Summary
Background
Understanding the genetic basis of airflow obstruction and smoking behaviour is key to determining the pathophysiology of chronic obstructive pulmonary disease (COPD). We used UK Biobank data to study the genetic causes of smoking behaviour and lung health.
Methods
We sampled individuals of European ancestry from UK Biobank, from the middle and extremes of the forced expiratory volume in 1 s (FEV1) distribution among heavy smokers (mean 35 pack-years) and never smokers. We developed a custom array for UK Biobank to provide optimum genome-wide coverage of common and low-frequency variants, dense coverage of genomic regions already implicated in lung health and disease, and to assay rare coding variants relevant to the UK population. We investigated whether there were shared genetic causes between different phenotypes defined by extremes of FEV1. We also looked for novel variants associated with extremes of FEV1 and smoking behaviour and assessed regions of the genome that had already shown evidence for a role in lung health and disease. We set genome-wide significance at p<5 × 10−8.
Findings
UK Biobank participants were recruited from March 15, 2006, to July 7, 2010. Sample selection for the UK BiLEVE study started on Nov 22, 2012, and was completed on Dec 20, 2012. We selected 50 008 unique samples: 10 002 individuals with low FEV1, 10 000 with average FEV1, and 5002 with high FEV1 from each of the heavy smoker and never smoker groups. We noted a substantial sharing of genetic causes of low FEV1 between heavy smokers and never smokers (p=2·29 × 10−16) and between individuals with and without doctor-diagnosed asthma (p=6·06 × 10−11). We discovered six novel genome-wide significant signals of association with extremes of FEV1, including signals at four novel loci (KANSL1, TSEN54, TET2, and RBM19/TBX5) and independent signals at two previously reported loci (NPNT and HLA-DQB1/HLA-DQA2). These variants also showed association with COPD, including in individuals with no history of smoking. The number of copies of a 150 kb region containing the 5′ end of KANSL1, a gene that is important for epigenetic gene regulation, was associated with extremes of FEV1. We also discovered five new genome-wide significant signals for smoking behaviour, including a variant in NCAM1 (chromosome 11) and a variant on chromosome 2 (between TEX41 and PABPC1P2) that has a trans effect on expression of NCAM1 in brain tissue.
Interpretation
By sampling from the extremes of the lung function distribution in UK Biobank, we identified novel genetic causes of lung function and smoking behaviour. These results provide new insight into the specific mechanisms underlying airflow obstruction, COPD, and tobacco addiction, and show substantial shared genetic architecture underlying airflow obstruction across individuals, irrespective of smoking behaviour and other airway disease.
Funding
Medical Research Council.
Introduction
Chronic obstructive pulmonary disease (COPD) is a global public health concern and is currently the third leading cause of death worldwide.1 Smoking and indoor air pollution are major environmental risk factors for development of COPD, but heritability studies also suggest a strong genetic component in smoking behaviour and in risk of COPD.1–4 Spirometry, particularly measurements of forced expiratory volume in 1 s (FEV1) and forced vital capacity (FVC), is used to measure airflow obstruction and helps in the diagnosis and grading of severity of COPD. Previous large genome-wide association studies (GWAS) of general population cohorts have identified 32 common genetic variants (minor allele frequency [MAF] >5%) associated with lung function,5–9 12 of which have also shown association with airflow obstruction and risk of COPD.10–15 However, these findings only explain a small proportion of the phenotypic variance (∼1·5% for FEV1).8
Research in context.
Evidence before this study
UK Biobank had completed its recruitment, including its baseline phenotyping and biobanking of samples, before our study began. The DNA had not yet been extracted from the biobanked samples, and the spirometry data quality had not yet been analysed across all UK Biobank participants. We searched for evidence of other large biobanks with spirometry data, including the P3G Catalogue. We did not identify any other biobank with spirometry data and DNA as large as UK Biobank. Evidence regarding the global burden of disease due to smoking or chronic obstructive pulmonary disease (COPD) was obtained from the WHO Global Health Risks Report and a systematic analysis of the Global Burden of Disease Study 2010. Tobacco smoking accounted for about 5·1 million deaths globally in 2004; because of recent increases in smoking prevalence in developing countries, the full global effect of smoking is yet to occur. COPD is the third leading cause of death globally. For previous evidence of genetic associations, we gave the highest ranking to associations reaching genome-wide significance in genome-wide association studies, a lower ranking to associations not reaching genome-wide significance in genome-wide association studies, and the lowest ranking to associations reported in candidate gene studies. To assess evidence of loci associated with lung function, COPD, and smoking behaviour, we queried the Catalog of Published Genome-Wide Association Studies. We used this evidence to report our known findings for genetic variants shown to be associated with forced expiratory volume in 1 s (FEV1; eight loci) and smoking behaviour (seven loci). We report candidate gene associations only for variants for which we found genome-wide evidence of association.
Added value of this study
We describe, to our knowledge, the first genetic study using the UK Biobank resource and show the quality of the phenotype and genotype data. Additionally, we describe an advance in imputation quality afforded by the use of a newly designed genotyping array used in conjunction with the largest reference panel available so far. A slightly modified version of this array is being used to genotype the remaining samples in UK Biobank. As evidence of the usefulness of these data, we describe novel insights into the genetic architecture of airflow obstruction and smoking. Specifically, we show that there are shared genetic causes of airflow obstruction between smokers and non-smokers, consistent with the limited evidence for gene–smoking interactions described so far. We show that the genetic determinants of low FEV1 in individuals without asthma are also informative in individuals with asthma. We report new loci associated with extremes of FEV1 and COPD, including evidence that a genomic region of complex structural variation has an effect on lung function and airflow obstruction in the general population. Our novel signals implicate epigenetic mechanisms as contributors to lung health. These findings, taken together with previous findings, will help define pathways underlying predisposition to development of COPD and smoking behaviours. A full understanding of the biological mechanisms underlying these genetic associations will improve our understanding of the pathophysiology of COPD and smoking behaviour, and potentially give rise to novel therapeutic strategies for the management of airway disease and prevention of nicotine addiction.
Implications of all the available evidence
This study has improved our understanding of the genetic and molecular basis of smoking behaviour and lung function and provided potential targets for therapeutic intervention. It has also shown the value of sampling from the extremes using a large biobank such as UK Biobank. A similar approach could be adopted for genetic studies of other health-related traits in UK Biobank, using either new genetic assays or the extensive genome-wide data that we and UK Biobank have generated.
Tobacco smoking accounted for about 5·1 million deaths globally in 2004 and for 18% of deaths in high-income countries.16 Large GWAS of smoking behaviour17–19 have identified up to eight associated loci; the strongest association reported is at the 15q25 locus.17–19 Further insight into the genetic factors affecting lung function, smoking behaviour, and COPD could lead to new approaches for smoking cessation and prevention and treatment of COPD.
UK Biobank is the largest European biobank available at present and represents an extensive resource from which to sample phenotypic extremes in the UK population.20 UK Biobank contains data from 502 682 individuals (94% of self-reported European ancestry), with extensive health and lifestyle questionnaire data, physical measures (including spirometry), and DNA.
In the UK Biobank Lung Exome Variant Evaluation (UK BiLEVE) study, we undertook nested case-control studies in individuals of European ancestry from UK Biobank to: (1) identify whether there are shared genetic causes underlying low FEV1 and high FEV1, and a shared genetic cause of low FEV1 between never smokers and heavy smokers and between individuals with and without a doctor diagnosis of asthma; (2) identify novel variants associated with extremes of FEV1 and smoking behaviour; and (3) provide further insight into regions of the genome that had already shown evidence for a role in lung health and disease.
Methods
Study design
We defined case and control groups by selecting individuals from the middle and extremes of the FEV1 distribution among both heavy smokers (mean 35 pack-years) and never smokers. We developed a custom array to provide optimum genome-wide coverage of common and low frequency (MAF 1–5%) coding variants and rare (MAF <1%) coding variants relevant to the UK population; this platform also provided dense coverage of genomic regions implicated in lung health and disease. Spirometry data in UK Biobank were obtained using a Vitalograph Pneumotrac 6800 (Buckingham, UK) on at least two occasions. Sampling was undertaken such that equal numbers of males and females were selected in total and the numbers of individuals selected from each age–sex band were proportional to the number of individuals in the band being sampled (appendix pp 3–5). One consequence of this approach is that we enriched our sample for non-smoking individuals with airflow obstruction.
To assess whether the novel regions that we identified as associated with FEV1 extremes are also associated with COPD, we defined individuals fulfilling spirometric criteria for the Global Initiative for Chronic Obstructive Lung Disease (GOLD) Stage 2+ COPD (FEV1:FVC ratio <0·7 and percent predicted FEV1<80%) as COPD cases and we defined individuals with FEV1:FVC ratio >0·7 and percent predicted FEV1 in excess of 80% from the high FEV1 strata as controls. Post-bronchodilator spirometry was not available, although drug treatment was not withheld before spirometry.
To assess the extent of the shared genetic causes of low FEV1 between individuals with and without reported or doctor-diagnosed asthma, we identified individuals within our study selection who were also asthma cases as participants who either (1) answered “asthma” to the touch-screen question “Has a doctor ever told you that you have had any of the following conditions?” or (2) reported asthma in a verbal interview at the time of recruitment to UK Biobank.
UK Biobank has received ethics approval from the National Health Service National Research Ethics Service (Ref 11/NW/0382).
Procedures
We undertook genome-wide genotyping of variants using a new custom Affymetrix Axiom array (UK BiLEVE array; Santa Clara, CA, USA; appendix pp 5–8) that was designed to (1) measure rare coding variation; (2) provide a framework for optimum imputation of non-genotyped variants that are common (MAF >5%) or of low frequency (MAF 1–5%) in the European population, when used in conjunction with a large imputation reference panel of individuals with whole-genome sequence data;21 and (3) optimise coverage of genes and genomic regions with established or putative roles in lung health and disease to enable fine mapping. After thorough sample and variant quality control (appendix pp 8–15), we imputed non-genotyped variants using a combined 1000 Genomes Project Phase 122 and UK10K Project23,24 reference panel (appendix pp 15–16). The data were used to finalise the design of the UK Biobank array, which is being used for genome-wide genotyping and imputation of the remaining UK Biobank participants.
Using data from previously published studies of whole-genome gene expression and genome-wide genotyping,25–29 we assessed whether variants at associated loci (identified as described in the Statistical analysis) regulate levels of mRNA. These expression quantitative trait loci (eQTL) studies included non-tumour lung tissue, blood, and, for variants associated with smoking behaviour, brain. For genes close to peaks of novel signals or genes implicated through eQTL, we assessed differential expression in the lungs of individuals with and without COPD and differential expression in the pseudoglandular and canalicular stages of development of the fetal lung.30,31 Additionally, we generated RNA sequencing data to discover novel transcripts of these genes in human bronchial epithelial cells. We tested all genome-wide meta-analysis p values for enrichment in biological pathways defined in publicly available databases. All functional analyses are described in detail in the appendix (pp 21–23).
Statistical analysis
Case-control comparisons of low FEV1 versus high FEV1, low FEV1 versus average FEV1, and high FEV1 versus average FEV1 were done within each of the heavy and never smokers subsets separately (appendix p 17). To identify whether any individual variants had a significantly different effect on the risk of airflow obstruction in heavy smokers compared with never smokers, we tested for interaction with smoking (appendix p 17). We calculated the proportion of the variance in FEV1 explained by genetic variants (appendix p 17). We compared heavy versus never smokers to identify loci associated with smoking behaviour. Association testing was done using a Score test (and Firth test for variants with minor allele count <400)32 with imputed marker doses, adjusting for pack-years in smokers and ten principal components. Full genome-wide association results are available via UK Biobank (appendix p 17). For genome-wide association analyses, we set genome-wide significance as p<5 × 10−8 and suggestive significance as 5 × 10−8<p<5 × 10−7. For other analyses, we used a Bonferroni correction for multiple testing. The appendix (p 17) describes quality control after association testing. For the lead single nucleotide polymorphism (SNP) at each of our novel signals of association with FEV1 extremes, we tested for association with COPD risk using the aforementioned definition (appendix p 18). We did a meta-analysis across smoking strata using inverse variance weighting. We assessed evidence for polygenic architecture of FEV1-defined phenotypes (appendix p 18).33 For this analysis, we created discovery and target subpopulations, each of which comprised cases and control groups created by randomly splitting the low FEV1 and average FEV1 groups (appendix pp 18–20). Variants of MAF of at least 1% associated with low FEV1 below given p value thresholds in the discovery population were incorporated into an aggregate score, and the association with the aggregate score was tested in the independent target population. A similar approach (appendix pp 18–20), in each case using independent discovery and target populations, was used to test for a shared polygenic component between high FEV1 and low FEV1, low FEV1 in heavy smokers and in never smokers, and low FEV1 in participants who did and those who did not report a history of doctor-diagnosed asthma. To show the reliability of the doctor diagnosis of asthma variable, we showed association with asthma at ten previously reported genome-wide significant loci (appendix pp 21, 29).
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the UK BiLEVE study and had final responsibility for the decision to submit for publication.
Results
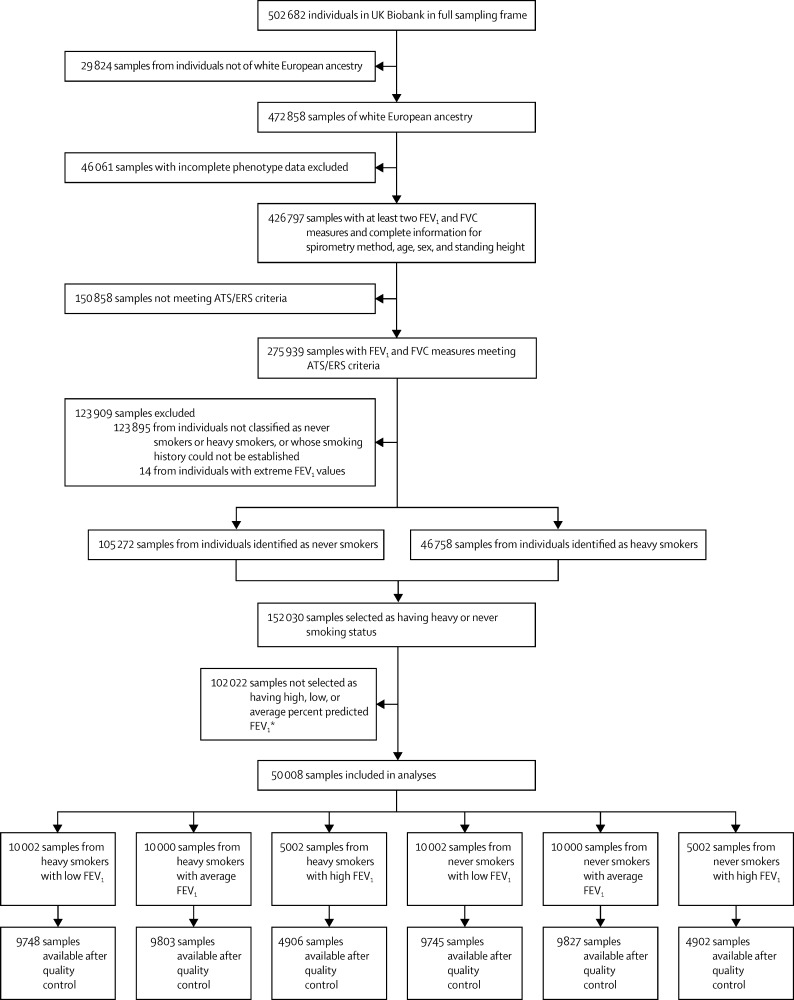
UK Biobank participants were recruited from March 15, 2006, to July 7, 2010. Sample selection for the UK BiLEVE study started on Nov 22, 2012, and was completed on Dec 20, 2012. We initially selected 50 008 unique samples representing the extremes and middle of the percent predicted FEV1 distributions; this comprised 10 002 individuals with low percent predicted FEV1, 10 000 individuals with average percent predicted FEV1, and 5002 individuals with high percent predicted FEV1 from each of the heavy smoker and never smoker groups (figure 1). Within this dataset, 48 931 unrelated individuals passed quality control and were included in subsequent analyses.
Figure 1.
Sample selection strategy
ATS=American Thoracic Society. ERS=European Respiratory Society.34 FEV1=forced expiratory volume in 1 s. FVC= forced vital capacity. *See appendix (pp 3–5) for more details of sample selection.
We undertook genome-wide genotyping of 807 411 variants. After filtering, genome-wide imputation using the 1000 Genomes Project Phase 1 and UK10K Project reference panel resulted in 42 795 484 variants. Our final dataset for analysis, after excluding variants with information quality less than 0·5 or minor allele count less than three, comprised 28 509 962 imputed or genotyped variants in 48 931 unrelated individuals (table 1 and appendix pp 16 and 95).23
Table 1.
Absolute and percent predicted forced expiratory volume in 1 s in each subgroup in heavy and never smokers
|
Heavy smokers |
Never smokers |
|||||
|---|---|---|---|---|---|---|
| Number of individuals (n=24 457) | Absolute FEV1 (L) | Predicted FEV1 (%) | Number of individuals (n=24 474) | Absolute FEV1 (L) | Predicted FEV1 (%) | |
| Low FEV1 | 9748 | 1·93 (0·55) | 65·6% (11·8) | 9745 | 2·05 (0·54) | 69·3% (10·0) |
| Average FEV1 | 9803 | 2·68 (0·56) | 90·6% (3·9) | 9827 | 2·92 (0·57) | 98·7% (1·3) |
| High FEV1 | 4906 | 3·49 (0·72) | 118·0% (8·1) | 4902 | 3·83 (0·73) | 130·3% (8·3) |
Data are mean (SD), unless otherwise specified. See appendix (pp 3–5) for details of sample selection. FEV1=forced expiratory volume in 1 s.
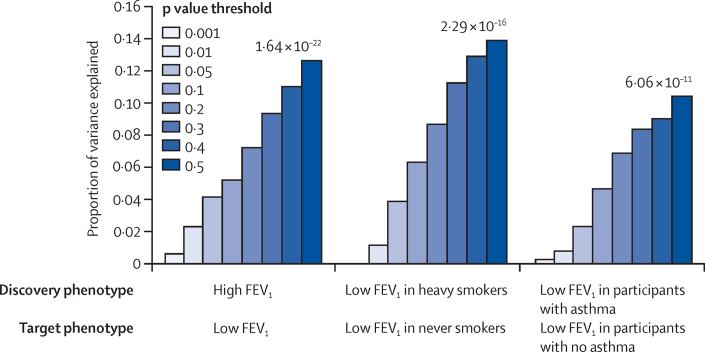
Using independent discovery and target subpopulations to generate and test risk scores, we found that the association of low FEV1 versus average FEV1 with the risk score became stronger for increasingly liberal p value thresholds of association in the discovery population (p=6·24 × 10−16 for a p value threshold of 0·5). This finding suggests a polygenic component to low FEV1, in which many variants of individually small effect size contribute to the risk of low FEV1. We found substantial sharing of genetic causes across thousands of genetic variants between low FEV1 in heavy smokers and low FEV1 in never smokers (p=2·29 × 10−16; p value threshold <0·5; figure 2; appendix p 30). Similarly, we found substantially overlapping genetic causes for low FEV1 in participants reporting a history of doctor-diagnosed asthma and low FEV1 in those without asthma (p=6·06 × 10−11; p value threshold <0·5; figure 2; appendix p 31). Finally, overlapping genetic causes were shown for high FEV1 and low FEV1 (p=1·64 × 10−22; p value threshold <0·5; figure 2; appendix p 30).
Figure 2.
Polygenic component of low forced expiratory volume in 1 s and shared polygenic component of different phenotypes defined by forced expiratory volume in 1 s, smoking, and doctor diagnosis of asthma
The p value in the target population shown above the bars is for the p value threshold <0·5. The sample sizes differed between the comparisons; details of these and the assumptions used in the analyses are described in the appendix (pp 18–20). FEV1=forced expiratory volume in 1 s.
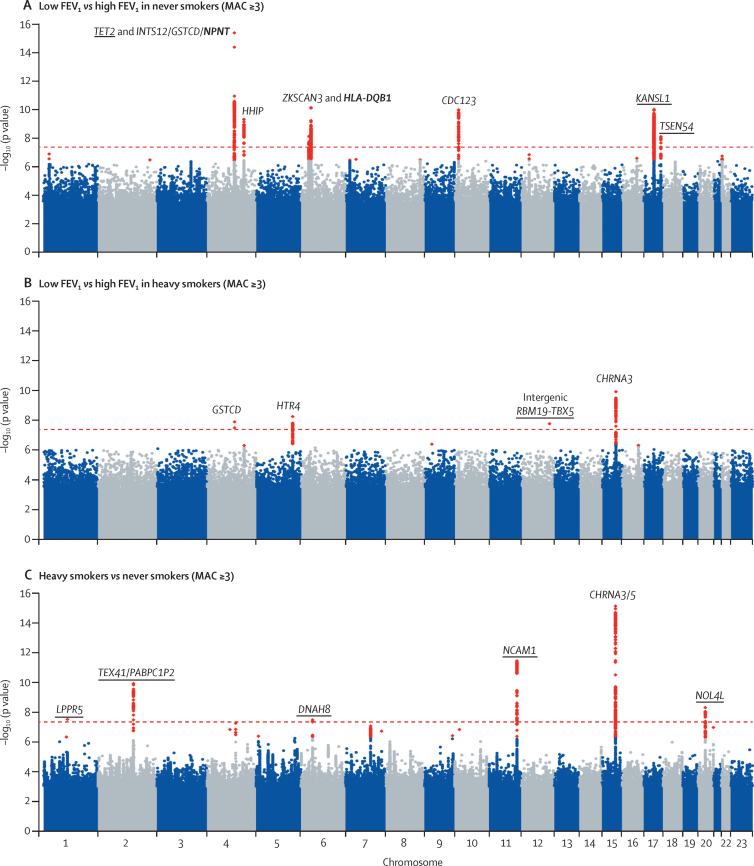
In addition to detecting signals of association previously reported by studies of quantitative lung function (appendix p 32–37), in our case-control analysis of FEV1 extremes we identified six novel signals of association (p<5 × 10−8) with low FEV1 versus high FEV1 (table 2; figure 3; appendix pp 38–53, 96–98, 102). The sentinel SNPs at five of these six signals, in or near TET2, NPNT, HLA-DQB1/HLA-DQA2, KANSL1, and TSEN54, were common (MAF ≥5%) and showed a stronger association with low FEV1 in never smokers than heavy smokers. The sentinel SNP at an intergenic signal between RBM19 and TBX5 was a rare variant (MAF=0·13%) that showed strongest association with low FEV1 in heavy smokers. The lead SNPs at each of these loci showed association with COPD (table 2; appendix p 55). The 26 previously reported SNPs (associated with FEV1, FEV1:FVC ratio, or both)5,7–9 explained 2·33% of the variance of FEV1 in our data; adding in the SNPs representing our six novel signals of association with FEV1 extremes, we explained 3·63% of the variance of FEV1 (appendix p 17).
Table 2.
Novel genome-wide significant signals of association with extremes of forced expiratory volume in 1 s or smoking behaviour
| Locus | Non-coded/coded allele (minor allele) | Imputation info score* | Smoking status | MAF (MAC)† | OR (95% CI) | p value (genomic control corrected) |
Association with COPD‡ |
Effect on FEV1 |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p value | Beta (SE)§ | p value | |||||||||
| Extremes of FEV1: low vs high FEV1 | ||||||||||||
| Genome-wide significant in heavy smokers | ||||||||||||
| Chr12:114743533 | RBM19/TBX5 | T/C (T) | 0·737 | Never | 0·002 (60) | 0·97 (0·57–1·67) | 0·90 | 1·16 (0·54–2·51) | 0·71 | 0·101 (0·118) | 0·39 | |
| .. | .. | .. | .. | Heavy | 0·001 (39) | 11·73 (5·03–27·32) | 1·16 × 10−8 | 6·44 (2·89–14·37) | 5·40 × 10−6 | −0·728 (0·151) | 1·31 × 10−6 | |
| Genome-wide significant in never smokers | ||||||||||||
| rs34712979 ¶‖, Chr4:106819053 | NPNT | G/A (A) | 1·000 | Never | 0·268 (7842) | 1·27 (1·20–1·34) | 9·62 × 10−16 | 1·36 (1·27–1·46) | 2·10 × 10−18 | −0·087 (0·010) | 2·27 × 10−17 | |
| .. | .. | .. | .. | Heavy | 0·261 (7636) | 1·18 (1·11–1·25) | 1·10 × 10−8 | 1·26 (1·18–1·34) | 5·43 × 10−13 | −0·056 (0·010) | 4·22 × 10−8 | |
| rs9274600¶, Chr6:32635592 | HLA-DQB1/HLA-DQA2 | A/G (G) | 0·962 | Never | 0·472 (13 838) | 1·18 (1·13–1·25) | 1·26 × 10−10 | 1·24 (1·16–1·32) | 1·95 × 10−11 | −0·057 (0·009) | 6·72 × 10−10 | |
| .. | .. | .. | .. | Heavy | 0·468 (13 719) | 1·05 (1·00–1·10) | 0·096 | 1·08 (1·02–1·14) | 8·58 × 10−3 | −0·019 (0·009) | 0·037 | |
| rs2532349, Chr17:44339473 | KANSL1 | A/G (G) | 0·976 | Never | 0·242 (7088) | 1·22 (1·15–1·29) | 1·66 × 10−10 | 1·24 (1·16–1·34) | 3·97 × 10−9 | −0·063 (0·011) | 3·22 × 10−9 | |
| .. | .. | .. | .. | Heavy | 0·233 (6832) | 1·15 (1·08–1·21) | 1·47 × 10−5 | 1·14 (1·07–1·22) | 9·56 × 10−5 | −0·050 (0·011) | 3·64 × 10−6 | |
| rs7218675, Chr17:73513185 | TSEN54 | C/A (C) | 0·997 | Never | 0·291 (8538) | 1·18 (1·11–1·25) | 1·18 × 10−8 | 1·22 (1·14–1·31) | 4·56 × 10−9 | −0·052 (0·010) | 1·94 × 10−7 | |
| .. | .. | .. | .. | Heavy | 0·290 (8503) | 1·04 (0·98–1·09) | 0·23 | 1·06 (1·00–1·13) | 0·059 | −0·017 (0·010) | 0·080 | |
| rs2047409, Chr4:106137033 | TET2 | G/A (G) | 0·998 | Never | 0·345 (10 117) | 1·17 (1·11–1·23) | 1·31 × 10−8 | 1·17 (1·10–1·25) | 1·64 × 10−6 | −0·056 (0·009) | 4·19 × 10−9 | |
| .. | .. | .. | .. | Heavy | 0·356 (10 440) | 1·07 (1·02–1·13) | 8·01 × 10−3 | 1·09 (1·03–1·16) | 2·92 × 10−3 | −0·023 (0·009) | 0·014 | |
| Smoking behaviour: heavy vs never smokers | ||||||||||||
| rs4466874, Chr11:112861434 | NCAM1 | T/C (C) | 0·998 | NA | 0·385 (37 709) | 1·10 (1·07–1·13) | 3·22 × 10−12 | NA | NA | NA | NA | |
| rs10193706, Chr2:146316319 | TEX41/ PABPC1P2 | A/C (A) | 0·983 | NA | 0·473 (46 280) | 1·09 (1·06–1·12) | 1·10 × 10−10 | NA | NA | NA | NA | |
| rs143125561; rs57342388, Chr20:31162590 | NOL4L | C/CACGG (CACGG) | 0·983 | NA | 0·233 (22 820) | 1·10 (1·07–1·13) | 4·65 × 10−9 | NA | NA | NA | NA | |
| rs61784651‖, Chr1:99445471 | LPPR5 | C/T (T) | 1·000 | NA | 0·170 (16 609) | 1·10 (1·07–1·14) | 2·89 × 10−8 | NA | NA | NA | NA | |
| rs10807199, Chr6:38901867 | DNAH8 | C/T (T) | 1·000 | NA | 0·473 (46 286) | 1·08 (1·05–1·11) | 3·17 × 10−8 | NA | NA | NA | NA | |
For variants that showed association with extremes of FEV1 in either heavy smokers or never smokers, the results from both the never smokers and heavy smokers are presented. For variants that had genome-wide significant evidence of association for smoking behaviour (and not for extremes of FEV1), association with COPD and effect on FEV1 were not assessed. Chromosome and position relate to National Center for Biotechnology Information build 37 (hg19). ..=as above. COPD=chronic obstructive pulmonary disease. FEV1=forced expiratory volume in 1 s. MAC=minor allele count. MAF=minor allele frequency. NA=not applicable. OR=odds ratio. SE=standard error.
Indication of certainty of imputation for this variant; an imputation info score of 1 suggests a variant imputed with the highest certainty or a directly genotyped variant.
In samples included in the comparison.
Analysis of Global Initiative for Chronic Obstructive Lung Disease stage 2+ COPD cases versus controls (heavy smokers: 5803 cases vs 4661 controls; never smokers: 3761 cases vs 4792 controls).
Effect on FEV1 beta values are effect-size estimates on an inverse-normal transformed scale after adjustments for age, age2, sex, height, and ancestry principal components (appendix p 21).
Novel signals of association within previously reported loci.
Directly genotyped.
Figure 3.
Manhattan plots for low versus high forced expiratory volume in 1 s in never smokers and heavy smokers and for heavy versus never smokers
p values are from a Score test and have genomic control applied unless the MAC was less than 400 and Score test p<1·00 × 10−6, in which case p values are from a Firth test with no genomic control. Novel loci are underlined and novel signals at previously reported loci are shown in bold. The dashed red line shows the threshold for genome-wide significance (p<5 × 10−8). Variants with suggestive evidence of association (p<5 × 10−7) are coloured red. Quantile–quantile plots for these analyses are shown in the appendix (pp 113–116). FEV1=forced expiratory volume in 1 s. MAC=minor allele count.
Although association with lung function at 4q24 is well established,5,7 we report two further independent signals of association at this locus (table 2; appendix pp 56–57, 99–101). The first (rs34712979) was localised to NPNT but independent of the previously reported signal, which spanned INTS12, GSTCD, and NPNT (appendix p 102). The second (rs2047409) was 552 kb from the signals at INTS12, GSTCD, and NPNT and was localised to TET2. The signal of association for rs34712979 was strongest in never smokers (p=9·62 × 10−16; table 2) and was weakly correlated (linkage disequilibrium r2=0·31) with another SNP in NPNT—rs6856422—which has also been identified as a novel secondary signal of association at 4q24 by an independent concurrent study of lung function in the general population.35 When rs6856422 was included as a covariate in our analysis, the signal for rs34712979 (p=9·62 × 10−16) was only slightly attenuated (p=4·66 × 10−11). The novel signal at TET2 (rs2047409) also showed strongest association in never smokers (p=1·31 × 10−8). rs2047409 is separated from the previously reported association of rs10516526 with FEV1 (GSTCD)5,7 by a recombination hotspot and is statistically independent (rs105165267 included as a covariate; rs2047409, p=9·8 × 10−9). TET2 encodes tet methylcytosine dioxygenase 2, which has a role in myelopoiesis, and SNPs in TET2 have shown association with height.36 TET2 was differentially expressed during fetal lung development (appendix pp 58–59).
We detected a signal of association with FEV1 extremes within the HLA region on chromosome 6 that was correlated with a previously reported signal of association with asthma.37 The signal we report was strongest in never smokers (rs9274600, p=1·26 × 10−10; table 2; appendix p 102). With an imputed proxy (rs17843604) of the asthma-associated SNP rs927334937—included as a covariate in the analysis of rs9274600—the signal for rs9274600 was attenuated (p=5·66 × 10−4), confirming that rs9274600 and rs9273359 are correlated. After exclusion of individuals with doctor-diagnosed asthma, the odds ratio for rs9274600 decreased from 1·18 (95% CI 1·11–1·25) to 1·14 (1·08–1·20), but remained significant (p=3·25 × 10−6; appendix p 103). This signal is independent of nearby signals reported for lung function,5,7,8 including rs7764819,38 which is 45 kb from rs9274600 (association for rs9274600 conditioned on rs7764819; p=6·71 × 10−11).
We identified a rare SNP that was associated with FEV1 extremes in heavy smokers only, after adjusting for pack-years of smoking (p=1·16 × 10−8; table 2). This intergenic SNP on chromosome 12 (chr12:114743533, MAF=0·13%) also showed weak evidence of association with smoking behaviour (p=6·12 × 10−3; appendix pp 60–62). We noted evidence for change in expression levels with increasing fetal lung age (p=0·04) for one or more probes after adjustment for multiple testing for the nearby gene TBX5 (appendix pp 58–59).
We noted a broad signal of association (∼1·5 Mb) in an inversion locus at 17q21.31 (rs2532349, near KANSL1; appendix p 97). This signal was strongest in never smokers (rs2532349, p=1·66 × 10−10), but was also detected in heavy smokers (p=1·47 × 10−5). Genes in this locus, which include MAPT and CRHR1, have previously been associated with pulmonary fibrosis39,40 and inhaled corticosteroid response in asthma.41 SNP rs2532349 (and SNPs in strong linkage disequilibrium [r2>0·8]) was associated with mRNA expression levels of at least 15 genes in lung and blood (appendix pp 63–70). We identified differential expression for six genes at 17q21.31 during fetal lung development (and for four genes on different chromosomes regulated by trans eQTLs at 17q21.41; appendix pp 58–59). Relatively abundant novel transcripts (ie, compared with other transcripts detected) were identified by RNA sequencing in human bronchial epithelial cells for WNT3 and LRRC37A4P; expression of both genes was associated with rs2532349 in lung and blood (appendix pp 104–109). The SNP rs2532349 (MAF=24%) was in linkage disequilibrium with the inversion (r2>0.9); the allele associated with low FEV1 was positively correlated with the inverted haplotype.42,43 The inversion locus contains structural variation resulting from three duplication events (150–300 kb).42,43 We imputed the nine common structural haplotypes (appendix pp 23–24)42 and found that the number of copies of the 150 kb region containing the 5′ end of KANSL1 (the entire 150 kb duplication region found only in individuals who carry the inversion and a nested region of the 300 kb duplication region found only in individuals who do not carry the inversion) was associated with extremes of FEV1 (p=2·40 × 10−6; appendix p 71). The sentinel SNP rs2532349 lies within this region.
A second signal of association with FEV1 extremes on chromosome 17 (17q25.1) was within TSEN54 and occurred only in never smokers (rs7218675; p=1·18 × 10−8; table 2). TSEN54 encodes a subunit of the tRNA splicing endonuclease complex, and rs7218675 was associated with expression of KIAA0195, TSEN54, and GRB2 in blood and expression of GRB2 in lung tissue (appendix pp 63–66). GRB2 is a ligand of the epidermal growth factor receptor, which links signalling by epidermal growth factor with the MAPK/ERK signalling pathway, triggering cell proliferation. RNA sequencing in human bronchial epithelial cells identified a relatively abundant novel fusion transcript of TSEN54 and LLGL2 (appendix p 108).
To corroborate the new signals we identified in TET2 and TSEN54, we present evidence of association with FEV1 in a previously reported study8 of 48 201 individuals (p=9·9× 10−5 and p=0·006, respectively; appendix pp 24, 82).
We identified a further 21 loci with suggestive (5 × 10−8<p<5 × 10−7) evidence of association with FEV1 extremes (appendix pp 72–73), including six rare variants with a minor allele count less than 400. These included signals in CCDC91 and RSRC1, both of which showed genome-wide significant association with lung function in an independent concurrent study of lung function in the general population.35
By comparing heavy smokers and never smokers, we identified five novel regions of association with smoking behaviour and confirmed four previously reported loci (15q25, 7p14, DBH, and BDNF; table 2; appendix pp 32–37).17–19 The novel signals included rs4466874, in an intron of NCAM1 (chromosome 11), and rs10193706, an intergenic SNP on chromosome 2 downstream of TEX41 and upstream of PABPC1P2 (table 2; appendix pp 110–112). Uncorrelated (r2<0·0001 with rs4466874) SNPs in TTC12 and ANKK1, near to NCAM1, have also previously shown association with nicotine dependence (appendix pp 74–75).44 A proxy of rs10193706 on chromosome 2 (rs953246, r2=0·48) is a trans eQTL for NCAM1 on chromosome 11 in brain tissue (appendix pp 76–77). Another proxy of rs10193706 on chromosome 2 (rs12622738, r2=0·86) is a trans eQTL in the substantia nigra for WDR61 on chromosome 15, 300 kb from the established 15q25 smoking locus (appendix pp 76–77).
We also noted novel genome-wide significant signals of association with smoking behaviour in NOL4L, LPPR5, and DNAH8 (table 2, appendix pp 110–112). A SNP in C20orf203, near to NOL4L, but independent of our sentinel variant, has previously been implicated in nicotine dependence.45 We identified secondary independent signals, which did not reach genome-wide significance, at three of the loci associated with smoking behaviour (appendix pp 56–57, 99–101),46 including a novel rare (MAF=0·09%) intergenic SNP near NCAM1. For novel signals for smoking behaviour, we did a meta-analysis of summary statistics from two previous, less powerful studies17,19 and found corroborative evidence for NCAM1, TEX41/PABPC1P2, and NOL4L (eg, for smoking initiation p=0·0003, p=0·017, and p=0·0006, respectively; appendix pp 24, 83–84). We identified a further eight loci with suggestive (5 × 10−8<p<5 × 10−7) evidence of association with smoking behaviour (appendix p 72–73), including CHRNA4 at 20q13.33 (p=1·01 × 10−7).47
In a genome-wide gene–smoking interaction analysis, although common SNPs on chromosomes 6 and 19 showed suggestive SNP–smoking interactions (p<5 × 10−7; appendix p 78), no gene–smoking interactions were detected at genome-wide significance (p<5 × 10−8). Three of the six variants associated with FEV1 extremes (table 2) showed weak evidence of interaction with smoking (Bonferroni correction for six tests p=0·0083; appendix pp 60–62), including common variants at the HLA-DQB1/HLA-DQA2 and TSEN54 loci and the rare variant at the RBM19/TBX5 locus. In a meta-analysis of the genome-wide association test statistics for low FEV1 versus high FEV1 across heavy and never smokers, motivated by our finding of shared genetic causes between heavy and never smokers and to increase the sample size, we identified an additional six novel genome-wide significant signals of association with FEV1 extremes. These included CCDC91, reported as a novel signal of association with lung function in the general population by a concurrent study,35 and SLMAP, for which there is corroborative evidence of association with lung function (appendix pp 79–80).8 Our pathway analysis identified a novel signal of enrichment of the histone subset of the chromatin packaging and remodelling process gene set, which was independently replicated in a concurrent GWAS of lung function in the general population (appendix p 81).35 Replication of a previously reported8 enrichment of the systemic lupus erythematosus pathway was also noted (appendix p 81).
Discussion
We describe, to our knowledge, the first genetic association analyses in UK Biobank, targeting the genetic architecture of smoking behaviour and lung function phenotypes. By sampling from the extremes of the FEV1 and smoking phenotype distributions, we identified novel associations for FEV1 and smoking behaviour. We show genome-wide evidence for shared genetic causes of low FEV1 between heavy smokers and never smokers. Furthermore, our analyses suggest that smoking is only likely to interact with a small proportion of the genetic effects we have identified on lung function—that is, smoking and genetic effects generally act separately. We also show shared genetic causes of airflow obstruction between participants who reported doctor-diagnosed asthma and those who did not.
Two of our novel signals of association with smoking behaviour implicate NCAM1; one SNP lies within an intron of NCAM1 and a second variant, located distantly on chromosome 2, is a trans eQTL for NCAM1 in brain tissue (medulla)—ie, it is associated with the level of expression of NCAM1. This second SNP is also a trans eQTL in substantia nigra tissue for another gene called WDR61, which is close to the genes CHRNA3 and CHRNA5 at 15q25—a locus strongly associated with smoking behaviour.17–19 The substantia nigra plays an important part in reward and addiction,48 but little is known about WDR61 other than that expression can be induced by mechanical strain in mesenchymal stem cells.49
We describe six new signals of association with FEV1 extremes, all of which were also associated with COPD using our definition based on spirometry. Five of these signals were most strongly associated with extremes of FEV1 (low vs high) in never smokers. The signal at 17q21.31 for extremes of FEV1 suggests a role for structural variation and epigenetic regulation in lung health. We found that the number of copies of the 5′ end of KANSL1—a gene disrupted by duplication events—is associated with FEV1 extremes. KANSL1 encodes a protein that is a key component of the NSL1 (histone acetyltransferase) complex.50 The disruption of the gene gives rise to a novel truncated transcript,42 which encodes a protein missing a domain essential for key interactions with other proteins important for NSL1 function.51 Therefore, widespread effects on gene regulation through altered histone acetylation could underlie this association. Reduced expression of KANSL1 causes a rare multisystem disorder,52,53 suggesting an essential role for KANSL1 in epigenetic regulation. In a genome-wide pathway analysis, we identified the histone gene set, further implicating a role for epigenetic regulation in lung health.
We maximised the power of our study by sampling from the extremes of a large biobank. No other similar resources of a comparable size were available for replication studies. Nevertheless, the novel genome-wide significant signals of association with FEV1 extremes in NPNT and KANSL1 in never smokers were also significantly associated in the independent set of heavy smokers. Furthermore, to corroborate the new signals we identified in TET2 and TSEN54, we present evidence of association with FEV1 in a previously reported large study of FEV1 in the general population.8 However, the rare SNP on chromosome 12 for which we found association with extremes of FEV1 was exclusive to the recently released UK10K Project component of the imputation panel and has not yet been measured in suitably large studies. Our comparison of smokers and never smokers represents a powerful approach because of the restriction to heavy smokers rather than ever smokers. For novel signals for smoking behaviour, we present additional evidence of association with smoking behaviour for NCAM1, TEX41/PABPC1P2, and NOL4L in independent populations.17,19 Although these independent datasets have limited power, they provide corroboration of key genome-wide significant findings in UK BiLEVE.
One of the strengths of our study design was that the genotyping platform we used allowed for fine mapping of regions already known to contain genetic variants that affect lung function. For example, we were able to identify a novel signal in NPNT that was independent of the previously reported signal of association at this locus (spanning GSTCD, INTS12, and NPNT). The independent NPNT signal captured by the genotyped variant rs34712979 was not detected in previous or concurrent studies because it was neither directly genotyped nor imputed with sufficient quality; this finding highlights a further advantage of the UK BiLEVE and UK Biobank array design.
The design of this genotyping array combined the best features of existing genome-wide platforms targeting common SNPs (MAF ≥5%) and putative functional exome chip content, plus additional content to improve imputation of low-frequency variants (MAF 1–5%). In combination with a new large UK-specific imputation reference panel (UK10K Project), these features increase the potential to discover novel signals. In our study, more than 28·5 million variants were imputed; current large meta-analyses combining data from several studies with older arrays and using equivalent quality control filters after imputing to 1000 Genomes Project Phase 1 alone typically measure about 10·6 million variants.35 The genome-wide genotype data for these 50 008 individuals have been deposited in UK Biobank to be made available to other approved research projects across many disease areas. The UK BiLEVE array was used as a prototype for the array that is being used in the remaining roughly 450 000 UK Biobank participants. The UK Biobank array shares more than 95% of its content with the UK BiLEVE array. When genotyping of all UK Biobank participants is complete, UK Biobank will provide a unique resource for genome-wide studies of quantitative traits, nested case-control studies, and studies in which longitudinal outcomes can be studied.
Despite the strengths of using a large resource such as UK Biobank, this study has some limitations. In particular, there is a trade-off between obtaining the large sample sizes generally needed for genetic studies and the depth of phenotyping that is practicable in such large populations. Our spirometric definition of COPD was not based on bronchodilator reversibility testing, although we have shown previously that by limiting inclusion to individuals with GOLD stage 2+ spirometry, most of these individuals are likely to have COPD according to more rigorous criteria.13 Similarly, our definition of asthma was based on self-reporting of doctor-diagnosed disease. These limitations might have reduced our ability to identify some novel disease associations, although we were able to replicate many known associations using this approach.
In summary, we show the usefulness in sampling from the extremes of UK Biobank data to identify novel genetic signatures underlying phenotypes important in the development of airway disease and smoking behaviour. The ongoing genotyping, and further phenotyping, of the rest of the UK Biobank resource will facilitate further GWAS, which will undoubtedly improve our understanding of the genetic and molecular basis of common disease.
Acknowledgments
Acknowledgments
This work was funded by a Medical Research Council (MRC) strategic award to MDT, IPH, DPS, and LVW (MC_PC_12010). This research was done using the UK Biobank resource. MDT was supported by MRC fellowships G0501942 and G0902313. IPH is supported by an MRC programme grant (G1000861). This Article presents independent research funded partially by the National Institute for Health Research (NIHR). The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, or the Department of Health. We thank Affymetrix for their role in array design and for undertaking genotyping and genotype calling. We thank all members of the UK Biobank Array Design Group: Peter Donnelly (chair), Jose Bras, Adam Butterworth, Richard Durbin, Paul Elliott, Ian Hall, John Hardy, Mark McCarthy, Gil McVean, Tim Peakman, Nazneen Rahman, Nilesh Samani, Martin Tobin, and Hugh Watkins. This study makes use of data generated by the UK10K Consortium, derived from samples from TwinsUK and ALSPAC. A full list of the investigators who contributed to the generation of the data is available from the UK10K website. Funding for UK10K was provided by the Wellcome Trust under award WT091310. JM is funded by an ERC Consolidator Grant (617306). APM is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science (grant number WT098017). The lung eQTL study at Laval University was supported by the Chaire de pneumologie de la Fondation JD Bégin de l'Université Laval, the Fondation de l'Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP - 123369), and the Cancer Research Society and Read for the Cure. YB is the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec – Santé (FRQS). EGCUT received financing by FP7 grants (278913, 306031, 313010), Center of Excellence in Genomics (EXCEGEN), and University of Tartu (SP1GVARENG). We thank EGCUT technical personnel, especially V Soo and S Smit. Data analyses were done in part in the High Performance Computing Center of University of Tartu. We thank Paul Jennings for assistance with generation of code to extract expression data. MO is a Postdoctoral Fellow of the Michael Smith Foundation for Health Research and the Canadian Institute for Health Research Integrated and Mentored Pulmonary and Cardiovascular Training program. This research used the ALICE and SPECTRE High Performance Computing Facilities at the University of Leicester.
Contributors
LVW, DPS, MDT, and IPH conceived and designed the study. LVW, NS, VEJ, IN, MSA, RA, JPC, CF, DP, JM, DPS, MDT, and IPH analysed data. SM, CKB, AKK, KP, MO, AR, RM, EMi, and ER undertook functional follow-up analyses. YB, KH, DSP, PDP, EMe, JO'C, EF, OD, IS, and JM provided data for follow-up of signals. NS, CF, MM, EZ, APM, JM, and DPS provided statistical support and advice. LVW, IS, PD, RH, IP, ALH, NCT, DPS, MDT, and IPH provided advice on study conduct. All authors discussed the results and implications and commented on the manuscript at all stages.
UK Brain Expression Consortium (UKBEC)
John A Hardy (UCL Institute of Neurology, London, UK), Michael E Weale (King's College London, London, UK), Mina Ryten (UCL Institute of Neurology, London, UK; and King's College London, London, UK), Colin Smith (The University of Edinburgh, Edinburgh, UK), Robert Walker (The University of Edinburgh, Edinburgh, UK), Juan Botía (UCL Institute of Neurology, London, UK; and King's College London, London, UK), Jana Vandrocova (UCL Institute of Neurology, London, UK; and King's College London, London, UK), Sebastian Guelfi (UCL Institute of Neurology, London, UK; and King's College London, London, UK), Karishma D'Sa (UCL Institute of Neurology, London, UK; and King's College London, London, UK), Mar Matarin (UCL Institute of Neurology, London, UK), Vibin Varghese (King's College London, London, UK), Daniah Trabzuni (UCL Institute of Neurology, London, UK), Adaikalavan Ramasamy (UCL Institute of Neurology, London, UK; King's College London, London, UK; and Jenner Institute, University of Oxford, Oxford, UK), and Paola Forabosco (King's College London, London, UK; and Cittadella Universitaria di Cagliari, Monserrato, Sardinia, Italy).
OxGSK Consortium
Jason Z Liu (University of Oxford, Oxford, UK), Federica Tozzi (GlaxoSmithKline, Verona, Italy), Dawn M Waterworth (GlaxoSmithKline, Upper Merion, PA, USA), Sreekumar G Pillai (GlaxoSmithKline, Upper Merion, PA, USA), Pierandrea Muglia (GlaxoSmithKline, Verona, Italy), Lefkos Middleton (Imperial College London, London, UK), Wade Berrettini (University of Pennsylvania School of Medicine, Philadelphia, PA, USA), Christopher W Knouff (GlaxoSmithKline, Research Triangle Park, NC, USA), Xin Yuan (GlaxoSmithKline, Upper Merion, PA, USA), Gérard Waeber (University of Lausanne, Lausanne, Switzerland; and University Hospital of Lausanne, Lausanne, Switzerland), Peter Vollenweider (University of Lausanne, Lausanne, Switzerland; and University Hospital of Lausanne, Lausanne, Switzerland), Martin Preisig (University of Lausanne, Lausanne, Switzerland; and University Hospital of Lausanne, Lausanne, Switzerland), Nicholas J Wareham (Institute of Metabolic Science, Cambridge, UK), Jing Hua Zhao (Institute of Metabolic Science, Cambridge, UK), Ruth J F Loos (Institute of Metabolic Science, Cambridge, UK), Inês Barroso (Wellcome Trust Sanger Institute, Hinxton, UK), Kay-Tee Khaw (University of Cambridge, Cambridge, UK), Scott Grundy (University of Texas Southwestern Medical Center, Dallas, TX, USA), Philip Barter (The Heart Research Institute, Sydney, NSW, Australia), Robert Mahley (Gladstone Institute of Cardiovascular Disease, University of California, San Francisco, CA, USA; and American Hospital, Istanbul, Turkey), Antero Kesaniemi (University of Oulu, Oulu, Finland), Ruth McPherson (University of Ottawa Heart Institute, Ottawa, ON, Canada), John B Vincent (University of Toronto, Toronto, ON, Canada), John Strauss (University of Toronto, Toronto, ON, Canada), James L Kennedy (University of Toronto, Toronto, ON, Canada), Anne Farmer (King's College London, London, UK), Peter McGuffin (King's College London, London, UK), Richard Day (University of Dundee, Dundee, UK), Keith Matthews (University of Dundee, Dundee, UK), Per Bakke (University of Bergen, Bergen, Norway), Amund Gulsvik (University of Bergen, Bergen, Norway), Susanne Lucae (Max Planck Institute of Psychiatry, Munich, Germany), Marcus Ising (Max Planck Institute of Psychiatry, Munich, Germany), Tanja Brueckl (Max Planck Institute of Psychiatry, Munich, Germany), Sonja Horstmann (Max Planck Institute of Psychiatry, Munich, Germany), H-Erich Wichmann (Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Ludwig-MaximiliansUniversität, Munich, Germany; and Klinikum Grosshadern, Munich, Germany), Rajesh Rawal (Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany), Norbert Dahmen (University of Mainz, Mainz, Germany), Claudia Lamina (Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; and Innsbruck Medical University, Innsbruck, Austria), Ozren Polasek (University of Zagreb, Zagreb, Croatia), Lina Zgaga (University of Edinburgh, Edinburgh, UK), Jennifer Huffman (MRC Human Genetics Unit, Edinburgh, UK), Susan Campbell (MRC Human Genetics Unit, Edinburgh, UK), Jaspal Kooner (Imperial College London, London, UK), John C Chambers (Imperial College London, London, UK), Mary Susan Burnett (Washington Hospital Center, Washington, DC, USA), Joseph M Devaney (Washington Hospital Center, Washington, DC, USA), Augusto D Pichard (Washington Hospital Center, Washington, DC, USA), Kenneth M Kent (Washington Hospital Center, Washington, DC, USA), Lowell Satler (Washington Hospital Center, Washington, DC, USA), Joseph M Lindsay (Washington Hospital Center, Washington, DC, USA), Ron Waksman (Washington Hospital Center, Washington, DC, USA), Stephen Epstein (Washington Hospital Center, Washington, DC, USA), James F Wilson (University of Edinburgh, Edinburgh, UK), Sarah H Wild (University of Edinburgh, Edinburgh, UK), Harry Campbell (University of Edinburgh, Edinburgh, UK), Veronique Vitart (MRC Human Genetics Unit, Edinburgh, UK), Muredach P Reilly (University of Pennsylvania, Philadelphia, PA, USA), Mingyao Li (University of Pennsylvania, Philadelphia, PA, USA), Liming Qu (University of Pennsylvania, Philadelphia, PA, USA), Robert Wilensky (University of Pennsylvania, Philadelphia, PA, USA), William Matthai (University of Pennsylvania, Philadelphia, PA, USA), Hakon H Hakonarson (Children's Hospital of Philadelphia, Philadelphia, PA, USA), Daniel J Rader (University of Pennsylvania, Philadelphia, PA, USA), David Ellinghaus (ChristianAlbrechts-University of Kiel, Kiel, Germany), Wolfgang Lieb (ChristianAlbrechts-University of Kiel, Kiel, Germany), Andre Franke (ChristianAlbrechts-University of Kiel, Kiel, Germany), Manuela Uda (Consiglio Nazionale delle Ricerche, Monserrato, Cagliari, Italy), Antonio Terracciano (National Institute on Aging, Baltimore, MD, USA), Xiangjun Xiao (University of Texas MD Anderson Cancer Center, Houston, TX, USA), Fabio Busonero (Consiglio Nazionale delle Ricerche, Monserrato, Cagliari, Italy), Paul Scheet (University of Texas MD Anderson Cancer Center, Houston, TX, USA), David Schlessinger (National Institute on Aging, Baltimore, MD, USA), David St Clair (University of Aberdeen, Aberdeen, UK), Dan Rujescu (Ludwig-Maximilians-University, Munich, Germany), Gonçalo R Abecasis (University of Michigan, Ann Arbor, MI, USA), Hans Jörgen Grabe (University of Greifswald, Greifswald, Germany), Alexander Teumer (University of Greifswald, Greifswald, Germany), Henry Völzke (University of Greifswald, Greifswald, Germany), Astrid Petersmann (University of Greifswald, Greifswald, Germany), Ulrich John (University of Greifswald, Greifswald, Germany), Igor Rudan (University of Split, Split, Croatia; and University of Edinburgh, Edinburgh, UK), Caroline Hayward (MRC Human Genetics Unit, Edinburgh, UK), Alan F Wright (MRC Human Genetics Unit, Edinburgh, UK), Ivana Kolcic (University of Zagreb, Zagreb, Croatia), Benjamin J Wright (University of Leicester, Leicester, UK), John R Thompson (University of Leicester, Leicester, UK), Anthony J Balmforth (University of Leeds, Leeds, UK), Alistair S Hall (University of Leeds, Leeds, UK), Nilesh J Samani (University of Leicester, Glenfield Hospital, Leicester, UK), Carl A Anderson (Wellcome Trust Sanger Institute, Hinxton, UK), Tariq Ahmad (Peninsula College of Medicine and Dentistry, Exeter, UK), Christopher G Mathew (King's College London School of Medicine, Guy's Hospital, London, UK), Miles Parkes (Addenbrooke's Hospital, Cambridge, UK), Jack Satsangi (University of Edinburgh, Western General Hospital, Edinburgh, UK), Mark Caulfield (Barts and the London School of Medicine, Queen Mary University of London, London, UK), Patricia B Munroe (Barts and the London School of Medicine, Queen Mary University of London, London, UK), Martin Farrall (University of Oxford, Wellcome Trust Centre for Human Genetics, Oxford, UK), Anna Dominiczak (British Heart Foundation Glasgow Cardiovascular Research Centre, University of Glasgow, Western Infirmary, Glasgow, UK), Jane Worthington (Arthritis Research UK Centre for Genetics and Genomics, University of Manchester, Manchester, UK), Wendy Thomson (Arthritis Research UK Centre for Genetics and Genomics, University of Manchester, Manchester, UK), Steve Eyre (Arthritis Research UK Centre for Genetics and Genomics, University of Manchester, Manchester, UK), Anne Barton (Arthritis Research UK Centre for Genetics and Genomics, University of Manchester, Manchester, UK), The Wellcome Trust Case Control Consortium*, Vincent Mooser (GlaxoSmithKline, Upper Merion, PA, USA), Clyde Francks (Max Planck Institute for Psycholinguistics, Nijmegen, Netherlands; and Radboud University Nijmegen, Nijmegen, Netherlands), Jonathan Marchini (University of Oxford, Oxford, UK). *Members listed in appendix pp 128–131.
Declaration of interests
We declare no competing interests.
Contributor Information
Martin D Tobin, Email: mt47@leicester.ac.uk.
Ian P Hall, Email: ian.hall@nottingham.ac.uk.
Supplementary Material
References
- 1.Lozano R, Naghavi M, Foreman K. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClearn GE, Svartengren M, Pedersen NL, Heller DA, Plomin R. Genetic and environmental influences on pulmonary function in aging Swedish twins. J Gerontol. 1994;49:264–268. doi: 10.1093/geronj/49.6.m264. [DOI] [PubMed] [Google Scholar]
- 3.Palmer LJ, Knuiman MW, Divitini ML. Familial aggregation and heritability of adult lung function: results from the Busselton Health Study. Eur Respir J. 2001;17:696–702. doi: 10.1183/09031936.01.17406960. [DOI] [PubMed] [Google Scholar]
- 4.Zhai G, Valdes AM, Cherkas L, Clement G, Strachan D, Spector TD. The interaction of genes and smoking on forced expiratory volume: a classic twin study. Chest. 2007;132:1772–1777. doi: 10.1378/chest.07-1438. [DOI] [PubMed] [Google Scholar]
- 5.Hancock DB, Eijgelsheim M, Wilk JB. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loth DW, Artigas MS, Gharib SA. Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nat Genet. 2014;46:669–677. doi: 10.1038/ng.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Repapi E, Sayers I, Wain LV. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soler Artigas M, Loth DW, Wain LV. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilk JB, Chen TH, Gottlieb DJ. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castaldi PJ, Cho MH, Litonjua AA. The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. Am J Respir Cell Mol Biol. 2011;45:1147–1153. doi: 10.1165/rcmb.2011-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho MH, Boutaoui N, Klanderman BJ. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillai SG, Ge D, Zhu G. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soler Artigas M, Wain LV, Repapi E. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184:786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilk JB, Shrine NR, Loehr LR. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho MH, McDonald ML, Zhou X. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO . Global health risks: mortality and burden of disease attributable to selected major risks. World Health Organization; Geneva: 2009. [Google Scholar]
- 17.Liu JZ, Tozzi F, Waterworth DM. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorgeirsson TE, Gudbjartsson DF, Surakka I. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudlow C, Gallacher J, Allen N. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Altshuler D, Auton A, for the The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Howie B, McCarthy S. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015 doi: 10.1038/nature14962. published online Sept 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao K, Bosse Y, Nickle DC. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamontagne M, Couture C, Postma DS. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eQTLs. PLoS One. 2013;8:e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obeidat M, Miller S, Probert K. GSTCD and INTS12 regulation and expression in the human lung. PLoS One. 2013;8:e74630. doi: 10.1371/journal.pone.0074630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westra HJ, Peters MJ, Esko T. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramasamy A, Trabzuni D, Guelfi S. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiling K, van den Berge M, Hijazi K. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med. 2013;187:933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melen E, Kho AT, Sharma S. Expression analysis of asthma candidate genes during human and murine lung development. Respir Res. 2011;12:86. doi: 10.1186/1465-9921-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma C, Blackwell T, Boehnke M, Scott LJ, for the GoT2D investigators Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37:539–550. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell SM, Wray NR, Stone JL, for the International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MR, Hankinson J, Brusasco V. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 35.Soler Artigas M, Wain L, Miller S, et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun (in press). [DOI] [PMC free article] [PubMed]
- 36.Lango Allen H, Estrada K, Lettre G. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moffatt MF, Gut IG, Demenais F. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hancock DB, Artigas MS, Gharib SA. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noth I, Zhang Y, Ma SF. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fingerlin TE, Murphy E, Zhang W. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tantisira KG, Lazarus R, Litonjua AA, Klanderman B, Weiss ST. Chromosome 17: association of a large inversion polymorphism with corticosteroid response in asthma. Pharmacogenet Genomics. 2008;18:733–737. doi: 10.1097/FPC.0b013e3282fe6ebf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet. 2012;44:881–885. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinberg KM, Antonacci F, Sudmant PH. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet. 2012;44:872–880. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gelernter J, Yu Y, Weiss R. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- 45.Wang D, Ma JZ, Li MD. Mapping and verification of susceptibility loci for smoking quantity using permutation linkage analysis. Pharmacogenomics J. 2005;5:166–172. doi: 10.1038/sj.tpj.6500304. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Ferreira T, Morris AP, for the Genetic Investigation of ANthropometric Traits (GIANT) Consortium. the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Niu T, Xing H. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. Am J Hum Genet. 2004;75:112–121. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faure A, Haberland U, Condé F, El Massioui N. Lesion to the nigrostriatal dopamine system disrupts stimulus-response habit formation. J Neurosci. 2005;25:2771–2780. doi: 10.1523/JNEUROSCI.3894-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rathbone SR, Glossop JR, Gough JE, Cartmell SH. Cyclic tensile strain upon human mesenchymal stem cells in 2D and 3D culture differentially influences CCNL2, WDR61 and BAHCC1 gene expression levels. J Mech Behav Biomed Mater. 2012;11:82–91. doi: 10.1016/j.jmbbm.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 50.Mendjan S, Taipale M, Kind J. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koolen DA, Kramer JM, Neveling K. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat Genet. 2012;44:639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- 53.Zollino M, Orteschi D, Murdolo M. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat Genet. 2012;44:636–638. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.