Significance
The process by which cells differentiate is central to multicellular development and cancer. Dramatic gene expression changes mediate this complex process, which involves the termination of proliferation and the acquisition of distinct cell-specific features. We identified a transcription factor, MYB DOMAIN PROTEIN 36 (MYB36), that regulates this developmental transition in the Arabidopsis thaliana root endodermis. Differentiated endodermis forms a protective waxy barrier called the Casparian strip. We found that MYB36 activates genes involved in Casparian strip formation and represses genes involved in proliferation. Our results suggest that MYB36 is a critical regulator of developmental timing in the root endodermis.
Keywords: differentiation, Casparian strip, MYB36, endodermis, proliferation
Abstract
Stem cells are defined by their ability to self-renew and produce daughter cells that proliferate and mature. These maturing cells transition from a proliferative state to a terminal state through the process of differentiation. In the Arabidopsis thaliana root the transcription factors SCARECROW and SHORTROOT regulate specification of the bipotent stem cell that gives rise to cortical and endodermal progenitors. Subsequent progenitor proliferation and differentiation generate mature endodermis, marked by the Casparian strip, a cell-wall modification that prevents ion diffusion into and out of the vasculature. We identified a transcription factor, MYB DOMAIN PROTEIN 36 (MYB36), that regulates the transition from proliferation to differentiation in the endodermis. We show that SCARECROW directly activates MYB36 expression, and that MYB36 likely acts in a feed-forward loop to regulate essential Casparian strip formation genes. We show that myb36 mutants have delayed and defective barrier formation as well as extra divisions in the meristem. Our results demonstrate that MYB36 is a critical positive regulator of differentiation and negative regulator of cell proliferation.
Progression from stem cell through specification to differentiation is central to multicellular development. This process involves both the loss of proliferative potential and the acquisition of characteristic features that enable cells to perform specific functions. In plant roots, differentiated endodermis surrounds the central vasculature, which transports water and nutrients throughout the plant. The endodermis protects the root vasculature from the entry of harmful toxins and pathogens by means of a cell-wall modification called the Casparian strip (1). This lignin-composed extracellular barrier prevents the passive flow of water and solutes into and out of the vasculature (2). To enter the vasculature, ions must pass through the endodermis, which selectively filters nutrients.
Given this crucial cellular function, endodermal specification has been extensively studied. The cortex/endodermal initial cell divides asymmetrically to regenerate itself and produce a daughter cell. This daughter cell divides to produce the first cells of the cortex and endodermal lineages, which we will refer to as progenitors. Several transcription factors have been identified as essential regulators of the daughter cell division and of endodermal cell-fate specification. SHORTROOT (SHR) is expressed in the vasculature and moves into the daughter cell and endodermis, where it induces the expression of SCARECROW (SCR) (3) and regulates daughter cell division (4). Together, SHR and SCR regulate the expression of JACKDAW (JKD), MAGPIE (MGP), and NUTCRACKER (NUC), several members of the C2H2 transcription factor family (5–8). Collectively, these proteins pattern the cortical and endodermal progenitors. SCR, JKD, MGP, and NUC regulate SHR movement and nuclear localization in the daughter cell, thus regulating its asymmetric division (5, 6, 8). The molecular pathways that subsequently induce differentiation of these progenitors are not known.
Differentiated endodermis is marked by Casparian strip formation. Although the molecular mechanisms driving differentiation are not known, it has been shown that ectopic SHR expression results in ectopic Casparian strip formation (3, 9). Five functionally redundant Casparian strip proteins (CASP1–5) are required for Casparian strip formation (10). CASPs are transmembrane proteins that are localized to a narrow zone within endodermal cells, where they anchor peroxidase and oxidase enzymes that polymerize lignin, the primary component of the Casparian strip (10, 11). Due to the irreversibility of lignin polymerization, Casparian strip formation can be used as a proxy for endodermal differentiation. Thus, understanding how CASP genes are regulated should provide insights into the mechanistic basis of differentiation.
We performed a genetic screen for mutants with altered pCASP2::GFP expression and identified a gene that codes for the transcription factor MYB DOMAIN PROTEIN 36 (MYB36). myb36 mutant seedlings have no visible pCASP2::GFP expression and Casparian strip barrier formation is dramatically delayed. Both SHR and SCR have been shown to activate MYB36 expression, and we demonstrate that SCR activation is direct. Additionally, we show that MYB36 activates genes involved in Casparian strip formation, but this activation, although rapid, requires protein synthesis. RNA-sequencing (RNA-seq) transcriptional profiling from the endodermis of myb36 mutants reveals that genes involved in daughter cell division and specification, including SCR, JKD, and MGP, are repressed by MYB36. These results demonstrate that MYB36 regulates essential genes for endodermal differentiation and represses transcription factors that regulate cell division and specification. We propose that MYB36 regulates the developmental switch from a proliferative state toward differentiation.
Results
MYB36 Is a Key Regulator of Endodermal Differentiation.
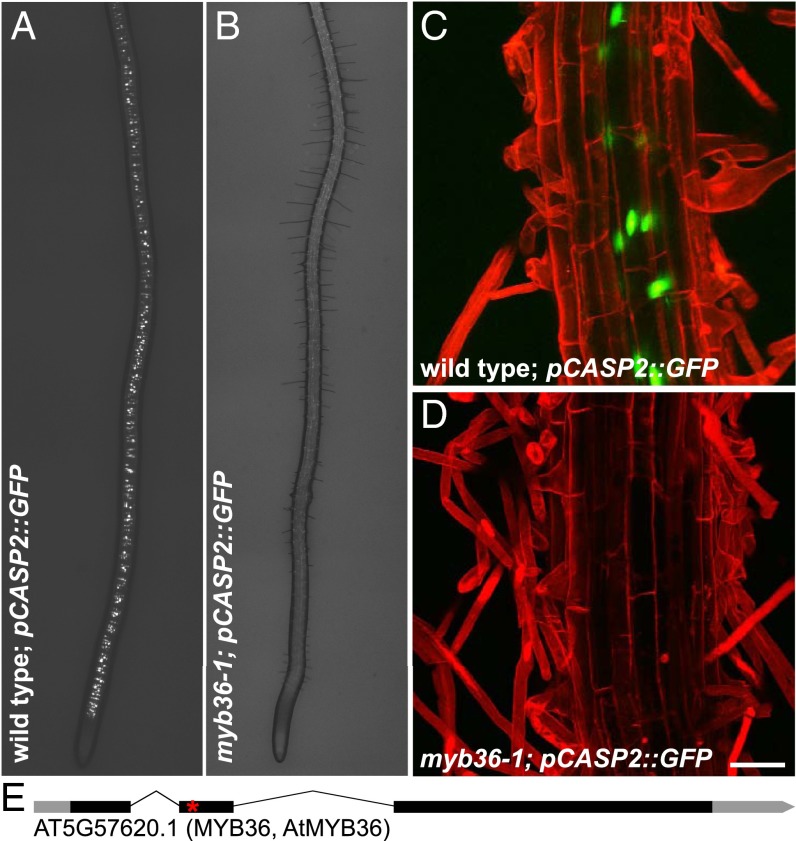
To identify regulators of endodermal differentiation, we conducted a genetic screen for mutations that alter the expression of a CASP2 transcriptional reporter (Fig. 1 A and C). Among mutagenized seedlings with a pCASP2::GFP reporter, we identified one recessive mutant that lacked any detectable GFP expression (Fig. 1 B and D). Using bulk segregant analysis and whole-genome resequencing, we identified a point mutation in the MYB36 locus (At5g57620) resulting in a premature STOP codon in the predicted DNA-binding domain (Fig. 1E). Due to the nature and location of the mutation and its recessivity, we presume our allele is a genetic null. MYB36 encodes an R2R3-MYB class transcription factor (12), which had previously been identified as a core expression marker for endodermal cell identity (13). We confirmed that our mutant is allelic to two other myb36 alleles, FLAG_412E06 and WiscDsLox442H5, which both fail to complement pCASP2::GFP expression when crossed to myb36-1 mutants [n = 20; F1 (first generation progeny) seedlings examined from each cross]. These experiments collectively suggest that MYB36 is the causal gene.
Fig. 1.
Identification of a mutant lacking pCASP2::GFP expression. (A and B) Stitched dissecting scope image of (A) wild-type plants expressing the pCASP2::GFP transcriptional reporter and (B) myb36-1 mutants that lack pCASP2::GFP expression. (C and D) Maximum-projection confocal images of (C) wild-type plants expressing the pCASP2::GFP transcriptional reporter and (D) myb36-1 mutants that lack pCASP2::GFP expression. (E) The mutation was mapped by sequencing to reveal a premature STOP (asterisk) at W58 in the coding region of MYB36, a putative transcription factor. (Scale bar, 50 μm.)
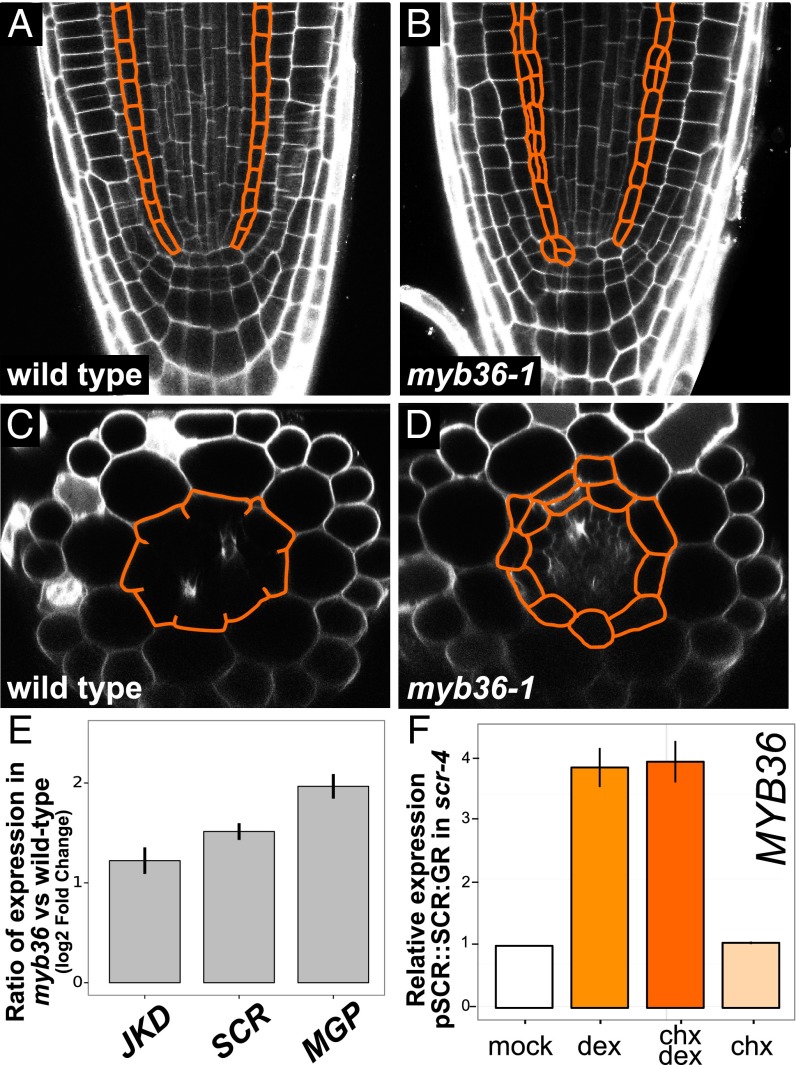
It has been shown that the intercalating agent propidium iodide (PI) is blocked from entering the vasculature by the Casparian strip (Fig. 2A) (14). We used this assay to determine Casparian strip function in myb36-1 (Methods). In wild-type roots, the Casparian strip restricts PI penetration into the vasculature ∼14 cells after the onset of elongation (Fig. 2C), coincident with the onset of differentiation. In myb36-1 mutants, despite evidence of differentiation, such as root hairs (Fig. 1B), PI was able to penetrate into the vasculature (Fig. 2B). Restriction of PI occurred ∼30 cells after the onset of elongation, indicating delayed barrier formation (Fig. 2C). Similar delays are observed in the casp1/casp3 double mutant (10) (Fig. 2C). These results indicate that MYB36 regulates genes involved in Casparian strip formation, a proxy for endodermal differentiation.
Fig. 2.
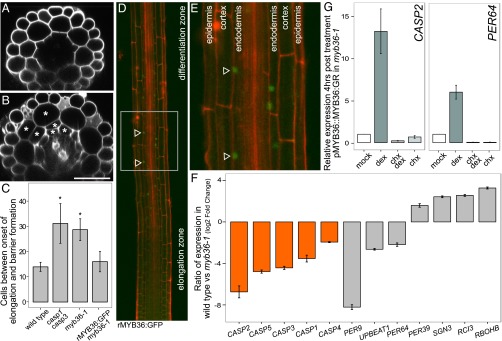
myb36-1 mutant seedlings exhibit delayed barrier formation and dramatic changes in gene expression. (A) Optical cross-section of wild-type root in the differentiation zone. (B) Optical cross-section of myb36-1 mutant root displays Casparian strip defect and extra cell divisions (asterisks). (Scale bar, 50 μm.) (C) Quantification of cells after onset of differentiation in wild type, casp1 casp3, and myb36-1 and genetic complementation of myb36-1 with rMYB36:GFP seedlings. Error bars are SD of the mean from three biological replicates. Significance was determined by Student’s t test, *P < 0.01 (for source data see Dataset S5). (D) Confocal images (25×) stitched together to reveal rMYB36:GFP expression. The fusion is most strongly detected in the elongation zone, before the onset of differentiation (arrowheads). (E) rMYB36:GFP is expressed specifically in the nuclei in the endodermis (arrowheads). Epidermis, cortex, and endodermis are labeled for orientation. (F) myb36-1 mutant endodermis has reduced expression of CASP genes (orange) and altered expression of many genes involved in oxidoreductase activity (gray) compared with wild-type endodermis. Error bars are SEM from three biological replicates. Adjusted P value ≤ 0.01 (Methods) (for source data see Dataset S6). (G) qRT-PCR of two putative MYB36 targets 4 h after induction of pMYB36::MYB36:GR. Mean expression values of three biological replicates are reported after normalization with PP2A expression. Error bars are SEM (for source data see Dataset S7).
We found that MYB36 expression is enriched in the endodermis compared with both whole root and cortex in cell type-specific RNA expression analysis (15) as well as RNA-seq data (16). To examine the expression pattern of MYB36 protein, we generated a translational fusion to GFP using recombineering to maintain the flanking DNA 5′ and 3′ of the MYB36 genomic locus (rMYB36:GFP). After crossing myb36-1 with plants expressing this construct, we found that rMYB36:GFP complements barrier function (Fig. 2C) and pCASP2::GFP expression (50/50 seedlings), providing further evidence that MYB36 is the causal gene for both phenotypes. rMYB36:GFP is most highly expressed before the onset of differentiation (Fig. 2D) and is expressed exclusively in endodermal cells (Fig. 2E).
MYB36 Activates Genes Required for Endodermal Differentiation.
To identify genes that are regulated by MYB36, we used RNA-seq to assess gene expression in myb36-1 and wild-type roots. From RNA-seq libraries of whole roots, we found 178 genes down-regulated and 324 genes up-regulated in myb36-1 (Dataset S1). Down-regulated genes are enriched for Gene Ontology (GO) terms associated with cell-wall organization, including all five CASP genes. Up-regulated genes are enriched for GO terms associated with extracellular region and peroxidase activity (Dataset S2).
Considering the cell-type specificity of MYB36 expression, we wanted to investigate changes in gene expression specific to the endodermis. To this end, we used the pSCR::ER:GFP transcriptional reporter and fluorescence-activated cell sorting to enrich for endodermal cells (17) from wild-type (pSCR::ER:GFP) and myb36-1 (pSCR::ER:GFP) seedlings. The endodermis-enriched RNA-seq data revealed 1,159 down-regulated genes and 949 up-regulated genes (Dataset S3). Down-regulated genes are associated with GO terms related to differentiation, including “peroxidase activity” and “Casparian strip.” Consistent with the whole-root data, all five CASP genes are significantly down-regulated in the mutant endodermis (Fig. 2F). Up-regulated genes are associated with GO terms related to response to stimulus, oxidoreductase activity, and binding, including “innate immune response,” “peroxidase activity,” and “RNA binding” (Dataset S4). Because peroxidase activity-related genes are enriched in both groups, MYB36 may regulate reactive oxygen species (ROS) homeostasis, which has been shown to play a critical role in the transition from proliferation to differentiation (18). Expression of a key regulator of this transition, UPBEAT1, is down-regulated in myb36-1 mutant plants (Fig. 2F).
CASP proteins are required to localize the lignin polymerization machinery enabling localized Casparian strip deposition (11). The NADPH oxidase respiratory burst oxidase homolog F (RBOHF) enables the production of H2O2, which is used by peroxidase 64 (PER64) to polymerize monolignol subunits into lignin (11). Although PER64 is significantly down-regulated in myb36-1 (Fig. 2F), RBOHF did not meet our significance threshold. However, other genes involved in ROS homeostasis are significantly differentially expressed (gray bars, Fig. 2F), as are numerous other peroxidases and oxidases (Dataset S3). Additionally, SCHENGEN3 (SGN3), a receptor-like kinase involved in localizing the CASP proteins (19), is significantly up-regulated in myb36-1 (Fig. 2F). Considering that expression of ROS-related genes both increases and decreases in myb36-1 mutants, MYB36 may play a general role in maintaining ROS homeostasis during differentiation. Alternatively, disruption of Casparian strip formation may cause ROS imbalance.
To assess whether regulation of CASP genes or ROS-related genes is direct or indirect, we performed an induction experiment using a MYB36 construct under its native promoter fused to the glucocorticoid receptor (GR) hormone-binding domain (pMYB36::MYB36:GR). GR fusions are sequestered in the cytoplasm and translocated to the nucleus upon induction by dexamethasone (dex) (20). Following dex treatment, myb36-1 mutants harboring this construct expressed pCASP2::GFP 4 h after induction (n = 100/100). Quantitative reverse transcription PCR (qRT-PCR) of CASP2 and PER64 confirmed that pMYB36::MYB36:GR induction resulted in increased expression of these genes (Fig. 2G). To assess whether this increased expression was the result of direct activation by MYB36, independent of protein synthesis, we induced with dex and added cyclohexamide (chx), a potent protein synthesis inhibitor. Surprisingly, there was no significant increase in expression of either CASP2 or PER64, suggesting that the activation is indirect (Fig. 2G). Alternatively, because protein synthesis is required for expression, MYB36 may induce a cofactor required for activation of CASP and PER64 in a feed-forward loop.
MYB36 Represses Genes That Are Involved in Proliferative Divisions.
In myb36-1 mutants there is evidence of extra divisions in both the meristematic and differentiation zones of the endodermis (Fig. 3 B and D). Extra divisions were previously observed for the WiscDsLox442H5 allele of myb36 at low penetrance (6/20 roots) (13), but we observed extra meristematic divisions at a higher penetrance (15/20 roots) in myb36-1, suggesting that our allele is more severe. These results suggest that MYB36 suppresses meristematic cell divisions. Because MYB36 is most highly expressed in the elongation zone before the onset of differentiation, MYB36 regulation of meristematic cell divisions is likely indirect, either by feedback on meristematic regulators or possibly by misregulation of ROS homeostasis. There is a growing body of literature that implicates ROS in cell division and differentiation (21, 22).
Fig. 3.
myb36-1 has more divisions in the meristem and in the differentiation zone than wild type. (A) Wild-type root meristem; the endodermis is outlined in orange (n = 20). (B) myb36-1 mutant root meristem displays additional divisions (outlined in orange) with 75% penetrance (n = 15/20). (C) Optical cross-section of wild-type seedling at the onset of the differentiation zone. Note the stereotypical eight cells present in both the endodermis and cortex. (D) Optical cross-section of myb36-1 at the onset of the differentiation zone. Additional divisions are outlined in orange. (E) myb36-1 has increased endodermal expression of JKD, SCR, and MGP compared with wild type (for source data see Dataset S8). (F) qRT-PCR of MYB36 expression 3 h after induction of pSCR::SCR:GR. Mean expression values of three biological replicates are reported after normalization with PP2A expression. Error bars are SEM (for source data see Dataset S9).
To investigate whether MYB36 regulates transcription factors involved in proliferative divisions, we reexamined the endodermis-enriched RNA-seq data. We found that SCR as well as MGP and JKD genes encoding the C2H2 transcription factors are significantly up-regulated in the endodermis of myb36-1 mutants (Fig. 3E). The change in expression from the closely related gene NUC did not satisfy our significance threshold. These BIRD transcription factors have been shown to play a role in early endodermal specification and proliferation (5, 6, 8). Thus, we speculate that MYB36 represses genes regulating proliferation as cells transition to a differentiated state.
SCARECROW Directly Activates MYB36 Expression in the Endodermis.
Based on the endodermal specificity of MYB36 expression, we hypothesized that SHR and/or SCR could be involved in its regulation. Time course gene expression data from inducible SHR and SCR proteins in their respective mutant backgrounds revealed that MYB36 is significantly up-regulated after induction of either SCR or SHR (4). To verify that SCR activates MYB36 expression, we performed an induction experiment using an inducible SCR construct under its native promoter fused to the GR hormone-binding domain (pSCR::SCR:GR) in the scr-4 mutant (4, 5, 7). We found that MYB36 expression is induced 3 h after induction, consistent with previous results (4). Treatment with dex and chx induced MYB36 expression as much as dex alone, providing strong evidence that SCR directly activates MYB36 expression (Fig. 3F).
A Key Link Between Proliferation and Differentiation.
The progression from stem cell to differentiated tissue involves transitions through different gene expression states. We have identified and characterized MYB36 as a key mediator of the transition from proliferation to end-stage differentiation. When mutated, critical genes for Casparian strip formation are not expressed, indicating that MYB36 controls this hallmark of differentiation. We find it interesting that MYB36 activates CASPs but represses the gene encoding the receptor-like kinase SGN3. This provides evidence for a more complex role of SGN3 in Casparian strip deposition. Although a barrier to small molecules forms eventually, future work will reveal whether this barrier is a fully functional Casparian strip.
Loss of MYB36 activity also results in increased cell proliferation and expression of SCR and certain genes encoding C2H2 transcription factors, suggesting a dual role of damping down cell division while promoting differentiation. This is analogous to the retinoblastoma (Rb) tumor suppressor protein in animals. Rb normally regulates progression through the cell cycle; however, when mutated or hyperphosphorylated, errant cell divisions result (23). Why ectopic cell divisions in plants appear to be more circumscribed than in animals is an interesting question for future research.
Expression of rMYB36:GFP is highest in the region just below the differentiation zone where the CASP proteins are expressed. The protein localization pattern mimics this expression (Fig. 2D). Considering that SCR and SHR are expressed throughout the endodermis, the expression pattern of MYB36 suggests that either there is an additional activator, which is also spatially restricted, or a repressor localized to the meristematic zone. Further studies will reveal additional upstream regulators of MYB36 and how network perturbation alters differentiation.
Our expression and direct activation data combined with results from the literature allowed us to construct a model describing the role of MYB36 in regulating the transition from proliferation to differentiation in the endodermis (Fig. 4). SCR and SHR activate MYB36 expression, which promotes expression of genes essential for localized Casparian strip formation. Additionally, MYB36 represses the genes encoding JKD and MGP, which regulate periclinal divisions in the endodermis and cortex (5, 6, 8). This repression by MYB36 results in restriction of proliferative divisions once the endodermis starts to terminally differentiate. These interactions generate an incoherent feed-forward loop (Fig. 4 D and E), which could provide a mechanism for the transitions between proliferation and differentiation (Fig. 4F).
Fig. 4.
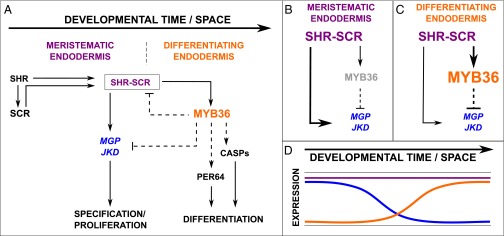
Model of the transition from specification to differentiation. (A) Wiring diagram describing the transition from proliferation to differentiation based on this work and published data discussed in the text. SHR activates expression of SCR. SHR and SCR form a complex that regulates expression of MGP and JKD, which regulate proliferation. At the transition between specification and differentiation, SHR/SCR, MYB36, and MGP/JKD form an incoherent feed-forward loop predicting two behaviors, as follows. (B) MGP and JKD gene expression should be high in the meristem in the absence of MYB36 expression. (C) Increased expression of MYB36 in the differentiation zone rapidly reduces MGP and JKD expression. (D) Predicted expression of proliferation-inducing genes (MGP and JKD) (blue) and MYB36 (orange) throughout developmental time. Thickness of lines represents the strength of interaction.
Methods
Plant Materials, Growth Conditions, and Steroid Treatment.
Arabidopsis accession Columbia-0 (Col-0) or Col-0 with the pCASP2::NLS:GFP:GUS (pCASP2::GFP) transgene was used as wild type for all experiments. The myb36-1 ethylmethane sulfonate (EMS)-generated line was used for all mutant experiments unless otherwise noted. For all experiments, seeds were sterilized, stratified, and imbibed 48 h at 4 °C. For the barrier assay, seedlings were plated on 0.5× Murashige and Skoog (MS) media, 1% agar, with no sucrose, and sealed with Micropore tape. Plates were grown vertically on square plates under long-day conditions and assayed at 6 d post exposure to light.
Induction and cyclohexamide experiments were performed as described (24) with modifications for younger plants. Briefly, pMYB36::MYB36:GR myb36-1 seeds were plated on sterile mesh, 0.5× MS media, with no sucrose. Seedlings were transferred 6 d post exposure to light to fresh 0.5× MS media (mock), 10 mM dexamethasone-containing media (dex), 10 mM cyclohexamide/10 mM dexamethasone-containing media (chx dex), or 10 mM cyclohexamide-containing media (chx). After 4 h of treatment, root tissue was harvested with a scalpel and frozen in liquid nitrogen. pSCR::SCR:GR scr-4 seeds were plated on sterile mesh, 1× MS, 1% sucrose media. Seedlings were transferred 6 d post exposure to light to fresh 1× MS, 1% sucrose media (mock), 10 mM dexamethasone-containing media, 10 mM dexamethasone/10 mM cyclohexamide-containing media, or 10 mM cyclohexamide-containing media. After 3 h of treatment, root tissue was harvested with a scalpel and frozen in liquid nitrogen.
Microscopy.
Barrier assays assessing Casparian strip function were performed as previously described (14). Briefly, seedlings were incubated in the dark for 10 min in fresh 10 μg/mL propidium iodide (Invitrogen) and rinsed two times in water. Stock solution (100×) was stored at 4 °C in water. The onset of elongation was defined as the first endodermal cell. Cortical cells were counted from the onset of elongation in a median optical section that was twice the size of its younger neighbor until the neighboring endodermal cell restricted PI penetration. The barrier assay and aberrant meristematic cell divisions were imaged and quantified using a Zeiss 510 upright confocal microscope.
Dissecting scope images were acquired with an Axio Zoom.V16 fluorescence dissecting scope (Zeiss) at 25× magnification. Images were stitched using Fiji (25) with the Pairwise Stitching plugin (26).
Mutagenesis Screen.
Seeds were mutagenized as previously described (27). Briefly, ∼7,500 seeds expressing homozygous pCASP2::GFP were imbibed overnight, mutagenized with 0.2% EMS for 15 h, washed with sterile water 10 times for a total of 4 h, and sown on soil. Approximately 5,000 mutagenized (M0) seeds germinated and allowed to self-cross to produce M1 seeds. Seeds were collected as bulks with 5–10 M1 plants per bulk. Nine hundred thirty bulks were collected and screened. In the next generation, ∼60 M2 seedlings per bulk were screened for changes in pCASP2::GFP expression intensity using a fluorescence stereo dissecting scope (Leica).
Mapping.
Bulk segregant analysis and whole-genome resequencing were used to map the EMS-generated myb36-1 mutant as previously described (28). Briefly, mutant plants were back-crossed to the pCASP2::GFP parental line and map-crossed to Landsberg erecta (Ler) to generate F1 heterozygotes. These plants were self-crossed, producing two populations of segregating F2 plants. Approximately 50 mutant plants were bulked from each cross. Unmutagenized pCASP2::GFP and Ler parental plants were grown and 100 seedlings were sequenced. Four pools were used to make DNA libraries using a TruSeq Library Prep Kit (Illumina). SNPtrack genetics.bwh.harvard.edu/snptrack/ was used to analyze the data and determine the causative mutation (28). dCAPS genotyping primers were designed (helix.wustl.edu/dcaps/dcaps.html) (29) to screen for a mutant allele: genotype forward (F)/reverse (R): 5′-AGATGTGGTAAGAGTTGCAGACTGAGATA-3′, 5′-caagagagatagatcaccgacCG-3′. Product size is 144 bp; wild type is cut into 114- and 31-bp fragments with AluI enzyme (New England Biolabs; R0137S). Whole-genome resequencing data have been uploaded to NCBI BioProject (www.ncbi.nlm.nih.gov/bioproject) accession no. PRJNA289027.
RNA-Seq Library Preparation.
Seeds were sterilized with 3% (vol/vol) sodium hypochlorite and 0.1% Tween for 7 min and rinsed five times in sterile water. Seeds were plated on sterile mesh, 1× MS, 1% agar, 1% sucrose media, and sealed with parafilm. Plates were grown under long-day conditions for 6 d. Whole-root samples from wild type (Col-0) and myb36-1 were harvested and frozen in liquid nitrogen. Endodermal-enriched samples expressing the transcriptional reporter pSCR::ER:GFP (30) in the Col-0 and myb36-1 backgrounds were obtained by fluorescence-activated cell sorting as described previously (17). Total RNA from whole roots and sorted cells was extracted using a Plant RNeasy Kit (Qiagen). rRNA was depleted from total RNA using a RiboMinus Plant Kit (Invitrogen). RNA quantity and integrity were assessed by Qubit (ThermoFisher Scientific) and Agilent Bioanalyzer. A ScriptSeq v2 RNA-Seq Library Prep Kit (Epicentre) was used to generate strand-specific RNA-seq libraries. Paired-end reads (50-bp) were obtained using the Illumina HiSeq 2000 platform at the Duke University Genome Sequencing & Analysis Core.
RNA-Seq Data Analysis.
Reads were quality-filtered and mapped to the Arabidopsis genome version TAIR10 using TopHat (31), and read counts were determined by HTSeq (32). Differentially expressed genes were determined using the R package DESeq (33). Significance was assessed using a threshold of log2 fold change ≥1 and padj ≤0.01 [padj: P value adjusted for multiple testing and controlled for false discovery rate with the Benjamini–Hochberg statistical procedure (33)]. Libraries were also run through Cufflinks (34) to determine fragments per kilobase of transcript per million mapped reads (FPKM) values for whole-root (Dataset S5) and sorted libraries (Dataset S6). All RNA-seq data have been uploaded to the GEO accession no. GSE70584. AGI numbers or locus identifiers for selected genes are as follows: AT2G36100 CASP1; AT3G11550 CASP2; AT2G27370 CASP3; AT5G06200 CASP4; AT5G15290 CASP5; AT5G03150 JACKDAW; AT1G03840 MAGPIE; AT5G44160 NUTCRACKER; AT4G11290 PER39; AT5G42180 PER64; AT1G09090 RBOHB; AT1G64060 RBOHF; AT1G05260 RCI3; AT4G20140 SGN3/GSO1; AT4G37650 SHR; AT3G54220 SCR; AT2G47270 UPBEAT1.
Functional categories were assigned to differentially expressed genes using the GOrilla online tool (35). A background set of detected genes was defined for whole roots and sorted endodermis independently. A gene was called detected if it was found in at least two biological replicates with an arbitrary cutoff of two normalized counts per replicate. A P value of 1E-04 was used for both conditions; only the most descendant Gene Ontology terms are reported for the sorted endodermis analysis.
Plasmid Construction and Transformation.
The JaTY clone JaTY60A10 (64,154 bp) was used to generate the rMYB36:GFP recombineering line as described (36). The oligos MYB36_Rec_F/R were designed to remove the STOP codon upon recombination with the GFP cassette. Homology to the MYB36 locus is in capital letters: MYB36_Rec_F/R: 5′-TGGTTATGCTTCAAGATTACGCTCAGATGAGCTACCACAGTGTTggaggtggaggtgg-3′, 5′-ATCCATCCCTATAGTTACGCATTTATATATATGCATGATATAACTTActaagcgtaatcaggaacatcgtaagggtaggccccagcggcc-3′. This construct was transformed into Col-0 and crossed to myb36-1.
Standard molecular biology procedures and the Gateway Cloning System (Invitrogen) were used for cloning. The 3.0-kb region upstream of the MYB36 transcription start site and the genomic region of the MYB36 gene were used to generate the pMYB36::MYB36:GR construct. pMYB36::MYB36:GR was transformed into the myb36-1 mutant.
qRT-PCR.
RNA was prepared from whole roots as described for RNA-seq libraries. Five hundred nanograms of RNA was used to generate cDNA with SuperScript III (Invitrogen) and diluted 1:10 for assays. Primers were designed and tested for amplification efficiency. qRT-PCR was preformed using FastStart Universal SYBR Green Master Mix (Roche) on a StepOnePlus instrument (Applied Biosciences). Three biological replicates and three technical replicates were used for each experiment. Standard curves were run for each primer pair. Values reported are the means of three biological replicates after efficiency corrected quantification with PP2A as the reference. The following pairs were used: CASP2exon1F/R: 5′-TACAACAGAGGACTCGCCATC-3′, 5′-TGGAAAGTTGGTAGATCGTCG-3′; PER64exon2F/R: 5′-ACAAAGCAGAGAAAGATGGACC-3′, 5′-CTCCTGAGAGAGCGACAGCATC-3′; MYB36exon3F/R: 5′-GGTCCATAATTGCAGCTCAG-3′, 5′-AATCGGTTATGGAGTCTTGACG-3′; PP2AF/R: 5′-TAACGTGGCCAAAATGATGC-3′, 5′-GTTCTCCACAACCGCTTGGT-3′.
Note.
While our paper was in revision, another paper was published in PNAS that describes the cell biology of MYB36 (37). Our paper demonstrates how MYB36 regulates the transition from proliferation to differentiation. We find the two papers to be complementary.
Supplementary Material
Acknowledgments
We thank the Duke University Genome Sequencing & Analysis Core Resource for sequencing the DNA and RNA libraries. The Biology Department Phytotron space was used for growing plants. We thank Cara Winter for insightful comments on the manuscript and helpful experimental suggestions, Eline Verbon for providing useful feedback on the manuscript, and Jingyuan Zhang and Heather Belcher for technical assistance. We thank Lindsay Marjoram and Ignaty Leshchiner for assistance with SNPtrack, and Niko Geldner for sharing the pCASP2::NLS:GFP:GUS (pCASP2::GFP) construct. This work was funded by a grant to P.N.B. from the National Science Foundation Arabidopsis 2010 Program (IOS-1021619) from the NIH (R01-GM043778) and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through Grant GBMF3405). L.M.L. was funded in part by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. J.J.P. was supported by an NIH Ruth L. Kirschstein National Research Service Award (F32 GM086976) fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: All whole-genome sequencing data and RNA-seq data reported in this paper have been deposited in the Sequence Read Archive (SRA) and Gene Expression Omnibus (GEO) database under the umbrella BioProject, www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA292308). Seed lines generated for this paper have been sent to the Arabidopsis Biological Resource Center, abrc.osu.edu/order-stocks (ABRC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515576112/-/DCSupplemental.
References
- 1.Caspary R. Bemerkungen über die Schutzscheide und die Bildung des Stammes und der Wurzel. Methods Mol Biol. 1865;4:24. [Google Scholar]
- 2.Naseer S, et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA. 2012;109(25):10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413(6853):307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 4.Sozzani R, et al. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466(7302):128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levesque MP, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4(5):e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch D, et al. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21(17):2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui H, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316(5823):421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 8.Long Y, et al. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell. 2015;27(4):1185–1199. doi: 10.1105/tpc.114.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sena G, Jung JW, Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131(12):2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- 10.Roppolo D, et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473(7347):380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153(2):402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 12.Dubos C, et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Iyer-Pascuzzi AS, et al. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell. 2011;21(4):770–782. doi: 10.1016/j.devcel.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alassimone J, Naseer S, Geldner N. A developmental framework for endodermal differentiation and polarity. Proc Natl Acad Sci USA. 2010;107(11):5214–5219. doi: 10.1073/pnas.0910772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Liberman LM, Mukherjee N, Benfey PN, Ohler U. Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res. 2013;23(10):1730–1739. doi: 10.1101/gr.149310.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 18.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143(4):606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Pfister A, et al. A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife. 2014;3:e03115. doi: 10.7554/eLife.03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalman FC, Scherrer LC, Taylor LP, Akil H, Pratt WB. Localization of the 90-kDa heat shock protein-binding site within the hormone-binding domain of the glucocorticoid receptor by peptide competition. J Biol Chem. 1991;266(6):3482–3490. [PubMed] [Google Scholar]
- 21.Livanos P, Apostolakos P, Galatis B. Plant cell division: ROS homeostasis is required. Plant Signal Behav. 2012;7(7):771–778. doi: 10.4161/psb.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt R, Schippers JHM. ROS-mediated redox signaling during cell differentiation in plants. Biochim Biophys Acta. 2015;1850(8):1497–1508. doi: 10.1016/j.bbagen.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 23.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 24.Wagner D, Sablowski RW, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285(5427):582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- 25.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25(11):1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigel D, Glazebrook J. 2006. EMS mutagenesis of Arabidopsis seed. CSH Protoc 2006(5):pdb.prot4621.
- 28.Leshchiner I, et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22(8):1541–1548. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18(12):613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 30.Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127(3):595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):516–520. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou R, Benavente LM, Stepanova AN, Alonso JM. A recombineering-based gene tagging system for Arabidopsis. Plant J. 2011;66(4):712–723. doi: 10.1111/j.1365-313X.2011.04524.x. [DOI] [PubMed] [Google Scholar]
- 37.Kamiya T, et al. The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci USA. 2015;112(33):10533–10538. doi: 10.1073/pnas.1507691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.