sequence reads to the D. melanogaster mitochondrial reference genome (NC_001709). We allowed up to five gaps, five gap extensions, and five mismatches per aligned read, but few reads needed such flexibility and most were filtered out in later steps. Using SAMtools, we postprocessed the alignments to filter out low-quality alignments and to detect single-nucleotide polymorphisms (SNPs) (Li et al. 2009). SNPs with a quality score greater than 20 and indels with a quality score greater than 50 were kept for further analyses. We then generated a consensus sequence for each of the D. melanogaster mtDNAs listed in Supporting Information, Table S1. Because of the high variance in coverage across the hypervariable region, we did not include this region in our final assemblies or analyses.
We annotated the consensus sequence for each mtDNA using the GenBank annotation of the D. melanogaster reference sequence (NC_001709). Using ClustalW (Larkin et al. 2007), we performed a whole-genome alignment, as well as gene-specific alignments, of each consensus sequence to the reference sequence and to the outgroup species Drosophila yakuba (NC_001322). There are very few indels in the protein-coding regions of Drosophila mtDNA (Montooth et al. 2009), making alignment straightforward. From these alignments we calculated expected heterozygosity (π), the number of segregating sites (S), and Watterson’s θW (Watterson 1975) as measures of sequence diversity. The mitochondrial haplotype network was inferred from 80 segregating sites in the coding region of the mtDNA for which there were no missing or ambiguous data using TCS version 1.21 (Clement et al. 2000).
Tests for recombination in the D. melanogaster mtDNA
We estimated linkage disequilibrium (LD) between all pairs of mitochondrial SNPs using the statistic D′ (Lewontin 1964), where D′ = 0 indicates no LD and |D′| = 1 indicates perfect LD. Because recombination erodes LD as a function of distance, a negative correlation between |D′| and genetic distance between pairs of SNPs has been used as evidence for recombination in mtDNA (Awadalla et al. 1999). To test this prediction, we looked for significant negative correlations between |D′| and genetic distance. We also conducted these same tests using another statistical measure of association, r2 (Hill and Robertson 1966), which is more robust to variation in mutation rates (Awadalla et al. 1999; Meunier and Eyre-Walker 2001; Innan and Nordborg 2002). We calculated these correlations by using a variety of minor allele frequency cutoffs. We also tested for the presence of all four genotypes at pairs of SNPs (the “four-gamete test”; Hudson and Kaplan 1985) using DNAsp version 5 (Rozas et al. 2003).
Neutrality tests
Using π and θW, we calculated Tajima’s D (Tajima 1989), which is expected to be 0 for a neutrally evolving locus. Demographic effects will skew the site-frequency spectrum of both synonymous and nonsynonymous polymorphisms at a locus. Contrasting Tajima’s D between nonsynonymous and synonymous polymorphisms therefore tests whether nonsynonymous alleles experience a greater skew in frequency relative to putatively neutral synonymous alleles, indicative of selection (Rand and Kann 1996). We implemented this analysis using the heterogeneity test (Hahn et al. 2002), which simulates 10,000 genealogies with no recombination by using the values of synonymous and nonsynonymous S calculated from the data and compares the estimated difference in Tajima’s D to the random distribution of differences between synonymous and nonsynonymous polymorphisms. We calculated several other summaries of the site-frequency spectrum, including Fu and Li’s D, which characterizes the proportion of mutations on external and internal branches of a genealogy (Fu and Li 1993) and Fay and Wu’s H, which tests for an excess of high-frequency, derived alleles in a sample relative to the neutral expectation (Fay and Wu 2000). These latter statistics were calculated using a set of 80 segregating sites in the coding region of the mtDNA for which there were no missing or ambiguous data. Significance was determined using 10,000 coalescent simulations as implemented in DNAsp version 5 (Rozas et al. 2003).
We constructed MK (McDonald and Kreitman 1991) two-by-two contingency tables of counts of nonsynonymous and synonymous polymorphisms (PN and PS) within D. melanogaster and nonsynonymous and synonymous fixed differences (DN and DS) between D. melanogaster and either D. yakuba or Drosophila simulans. Polymorphic sites within D. melanogaster only contributed to fixed differences if the allele in the outgroup sequence was not present in D. melanogaster. We tested for significant deviations from neutrality by using the Fisher’s exact tests of the MK table in R version 2.15.1 (R Core Team 2012). We calculated NI—the ratio of PN/PS to DN/DS—as a summary statistic of the MK table (Rand and Kann 1996). Assuming that selection is constant, the neutral expectation is that DN/Ds will equal PN/PS (Kimura 1983; McDonald and Kreitman 1991), and NI is expected to be 1. When calculating NI for any gene with a count of 0 in any cell of the MK table, we added a count of 1 to all cells (Sheldahl et al. 2003; Presgraves 2005). Twenty-three percent of 13 D. melanogaster mitochondrial genes, 9.5% of 6113 D. melanogaster nuclear genes, 0% of 13 H. sapiens mitochondrial genes, and 73% of 11,624 H. sapiens nuclear genes required these additional counts. If the MK test is significant, an NI value less than 1 indicates a significant excess of nonsynonymous fixed differences, whereas an NI value greater than 1 indicates a significant excess of nonsynonymous polymorphisms. We also calculated , as in Presgraves (2005), the sign of which is more intuitive; negative values are consistent with an excess of weakly deleterious (negatively selected) polymorphisms and positive values are consistent with an excess of advantageous (positively selected) substitutions.
The short length and low DN values for mitochondrial genes upwardly biases NI (Weinreich and Rand 2000), and we initially used D. yakuba as the outgroup to increase the amount of divergence. However, using D. yakuba to calculate divergence also increases the potential for multiple substitutions at silent sites. Because of this, we constructed MK tables for the 13 mitochondrial protein-coding genes using a range of taxa and methods that capture different amounts of sequence divergence (Table S2 and Table S3). We used both D. simulans and D. yakuba to polarize changes on the branch leading to D. melanogaster, which resulted in very few nonsynonymous substitutions and highly variable NI values (Table S2). We also used either D. simulans or D. yakuba singly to calculate divergence with D. melanogaster in two ways. The “more-inclusive” approach included codons that were missing data in some lines, and averaged across all possible mutational pathways between codons with multiple substitutions to estimate DN and DS (Nei and Gojobori 1986). The “less-inclusive” method omitted codons with any missing data, omitted mtDNA SRX022291 (which contained more missing data than any other mtDNA) and calculated divergence using the mutational path that minimized DN between codons with multiple substitutions. Unless otherwise noted, the more-inclusive method using D. yakuba as outgroup is presented and discussed.
In addition to using counts for single genes, we also analyzed MK tables of the summed counts of polymorphic and divergent sites for each of the mitochondrial-encoded OXPHOS complexes: Complex I (NADH dehydrogenase, ND), Complex IV (cytochrome c oxidase), and Complex V (ATP synthase). Cytochrome B is the only Complex III gene encoded by the mtDNA. Stoletzki and Eyre-Walker (2011) emphasize that contingency data generally should not be summed, particularly when there is heterogeneity among contingency tables, and they provide an unbiased estimator of overall NI for combining counts across genes, . We calculated this statistic and used the DoFE software package (Stoletzki and Eyre-Walker 2011) to calculate bootstrap confidence intervals and to conduct Woolf’s tests of homogeneity (Woolf 1955). The only data set with significant heterogeneity was the D. melanogaster nuclear gene set (P < 0.0001). Similar statistics were used to analyze polymorphism and divergence in the human data sets, as well as in a subset of the mitochondrial haplotypes reported in our study that were independently sequenced and assembled by Richardson et al. (2012) (Table S4).
Comparisons of mitochondrial and nuclear NI in flies and humans
To compare patterns of polymorphism and divergence between mitochondrial and nuclear genomes, we obtained existing data for nuclear genes in D. melanogaster and for nuclear and mitochondrial genes in Homo sapiens. Counts of polymorphism and divergence for D. melanogaster nuclear genes were obtained from the Drosophila Population Genomics Project analysis of the same 38 genomes from Raleigh, North Carolina, with divergence polarized along the D. melanogaster lineage and using the mutational path that minimized DN between codons with multiple substitutions (Langley et al. 2012). The human nuclear data from Bustamante et al. (2005) included counts of polymorphic and divergent sites from 19 African Americans and 20 European Americans, using the chimpanzee Pan troglodytes as an outgroup. We calculated the number of polymorphic and divergent sites for human mitochondrial genes by using mtDNA sequences from 19 African Americans (Just et al. 2008), 20 European Americans (Rubino et al. 2012), and the chimpanzee mitochondrial reference genome D38113.1 (Horai et al. 1995) using the “more-inclusive” method described previously. We also analyzed subsets of the human mitochondrial data to illustrate the sensitivity of NI to sampling (Table S5 and Table S6). Comparisons of the distributions of NI and Z* between gene sets were performed using Mann−Whitney U-tests in R version 2.15.1 (R Core Team 2012).
Data availability
File S1 contains the 38 D. melanogaster assembled mtDNA genomes used in this study aligned with D. yakuba (NC_001322). Human mtDNA sequence data are available in GenBank, and the accession numbers are listed in Table S6.
Results
An excess of low- and high-frequency, derived mitochondrial polymorphisms
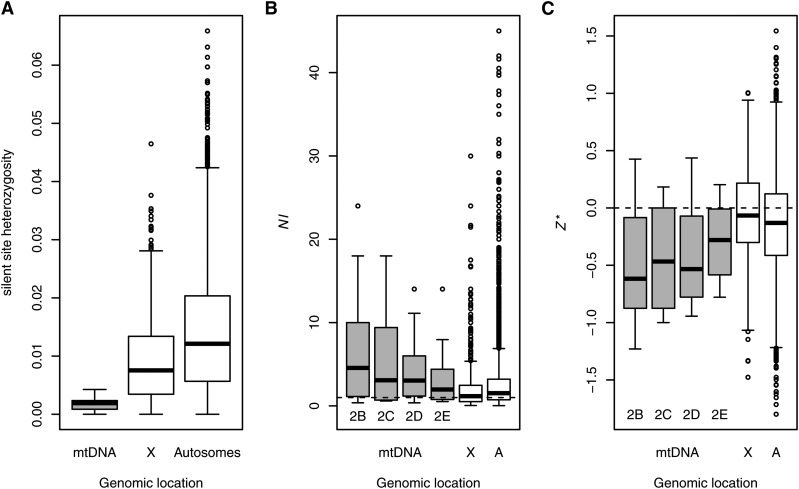
We assembled 14,916 bp of sequence containing the transcribed regions of the mtDNA with a median coverage of 32x for 38 genetic lines sampled from a single population of D. melanogaster in Raleigh, North Carolina (Langley et al. 2012; Mackay et al. 2012) (Table S1). More than 98% of these nucleotides encode the 13 protein-coding genes, 22 tRNAs, and two ribosomal RNAs. The per-site expected heterozygosity in this region (π) of the mtDNA was 0.0008. We identified 137 segregating sites in this population sample, 103 of which were in protein-coding genes. Median heterozygosity in protein-coding genes was 0.0023 per synonymous site and 0.0002 per nonsynonymous site. Silent site heterozygosity was significantly lower in mitochondrial genes relative to nuclear genes (Mann−Whitney U, mtDNA vs. X chromosome, PMWU = 0.00002; mtDNA vs. autosomes, PMWU < 0.00001) and was only 0.16 times that of the autosomes (Figure 1A), lower than what is expected if the mtDNA has an effective population size that is one-quarter that of the autosomes.
Figure 1.
Effect of genomic location on silent-site heterozygosity and on summary statistics of polymorphism and divergence in D. melanogaster. (A) Genomic location has a significant effect on per-site silent-site heterozygosity (PMWU < 0.001 for all pairwise contrasts), consistent with predicted differences in the effective population size (Ne). The ratio of median mitochondrial to autosomal silent site heterozygosity was 0.157, less than predicted for neutral sites if mitochondrial Ne is one quarter that of the autosomes. mitochondrial DNA (mtDNA), X-chromosome, and autosome data sets contained 12, 1255, and 8073 genes, respectively. (B, C) Distributions of neutrality index (NI) and Z* are similar between mitochondrial and autosomal (abbreviated as A) genes, with moderately significant differences between mitochondrial and X-linked (abbreviated as X) genes. The four mtDNA boxes represent estimates from the corresponding MK tables in Table S2, B−E that used either D. simulans (Table S2, B−C) or D. yakuba (Table S2, D−E) as the outgroup. Dashed lines represent the neutral expectations for these statistics. Three nuclear loci for which NI exceeded 50 were excluded from (B) to improve visualization. Statistical results are presented in Table S2. mtDNA, X-chromosome, and autosome data sets contained 13, 712, and 5401 genes, respectively.
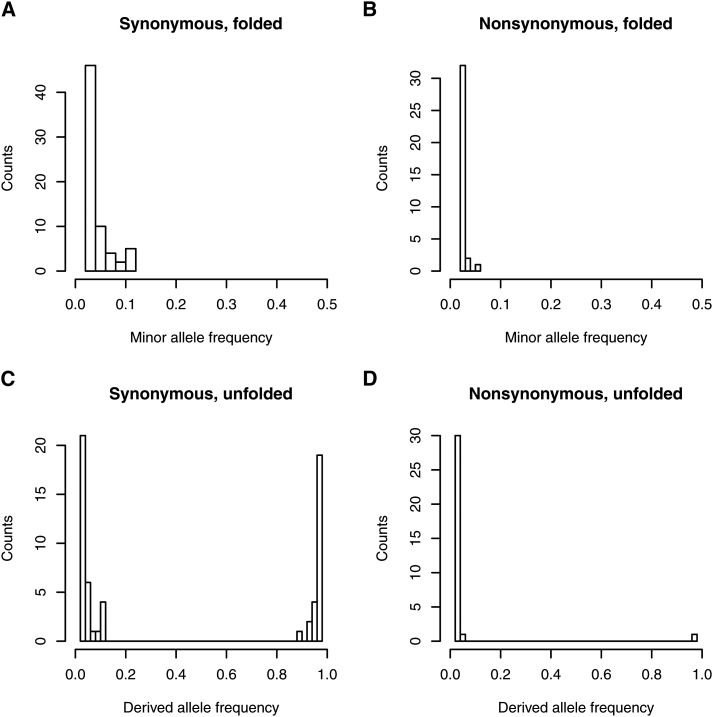
In addition to the scarcity of segregating sites in the D. melanogaster mtDNA, polymorphisms at these sites were skewed toward low frequencies (Figure 2, A and B), as evidenced by consistently negative values of Tajima’s D (Table 1). Tajima’s D across the mtDNA was −2.607 and differed significantly from the neutral expectation of 0 (S = 80 for coding sites with no missing data, P < 0.0001), as did Fu and Li’s D (D = −2.67, P < 0.05). The minor allele frequency for unpolarized synonymous polymorphisms was always less than 11%, and all but one of the nonsynonymous polymorphisms were singletons (Figure 2, A and B). Using D. yakuba as an outgroup revealed that the derived allele was nearly fixed for 44% of segregating synonymous sites, whereas there was only a single derived nonsynonymous polymorphism at high frequency (Figure 2, C and D). Using both D. simulans and D. yakuba to polarize mutations did not qualitatively change this result (Figure S1). Thus, the mitochondrial genome was essentially devoid of intermediate-frequency polymorphisms, with derived synonymous mutations at either very high (greater than 89%) or very low (less than 11%) frequencies and nearly all derived nonsynonymous polymorphisms at frequencies less than 5%. This skew toward high-frequency, derived alleles resulted in a significant negative value of Fay and Wu’s H statistic (H = −41.2, P = 0.005).
Figure 2.
Site-frequency spectra of synonymous and nonsynonymous polymorphisms in the D. melanogaster mitochondrial DNA. (A, B) Folded site-frequency spectra for synonymous and nonsynonymous segregating sites across the mitochondrial protein-coding region reveal that mitochondrial polymorphisms are skewed to low frequencies. (C, D) Unfolded site-frequency spectra reveal that derived, synonymous polymorphisms are almost equally likely to be at low frequency (56% of 59 sites at frequencies less than 0.11) or nearly fixed (44% of 59 sites at frequencies greater than 0.89), while derived, nonsynonymous polymorphisms are nearly always present as singletons (94% of 32 sites). There are essentially no mitochondrial polymorphisms at intermediate frequencies. Sites were omitted from the unfolded site frequency spectra if neither allelic state was shared with D. yakuba. The number of sites included in each distribution is 67 (A), 35 (B), 59 (C), and 32 (D).
Table 1. Synonymous and nonsynonymous variation in D. melanogaster mitochondrial genes and OXPHOS complexes.
| Gene/Complexa | Synonymous Sitesb | Nonsynonymous Sites | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (bp) | S | π | ӨW | D | n (bp) | S | π | ӨW | D | |
| COI | 347 | 16 | 1.57 | 3.81 | −1.91 | 1183 | 0 | 0 | 0 | Undef |
| COII | 142 | 5 | 0.31 | 1.19 | −1.90 | 539 | 1 | 0.05 | 0.24 | −1.13 |
| COIII | 169.33 | 9 | 0.72 | 2.14 | −1.96 | 613.67 | 1 | 0.05 | 0.24 | −1.13 |
| ND1 | 194 | 3 | 0.16 | 0.71 | −1.72 | 739 | 3 | 0.16 | 0.71 | −1.72 |
| ND2 | 198 | 5 | 0.41 | 1.19 | −1.69 | 822 | 6 | 0.34 | 1.43 | −2.07 |
| ND3 | 68 | 1 | 0.05 | 0.24 | −1.12 | 280 | 1 | 0.05 | 0.24 | −1.12 |
| ND4 | 273.33 | 7 | 0.51 | 1.67 | −1.95 | 1061.67 | 7 | 0.38 | 1.67 | −2.17 |
| ND4L | 56 | 1 | 0.05 | 0.24 | −1.13 | 229 | 2 | 0.11 | 0.48 | −1.49 |
| ND5 | 351 | 10 | 0.68 | 2.38 | −2.16 | 1365 | 4 | 0.21 | 0.95 | −1.88 |
| ND6 | 103.33 | 4 | 0.26 | 0.95 | −1.75 | 415.67 | 2 | 0.11 | 0.48 | −1.49 |
| Complex I | 1243.66 | 31 | 2.12 | 7.38 | −2.48 | 4912.34 | 25 | 1.35 | 5.95 | −2.64 |
| Complex III | 239 | 5 | 0.55 | 1.19 | −1.38 | 892 | 2 | 0.16 | 0.48 | −1.29 |
| Complex IV | 658.33 | 30 | 2.60 | 7.14 | −2.21 | 2335.67 | 2 | 0.11 | 0.48 | −1.49 |
| Complex V | 174.67 | 2 | 0.16 | 0.71 | −1.72 | 650.33 | 6 | 0.32 | 1.43 | −2.10 |
| Total | 2315.66 | 68 | 5.43 | 16.42 | −2.44 | 8790.34 | 35 | 1.93 | 8.33 | −2.70 |
OXPHOS, oxidative phosphorylation; Undef, undefined; CO, cytochrome c oxidase; ND, NADH dehydrogenase; ATPase, ATP synthase.
Complex I (ND), 7 loci; Complex III (Cytochrome B), 1 locus; Complex IV (CO), 3 loci; Complex V (ATPase), 2 overlapping loci.
For synonymous and nonsynonymous sites, we calculated the number of segregating sites (S), heterozygosity (π), Watterson’s ӨW, and Tajima’s D. The heterogeneity test for differences between synonymous and nonsynonymous D was never significant (P > 0.35 for all comparisons).
A partial sweep in the D. melanogaster mtDNA
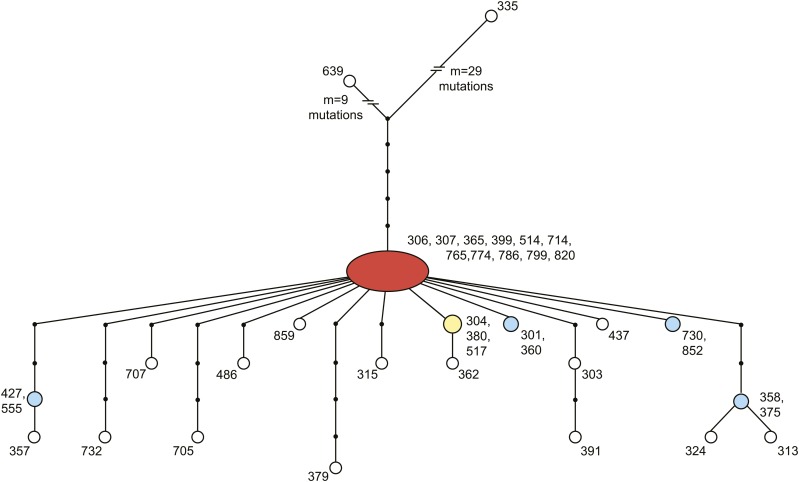
The large fraction of derived alleles at high frequencies is a consequence of the haplotype structure of this sample (Figure 3). Nearly 30% of individuals in this population shared an identical mitochondrial haplotype, and an additional 66% of individuals differed from this haplotype by only one to five mutations. The two remaining haplotypes (RAL-639 and RAL-335) were highly divergent from this common haplotype group, contributing nearly half of the segregating sites to the population sample. These two haplotypes shared the ancestral state with D. yakuba at 17 of the 23 derived high-frequency synonymous polymorphisms (i.e., they have the low-frequency ancestral allele). When these two haplotypes were removed from the analysis, there remained a strong skew toward rare alleles (Tajima’s D = −2.31, P < 0.01; Fu and Li’s D = −3.14, P < 0.02), but Fay and Wu’s H, which is sensitive to the number of high-frequency derived alleles, was only weakly significant (H = −10.34, P = 0.043). The remaining six derived, high-frequency synonymous polymorphisms, as well as the single derived, high-frequency nonsynonymous polymorphism, were the result of single mtDNAs within the common haplotype group having the same allelic state as D. yakuba. Given the lack of recombination in the mtDNA, these are likely new, rather than ancestral, mutations. Six of these seven mutations would have changed a C or G to a T or A, consistent with the mutation bias in the D. melanogaster mtDNA (Haag-Liautard et al. 2008).
Figure 3.
Haplotype network for 38 D. melanogaster mitochondrial DNAs (mtDNAs) sampled from Raleigh, North Carolina. The network, inferred from 80 coding region single-nucleotide polymorphisms (SNPs) with no missing information, reveals that nearly 30% of individuals sampled (11/38) share the same common haplotype (red) and an additional 65% of individuals carry a haplotype only a few mutations away from this haplotype. This common set of mitochondrial haplotypes is highly diverged from the two other mtDNAs sampled in the population; lines RAL-639 and RAL-335 differ from the common haplotype at 14 and 34 SNPs, respectively. At least one of these two haplotypes carries the ancestral state (shared with D. yakuba) at 38% of these SNPs. Numbers represent the Raleigh line carrying the haplotype. Red, yellow, blue, and white nodes were present in 11, 3, 2, and 1 lines, respectively.
No evidence for recombination in the D. melanogaster mtDNA
We tested for a negative correlation between LD and the distance between each pair of polymorphic sites in the D. melanogaster mitochondrial genome, as a signature of the decay of LD over distance via recombination (Awadalla et al. 1999). There was no evidence for a decrease in LD with increasing distance between sites, regardless of the measure of LD or the minor allele cutoff used (Table S7). There were no pairs of polymorphic sites for which all four gametes were present (Hudson and Kaplan 1985; Bruen et al. 2006), further supporting an absence of effective recombination.
Weakly deleterious polymorphism in the D. melanogaster mtDNA
The skew in the site-frequency spectrum toward rare alleles (Figure 2) resulted in negative values of Tajima’s D across the entire mtDNA (Table 1). However, there was no evidence that the skew toward rare alleles differed between synonymous and nonsynonymous polymorphisms (Figure 2, A and B)—heterogeneity tests (Hahn et al. 2002) of Tajima’s D between synonymous and nonsynonymous sites were never significant (P > 0.35 for all genes and complexes). However, unfolding the site frequency spectra revealed that the large number of high-frequency, derived sites were nearly all synonymous (Figure 2, C and D), suggesting that the haplotype that has increased in frequency carried many more synonymous than nonsynonymous polymorphisms. Given that the mutation bias in the D. melanogaster mtDNA greatly favors nonsynonymous mutations (Haag-Liautard et al. 2008), this pattern suggests a history of effective purifying selection removing mitochondrial haplotypes that contain nonsynonymous polymorphisms. Furthermore, all nonsynonymous polymorphisms that have arisen on the common mitochondrial haplotype are present at very low frequencies.
The current distribution of polymorphisms relative to divergence in D. melanogaster showed little evidence for a large and significant excess of segregating deleterious polymorphisms. Across MK tables, no single gene departed significantly from the neutral expectation after Bonferroni correction (P < 0.05/13) (Table 2 and Table S2). For the entire set of protein-coding mitochondrial genes, there was a slight excess of nonsynonymous polymorphism relative to the neutral expectation, as indicated by moderately significant MK tests [Fisher’s exact test, PFET ranged from 0.0004 to 0.041 across methods (Table S2)] and values of NITG that ranged from 1.67 to 2.57 across methods, with confidence intervals that did not include the neutral expectation of 1 (Table S2). There was some OXPHOS-complex specificity to this result—Complexes I (ND) and V (ATPase) tended to deviate significantly from neutrality with NI values greater than 1, whereas Complex IV (CO) was consistent with the neutral expectation (Table 3 and Table S3).
Table 2. Counts of polymorphic (P) and divergent (D) nonsynonymous (N) and synonymous (S) sites along with summary statistics of the MK table for D. melanogaster mitochondrial genes.
| Gene | PNa | PSa | DNa | DSa | NIb | Z*c | PFETd |
|---|---|---|---|---|---|---|---|
| ATPase6 | 5 | 2 | 11 | 35 | 7.955 | −0.778 | 0.021 |
| ATPase8 | 1 | 0 | 2 | 8 | 6.000 | −0.778 | 0.273 |
| COI | 0 | 16 | 8 | 101 | 0.667 | 0.176 | 0.595 |
| COII | 1 | 5 | 6 | 39 | 1.300 | −0.280 | 1.000 |
| COIII | 1 | 9 | 8.5 | 47.5 | 0.621 | −0.009 | 1.000 |
| Cyt-b | 2 | 5 | 17.5 | 67.5 | 1.543 | −0.267 | 0.641 |
| ND1 | 3 | 3 | 11 | 45 | 4.091 | −0.584 | 0.122 |
| ND2 | 6 | 5 | 25 | 41 | 1.968 | −0.275 | 0.334 |
| ND3 | 1 | 1 | 5 | 22 | 4.400 | −0.584 | 0.377 |
| ND4 | 7 | 7 | 24 | 63 | 2.625 | −0.408 | 0.120 |
| ND4L | 2 | 1 | 1 | 7 | 14.00 | −0.778 | 0.152 |
| ND5 | 4 | 10 | 55.833 | 107.167 | 0.768 | 0.063 | 0.775 |
| ND6 | 2 | 4 | 21.5 | 22.5 | 0.523 | 0.203 | 0.669 |
MK, McDonald-Kreitman; NI, neutrality index; ATPase, ATP synthase; CO, cytochrome c oxidase; Cyt-b, Cytochrome B; ND, NADH dehydrogenase.
MK counts from the “more-inclusive” method. Values from other methods are in Table S2.
A count of 1 was added to each cell when calculating for any gene with a zero count in any cell.
, as in Presgraves (2005).
P-value from Fisher’s exact test of the MK table.
Table 3. Summary statistics of the MK table for mitochondrially encoded OXPHOS complexes and nuclear genes.
| Species | Genome | Gene Seta | NIb | NITGc | Zb | PFETd |
|---|---|---|---|---|---|---|
| D. mel | mtDNAe | Complex I | 1.73 | 1.59 (0.94, 3.17) | −0.238 | 0.070 |
| mtDNA | Complex IV | 0.56 | 0.55 (0, 1.30) | 0.255 | 0.750 | |
| mtDNA | Complex V | 9.92 | 9.64 (undef) | −0.997 | 0.007 | |
| mtDNA | All coding | 1.59 (1.97,3.57,3.89) | 1.67 (1.03, 2.86) | −0.201 (-0.28,-0.33,0.36) | 0.041 | |
| Nuclear | Autosomes | 1.16 (1.52,2.83,4.25) | 1.39 (1.35, 1.43) | −0.064 (-0.13,-0.15,0.42) | <1e-6 | |
| Nuclear | X chrom | 0.91 (1.17,2.21,3.24) | 1.02 (0.94, 1.10) | 0.041 (-0.07,-0.06,0.41) | 0.001 | |
| H. sap | mtDNA | Complex I | 1.19 | 1.20 (0.56, 2.40) | −0.074 | 0.475 |
| mtDNA | Complex IV | 1.98 | 2.00 (0.86, 3.61) | −0.296 | 0.402 | |
| mtDNA | Complex V | 1.62 | 1.66 (0.79, 2.42) | −0.209 | 0.127 | |
| mtDNA | All coding | 1.46 (1.38,1.95,1.38) | 1.48 (0.90, 2.24) | −0.165 (-0.23,-0.23,0.32) | 0.034 | |
| Nuclear | All coding | 1.51 (1.39,2.28,2.85) | 1.57 (1.51, 1.63) | −0.180 (-0.12,-0.13,0.40) | <1e-6 |
MK, McDonald-Kreitman; OXPHOS, oxidative phosphorylation; NI, neutrality index; D. mel, D. melanogaster; mtDNA, mitochondrial DNA; undef, undefined; X chrom, X chromosomes; H. sap, H. sapiens.
Complex I (ND) seven loci; Complex IV (CO) three loci; Complex V (ATPase), two overlapping loci. Complex II is nuclear encoded and Complex III has only a single mitochondrial locus, Cyt-b.
NI and were calculated using counts of PN, PS, DN, and DS summed across genes within gene sets. Median, mean, and SD provided for whole genome.
with confidence intervals from 5000 bootstrap samples (Stoletzki and Eyre-Walker 2011).
P-value from Fisher’s exact test of the MK table.
D. melanogaster mtDNA data from the “more-inclusive” method. Values from other methods are in Table S3.
Analysis of 36 of the 38 mitochondrial haplotypes in our sample that were independently sequenced and assembled by Richardson et al. (2012) confirmed these patterns (Table S4). When counts of polymorphism and divergence differed between datasets, they typically differed by a single count. The exception was in several Complex I (ND) genes, for which our assembled mtDNAs had a small number of additional nonsynonymous polymorphisms relative to the Richardson et al. (2012) data set (ND genes, df = 6, PMWU, paired = 0.021; all other genes, df = 5, PMWU, paired = 1) that resulted in slightly greater values of NI (ND included, df = 12, PMWU, paired = 0.016; all other genes, df = 5, PMWU, paired = 1). This was not due to absence of two mitochondrial haplotypes in the Richardson et al. (2012) data set, and the additional polymorphisms in our data were not clustered on any single mitochondrial haplotype. The reduced number of nonsynonymous polymorphisms in the Richardson et al. (2012) data provided even less support for an excess of nonsynonymous segregating variation in the mitochondrial genome. Summed counts of polymorphism and divergence for the entire set of mitochondrial-encoded proteins in this dataset did not deviate from the neutral expectation (PFET = 0.423), and the confidence intervals on NITG for mitochondrial-encoded proteins contained the neutral expectation of 1 (NITG = 0.821, 95% confidence interval = 0.386 to 1.90).
On average, NI is similar for mitochondrial and nuclear genes in flies
Although there is a weak signature of an excess of nonsynonymous segregating variation in the D. melanogaster mitochondrial genome, both mitochondrial and nuclear gene sets have median NI, Z*, or NITG values that deviate in the same manner from the neutral expectation, indicative of both genomes harboring weakly deleterious polymorphisms. Furthermore, the distribution of D. melanogaster mitochondrial gene NI values was contained within that of the nuclear genes, with many nuclear genes having both more positive and more negative values of NI and Z* (Figure 1, B and C). Weakly significant differences between mitochondrial and nuclear gene NI were affected by the genomic location of nuclear genes (Table S2), as X-linked genes had significantly lower values of NI and more positive Z* values relative to autosomal genes (PMWU < 1e-6 for both statistics) (Figure 1, B and C). Because of this, mitochondrial gene NI differed significantly from X-linked genes (PMWU ranged from 0.007 to 0.065) but not from autosomal genes (PMWU ranged from 0.047 to 0.325), with similar patterns for Z* (mtDNA vs. X, PMWU ranged from 0.002 to 0.019; mtDNA vs. autosomes, PMWU ranged from 0.012 to 0.104). The lower counts of polymorphic sites in the assembled mtDNAs from Richardson et al. (2012) provided less support for genomic differences in MK statistics. Neither NI nor Z* differed significantly between the mitochondria and either the X or the autosomes (NI, PMWU > 0.441 for both comparisons; Z*, PMWU > 0.471 for both comparisons).
The moderate levels of significance associated with some of these contrasts, and the sensitivity of these contrasts to small differences in MK counts and methods, suggest that although there is a trend for mitochondrial genes to have larger NI (and more negative Z*) values relative to nuclear genes, the differences between genomes are not large. Contrasts with mitochondrial genomes may have low power due to the smaller number of genes and low levels of nonsynonymous polymorphism and divergence, relative to nuclear genomes. However, the mitochondrial data are not a sample of genes, as they represent the complete protein-coding complement of this genome. Nevertheless, a traditional power analysis suggests that we would require an 18-fold increase in the number of mitochondrial genes for the smallest effect size (Table 3) to reach statistical significance. To provide biological context for the small differences in MK summary statistics that we observed between genomes, we calculated and contrasted effect sizes as the difference in means between genomes divided by the root mean square of the SD for NI and Z*. Across MK tables, the differences in NI between mitochondrial and autosomal genes yielded effect sizes that range from 0.18 to 0.71, smaller than those reported in the meta-analyses of Weinreich and Rand (2000), where the difference in mean NI between mitochondrial and nuclear genes was 3.2, with an effect size of 0.96. In an analysis of 98 nuclear loci in D. melanogaster, Presgraves (2005) reported significant differences in Z* for genes located in regions of high and low recombination for which the effect size was 0.96, over twice that which we observed between mitochondrial and autosomal gene Z* (Table 3).
NI does not differ between mitochondrial and nuclear genes in humans
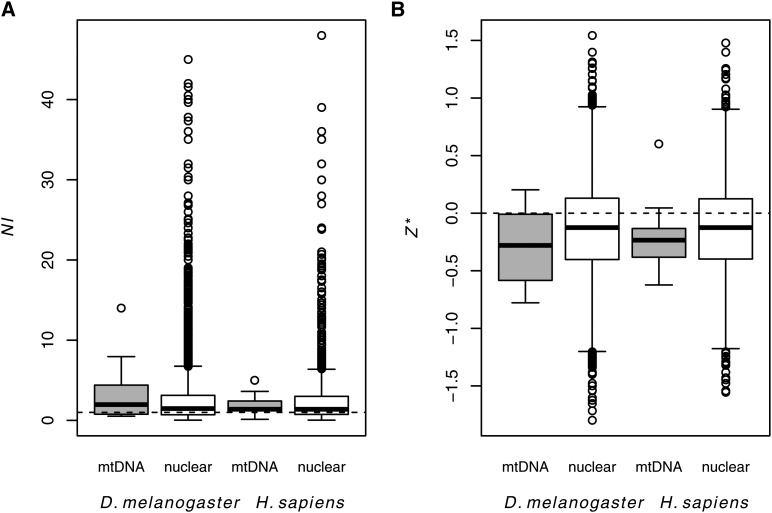
Summary statistics of the MK table also did not differ between mitochondrial and nuclear genes in H. sapiens (NI, PMWU = 0.657; Z*, PMWU = 0.243), nor did the site-frequency spectrum differ between nonsynonymous and synonymous mitochondrial polymorphisms in humans (heterogeneity test, P > 0.36 for all genes). Values of NI and Z* for mitochondrial genes in humans were well within the distribution of these statistics for nuclear genes (Figure 4), and the confidence intervals around NITG for the mitochondrial and nuclear genomes were overlapping (Table 3). Similar to the fly mtDNA and nuclear genome, the median values of NI and Z* for both the human mtDNA and nuclear genome were consistent with a slight excess of nonsynonymous polymorphism (Table 3). The distributions of NI and Z* were also largely overlapping and did not differ significantly between D. melanogaster and H. sapiens mitochondrial genes (NI, PMWU = 0.545; Z*, PMWU = 0.441) (Figure 4), despite differing nuclear Ne between these species. This further supports the idea that the efficacy of purifying selection in these mitochondrial genomes is largely independent of Ne.
Figure 4.
Distributions of (A) neutrality index (NI) and (B) Z* for mitochondrial and nuclear genes in D. melanogaster and H. sapiens. Three nuclear genes in flies and two nuclear genes in humans that had NI values greater than 50 were removed to improve visualization. Dashed lines represent the neutral expectation for each statistic. The D. melanogaster mitochondrial DNA (mtDNA) and nuclear sets contained 13 and 6113 genes, respectively. The H. sapiens mtDNA and nuclear sets contained 13 and 11,624 genes, respectively. See Table S2 and Table 3 and main text for statistical results.
Using data from flies and humans, we tested whether contrasts between nuclear and mitochondrial genes with similar function in OXPHOS and putatively similar selective effects of mutations (s) would reveal greater differences in NI between mitochondrial and nuclear genomes as a function of differing Ne. For humans, there was no difference in NI or Z* between OXPHOS genes encoded in the mitochondrial and nuclear genomes (PMWU > 0.46 for both statistics), whereas for flies there was a weakly significant difference that was driven by the fact that the nuclear OXPHOS genes in our sample had values of NI and Z* that were more consistent with an excess of nonsynonymous substitutions (NI, PMWU = 0.026; Z*, PMWU = 0.022) (Figure S2). However, these data should be treated with some caution, as there were only 11 genes in our nuclear data set annotated to have OXPHOS function, and nine of these genes are part of Complex I (ND). Mitochondrial ND genes accumulate more amino acid substitutions than do other OXPHOS-complex genes in Drosophila (Ballard 2000; Montooth et al. 2009), potentially reflecting differences in functional constraint among complexes that are consistent with the OXPHOS-complex differences in NITG that we observed in this study (Table 3 and Table S3).
Finally, we used the human data to illustrate the sensitivity of NI to sampling. When only a few individuals are sampled, the choice of genomes can lead to high variability and extreme values in NI, potentially as a result of single haplotypes that may carry multiple polymorphisms, as appears to be the case for human ND6 (Table 4 and Table S5). For example, depending on which Japanese individuals we included in our analyses, NI for ND6 takes on values of 30.71 (MK test, PFET = 0.001), 5.50 (PFET = 0.308), or 1.79 (PFET = 0.522) when sampling only three mtDNAs. As more mtDNAs are sampled, NI and Z* for each mitochondrial gene become more similar to the neutral expectation (Table 4 and Table S5). Overall, these analyses using D. melanogaster and H. sapiens mitochondrial genomes highlight the sensitivity of these MK statistics to the number of genomes sampled, the amount of divergence between species, and the low levels of polymorphism in these genes.
Table 4. The sensitivity of NI to sampling.
| Sample | 1 African | 1 African | 1 African | 19 African-American | 30 African-American | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 European | 1 European | 1 European | 20 European-Americanc | 30 European-Americanc | ||||||
| 1 Japanesea | 1 Japaneseb | 1 Japaneseb | ||||||||
| NId | Z* | NI | Z* | NI | Z* | NI | Z* | NI | Z* | |
| ATPase | 2.27 | −0.37 | 2.78 | −0.44 | 2.84 | −0.45 | 1.62 | −0.22 | 1.65 | −0.22 |
| COI | 12.50 | -1.08 | 6.89 | −0.88 | 10.33 | -1.03 | 3.61 | -0.56 | 3.68 | -0.56 |
| COII | 3.33 | −0.52 | 5.80 | −0.82 | 3.28 | −0.52 | 0.86 | −0.13 | 0.64 | −0.01 |
| COIII | 2.36 | −0.53 | 1.38 | −0.14 | 1.60 | −0.39 | 1.38 | −0.23 | 0.95 | −0.08 |
| Cyt-b | 3.78 | -0.57 | 3.64 | −0.55 | 3.64 | −0.55 | 2.29 | −0.36 | 3.23* | -0.50 |
| ND1 | 1.58 | −0.28 | 2.51 | −0.43 | 2.63 | −0.45 | 1.49 | −0.20 | 1.49 | −0.19 |
| ND2 | 5.86 | -0.76 | 5.86 | -0.76 | 7.73 | -0.86 | 3.59 | −0.55 | 2.95 | -0.46 |
| ND3 | 2.83 | −0.52 | 6.80 | −0.77 | 2.83 | −0.52 | 1.33 | −0.28 | 1.52 | −0.25 |
| ND4 | 0.93 | −0.09 | 0.52 | 0.05 | 0.52 | 0.05 | 0.13 | 0.60 | 0.11 | 0.70 |
| ND4L | 2.20 | −0.34 | 2.63 | −0.42 | 2.63 | −0.42 | 5.00 | −0.62 | 4.00 | −0.54 |
| ND5 | 2.00 | −0.32 | 2.38 | −0.39 | 2.23 | −0.37 | 1.20 | −0.09 | 1.44 | −0.16 |
| ND6 | 30.71* | -1.22 | 5.50 | −0.74 | 1.79 | −0.25 | 1.30 | −0.20 | 1.17 | −0.16 |
| All coding | 2.88* | -0.46 | 2.50* | -0.40 | 2.41* | -0.39 | 1.46 | -0.17 | 1.55 | -0.19 |
| NITG (C.I.)e | 2.85 (1.83, 4.99) | 2.60 (1.65, 3.91) | 2.56 (1.59, 3.81) | 1.48 (0.90, 2.24) | 1.59 (0.93, 2.36) | |||||
NI, neutrality index; ATPase, ATP synthase; CO, cytochrome c oxidase; Cyt-b, Cytochrome B; ND, NADH dehydrogenase; C.I., confidence interval; MK, McDonald-Kreitman.
MK table counts from Nachman et al. (1996).
MK table counts as mentioned previously, but substituting two different, randomly chosen Japanese samples.
MK table counts from African-American and European-American sequences sampled from (Just et al. 2008) and (Rubino et al. 2012) with the chimpanzee mitochondrial reference genome as an outgroup (Horai et al. 1995).
A count of 1 was added to each cell when calculating NI for any locus with a zero count in any cell. Values in bold indicate P ≤ 0.05; * indicates significant sample-wise Bonferroni-corrected P-value of less than 0.004 for Fisher’s exact test of the MK table.
Calculated as in Table 3. No sample rejected Woolf’s test of homogeneity (P > 0.19 for all samples). Values in bold indicate that the confidence intervals do not overlap the neutral expectation of 1.
Discussion
Using a large sample of whole-genome sequence data, we have tested a number of hypotheses about mtDNA evolution, and about differences in the efficacy of selection on mitochondrial vs. nuclear genes. Our data confirm that mtDNA do not have a signature of recombination and have lower silent-site diversity than do nuclear genes in D. melanogaster, which supports the prediction that the mitochondrial genome has a lower Ne than the nuclear genome. We also show a skew in the site-frequency spectrum toward rare alleles in D. melanogaster that likely has two sources: 1) the accumulation of new mutations on what appears to be a mtDNA haplotype that has swept to high frequency in the recent past, and 2) the ancestral polymorphisms contained on migrant or remnant haplotypes that are now rare in this population. Despite the apparent reduction in Ne for mtDNA, our findings indicate that selection is similarly effective at purging deleterious polymorphisms from the mitochondrial and nuclear genomes of D. melanogaster, and that the same is true in H. sapiens. Although all genomes that we analyzed showed some evidence of an excess of nonsynonymous polymorphism relative to the neutral expectation, the only significant differences in NI and Z* were between D. melanogaster mitochondrial genes and X-linked genes. X-linked genes in Drosophila have a greater proportion of beneficial substitutions than do autosomes (Langley et al. 2012; Mackay et al. 2012; Meisel and Connallon 2013; Garrigan et al. 2014), suggesting that what differs between mitochondrial genes and nuclear genes is likely the fraction of beneficial substitutions rather than the efficacy of purifying selection, which appears to be largely independent of Ne in the D. melanogaster and H. sapiens mitochondrial genomes that we have analyzed.
Given its uniparental and haploid transmission, the expectation under neutrality is that the mtDNA has one-quarter the population size of the autosomes. This reduced value of N (and subsequently Ne) matches that expected for the Y (or W) chromosome, and, like the Y chromosome, the mtDNA has little to no recombination. However, very much unlike the Y chromosomes that have been sequenced (e.g., Charlesworth and Charlesworth 2000; Carvalho et al. 2009; Carvalho and Clark 2013; Bellott et al. 2014), animal mtDNA genomes do not show an accumulation of transposable elements, and the gene content of the animal mitochondrial genome is remarkably stable, with few gene losses and even fewer pseudogenes (Boore 1999; Ballard and Rand 2005). Furthermore, dN/dS is two to 15 times lower for mitochondrial genes than for nuclear genes in mammals (Popadin et al. 2013), and average values of dN/dS for mitochondrial genes are well under 0.1 and are, on average, only 13% that of nuclear genes in Drosophila (Bazin et al. 2006; Montooth et al. 2009). This pattern of amino acid conservation is particularly striking, given that the mutation rate in the D. melanogaster mtDNA is an order of magnitude greater than the per-site mutation rate in the nuclear genome, with an extreme bias toward nonsynonymous mutations in the mitochondrial genome (Haag-Liautard et al. 2007; Haag-Liautard et al. 2008). Although heteromorphic Y chromosomes do show signatures of less effective purifying selection, such as proliferation of satellite repeats and reduced codon bias (Bachtrog 2013; Singh et al. 2014), the single copy, X-degenerate genes that have remained on the human Y chromosome experience effective purifying selection (Rozen et al. 2009; Bellott et al. 2014), as do the protein sequences of Drosophila Y-linked genes (Singh et al. 2014). Thus, despite early loss of many genes when heteromorphic Y chromosomes and mtDNA formed, both these nonrecombining chromosomes contain genes maintained by effective purifying selection in the presence of reduced Ne.
Many researchers have cited the early work on NI in Drosophila and mammals in support of the idea that mtDNA accumulate deleterious mutations (e.g., Meiklejohn et al. 2007; Green et al. 2008; Neiman and Taylor 2009; Akashi et al. 2012). In fact, this idea has become so engrained that it is regularly cited in reviews of mitochondrial gene evolution (e.g., Ballard and Whitlock 2004; Lynch 2007). What is perhaps surprising about this conversion of a small set of intriguing initial studies into dogma is that the early studies themselves were quite circumspect about the implications of their results. For instance, Nachman (1998), in noting that very few nuclear loci were available for comparison, stated “It is also unclear whether the patterns reported here are unique to mitochondrial DNA.” Data from the few nuclear genes that had been sequenced raised “the possibility that the patterns reported here for mtDNA may also be found at some nuclear loci” (Nachman 1998). Even studies that did have access to additional nuclear datasets were only able to calculate NI for 36 nuclear loci (Weinreich and Rand 2000), and the NI values that were available often did not deviate significantly from neutrality (Nachman 1998; Weinreich and Rand 2000). Those that did reject neutrality tended to do so weakly, perhaps due to the small number of polymorphisms in mitochondrial samples even when the number of individuals sampled is high (e.g., ND3, Nachman et al. 1996). Nevertheless, there are mitochondrial genes that do strongly reject neutrality, and some of these had NI values that greatly exceeded NI for the sampled nuclear loci. On the basis of these and similar comparisons, many authors have reached the conclusion that mtDNA evolves in a manner distinct from the nuclear genome. Our results using the whole genomes of flies and humans, combined with observations of low mitochondrial dN/dS, suggest that the mitochondrial genomes of flies and humans are not suffering less effective purifying selection relative to the nuclear genome, and that differences in selection between these genomes may lie in differing rates of adaptive evolution.
Reductions in Ne—due either to reductions in census population size or to the increased effect of linked, selected variants in regions of low recombination—are expected to result in a reduction in the efficacy of purifying selection. Indeed, comparisons of MK test results across a range of species with different values of Ne have revealed this expected relationship (e.g., Li et al. 2008; Wright and Andolfatto 2008; Gossmann et al. 2010), as have comparisons of NI across regions of the D. melanogaster nuclear genome with different recombination rates (Presgraves 2005; Langley et al. 2012). Therefore, all things being equal, mitochondrial loci would be expected to harbor an excess of nonsynonymous polymorphisms relative to nuclear loci due to reduced Ne. Our results suggest that all things are not equal between these two cellular compartments, and that there may be features of the mitochondrion that make it less likely to accumulate deleterious mutations. One such feature is the “bottleneck” that occurs in the number of mtDNAs that are passed from mother to offspring—this event makes it possible for selection to act within hosts, possibly increasing the power of selection to remove deleterious mutations (Bergstrom and Pritchard 1998; Rand 2011) and reducing variability in mitochondrial Ne among taxa, relative to nuclear genomes. The additional layers of selection imposed by mitochondrial inheritance, combined with stronger negative selective effects of amino acid changing mutations in mitochondrial genes (e.g., Popadin et al. 2013), may allow the mtDNA to escape the accumulation of deleterious mutation, resulting in relatively similar values of NI between nucleus and mitochondria. If the selective effects of mutations in mitochondrial genes are beyond the “horizon” where all mutations will behave similarly regardless of Ne (Nachman 1998; Eyre-Walker and Keightley 2007), then we expect patterns of mitochondrial polymorphism and divergence to be largely independent of Ne.
Our results come with several caveats. First, we have only studied two organisms—it may be that a more comprehensive review of NI in mtDNA and nuclear loci across many species will reveal a difference in the average efficacy of purifying selection or highlight lineage-specific patterns. The early meta-analyses of NI contained loci from a wide range of animals (Nachman 1998; Weinreich and Rand 2000), and using data from only Drosophila and humans may provide a limited perspective. Nevertheless, these are two model organisms for evolutionary biology that span a large range of mtDNA:nuclear substitution rates, and studies of these species have led the way for much of modern population genetics. Second, it is clear from our analysis of the D. melanogaster mtDNA that it is not at equilibrium, and may be recovering from a partial cytoplasmic sweep that may be associated with Wolbachia (Richardson et al. 2012). Much of the theory used to predict NI values from Ne and s assumes mutation-selection-drift balance (see, e.g., Nachman 1998), and deviations from this equilibrium can result in more complex relationships between Ne, s, and NI (Messer and Petrov 2013). Although nonequilibrium histories may mean that mtDNA NI values are not at equilibrium, it is equally likely that nuclear genes from D. melanogaster are not at mutation-selection-drift equilibrium (Hahn 2008; Langley et al. 2012). Whether or not the mtDNA is at equilibrium, and whether or not the NI values calculated from this snapshot of two species represent equilibrium values, our results still imply that there is little difference between nuclear and mitochondrial measures of the efficacy of purifying selection.
Despite the mitochondrial genome experiencing a distinct population genetic environment relative to the nuclear genome, our whole-genome analyses uncovered little evidence for an excess accumulation of slightly deleterious mutations in mitochondrial genomes, relative to nuclear genomes. In fact, the only strong evidence for a reduced efficacy of selection in animal mtDNA, relative to nuclear genomes, comes from comparative studies of nuclear and mitochondrial tRNAs (Lynch 1996; Lynch 1997). As discussed previously in this article, in the absence of a pattern in NI, there are few patterns of molecular evolution in animal mtDNA indicative of deleterious mutation accumulation [but see Osada and Akashi (2012)]. This pattern is in stark contrast to the patterns found in analogous nuclear regions with reduced Ne and low recombination, like the Y chromosome. Determining whether mtDNA accumulate deleterious polymorphisms and substitutions more readily than nuclear DNA in a larger sample of species (and what type of loci may be affected) will be a particularly fruitful avenue for future studies.
Acknowledgments
We thank Colin Meiklejohn and members of the Montooth and Hahn labs for constructive feedback. B.S.C. was supported on the Indiana University Genetics, Molecular and Cellular Sciences Training Grant T32-GM007757 funded by the National Institutes of Health and a Doctoral Dissertation Improvement Grant funded by the National Science Foundation. This research was supported by funding from Indiana University, award DBI-0845494, from the National Science Foundation to M.W.H. and a National Science Foundation CAREER award IOS-1149178 to K.L.M.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.016493/-/DC1
Communicating editor: S. I. Wright
Literature Cited
- Akashi H., Osada N., Ohta T., 2012. Weak selection and protein evolution. Genetics 192: 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla P., Eyre-Walker A., Smith J. M., 1999. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science 286: 2524–2525. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J. W., 2000. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 51: 48–63. [DOI] [PubMed] [Google Scholar]
- Ballard J. W. O., Kreitman M., 1994. Unraveling selection in the mitochondrial genome of Drosophila. Genetics 138: 757–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J. W. O., Rand D. M., 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36: 621–642. [Google Scholar]
- Ballard J. W. O., Whitlock M. C., 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13: 729–744. [DOI] [PubMed] [Google Scholar]
- Bazin E., Glemin S., Galtier N., 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572. [DOI] [PubMed] [Google Scholar]
- Bellott D. W., Hughes J. F., Skaletsky H., Brown L. G., Pyntikova T., et al. , 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom C. T., Pritchard J., 1998. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics 149: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Blanco-Martin B., Charlesworth B., 2012. The relation between the neutrality index for mitochondrial genes and the distribution of mutational effects on fitness. Evolution 66: 2427–2438. [DOI] [PubMed] [Google Scholar]
- Boore J. L., 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen T. C., Philippe H., Bryant D., 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M. T., et al. , 2005. Natural selection on protein-coding genes in the human genome. Nature 437: 1153–1157. [DOI] [PubMed] [Google Scholar]
- Carvalho A. B., Clark A. G., 2013. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 23: 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A. B., Koerich L. B., Clark A. G., 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 25: 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R., 1985. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 22: 252–271. [DOI] [PubMed] [Google Scholar]
- Clement M., Posada D., Crandall K. A., 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Fay J. C., Wu C. I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X., Li W. H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Vedanayagam J. P., Presgraves D. C., 2014. Genome diversity and divergence in Drosophila mauritiana: Multiple signatures of faster X evolution. Genome Biol. Evol. 6: 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 2000. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T. I., Song B.-H., Windsor A. J., Mitchell-Olds T., Dixon C. J., et al. , 2010. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E., Malaspinas A.-S., Krause J., Briggs A. W., Johnson P. L. F., et al. , 2008. A complete neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134: 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C., Coffey N., Houle D., Lynch M., Charlesworth B., et al. , 2008. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 6: e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., 2008. Toward a selection theory of molecular evolution. Evolution 62: 255–265. [DOI] [PubMed] [Google Scholar]
- Hahn M. W., Rausher M. D., Cunningham C. W., 2002. Distinguishing between selection and population expansion in an experimental lineage of bacteriophage t7. Genetics 161: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Horai S., Hayasaka K., Kondo R., Tsugane K., Takahata N., 1995. Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc. Natl. Acad. Sci. USA 92: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Nordborg M., 2002. Recombination or mutational hot spots in human mtDNA? Mol. Biol. Evol. 19: 1122–1127. [DOI] [PubMed] [Google Scholar]
- Just R. S., Diegoli T. M., Saunier J. L., Irwin J. A., Parsons T. J., 2008. Complete mitochondrial genome sequences for 265 African American and U.S. “Hispanic” individuals. Forensic Sci. Int. Genet. 2: e45–e48. [DOI] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R., et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lewontin R. C., 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Costello J. C., Holloway A. K., Hahn M. W., 2008. “Reverse ecology” and the power of population genomics. Evolution 62: 2984–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol. Biol. Evol. 13: 209–220. [DOI] [PubMed] [Google Scholar]
- Lynch M., 1997. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol. Biol. Evol. 14: 914–925. [DOI] [PubMed] [Google Scholar]
- Lynch M., 2007. Origins of Genome Architecture. Sinauer Associates, Inc., Sunderland, Massachusetts. [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Meiklejohn C. D., Montooth K. L., Rand D. M., 2007. Positive and negative selection on the mitochondrial genome. Trends Genet. 23: 259–263. [DOI] [PubMed] [Google Scholar]
- Meisel R. P., Connallon T., 2013. The faster-X effect: integrating theory and data. Trends Genet. 29: 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., Petrov D. A., 2013. Frequent adaptation and the McDonald–Kreitman test. Proc. Natl. Acad. Sci. USA 110: 8615–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J., Eyre-Walker A., 2001. The correlation between linkage disequilibrium and distance: implications for recombination in hominid mitochondria. Mol. Biol. Evol. 18: 2132–2135. [DOI] [PubMed] [Google Scholar]
- Montooth K. L., Rand D. M., 2008. The spectrum of mitochondrial mutation differs across species. PLoS Biol. 6: e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Abt D. N., Hofmann J., Rand D. M., 2009. Comparative genomics of Drosophila mtDNA: Novel features of conservation and change across functional domains and lineages. J. Mol. Evol. 69: 94–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman M. W., 1998. Deleterious mutations in animal mitochondrial DNA. Genetica 102–103: 61–69. [PubMed] [Google Scholar]
- Nachman M. W., Brown W. M., Stoneking M., Aquadro C. F., 1996. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142: 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Neiman M., Taylor D. R., 2009. The causes of mutation accumulation in mitochondrial genomes. Proc. Biol. Sci. 276: 1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N., Akashi H., 2012. Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol. Biol. Evol. 29: 337–346. [DOI] [PubMed] [Google Scholar]
- Popadin K. Y., Nikolaev S. I., Junier T., Baranova M., Antonarakis S. E., 2013. Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol. Biol. Evol. 30: 347–355. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15: 1651–1656. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2012 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria ISBN 3–900051–07–0: URL, Available from: http://www.R-project.org/. Accessed August 31, 2015.
- Rand D. M., 2011. Population genetics of the cytoplasm and the units of selection on mitochondrial DNA in Drosophila melanogaster. Genetica 139: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D. M., Kann L. M., 1996. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 13: 735–748. [DOI] [PubMed] [Google Scholar]
- Rand D. M., Kann L. M., 1998. Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA. Genetica 102–103: 393–407. [PubMed] [Google Scholar]
- Richardson M. F., Weinert L. A., Welch J. J., Linheiro R. S., Magwire M. M., et al. , 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8: e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Sanchez-Delbarrio J. C., Messeguer X., Rozas R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Rozen S., Marszalek J. D., Alagappan R. K., Skaletsky H., Page D. C., 2009. Remarkably little variation in proteins encoded by the Y chromosome’s single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 85: 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino F., Piredda R., Calabrese F. M., Simone D., Lang M., et al. , 2012. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res. 40: D1150–D1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldahl L. A., Weinreich D. M., Rand D. M., 2003. Recombination, dominance and selection on amino acid polymorphism in the Drosophila genome: contrasting patterns on the X and fourth chromosomes. Genetics 165: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker D. D., Dyer K. A., Ahrens M., Mcabee K., Jaenike J., 2004. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics 168: 2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Koerich L. B., Carvalho A. B., Clark A. G., 2014. Positive and purifying selection on the Drosophila Y chromosome. Mol. Biol. Evol. 31: 2612–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N., Eyre-Walker A., 2011. Estimation of the neutrality index. Mol. Biol. Evol. 28: 63–70. [DOI] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Weinreich D. M., Rand D. M., 2000. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics 156: 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. J., Wolff J. N., Pierson M., Gemmell N. J., 2008. Revealing the hidden complexities of mtDNA inheritance. Mol. Ecol. 17: 4925–4942. [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R., Clary D. O., 1985. Sequence evolution of Drosophila mitochondrial DNA. Genetics 109: 725–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf B., 1955. On estimating the relation between blood group and disease. Ann. Hum. Genet. 19: 251–253. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Andolfatto P., 2008. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annu. Rev. Ecol. Evol. Syst. 39: 193–213. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
File S1 contains the 38 D. melanogaster assembled mtDNA genomes used in this study aligned with D. yakuba (NC_001322). Human mtDNA sequence data are available in GenBank, and the accession numbers are listed in Table S6.