Abstract
Islets of Langerhans contain multiple hormone-producing endocrine cells controlling glucose homeostasis. Transcription establishes and maintains islet cellular fates and identities. Genetic and environmental disruption of islet transcription triggers cellular dysfunction and disease. Early transcriptional regulation studies of specific islet genes, including insulin (INS) and the transcription factor PDX1, identified the first cis-regulatory DNA sequences and trans-acting factors governing islet function. Here, we review how human islet “omics” studies are reshaping our understanding of transcriptional regulation in islet (dys)function and diabetes. First, we highlight the expansion of islet transcript number, form, and function and of DNA transcriptional regulatory elements controlling their production. Next, we cover islet transcriptional effects of genetic and environmental perturbation. Finally, we discuss how these studies’ emerging insights should empower our diabetes research community to build mechanistic understanding of diabetes pathophysiology and to equip clinicians with tailored, precision medicine options to prevent and treat islet dysfunction and diabetes.
Keywords: Genome-wide association study (GWAS), Promoter, Broad H3K4me3 Domain (BD), Enhancer, Stretch/Super Enhancer (SE), Chromatin Interaction Analysis by Paired End Tag sequencing (ChIA-PET), Chromatin Immunoprecipitation (ChIP)-seq, RNA-seq, islet, type 1/2 diabetes (T1D/T2D), chromatin, expression quantitative trait locus (eQTL), splicing quantitative trait locus (sQTL), allele-specific expression (ASE), single nucleotide polymorphism (SNP), inflammation, oxidative stress, endoplasmic reticulum (ER) stress
Introduction
The islets of Langerhans are clusters of at least 5 cell types--alpha, beta, delta, epsilon, and pancreatic polypeptide (PP)—that, together, comprise ~1-2% of the pancreas and execute pancreatic endocrine functions. The DNA in each of these cells is largely identical, yet they are wired to complete distinct and complementary functions to maintain tight glycemic control. Until recently, most of our understanding of islet composition, physiology, and pathophysiology has been driven by animal model studies (mostly mouse and rat). Recent comparative analyses have revealed species-specific differences in the cellular architecture/composition of islets [1], [2], gene expression programs [3], and insulin secretion properties [2], [3], emphasizing the importance of studying and understanding physiologic processes and pathophysiologic responses in human islets alongside model systems. Moreover, human genetic variants affecting islet (dys)function may not exist or be properly modeled in other species.
Islet endocrine cell type composition varies both between individuals and between pancreatic sub-regions of the same individual, but averages 54% (range 28-75%) insulin-producing beta cells, 35% (range 10-65%) glucagon-secreting alpha cells, 11% (1-22%) somatostatin-secreting delta cells, and very few epsilon and PP cells [1], [2]. As might be expected based on morphologic fluctuations, islet function also varies between individuals [4]. Many of the rare genetic differences that cause monogenic islet disorders, such as congenital hyperinsulinemia (CHI), permanent/transient neonatal diabetes mellitus (PNDM/TNDM), and maturity onset diabetes in the young (MODY), are protein-coding changes in islet transcription factors (TFs) or non-coding changes that affect islet transcriptional regulation. Islet transcriptional dysregulation is also implicated by type 1 and type 2 diabetes genetic susceptibility studies [5]–[8].
Transcription is a fundamental cellular process that governs cell fate choices in developing cells and myriad physiologic and pathophysiologic responses in mature cells. RNA Polymerase 2 (Pol2) transcribes genes encoded by the cell’s DNA into various messenger RNA (mRNA) molecules. Early transcriptional regulation studies demonstrated the importance of trans-acting factors (DNA binding proteins) binding to cis-acting DNA sequence motifs immediately prior to a gene’s transcriptional start site in transcriptional control. Recent studies indicate that sequences/sites distant from promoters also mediate cell type-specific transcription. Moreover, they indicate that molecular features not specifically predicted by DNA sequence motifs, such as local DNA shape and DNA accessibility and long-range folding/packing in the nucleus, also influence transcriptional regulation. Together, these features control the recruitment and/or activation of Pol2 transcriptional complexes at specific genes. Genetic (e.g., sequence variation) or epigenetic (e.g., chromatin remodeling) perturbation of these features can disrupt normal transcription, contributing to cellular dysfunction and disease.
Here, we review how next generation sequencing-based molecular profiling technologies performed on human islets over the past five years is transforming our understanding of transcriptional (dys)regulation in human islet (dys)function and disease. Islet transcriptome analyses by RNA sequencing (RNA-seq) have uncovered greater diversity of transcripts, in form, number, and function, than previously estimated. Epigenomic analyses have discovered tens of thousands of new transcriptional regulatory regions that coordinately control islet transcriptional output in resting, stimulated, and stressed states. Studies of islet genomes, epigenomes, and transcriptomes from multiple people are providing insights into genetic differences that alter transcriptional regulation to contribute to islet dysfunction and disease. Studies comparing diabetic and non-diabetic islet transcriptomes have identified abnormal transcriptional features; these have emphasized the need to better understand the transcriptional consequences of specific islet stresses, such as inflammation, endoplasmic reticulum stress, and oxidative stress. These studies have facilitated new insights into the genetics and molecular mechanisms of islet (dys)function and disease and are fueling new opportunities for preventative and therapeutic approaches to diabetes treatment.
The expanding islet transcriptome
How many genes are transcribed in the islets? Until recently, our understanding of the islet transcriptome was limited by the technology of the time. Microarray-based expression analysis required existing knowledge about the location and structure of genes to design the nucleotide probe sequences to interrogate gene expression. In contrast, RNA sequencing (RNA-seq) does not require such a priori knowledge or design limitations and has enabled agnostic interrogation of the entire transcriptomes of cultured cells and tissues covering a wider range of expression level than microarrays [9]. RNA-seq of human islets and sorted constituent cells has dramatically expanded islet transcriptome catalogue numbers, forms, and functions.
The first application of high throughput sequencing to catalogue the pancreatic islet transcriptome identified approximately 21,000 transcripts, corresponding to 7600 genes [10]. Deeper sequencing of islets from multiple individuals followed to characterize the transcriptomes of intact islets [11]–[17] and their dissociated, sorted, and purified constituent cells, notably insulin-producing beta cells and glucagon-producing alpha cells [11], [18]. These studies, performed on samples from different individuals and under different conditions, collectively suggest that as many as 50-60% of known genes are modestly expressed in islets (10,883 genes with reads per kilobase per million mapped reads (RPKM) >1 and 17,175 genes with RPKM > 0.5) [12], [13], [16]. These include several genes near DNA sequence variants (single nucleotide polymorphisms; SNPs) implicated by genome-wide association studies (GWAS) in genetic susceptibility to type 1 (T1D) and type 2 diabetes (T2D). These data support current views of the importance of islet (dys)regulation in T2D pathophysiology [19] and have rekindled interest in potential roles for islet transcriptional dysregulation in early or progressive pathophysiologic events leading to T1D [7], [20]. MicroRNA (miRNA) have also been profiled in human islets [21], [22]; a recent review describes their roles in post-transcriptional regulation and islet biology [23].
Initial RNA-seq analyses also hinted that several genes in islets and beta cells may undergo alternative splicing to form multiple, distinct transcripts (isoforms). Alternatively spliced transcripts contribute to cell type specific gene functions and have been implicated in both physiologic and pathophysiologic events in cells. For example, alternative splicing of TCF7L2 and G6PC2 was implicated as a molecular consequence of their respective GWAS SNP risk alleles [24]. Analysis of human islet transcriptomes from 11 individuals suggests that 1000-2000 genes may undergo alternative splicing in islets [11]. Analysis of ~90 human islet transcriptomes linked alternative splicing control of 371 islet transcripts to a specific SNP, termed splicing quantitative trait loci (sQTL) [17]. Thus, the islet transcriptome is quite diverse and this diversity is, to at least some extent, under genetic control. Better understanding of these mechanisms in islets should provide insights into islet dysfunction and diabetes.
Long non-coding RNA transcripts (lncRNAs) are a newly identified class of transcripts with implications in many diseases. LncRNAs mediate diverse developmental and pathophysiologic processes ranging from imprinting and X inactivation to tumorigenesis (reviewed in [26]), but lncRNA cell type specificity and low sequence conservation between species has made it difficult to predict lncRNA functions and modes of action [27]. Fueled by epigenome and transcriptome profiling of multiple cell types and tissues over the past 5-6 years, the catalog of lncRNAs is continuously and rapidly expanding. They were first systematically identified [25] using 3 criteria: (a) histone 3 lysine 4 trimethyl (H3K4me3)-histone 3 lysine 36 trimethyl (H3K36me3) “active transcription” epigenetic marks that occur over regions of the genome devoid of gene annotations; (b) minimum transcript size of 200 nucleotides; and (c) minimal protein-coding potential. They are exquisitely cell type-specific; each RNA-seq study of a new cell type or tissue identifies new lncRNAs. Morán et al [13] applied the above lncRNA criteria to islet and FACS-sorted beta cell RNA-seq data to identify 1128 lncRNAs. Independent RNA-seq studies of human islets have identified a similar number of lncRNA (1297 [15]); however, only 25-30% (n=349) of lncRNAs overlap between studies. It is unclear whether this discrepancy is due to technical differences in islet handling/data processing steps or to genetic differences between samples. Uniform and joint analysis of these datasets will help to rectify these apparent discrepancies and to identify the complete compendium of human lncRNAs in islet cells under baseline conditions.
Understanding molecular functions of newly identified islet lncRNAs is an important goal. The first islet lncRNAs were described in 2012, so it is still early to dissect their precise role(s) in islet cell development, identity, and (patho)physiology. Islet lncRNA expression patterns and genomic location suggest they may serve as important biomarkers or mediators of islet (dys)function and diabetes. Their expression is developmentally controlled, and a subset of them are glucose-responsive and associated with changes in HbA1c, a long-term measure of glucose control [13], [17]. Several are situated next to genes encoding islet TFs, including PDX1, HNF1A, NEUROD1, MAFB, FOXA2, ISL1, and NKX in the genome; others are co-expressed with islet TF and insulin secretion genes [17]. LncRNA knockouts in beta cells or whole animals, coupled with additional profiling of lncRNA behavior under different stimulatory and stress conditions, should shed light on the functions of these new and exciting RNA species.
Identification of islet transcriptional regulatory elements
Initial islet transcriptional regulation studies focused on regions immediately upstream of transcription start sites. They also relied almost exclusively on in vitro reporter assays for insights into in vivo control. These studies made at least two important contributions to understanding islet transcriptional regulation. First, they identified DNA sequence motifs that regulate islet gene expression, including the A-box, C1, E1, and CRE sequences in the insulin gene promoter [28] and areas I-IV upstream of the PDX1 gene [29], [30]. Second, they facilitated discovery of important islet TFs that bind these motifs, such as PDX1, MAFA, NeuroD, and FOXA2/HNF3Beta. However, detailed comparison of in vitro and in vivo Pdx1 binding to target promoters emphasized the importance of DNA accessibility and nuclear chromatin structure in dictating which sequences are actually TF-bound and used in vivo by islets and beta cells [31].
In the nucleus of each cell, DNA is wrapped around histone octamers to form chromatin. Inactive (heterochromatin) and actively transcribed (euchromatin) chromatin regions of the genome exhibit different features, including varying degrees of openness/accessibility and distinct histone protein covalent modifications [32]. Euchromatin is more loosely packed and accessible to specific and general TFs. As such, active regulatory elements are accessible to enzymes like DNase I [33]. Histone modification patterns are used to stratify them into promoter, enhancer, and insulator elements [32], [34]–[38] (Fig. 1A).
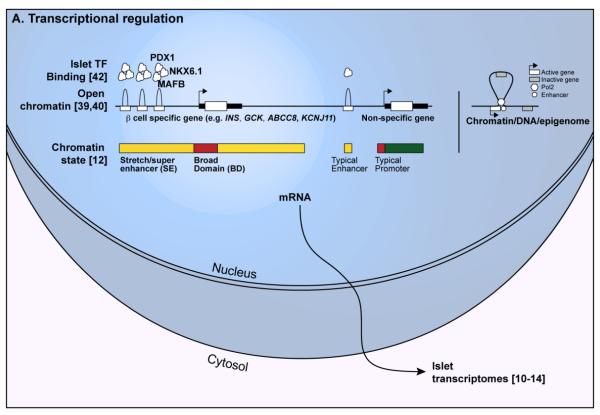
Figure 1. Transcriptomic and epigenomic features of normal and perturbed islets. (A) Transcriptional regulatory features in islets.
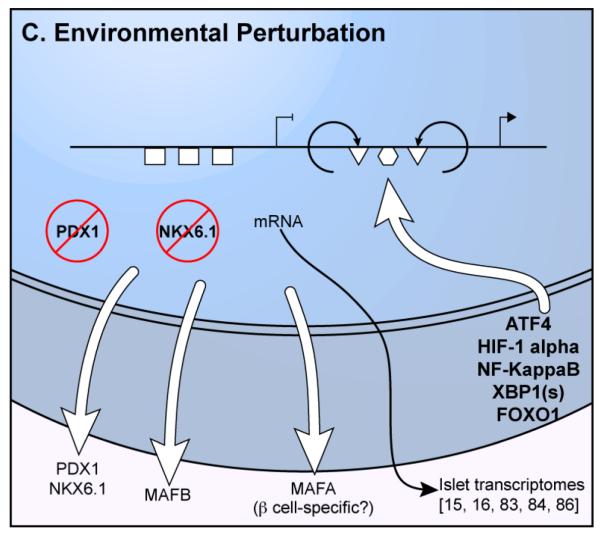
Left; Open chromatin, islet transcription factor (TF) binding, and combinations of histone modification patterns (chromatin state) identify regulatory features in islets. Genes important for islet/beta cell identity and function (e.g., INS, KCNJ11, ABCC8, GCK) exhibit important epigenetic features, such as clustered sites of open chromatin (humps) and multiple islet TF binding (cotton balls), and extended enhancer (yellow bars) and promoter (red bars) chromatin state compared to features around a typical gene. Green bar indicates a “transcription elongation” state typically observed over non-specific, expressed genes. Right; 3D epigenomic analyses identify enhancer-target gene links, which can involve looping out/exclusion of the nearest gene (gray rectangle) on the linear genome to mediate 3D interactions between the enhancer (white circle). (B) Some genetic variants disrupt TF binding motifs (red “X”), abrogating protein binding (e.g., PDX1), reducing chromatin accessibility, and inactivating the gene. (C) Islets respond to perturbations such as oxidative stress, inflammation, and oxidative stress with nuclear translocation of several stress-responsive TFs (e.g., NF-KappaB, ATF4, XBP1(s), HIF1-alpha). These factors bind to new islet regulatory elements (triangles, circle) to activate the appropriate stress response genes (arrowhead). Islet TFs are inactivated and/or exported from the nucleus (red circles with slashes), abandoning their binding sites (rectangles) and leading to gene inactivation.
Several techniques have been recently applied to functionally profile human pancreatic islets. DNase-seq and FAIRE-seq studies revealed approximately 100,000 [39] and 80,000 [40] open sites (referred to as “peaks”) in islets, respectively. These two studies expanded the catalogue of potential islet cis-regulatory elements from a few thousand promoters to tens of thousands of promoters and non-promoter (enhancer, insulator) elements. Together with genetic susceptibility studies (discussed below), these data are uncovering the complexity of islet transcriptional regulation and emphasizing the importance of enhancer control of islet transcription programs in physiologic and pathophysiologic states. Table 1 summarizes tools contributed by multiple studies [12], [39], [41], [42] to search, visualize, and retrieve the locations of epigenetic features, described below, in islets and other cell types.
Table 1.
Islet regulatory element databases
| Dataset | URL | Study |
|---|---|---|
| Islet open chromatin (DNAse-seq; “PanIslets” track) |
http://www.genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=405493199_538GgMsR3I61PZAYyET9dQcNHIVk&c=chr6&g=wgEncodeOpenChromDnase | [39] |
| Broad H3K4me3 Domains (BDs) | http://bddb.stanford.edu | [41] |
| Chromatin states from 10 cell types (including islets); Stretch Enhancers (SEs) |
http://research.nhgri.nih.gov/manuscripts/Collins/islet_chromatin/ | [12] |
| Islet enhancer clusters (Islet Regulome) |
http://www.isletregulome.org | [42] |
Promoters
Promoters are cis-regulatory elements located adjacent to the transcriptional start sites (TSSs) of genes (Fig. 1A). They are necessary for transcription of a gene to occur, but recent genome-wide surveys of cis-regulatory elements suggest they may not be sufficient to direct cell type-specific or robust gene transcription. Targeted promoter studies to determine the mechanisms governing INS transcription were instrumental to identify several important factors regulating gene expression in beta cells and islets, including PDX1, NEUROD1, and MAFA and to define the cis-regulatory sequences they bind to exert their control [28].
Genome-wide, active promoters are enriched for histone 3 lysine trimethylation (H3K4me3). H3K4me3 chromatin immunoprecipitation-sequencing (ChIP-seq) identified approximately 14,000-18,000 promoter sites in the genome that are potentially active or poised for use in islets [12], [18], [39], [42], [43]. In addition to cataloguing islet promoters, these studies contributed the following insights about transcriptional regulation in islets. First, very few promoters are islet-unique. When compared with multiple other cell types, only ~1.5% of promoters (n=256) were exclusively H3K4me3-positive in islets [39]. However, this set included several important genes for islet function such as the beta cell-specific hexokinase (GCK), the RNA binding protein, HuD, that regulates INS translation (ELAVL4) [44], and the zinc transporter, ZnT8, important for insulin granule assembly and secretion (SLC30A8). This observation is consistent with other studies [35], [45], [46] and suggests that cell type-specific (e.g., islet-specific) transcriptional control of gene expression is dictated by specific promoter use for only a small subset of genes. There is some discrepancy in the promoter architecture and putative transcriptional regulatory mechanisms at genes encoding major islet hormones such as insulin (INS), glucagon (GCG), and somatostatin (SST) [39], [42], [43], [47]. Data from multiple groups suggest that these promoters are not highly enriched for the typical punctate H3K4me3 active promoter mark despite the genes being highly expressed [39], [43], [47]. Detailed INS locus analysis defined this region as an islet “open chromatin domain”--an 80 kb region encompassing the INS/IGF2/TH genes that does not exhibit punctate promoter marks, but rather widespread general openness, active histone modification patterns, and evidence of pervasive transcription throughout the locus [48].
Enhancers
Enhancers are DNA sequences in the genome that amplify or “enhance” transcription of a gene above baseline levels. They confer spatial and temporal specificity to promoter activity and gene expression in developing and mature cells and tissues. Enhancer features and specific examples, including the limb-specific enhancer controlling SHH expression and the beta-globin locus control region (LCR) controlling expression of multiple hemoglobin genes at different stages of development, have been elegantly reviewed recently [49]. Extensive epigenomic surveys of open chromatin by DNase-seq and histone modifications by ChIP-seq in hundreds of cell types have enabled the genome-wide and systematic identification of these elements and elucidated some of their general features [34]–[37], [45], [47], [50], [51]. Enhancer sites are typically exhibit histone 3 lysine 4 monomethylation (H3K4me1) enrichment. Active and poised enhancers are distinguished by presence or absence of H3 lysine 27 acetylation (H3K27ac), respectively.
Islet ChIP-seq studies have identified 30,000-60,000 putative enhancers in islets [12], [39], [42], [43]. This is consistent with data from other cell types, such that enhancers outnumber promoters by two- to four-fold [34], [35], [47], and represents a dramatic expansion of the cis-regulatory landscape of islets,. Islet enhancers are more cell-specific than islet promoters, suggesting they are key mediators of islet-specific transcriptional responses. Moreover, sequence variants contributing to variation in islet expression, function (e.g., fasting glucose), or risk of type 2 diabetes are significantly and specifically enriched to overlap islet enhancers [12], [17], [42], [51].
Bigger is better
Independent studies of genome-wide histone modification patterns in multiple cell types have revealed that both promoter and enhancer chromatin marks range in length from hundreds to tens of thousands of nucleotides [12], [41]. Broad domains (BDs; Fig 1A) are the longest 5% of contiguous H3K4me3 promoter marks (>4 kilobases; kb) [41]. They mark promoters of genes of particular importance for cell-specific identity and function. For example, BDs mark promoters of genes encoding the pluripotency TFs (OCT4, SOX2, NANOG) specifically in embryonic stem cells. Approximately 1,000-3,000 genes are BD-marked in a given cell type. This long epigenetic mark seems to reflect or govern stable, consistent gene expression rather than solely high levels of expression [41]. Islets contain approximately 3,500 BD-marked genes. BD-marked genes in islets include major TFs (e.g., PDX1, MAFA/B, NKX6-1/2-2), genes encoding key enzymes for glucose processing (GCK, G6PC2), and those regulating insulin production and secretion (ELAVL4, KCNJ11, SLC30A8, PCSK1/2, CACNA1C/D). The BD mark has also been used as a screening tool to identify novel genes that play important roles in cell identity and function [41]. We expect that systematic screens of BD-marked genes in islets will uncover new gene(s) and pathway(s) controlling islet function.
Similar to Broad Domains, independent studies have identified a subset of enhancers in a cell that seem to govern the transcription of genes particularly important to cell identity and function. Stretch enhancers are the longest 5-10% of enhancer states (>3kb long) in each cell type and are located near to or overlapping genes important for cell type-specific functions. (Fig. 1A, left). Super enhancers were originally defined as single or clustered sites in the genome bound by a disproportionate amount of cell type-specific master TFs and/or coactivator proteins [52], [53]. Subsequently, surveys of multiple cells identified super enhancers based on long stretches of H3K27ac “active enhancer” modifications [54]. Although stretch and super enhancers (SEs) are not equivalent regulatory entities, they overlap at several loci and share important functional features: (i) they are highly cell type-specific and overlap locus control regions (LCRs)--complex regulatory regions dictating the developmental regulation of certain genes; (ii) they are associated with cell type-specific expression of genes important for cell type-specific functions; and (iii) they are enriched for SNPs associated with phenotypes and diseases affecting the relevant cell type (e.g., T2D or fasting glucose SNPs enriched in islet SEs) [12], [54]. In islets, they are comprised of clustered constituent open chromatin sites [39], [40] bound by multiple master islet TFs, such as PDX1, NKX6-1, FOXA2, and MAFB [42]. These complex regulatory sites may function as regulatory hubs or transcription factories to coordinate transcriptional activity.
Connecting the pieces
With transcriptional regulatory element “parts lists” in hand, a critical step is to assemble the components--enhancers, promoters, and insulators--into a detailed wiring diagram of the circuits that control transcriptional responses to stimulus and stress in the nucleus of islet cells (Fig. 1A, right). Connectivity maps provide mechanistic insights into cell type-specific transcriptional regulation [55]–[58] and link SNP-containing enhancers to their target genes [55], [59]. Felsenfeld and colleagues used 4C, a variation of the chromosome conformation capture technique (reviewed in [60]) to identify genes physically interacting or in close three-dimensional (3D) proximity with the INS promoter in islets. They discovered that the SYT8 and ANO1 genes, located approximately 300 kb and 68 megabases (Mb), respectively, from the INS promoter on the linear DNA, are close together in the 3D nucleus [61], [62]. Both interactions are islet-specific and functionally link INS transcription with that of two genes encoding a membrane protein (SYT8) and an ion channel (ANO1) important for insulin exocytosis. Glucose stimulation strengthened both interactions and enhanced SYT8 and ANO1 expression in islets. This suggests that these genes form glucose-responsive transcriptional co-regulatory units, supporting the transcription factory model introduced over 10 years ago [63].
The two- to four-fold excess of enhancers relative to promoters strongly suggests that this circuitry is more complex than a collection of single enhancer-promoter interactions. Clustered open chromatin sites [39], [40] and islet TF binding [42] in SEs suggest that these elements form complex regulatory hubs or co-regulatory units, with multiple enhancers coordinately regulating one or more promoters. Ferrer and colleagues used 4C to identify several putative enhancers interacting with specific promoters, including ISL1, PDX1, and MAFB [42]. They confirmed interaction between known regulatory elements and their target promoter (e.g., Regions I-IV upstream of PDX1 [29], [30]) and discovered novel interactions, some of which extended over 1 Mb from the promoter. Because these are bulk islet interactions, we do not know if they are present in all islet cell types or if all or a subset of these interactions is formed in a specific islet cell type (e.g., beta cells). Overall, however, these data support a model wherein multiple enhancers contact a gene promoter to coordinately regulate its activity in islets. These examples illustrate the utility and importance of 3D epigenome approaches to better understand transcriptional regulation in islets and to assign enhancers to their target genes. It will be important (1) to expand upon these targeted analyses to identify comprehensively all promoter-promoter, promoter-enhancer, and enhancer-enhancer interactions at high resolution using techniques such as ChIA-PET [64] or Capture-C [65]; and (2) to assign these interactions to their specific endocrine cell type to better understand cell type-specific (e.g., alpha, beta, delta cell) connectivity, especially those of BDs and SEs.
Perturbed transcriptional regulation in islet dysfunction and diabetes
Effects of individual genetic variation on islet transcription
DNA sequence variants that alter islet transcriptional programs lead to both rare and common forms of diabetes. Over half of the genes containing disease-causing mutations in patients with PNDM, TNDM, MODY, and CHI encode islet TFs. These include well-known genes such as PDX1, HNF1A, HNF1B, HNF4A, and NEUROD1. Recent exome sequencing of neonatal diabetes patients has identified mutations in two additional TF genes, GATA4 [66, p. -] and GATA6 [67]. Exome sequencing of PNDM, TNDM, MODY, and CHI patients with undiagnosed mutations is ongoing and is almost certain to identify mutations in additional, perhaps unexpected, TFs. Understanding the cis-regulatory elements bound by these factors and their target genes is necessary to better understand the pathophysiologic consequences of their disruption. Such an endeavor may define new sites that could contain disease-predisposing mutations.
Although less numerous than protein-coding mutations, rare variants altering islet cis-regulatory elements have been identified in families with monogenic islet disorders (Table 2). Promoter mutations in KCNJ11 and ABCC8 were identified in CHI patients [68]. Reporter assays indicate that these mutations each decreased promoter activity by ~60%. Similarly, inactivating promoter mutations in HNF4A P2 were identified for both MODY1, MODY-like, and gestational diabetes patients [69]–[71], and in the beta cell GCK promoter in patients with fasting hyperglycemia [72]. Intergenic mutations in the BLK locus, which decreased reporter gene activity, were identified as causative for MODY9 [73]. Inspection of islet chromatin maps [12], [39] indicate that the location of at least one of the described MODY9 mutations overlaps a putative islet enhancer also bound by CTCF, suggesting this rare variant could disrupt its function. Finally, six different recessive mutations in a developmental enhancer 25 kb downstream of the PTF1A gene (pancreas-specific transcription factor 1a) were identified in 10 families with pancreatic agenesis [74]. We expect that the recent documentation of hundreds of thousands of enhancer elements in pancreatic islets and anticipated identification of hundreds of thousands of enhancer elements in various islet developmental precursor cell types will lead to the discovery of new, rare enhancer mutations contributing to monogenic diabetes and islet dysfunction disorders.
Table 2.
Genetic variants altering islet transcriptional regulatory elements
| Target Gene (GWAS locus) |
SNP | Islet Regulator y element |
Alleles | Risk/ Effect allele |
eQTL/ aseQTL? |
Transcription al effect |
Molecula r Genetic Effect |
Physiologic Effect / Association |
Reference |
|---|---|---|---|---|---|---|---|---|---|
|
RARE
VARIANTS |
|||||||||
|
| |||||||||
| GCK | −71 G/C | Promoter | G/C | C | ND | Down | Loss of SP1 binding |
Fasting hyperglyce mia |
[72] |
| ABCC8 | −64 C/G | Promoter | C/G | G | ND | ND | ND | CHI | [68] |
| HFN4A | −146T>C | Promoter (P2) |
T/C | C | ND | ND | Mutated PDX1 binding site |
MODY | [70] |
| HFN4A | −192C>G | Promoter (P2) |
C/G | G | ND | ND | ND | Impaired GSIS; MODY-X; GDM; T2D |
[69] |
| HFN4A | −136A>G | Promoter (P2) |
A/G | G | ND | ND | ND | MODY-like | [71] |
| HFN4A | −169C>T | Promoter (P2) |
C/T | T | ND | ND | ND | MODY-like | [71] |
| BLK | chr8:11,45 9,364; chr8: 11,459,53 1 (NCBI Build 36.1) |
Putative Enhancer/ Insulator |
T/G;G/T | G;T | ND | ND | Decrease d reporter enhancer activity |
MODY | [73] |
| KCNJ11 | 88 G-T | Promoter | G/T | T | ND | ND | ND | CHI | [68] |
|
COMMON
VARIANTS |
|||||||||
|
| |||||||||
| TCF7L2 | rs7903146 | Enhancer | C/T | T | eQTL | Up | Increased open chromati n; increased reporter enhancer activity |
T2D | [14]; [39]; [40]; [77] |
| MTNR1B | rs1083096 3 |
Enhancer | C/G | G | eQTL | Up | ND | T2D | [17]; [81] |
| CAMK1D | rs1277979 0; rs1125765 5 |
Enhancer | A/G; C/T |
G;T | eQTL | Up | Increased FOXA1, FOXA2 binding |
T2D | [14]; [80] |
| ARAP1 | rs1160333 4 |
Promoter | C/T | T | eQTL | Up | Decrease d PAX6, PAX4 binding |
Decreased proinsulin; T2D |
[78] |
| G6PC2 | rs1343165 2 |
Promoter | G/A | A | eQTL | Up | Increased NF-Y binding |
Increased fasting plasma glucose |
[24] |
| G6PC2 | rs2232316 | Promoter | A/G | A | eQTL | Up | Increased FOXA2 binding |
Increased fasting plasma glucose |
[24] |
| G6PC2 | rs573225 | Promoter | A/G | A | eQTL | Down | Increased FOXA2 binding |
Increased fasting plasma glucose |
[24] |
| KCNJ11 | rs5912 | Promoter | C/T | T | aseQTL; eQTL |
Down | ND | T2D | [11]; [14] |
| CLEC16A | rs1270871 6 |
Enhancer (?) |
A/G | G | eQTL (Islets) |
Down | ND | T1D; decreased beta cell function (HOMA-B); increased HbA1c |
[20] |
| CTSH | rs3825932 | Promoter | T/C | T | eQTL | Down | ND | T1D; IGT | [76] |
| ZFAND3 | rs5869265 9 |
Enhancer | A/G | A | ND | ND | Disrupted NEUROD1 binding |
T2D | [42] |
| ADRA2A | rs553668 | 3’UTR | A/G | A | eQTL | Up | ND | Decrease insulin secretion; Decrease GSIS; T2D |
[4]; [78]; [82] |
ND = Not Determined; e/aseQTL = expression/allele-specific expression quantitative trait locus; GSIS=glucose stimulated insulin secretion; CHI = congenital hyperinsulinemia
MODY = maturity onset diabetes of the young; T2D = type 2 diabetes; T1D = type 1 diabetes; IGT = impaired glucose tolerance
Genome-wide association study (GWAS) results suggest that common variant effects on islet transcription are important for islet (dys)function, T1D, and T2D. Massive consortia efforts have identified >100 regions of the human genome (loci) containing DNA sequence variants (SNPs) associated with genetic variation in glycemic traits related to islet (dys)function and susceptibility to both T1D and T2D. Approximately 90% of these SNPs reside in noncoding regions of the genome, fueling the hypothesis that they disrupt transcriptional regulatory elements. T2D and glycemic trait GWAS SNPs are enriched in islet enhancers and several putative target genes are islet-expressed [12], [14]–[17], [42]. T1D GWAS SNPs are enriched in lymphoid enhancers [75]. However, detailed functional analysis of T1D susceptibility genes CLEC16A [20] and CTSH [76] and the observation that multiple T1D-associated genes are also expressed in islets [11], [16] warrant continued attention to potential roles for aberrant islet transcriptional control in T1D pathophysiology.
The non-coding location of GWAS SNPs present at least four challenges to understanding their effect(s): (1) identifying the variant(s) responsible for the association; (2) understanding the molecular effect(s) of these variants; (3) identifying the gene(s) affected by these perturbations; and (4) determining the direction of the effect, i.e., gain- or loss-of-function, on the target gene. Several studies have begun to address these challenges. Table 2 summarizes our collective knowledge and reflects the per-locus variability in our understanding of common variant effects on islet transcription. Both molecular genetic and islet transcriptional consequences of SNP risk alleles have been deciphered for a handful of T2D GWAS loci, including TCF7L2 [77], SLC30A8, ADRA2A [4], [78], G6PC2 [24], ARAP1 [79], and CAMK1D/CDC123 [14], [80]. For loci such as MTNR1B [81], CLEC16A [20], CTSH [76], and ZFAND3 [42], either the transcriptional or the molecular genetic consequence of the SNP allele has been determined.
Recently, Groop and colleagues conducted microarray and RNA-seq studies of islets from 63 and 89 organ donors, respectively [14], [17]. They found that expression of 640 genes is modulated by SNPs, including a subset associated with T2D and glycemic traits (Table 2). Additionally, they discovered ~1100 additional genes exhibiting allele-specific expression. Together, expression of approximately 1700 islet genes appears to be modified by genetic variation; this aligns with the median number of genes (1742) harboring at least one allele-specific expression SNP in an independent study [11]. Moreover, putative GWAS SNP target genes exhibited evidence of allelic islet expression (15/23 T1D, 20/28 T2D, and 15/18 glycemic trait genes) [11]. Further efforts to elucidate the molecular mechanisms controlling these islet expression and allelic expression differences should provide context to the pathophysiologic events and guide therapeutic strategies.
The motivation for these targeted and genome-wide analyses is to build more precise predictive risk models and prevention strategies, to identify diagnostic molecular markers, and to develop new and more precise therapeutic approaches to prevent and treat islet dysfunction and diabetes. Genome-informed modalities have been developed for a subset of MODY and neonatal diabetes patients and have impacted their prognosis and treatment [8]. MODY patients with HNF1A and HNF4A mutations respond particularly well to low-dose sulfonylurea therapy, whereas those with GCK mutations are best left untreated. Neonatal diabetes patients with activating KCNJ11/ABCC8 mutations can be effectively treated with high-dose sulfonylureas. Studies of rs553668, a GWAS SNP in ADRA2A associated with impaired beta cell function and T2D, provide an exciting example of translating common genetic variant association into molecular mechanism of action, physiologic consequences, and genotype-based treatment [4], [78], [82]. ADRA2A encodes an adrenergic receptor that mediates adrenergic suppression of glucose-stimulated insulin secretion (GSIS) in islets [78]. The rs553668 risk allele leads to ADRA2A overexpression in islets and impaired insulin secretion in risk allele carriers [4], [78]. Administration of the adrenergic receptor antagonist yohimbine did not affect insulin secretion in non-risk individuals, but it improved insulin secretion in risk allele carriers to levels seen in the non-risk individuals [82].
Environmental effects on islet transcription
Studies comparing T2D and non-diabetic islet and beta cell transcriptomes have detected differences in hundreds of mRNA and several miRNA [14], [83]–[85]. Few differentially expressed genes are overlapping between studies. This likely reflects a combination of biological differences (organ donor characteristics (e.g., race, genotype, sex, weight, age, and cause of death), duration of diabetes and degree of blood sugar control, and biological sample attainment and processing (enzymatic isolation of islets vs. LCM). However, on a pathway level, the studies consistently identified aberrant transcription of components of (1) core islet/beta cell function pathways, such as glucose sensing, insulin receptor signaling, glycolysis/beta oxidation, and glucose stimulated insulin secretion; (2) stress response pathways such as oxidative stress; and (3) islet TFs. These studies implicate transcriptional dysregulation of key pathways as a feature of islet dysfunction and T2D; more studies will be necessary to determine the causative nature of these changes.
Several studies have sought to understand the transcriptional consequences of early pathophysiologic events leading to islet dysfunction, T1D, and T2D. Stress response pathways postulated as early mediators of beta cell failure and death include inflammation, hypoxia/oxidative stress, and endoplasmic reticulum (ER) stress [8]. All of these responses lead to changes in localization and/or activity of TFs in the islet (Fig. 1C). Inflammation and ER stress mediate transcriptional changes in islets via induction and nuclear localization of TFs like NF-kappaB, CHOP, XBP1s, ATF4, and ATF6. Hypoxia leads to HIF1alpha/HIF1beta mediated transcription. Oxidative stress causes export or inactivation of islet TFs such as PDX1, NKX6.1, MAFA; these factors are also compromised in T2D islets [86]. Few studies have assessed the comprehensive transcriptome changes using RNA-seq. 1325 genes were differentially expressed and 3525 were alternatively spliced after acute (48 hour) exposure of human islets to the free fatty acid palmitate (modeling lipotoxicity), including 11/59 T2D GWAS candidate genes expressed in islets [15]. Similarly, exposure of islets to proinflammatory cytokines altered the expression and splicing of 3065 and 6875 genes, respectively [16]. Although these acute experimental exposures may not accurately reflect the precise changes in vivo, they provide a basis for understanding transcriptional consequences of islet damage and may identify pathways mediating pathophysiologic processes in T1D and T2D.
Conclusions
Islet “omics” studies have uncovered extensive diversity and complexity of the transcripts produced and also of the cis-regulatory elements controlling their production. Recent studies, highlighted in this review, have built a compendium of transcripts and regulatory elements and are working to assemble individual components into islet cis-regulatory transcriptional regulatory networks. They are building insights into the individual impact of either genetic variation or environmental perturbation of transcriptional control on islet physiology and pathophysiology. Diverse transcriptional features (regulatory element use, transcript levels, splicing) in islets are linked to genotype. Continued progress to identify the target gene(s) and direction of effect (gain-of-function or loss-of-function) of GWAS and other key regulatory SNPs is inevitable. Thus, we expect that additional stories akin to ADRA2A will emerge in coming years. Studies integrating both genetic and environmental contributions to islet dysfunction are needed to realize precision medicine (prevention, monitoring, and treatment) approaches to islet dysfunction and diabetes. Finally, we anticipate that epigenomic and transcriptomic analysis of single cells or stratified islet subpopulations will provide more precise understanding of the cell type-specific (e.g. alpha, beta, delta) effects of genetic and environmental perturbation, which should impact pathophysiologic understanding and therapeutic approaches for islet dysfunction and diabetes.
Acknowledgments
We apologize to colleagues whose work was not directly cited due to reference limitations. We thank Jesse Hammer in JAX Graphics Design & Production Services for the figure artwork. We would like to thank our colleagues at JAX-GM (Adam Williams), University of Michigan (Stephen C. J. Parker), and the National Human Genome Research Institute (Lori Bonnycastle and Brooke Wolford) for their helpful suggestions and critical review of this manuscript. This work was supported by US National Institutes of Health (NIH) grant R00DK092251-03 to M.L.S.
Footnotes
Conflict of Interest
Michael L. Stitzel, Ina Kycia, Romy Kursawe, and Duygu Ucar declare that they have no conflict of interest.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been
highlighted as:
• Of importance
•• Of major importance
- [1].Brissova M, Fowler MJ, Nicholson WE, Chu A, Hirshberg B, Harlan DM, Powers AC. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2005 Sep.53(no. 9):1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- [2].Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren P-O, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc. Natl. Acad. Sci. U. S. A. 2006 Feb.103(no. 7):2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dai C, Brissova M, Hang Y, Thompson C, Poffenberger G, Shostak A, Chen Z, Stein R, Powers AC. Islet-enriched gene expression and glucose-induced insulin secretion in human and mouse islets. Diabetologia. 2012 Mar.55(no. 3):707–718. doi: 10.1007/s00125-011-2369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rosengren AH, Braun M, Mahdi T, Andersson SA, Travers ME, Shigeto M, Zhang E, Almgren P, Ladenvall C, Axelsson AS, Edlund A, Pedersen MG, Jonsson A, Ramracheya R, Tang Y, Walker JN, Barrett A, Johnson PRV, Lyssenko V, McCarthy MI, Groop L, Salehi A, Gloyn AL, Renström E, Rorsman P, Eliasson L. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes. 2012 Jul.61(no. 7):1726–1733. doi: 10.2337/db11-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]••.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium, Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MCY, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JCN, Chang L-C, Chang T-J, Chang Y-C, Charpentier G, Chen C-H, Chen H, Chen Y-T, Chia K-S, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang L-M, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han B-G, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jöckel K-H, Jonsson A, Jowett JBM, Kadowaki T, Kang HM, Kanoni S, Kao WHL, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw K-T, Khor C-C, Kim H-L, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyövälti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee J-M, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim W-Y, Lind L, Lindgren CM, Lindholm E, Liu C-T, Liu JJ, Lobbens S, Long J, Loos RJF, Lu W, Luan J, Lyssenko V, Ma RCW, Maeda S, Mägi R, Männisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Mühleisen TW, Müller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng P-K, Nica AC, Nilsson PM, Njølstad I, Nöthen MM, Ohnaka K, Ong TH, Owen KR, Palmer CNA, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JRB, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segrè AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS, Shu XO, Shuldiner AR, Sigurđsson G, Sijbrands E, Silveira A, Sim X, Sivapalaratnam S, Small KS, So WY, Stančáková A, Stefansson K, Steinbach G, Steinthorsdottir V, Stirrups K, Strawbridge RJ, Stringham HM, Sun Q, Suo C, Syvänen A-C, Takayanagi R, Takeuchi F, Tay WT, Teslovich TM, Thorand B, Thorleifsson G, Thorsteinsdottir U, Tikkanen E, Trakalo J, Tremoli E, Trip MD, Tsai FJ, Tuomi T, Tuomilehto J, Uitterlinden AG, Valladares-Salgado A, Vedantam S, Veglia F, Voight BF, Wang C, Wareham NJ, Wennauer R, Wickremasinghe AR, Wilsgaard T, Wilson JF, Wiltshire S, Winckler W, Wong TY, Wood AR, Wu J-Y, Wu Y, Yamamoto K, Yamauchi T, Yang M, Yengo L, Yokota M, Young R, Zabaneh D, Zhang F, Zhang R, Zheng W, Zimmet PZ, Altshuler D, Bowden DW, Cho YS, Cox NJ, Cruz M, Hanis CL, Kooner J, Lee J-Y, Seielstad M, Teo YY, Boehnke M, Parra EJ, Chambers JC, Tai ES, McCarthy MI, Morris AP. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014 Mar.46(no. 3):234–244. doi: 10.1038/ng.2897. This large genetic meta-analysis study and references therein highlight the current knowledge about sequence variants contributing genetic risk for type 2 diabetes in multiple ethnic groups.
- [6].Dimas AS, Lagou V, Barker A, Knowles JW, Mägi R, Hivert M-F, Benazzo A, Rybin D, Jackson AU, Stringham HM, Song C, Fischer-Rosinsky A, Boesgaard TW, Grarup N, Abbasi FA, Assimes TL, Hao K, Yang X, Lecoeur C, Barroso I, Bonnycastle LL, Böttcher Y, Bumpstead S, Chines PS, Erdos MR, Graessler J, Kovacs P, Morken MA, Narisu N, Payne F, Stancakova A, Swift AJ, Tönjes A, Bornstein SR, Cauchi S, Froguel P, Meyre D, Schwarz PEH, Häring H-U, Smith U, Boehnke M, Bergman RN, Collins FS, Mohlke KL, Tuomilehto J, Quertemous T, Lind L, Hansen T, Pedersen O, Walker M, Pfeiffer AFH, Spranger J, Stumvoll M, Meigs JB, Wareham NJ, Kuusisto J, Laakso M, Langenberg C, Dupuis J, Watanabe RM, Florez JC, Ingelsson E, McCarthy MI, Prokopenko I, MAGIC Investigators Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014 Jun.63(no. 6):2158–2171. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Soleimanpour SA, Stoffers DA. The pancreatic β cell and type 1 diabetes: innocent bystander or active participant? Trends Endocrinol. Metab. TEM. 2013 Jul.24(no. 7):324–331. doi: 10.1016/j.tem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]••.Halban PA, Polonsky KS, Bowden DW, Hawkins MA, Ling C, Mather KJ, Powers AC, Rhodes CJ, Sussel L, Weir GC. β-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care. 2014 Jun.37(no. 6):1751–1758. doi: 10.2337/dc14-0396. This article summarizes progress in and recommendations for further understanding islet/beta cell failure in type 2 diabetes based on proceedings at the October 2013 Global Partnership to Accelerate Diabetes Research Conference.
- [9].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008 Jul.5(no. 7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- [10].Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, Eizirik DL, Welsh N, Goodman N, Hood L. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med. Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nica AC, Ongen H, Irminger J-C, Bosco D, Berney T, Antonarakis SE, Halban PA, Dermitzakis ET. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013 Sep.23(no. 9):1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]•.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, NISC Comparative Sequencing Program. Black BL, Visel A, Pennacchio LA, Collins FS, National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors. NISC Comparative Sequencing Program Authors Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. U. S. A. 2013 Oct.110(no. 44):17921–17926. doi: 10.1073/pnas.1317023110. This study identified long enhancer states as regulators of cell type-specific functions in islets and other cell types. Moreover, sequence variants associated with genetic risk for diseases (such as type 2 diabetes) were specifically enriched in stretch enhancers of disease-relevant cell types (such as islets).
- [13]•.Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, Pasquali L, Sauty-Colace C, Beucher A, Scharfmann R, van Arensbergen J, Johnson PR, Berry A, Lee C, Harkins T, Gmyr V, Pattou F, Kerr-Conte J, Piemonti L, Berney T, Hanley N, Gloyn AL, Sussel L, Langman L, Brayman KL, Sander M, McCarthy MI, Ravassard P, Ferrer J. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012 Oct.16(no. 4):435–448. doi: 10.1016/j.cmet.2012.08.010. This was the first study to systematically identify human islet lncRNAs.
- [14].Taneera J, Lang S, Sharma A, Fadista J, Zhou Y, Ahlqvist E, Jonsson A, Lyssenko V, Vikman P, Hansson O, Parikh H, Korsgren O, Soni A, Krus U, Zhang E, Jing X-J, Esguerra JLS, Wollheim CB, Salehi A, Rosengren A, Renström E, Groop L. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012 Jul.16(no. 1):122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- [15].Cnop M, Abdulkarim B, Bottu G, Cunha DA, Igoillo-Esteve M, Masini M, Turatsinze J-V, Griebel T, Villate O, Santin I, Bugliani M, Ladriere L, Marselli L, McCarthy MI, Marchetti P, Sammeth M, Eizirik DL. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014 Jun.63(no. 6):1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- [16].Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, Ortis F, Santin I, Colli ML, Barthson J, Bouwens L, Hughes L, Gregory L, Lunter G, Marselli L, Marchetti P, McCarthy MI, Cnop M. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(no. 3):e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]•.Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, Storm P, Osmark P, Ladenvall C, Prasad RB, Hansson KB, Finotello F, Uvebrant K, Ofori JK, Di Camillo B, Krus U, Cilio CM, Hansson O, Eliasson L, Rosengren AH, Renström E, Wollheim CB, Groop L. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. U. S. A. 2014 Sep.111(no. 38):13924–13929. doi: 10.1073/pnas.1402665111. This study of pancreatic islets from ~90 individuals identified hundreds of genetic variants that alter islet transcription by RNA-seq of pancreatic islets from ~90 individuals.
- [18]•.Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, Kaestner KH. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Invest. 2013 Mar.123(no. 3):1275–1284. doi: 10.1172/JCI66514. This study performed RNA-seq and ChIP-seq on dissociated, sorted islet cells to identify alpha and beta cell enriched transcripts and epigenetic promoter modifications.
- [19].Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell. 2012 Mar.148(no. 6):1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, Spruce LA, Kushner JA, Groop L, Seeholzer SH, Kaufman BA, Hakonarson H, Stoffers DA. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014 Jun.157(no. 7):1577–1590. doi: 10.1016/j.cell.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Klein D, Misawa R, Bravo-Egana V, Vargas N, Rosero S, Piroso J, Ichii H, Umland O, Zhijie J, Tsinoremas N, Ricordi C, Inverardi L, Domínguez-Bendala J, Pastori RL. MicroRNA expression in alpha and beta cells of human pancreatic islets. PloS One. 2013;8(no. 1):e55064. doi: 10.1371/journal.pone.0055064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].van de Bunt M, Gaulton KJ, Parts L, Moran I, Johnson PR, Lindgren CM, Ferrer J, Gloyn AL, McCarthy MI. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PloS One. 2013;8(no. 1):e55272. doi: 10.1371/journal.pone.0055272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, Regazzi R. Emerging roles of non-coding RNAs in pancreatic β-cell function and dysfunction. Diabetes Obes. Metab. 2012 Oct.14(Suppl 3):12–21. doi: 10.1111/j.1463-1326.2012.01654.x. [DOI] [PubMed] [Google Scholar]
- [24].O’Brien RM. Moving on from GWAS: functional studies on the G6PC2 gene implicated in the regulation of fasting blood glucose. Curr. Diab. Rep. 2013 Dec.13(no. 6):768–777. doi: 10.1007/s11892-013-0422-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009 Mar.458(no. 7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012 Feb.482(no. 7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014 Apr.24(no. 4):616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hay CW, Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006 Dec.55(no. 12):3201–3213. doi: 10.2337/db06-0788. [DOI] [PubMed] [Google Scholar]
- [29].Gerrish K, Van Velkinburgh JC, Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol. Endocrinol. Baltim. Md. 2004 Mar.18(no. 3):533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- [30].Gerrish K, Gannon M, Shih D, Henderson E, Stoffel M, Wright CV, Stein R. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 2000 Feb.275(no. 5):3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- [31].Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J. Biol. Chem. 2002 Apr.277(no. 15):13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- [32].Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011 Jan.12(no. 1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- [33].Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008 Jan.132(no. 2):311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007 Mar.39(no. 3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- [35].Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009 May;459(no. 7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011 Feb.470(no. 7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010 Dec.107(no. 50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007 May;129(no. 4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- [39].Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SCJ, Boyle AP, Scott LJ, NISC Comparative Sequencing Program. Margulies EH, Boehnke M, Furey TS, Crawford GE, Collins FS. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010 Nov.12(no. 5):443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, Berney T, Montanya E, Mohlke KL, Lieb JD, Ferrer J. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010 Mar.42(no. 3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]•.Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, Hitz BC, Gupta R, Rando TA, Baker JC, Snyder MP, Cherry JM, Brunet A. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014 Jul.158(no. 3):673–688. doi: 10.1016/j.cell.2014.06.027. Similar to stretch/super enhancers (SEs), this study identified long stretches of activating histone modifications as an important feature of cell type-specific promoters, termed Broad Domains (BDS)
- [42]•.Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Morán I, Gómez-Marín C, van de Bunt M, Ponsa-Cobas J, Castro N, Nammo T, Cebola I, García-Hurtado J, Maestro MA, Pattou F, Piemonti L, Berney T, Gloyn AL, Ravassard P, Gómez-Skarmeta JL, Müller F, McCarthy MI, Ferrer J. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014 Feb.46(no. 2):136–143. doi: 10.1038/ng.2870. Using ChIP-seq of islet TFs, this study found that islet-specific enhancer clusters are bound by multiple islet TFs. Consistent with earlier stretch/super enhancer studies [12, 54], GWAS SNPs for T2D and related traits are enriched in these islet enhancer clusters.
- [43].Bhandare R, Schug J, Le Lay J, Fox A, Smirnova O, Liu C, Naji A, Kaestner KH. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 2010 Apr.20(no. 4):428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee EK, Kim W, Tominaga K, Martindale JL, Yang X, Subaran SS, Carlson OD, Mercken EM, Kulkarni RN, Akamatsu W, Okano H, Perrone-Bizzozero NI, de Cabo R, Egan JM, Gorospe M. RNA-binding protein HuD controls insulin translation. Mol. Cell. 2012 Mar.45(no. 6):826–835. doi: 10.1016/j.molcel.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, Haugen E, Heimfeld S, Murry CE, Akey JM, Stamatoyannopoulos JA. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013 Aug.154(no. 4):888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep.489(no. 7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010 Oct.28(no. 10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mutskov V, Felsenfeld G. The human insulin gene is part of a large open chromatin domain specific for human islets. Proc. Natl. Acad. Sci. U. S. A. 2009 Oct.106(no. 41):17419–17424. doi: 10.1073/pnas.0909288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014 Mar.21(no. 3):210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- [50].Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012 Sep.337(no. 6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015 Feb.518(no. 7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013 Apr.153(no. 2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013 Apr.153(no. 2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]•.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013 Nov.155(no. 4):934–947. doi: 10.1016/j.cell.2013.09.053. This study extended earlier super enhancer (SE) studies into human cells, identifying SEs in 86 cell/tissue types and showing that sequence variants contributing to diseases overlap SEs of disease-relevant cell types.
- [55].Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei C-L, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung W-K, Snyder M, Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012 Jan.148(no. 1–2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kieffer-Kwon K-R, Tang Z, Mathe E, Qian J, Sung M-H, Li G, Resch W, Baek S, Pruett N, Grøntved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou J, Joung JK, Sartorelli V, Wei C-L, Ruan X, Hager GL, Ruan Y, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013 Dec.155(no. 7):1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, Mulawadi FH, Sung W-K, Nicolis S, Ahituv N, Ruan Y, Wei C-L. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013 Dec.504(no. 7479):306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009 Oct.326(no. 5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung H-K, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui C-C, Gomez-Skarmeta JL, Nóbrega MA. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014 Mar.507(no. 7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013 Jun.14(no. 6):390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat. Struct. Mol. Biol. 2011 Mar.18(no. 3):372–378. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu Z, Lefevre GM, Gavrilova O, Foster St Claire MB, Riddick G, Felsenfeld G. Mapping of long-range INS promoter interactions reveals a role for calcium-activated chloride channel ANO1 in insulin secretion. Proc. Natl. Acad. Sci. U. S. A. 2014 Nov.111(no. 47):16760–16765. doi: 10.1073/pnas.1419240111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cook PR. The organization of replication and transcription. Science. 1999 Jun.284(no. 5421):1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- [64].Zhang J, Poh HM, Peh SQ, Sia YY, Li G, Mulawadi FH, Goh Y, Fullwood MJ, Sung W-K, Ruan X, Ruan Y. ChIA PET analysis of transcriptional chromatin interactions. Methods San Diego Calif. 2012 Nov.58(no. 3):289–299. doi: 10.1016/j.ymeth.2012.08.009. [DOI] [PubMed] [Google Scholar]
- [65].Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014 Feb.46(no. 2):205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- [66].Shaw-Smith C, De Franco E, Lango Allen H, Batlle M, Flanagan SE, Borowiec M, Taplin CE, van Alfen-van der Velden J, Cruz-Rojo J, Perez de Nanclares G, Miedzybrodzka Z, Deja G, Wlodarska I, Mlynarski W, Ferrer J, Hattersley AT, Ellard S. GATA4 mutations are a cause of neonatal and childhood-onset diabetes. Diabetes. 2014 Aug.63(no. 8):2888–2894. doi: 10.2337/db14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].De Franco E, Shaw-Smith C, Flanagan SE, Shepherd MH, International NDM Consortium. Hattersley AT, Ellard S. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013 Mar.62(no. 3):993–997. doi: 10.2337/db12-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Tornovsky S, Crane A, Cosgrove KE, Hussain K, Lavie J, Heyman M, Nesher Y, Kuchinski N, Ben-Shushan E, Shatz O, Nahari E, Potikha T, Zangen D, Tenenbaum-Rakover Y, de Vries L, Argente J, Gracia R, Landau H, Eliakim A, Lindley K, Dunne MJ, Aguilar-Bryan L, Glaser B. Hyperinsulinism of infancy: novel ABCC8 and KCNJ11 mutations and evidence for additional locus heterogeneity. J. Clin. Endocrinol. Metab. 2004 Dec.89(no. 12):6224–6234. doi: 10.1210/jc.2004-1233. [DOI] [PubMed] [Google Scholar]
- [69].Ek J, Hansen SP, Lajer M, Nicot C, Boesgaard TW, Pruhova S, Johansen A, Albrechtsen A, Yderstraede K, Lauenborg J, Parrizas M, Boj SF, Jørgensen T, Borch-Johnsen K, Damm P, Ferrer J, Lebl J, Pedersen O, Hansen T. A novel - 192c/g mutation in the proximal P2 promoter of the hepatocyte nuclear factor-4 alpha gene (HNF4A) associates with late-onset diabetes. Diabetes. 2006 Jun.55(no. 6):1869–1873. doi: 10.2337/db05-1684. [DOI] [PubMed] [Google Scholar]
- [70].Thomas H, Jaschkowitz K, Bulman M, Frayling TM, Mitchell SM, Roosen S, Lingott-Frieg A, Tack CJ, Ellard S, Ryffel GU, Hattersley AT. A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum. Mol. Genet. 2001 Sep.10(no. 19):2089–2097. doi: 10.1093/hmg/10.19.2089. [DOI] [PubMed] [Google Scholar]
- [71].Wirsing A, Johnstone KA, Harries LW, Ellard S, Ryffel GU, Stanik J, Gasperikova D, Klimes I, Murphy R. Novel monogenic diabetes mutations in the P2 promoter of the HNF4A gene are associated with impaired function in vitro. Diabet. Med. J. Br. Diabet. Assoc. 2010 Jun.27(no. 6):631–635. doi: 10.1111/j.1464-5491.2010.03003.x. [DOI] [PubMed] [Google Scholar]
- [72].Gasperíková D, Tribble ND, Staník J, Hucková M, Misovicová N, van de Bunt M, Valentínová L, Barrow BA, Barák L, Dobránsky R, Bereczková E, Michálek J, Wicks K, Colclough K, Knight JC, Ellard S, Klimes I, Gloyn AL. Identification of a novel beta-cell glucokinase (GCK) promoter mutation (-71G>C) that modulates GCK gene expression through loss of allele-specific Sp1 binding causing mild fasting hyperglycemia in humans. Diabetes. 2009 Aug.58(no. 8):1929–1935. doi: 10.2337/db09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Borowiec M, Liew CW, Thompson R, Boonyasrisawat W, Hu J, Mlynarski WM, El Khattabi I, Kim S-H, Marselli L, Rich SS, Krolewski AS, Bonner-Weir S, Sharma A, Sale M, Mychaleckyj JC, Kulkarni RN, Doria A. Mutations at the BLK locus linked to maturity onset diabetes of the young and beta-cell dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2009 Aug.106(no. 34):14460–14465. doi: 10.1073/pnas.0906474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]•.Weedon MN, Cebola I, Patch A-M, Flanagan SE, De Franco E, Caswell R, Rodríguez-Seguí SA, Shaw-Smith C, Cho CH-H, Lango Allen H, Houghton JAL, Roth CL, Chen R, Hussain K, Marsh P, Vallier L, Murray A, International Pancreatic Agenesis Consortium. Ellard S, Ferrer J, Hattersley AT. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet. 2014 Jan.46(no. 1):61–64. doi: 10.1038/ng.2826. This very elegant study combined genetic analysis of patient samples with functional genomics to demonstrate that rare mutations likely cause pancreatic agenesis by disrupting a developmental enhancer.
- [75].Onengut-Gumuscu S, Chen W-M, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, Farber E, Bonnie JK, Szpak M, Schofield E, Achuthan P, Guo H, Fortune MD, Stevens H, Walker NM, Ward LD, Kundaje A, Kellis M, Daly MJ, Barrett JC, Cooper JD, Deloukas P, Type 1 Diabetes Genetics Consortium. Todd JA, Wallace C, Concannon P, Rich SS. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015 Mar. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fløyel T, Brorsson C, Nielsen LB, Miani M, Bang-Berthelsen CH, Friedrichsen M, Overgaard AJ, Berchtold LA, Wiberg A, Poulsen P, Hansen L, Rosinger S, Boehm BO, Ram R, Nguyen Q, Mehta M, Morahan G, Concannon P, Bergholdt R, Nielsen JH, Reinheckel T, von Herrath M, Vaag A, Eizirik DL, Mortensen HB, Størling J, Pociot F. CTSH regulates β-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc. Natl. Acad. Sci. U. S. A. 2014 Jul.111(no. 28):10305–10310. doi: 10.1073/pnas.1402571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjögren M, Ling C, Eriksson K-F, Lethagen A-L, Mancarella R, Berglund G, Tuomi T, Nilsson P, Del Prato S, Groop L. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 2007 Aug.117(no. 8):2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li D-Q, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L, Groop L, Rorsman P, Salehi A, Lyssenko V, Luthman H, Renström E. Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010 Jan.327(no. 5962):217–220. doi: 10.1126/science.1176827. [DOI] [PubMed] [Google Scholar]
- [79].Kulzer JR, Stitzel ML, Morken MA, Huyghe JR, Fuchsberger C, Kuusisto J, Laakso M, Boehnke M, Collins FS, Mohlke KL. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am. J. Hum. Genet. 2014 Feb.94(no. 2):186–197. doi: 10.1016/j.ajhg.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fogarty MP, Cannon ME, Vadlamudi S, Gaulton KJ, Mohlke KL. Identification of a regulatory variant that binds FOXA1 and FOXA2 at the CDC123/CAMK1D type 2 diabetes GWAS locus. PLoS Genet. 2014 Sep.10(no. 9):e1004633. doi: 10.1371/journal.pgen.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lyssenko V, Nagorny CLF, Erdos MR, Wierup N, Jonsson A, Spégel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat. Genet. 2009 Jan.41(no. 1):82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]••.Tang Y, Axelsson AS, Spégel P, Andersson LE, Mulder H, Groop LC, Renström E, Rosengren AH. Genotype-based treatment of type 2 diabetes with an α2A-adrenergic receptor antagonist. Sci. Transl. Med. 2014 Oct.6(no. 257):257ra139. doi: 10.1126/scitranslmed.3009934. This study, together with [4] and [78], is a compelling example translating a common genetic variant associated with diabetes risk into molecular function, pathophysiologic consequences, and a genotype-based, precision medicine approach to correcting these effects.
- [83].Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng Y-H, Roberson RS, Ricordi C, O’Connell PJ, Gonzalez FJ, Kahn CR. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005 Aug.122(no. 3):337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- [84].Marselli L, Thorne J, Dahiya S, Sgroi DC, Sharma A, Bonner-Weir S, Marchetti P, Weir GC. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PloS One. 2010;5(no. 7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, Chen Y, Choi I, Vourekas A, Won K-J, Liu C, Vivek K, Naji A, Friedman JR, Kaestner KH. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014 Jan.19(no. 1):135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]•.Guo S, Dai C, Guo M, Taylor B, Harmon JS, Sander M, Robertson RP, Powers AC, Stein R. Inactivation of specific β cell transcription factors in type 2 diabetes. J. Clin. Invest. 2013 Aug.123(no. 8):3305–3316. doi: 10.1172/JCI65390. This very thorough study demonstrates that oxidative stress alters islet transcription factor localization and activity. This phenomenon is also observed in islets from type 2 diabetics, implicating environmental perturbation of transcriptional elements/programs as an important pathophysiologic event in diabetes