Abstract
Molecular mechanisms underlying the functioning of central pattern generators (CPGs) are poorly understood. Investigations using genetic approaches in the model organism Drosophila may help to identify unknown molecular players participating in the formation or control of motor patterns. Here we report Drosophila genes as candidates for involvement in the neural mechanisms responsible for motor functions, such as locomotion and courtship song. Twenty-two Drosophila lines, used for gene identification, were isolated from a previously created collection of 1064 lines, each carrying a P element insertion in one of the autosomes. The lines displayed extreme deviations in locomotor and/or courtship song parameters compared with the whole collection. The behavioral consequences of CNS-specific RNAi-mediated knockdowns for 10 identified genes were estimated. The most prominent changes in the courtship song interpulse interval (IPI) were seen in flies with Sps2 or CG15630 knockdown. Glia-specific knockdown of these genes produced no effect on the IPI. Estrogen-induced knockdown of CG15630 in adults reduced the IPI. The product of the CNS-specific gene, CG15630 (a predicted cell surface receptor), is likely to be directly involved in the functioning of the CPG generating the pulse song pattern. Future studies should ascertain its functional role in the neurons that constitute the song CPG. Other genes (Sps2, CG34460), whose CNS-specific knockdown resulted in IPI reduction, are also worthy of detailed examination.
Keywords: candidate genes, central pattern generators, courtship song, Drosophila, locomotion, P element insertion, RNA interference
Abbreviations
- BDSC
Bloomington Drosophila Stock Center
- CNS
central nervous system
- CPG
central pattern generator
- CS
Canton-S strain
- FITC
fluorescein isothiocyanate
- Gal4
yeast transcription activator protein Gal4
- GFP
green fluorescent protein
- IPI
interpulse interval
- LA
locomotor activity
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PdL
Ponce de Leon P element construct
- RNAi
RNA interference
- shRNA
small hairpin RNA
- TRiP
Transgenic RNAi Project
- UAS
upstream activation sequence
- VDRC
Vienna Drosophila RNAi Center
Introduction
Motor output from animal nervous systems is formed by regulatory and trigger signals from sensory and integrative structures, and is dependent on the intrinsic properties of motor neurons and their interactions.1 Motor output may manifest as a brief burst of pulses (e.g., knee jerk) or a continuous rhythmically organized sequence of pulses (e.g., wing beats). The second type of activity is attributed to central pattern generators (CPGs) in the nervous system, either single-cell pacemakers or ensembles of interacting neurons creating rhythmic output. Pacemaker rhythmic activity is created by oscillations in membrane permeability for sodium, potassium and calcium ions, but elucidation of the molecular mechanisms controlling these oscillations is not yet complete.2-4
Drosophila melanogaster is a model organism with a simple nervous system. A powerful arsenal of genetic techniques has been assembled over the last hundred years of investigating the fruit fly. These advantages provide a system for discovering the molecular basis of rhythmic activity generation. Several new molecular components participating in motor functions, including CPG operation and regulation, have been described in Drosophila.5,6 However, the data in this field are fragmentary, and the approach of forward genetics, i.e., genetic screening for impairments of motor functions, may be very helpful in identifying genes involved in CPG functioning.
Many behaviors may be studied to investigate CPGs in Drosophila (e.g., walking, flight and sound production in adult flies or crawling in larvae). We chose walking and male courtship song because of the relative ease of performing mass testing of flies using simple automated techniques (see Methods).
The locomotor activity of single flies, as revealed by ‘open-field’ observations, is characterized by 2 different behavioral components, the ‘amount’ and ‘speed’ of movement.7 The ‘amount’ is determined by run initiation frequency and run duration. CPG malfunction can be reflected in both parameters. For example, mutation of the na gene, encoding a Na+ leak channel known to regulate pacemaker activity, makes flies walk in fits and starts, taking a few steps and then stopping before starting again.4,5,8 This motor phenotype would be characterized by high run initiation frequency and low run duration. Alteration of run speed may indicate changes in the motor pattern of leg coordination, which could be derived from defective CPG regulation. Mendes et al.9 reported that flies increasingly use tetrapod and non-canonical combinations instead of a tripod gait as they decrease their speed. Hence, an absent tripod gait in mutant lines could lead to a reduction in run speed.
The impulse component of the male D. melanogaster courtship song has a rhythmic repetitive structure of sound pulses varying from 2 to 50 pulses per train.10 The time between pulses is known as the interpulse interval (IPI), whose value is inversely correlated with the activation level of neurons of the putative song CPG.11 Thus, in contrast to locomotor parameters, the IPI pattern directly reflects the functioning of the song CPG.
Genetic studies of locomotor behavior in Drosophila have a long history starting with selection experiments12-17 and later with linkage analysis.7,18,19 To identify candidate genes affecting locomotor behavior, quantitative trait loci analysis21,22 and whole genome analysis of differences in expression levels between selection lines22 were used. With the same aim, Strauss23 performed a screen for ethyl methanesulfonate induced X-linked locomotor mutants. However, to date, Drosophila locomotor behavior in terms of CPG functioning has not been investigated and no genetic screen for autosomal genes affecting various locomotor parameters has been coupled with simultaneous investigation of courtship song parameters.
A great number of genetic studies of Drosophila courtship song, including genetic screens and analysis of numerous mutants, have been performed (for a review see ref. 24, and for recent advances in this field see ref. 25); however, the present paper shows that the list of genes having possible involvement in song CPG is not complete.
Here we report the identification of 22 Drosophila genes as candidates for involvement in the neural mechanisms responsible for motor functions. The 22 lines used for gene identification were isolated from a previously created collection of 1064 lines carrying a single random P element insertion (PdL transposon) in one of the autosomes. These 22 lines displayed extreme deviations in locomotor and/or courtship song parameters compared with the whole collection (the details and results of the behavioral screening will be published elsewhere).
The behavioral consequences of CNS-specific knockdowns of 10 identified genes were estimated using local RNA interference (RNAi) under the control of various GAL4 drivers (elav/nrv2/appl/tsh/repo-GAL4) in the Gal4/UAS system. Tissue-specificity and efficiency of RNAi were confirmed by GFP fluorescence localization in the embryo and quantitative assessment of target gene expression in imago (for genes CG15630, Sps2 and Mef2).
RNAi of each analyzed gene led to deviation of one or more parameters of locomotor behavior or courtship song. The results greatly depended on the suppressed gene and the CNS-specific driver used. The most prominent changes in the courtship song IPI, directly reflecting CPG functioning (motor pattern of wing vibration), were seen in flies with Sps2 and CG15630 RNAi. Absence of courtship song impairments in flies with glia-specific CG15630 knockdown and IPI reduction following estrogen-induced CG15630 RNAi at adult stage indicates that CG15630 codes for a product, which is necessary for the correct functioning of courtship song CPG neurons in adult flies.
Results
Identification of candidate genes in mutants with motor phenotypes
Location of the PdL transposon sequence and its direction were determined for selected lines with motor phenotypes. Of the 22 lines analyzed, each had a unique PdL transposon insertion site (Table 1). Insertion site locations were as follows: in 15 lines in non-coding regions of introns or exons of the genes, Cf2, Dgp-1, Ext2, lola, MESR4, Map205, Mef2, Sps2, Treh, wdp, yps, CG1943, CG5807, CG6746 and CG15630; in 2 lines in coding regions of the genes, jumu and CG8708; in one line in the cluster of transposons in the pericentromeric region (no gene); in 4 lines at various distances from the genes, drl, jing, Hsrω and CG34460.
Table 1.
Genomic location and orientation of PdL insertions in the mutant lines. Line ID - laboratory identifier of the mutant line. Designations in the second column are: XX:YYYYYYYY[+/-], where XX indicates the chromosome arm; YYYYYYYY indicates the Drosophila genome nucleotide number downstream of the PdL insertion; [+/-] indicates forward (5’-3’) or reverse (3’-5’) orientation of the PdL transposon in the chromosome. Genomic location and orientation are determined according to Flybase51 data released September 7th, 2010 (Dmel Release 5.30), sequenced strain y1; cn1 bw1 sp1 (Bloomington # 2057)
| Line ID | PdL genomic position and orientation | Candidate gene affected | Known or predicted gene products | Allele |
|---|---|---|---|---|
| 3724 | 2L:4882790[+] | Сf2 | transcription factor | Сf23724 |
| 663 | 2R:5842843[-] | Mef2 | transcription factor (predicted) | Mef2663 |
| 5282b | 2R:6429208[+] | lola | transcription factor (predicted) | lola5282b |
| 5493 | 3R:6182222[+] | jumu | transcription factor (predicted) | jumu5493 |
| 3494 | 2R:2389393[-] | jing | transcription factor | jing3494 |
| 2169 | 2R:13435745[+] | MESR4 | unknown | MESR42169 |
| 843К | 2R:14059049[+] | Dgp-1 | translation factor (predicted) | Dgp-1843К |
| 7081 | 3L:12118218[-] | yps | translation repressor | yps7081 |
| 3404а | 2L:4793956[+] | CG15630 | cell surface receptor (predicted) | CG156303404а |
| 3979a-s2 | 2R:18199553[-] | wdp | cell surface receptor (predicted) | wdp3979a-s2 |
| 2248 | 2L:19190341[-] | drl | receptor tyrosine kinase (predicted) | drl2248 |
| 3328b | 3R:20639718[-] | CG5807 | lipocalin-1 cell surface receptor (predicted) | CG58073328b |
| 5433-t3 | 3R:17122251[+] | Hsrω | regulatory RNA (predicted) | Hsrω5433-t3 |
| 6225a | 2L:20116133[+] | no gene | piRNA (predicted) | no gene |
| 3389 | 2L:12002261[-] | CG6746 | 3-hydroxyacyl-CoA dehydratases (predicted) | CG67463389 |
| 5769 | 2L:10342643[+] | Sps2 | monoselenphosphate synthetase (predicted) | Sps25769 |
| 5180-t3 | 3R:2901187[+] | CG1943 | unknown | CG19435180-t3 |
| 4653 | 2R:4047132[+] | CG8708 | β-1,3-galactosyltransferase | CG87084653 |
| 3290 | 2R:12716378[-] | CG34460 | unknown | CG344603290 |
| 5567a | 3R:27894136[-] | Map205 | microtubule binding protein | Map2055567a |
| 6387а-s2 | 2R:12036471[+] | Ext2/ CG10731 | glycosyltransferase (predicted)/ATP synthase subunit s | Ext26387а-s2/ CG107316387а-s2 |
| 4262с-s2 | 2R:16964321[+] | Treh | trehalase | Treh4262с-s2 |
Locomotor behavior in flies with CNS-specific knockdown of candidate genes
Parameters of locomotion were assessed in flies with suppressed CNS expression in 10 candidate genes (Sps2, CG15630, Dgp-1, CG6746, CG8708, CG34460, Mef2, lola, jing and drl). The selection of target genes was based on the availability of RNAi lines with attP landing sites from VDRC and TRiP collections. The GAL4/UAS system for post-transcriptional silencing of a target gene by means of local RNAi was used. The experimental flies were derived by crossing the VDRC (Vienna Drosophila RNAi Center) or TRiP (Transgenic RNAi Project) lines, which carry transgenes coding for target gene-specific interfering RNA under a UAS element, with lines carrying GAL4 CNS-specific drivers (elav/nrv2/appl/tsh-GAL4, Table 2). GAL4 control flies were derived from crosses of the same GAL4 lines with host strains for VDRC and TRiP RNAi lines, #60100 and #36303 respectively, which did not carry an interfering RNA transgene (see Materials and Methods). To exclude from analysis the motor deviations caused by genetic background differences between control and experimental lines and probable nonspecific leaking from RNAi transgene we tested UAS-RNAi control flies originated from crosses between wild-type strain Canton-S (CSBDSC) and RNAi line for each gene (Table2). The crosses between CSBDSC and host controls for RNAi lines (60100 and 36303) were used for comparison.
Table 2.
Collection and FlyBase identification numbers (ID) of transgenic flies used in experiments. w* - unspecified allele
| Stock type | Collection ID | Transgene | FlyBase ID | Genotype |
|---|---|---|---|---|
| Gal4 lines | 8760 | elav-GAL4 | FBst0008760 | w[*]; P{w[+mC] = GAL4-elav.L}3 |
| 8765 | elav-GAL4 | FBst0008760 | P{w[+mC] = GAL4-elav.L}2/CyO | |
| 6794 | nrv2-GAL4 | FBst0006794 | w[*]; P{w[+mC] = nrv2-GAL4.S}8 P{w[+mC] = UAS-GFP.S65T}eg[T10 | |
| 30546 | appl-GAL4 | FBst0030546 | P{w[+m*] = Appl-GAL4.G1a}1, y[1] w[*]; Mlf[Delta10]/CyO | |
| 3040 | tsh-GAL4 | FBst0003040 | y[1] w[1118]; P{w[+mW.hs] = GawB}tsh[md621]/CyO; P{w[+mC] = UAS-y.C}MC1/TM2 | |
| 25038 | GAL4.ER | FBst0025038 | w[*]; P{w[+mC] = hs-GAL4.ER}ER156 | |
| 7415 | repo-GAL4 | FBst0007415 | w[1118]; P{w[+m*] = GAL4}repo/TM3, Sb[1] | |
| TriP RNAi lines | 28699 | UAS-Mef2 | FBst0028699 | y1 v1; P{TRiP.JF03115}attP2 |
| 26714 | UAS-lola | FBst0026714 | y1 v1; P{TRiP.JF02254}attP2 | |
| 27024 | UAS-jing | FBst0027024 | y1 v1; P{TRiP.JF02345}attP2 | |
| 29602 | UAS-drl | FBst0029602 | y1 v1; P{TRiP.JF03281}attP2 | |
| VDRC RNAi KK lines | 105268 | UAS-Sps2 | FBst0477096 | y,w[1118];P{attP,y[+],w[3`]} |
| 107797 | UAS-CG15630 | FBst0479610 | y,w[1118];P{attP,y[+],w[3`]} | |
| 109410 | UAS-Dgp-1 | FBst0481099 | y,w[1118];P{attP,y[+],w[3`]} | |
| 103625 | UAS-CG6746 | FBst0475483 | y,w[1118];P{attP,y[+],w[3`]} | |
| 102288 | UAS-CG8708 | FBst0474157 | y,w[1118];P{attP,y[+],w[3`]} | |
| 110120 | UAS-CG34460 | FBst0481705 | y,w[1118];P{attP,y[+],w[3`]} | |
| host strain for TRiP RNAi lines | 36303 | No | FBst0036303 | y[1] v[1]; P{y[+t7.7] = CaryP}attP2 |
| host strain for VDRC KK RNAi lines | 60100 | No | No | y,w[1118];P{attP,y[+],w[3`]} |
| GFP line | 32202 | UAS-GFP | FBst0032202 | w[*]; P{w[+mC] = 10XUAS-IVS-GFP-WPRE}attP2 |
| CSBDSC | 1 | No | FBst0000001 | Wild type |
For VDRC and TRiP lines, comparison of UAS-RNAi controls with CSBDSC×60100 and CSBDSC×36303 host line hybrids respectively (see CS in Figs. 1, 2) showed own effect of UAS-RNAi transgene insertions. It was manifested as a decrease in run frequency (4 VDRC lines) and run speed (TRiP line for gene lola) and as an increase in run duration (2 other VDRC lines) and run speed (TRiP line for gene jing). For that reason, the knockdown effects coinciding with own effect of UAS-RNAi insertion (marked with asterisk in Figs. 1, 2) must be excluded from consideration. RNAi under tsh-GAL4 control caused lethality for Mef2 and lola and gross morphological wing defects for jing (Fig. 2).
Figure 1.
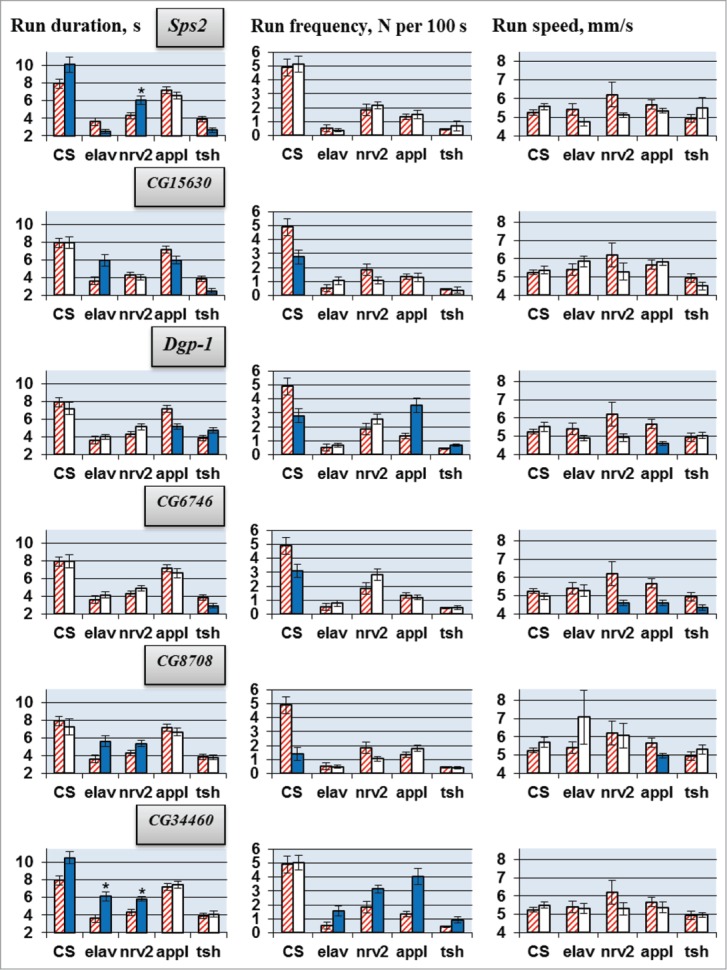
Locomotion parameters in flies with tissue-specific knockdown of the candidate genes (VDRC RNAi lines). GAL4/UAS-RNAi flies with spatially restricted knockdown (second column for each driver) derived by crossing elav/nrv2/appl/tsh-GAL4 drivers with VDRC RNAi lines. GAL4 controls without UAS-RNAi transgene (first, hatched columns) originate from cross of the same GAL4 drivers with host strain #60100 and are specific for each GAL4 driver. Also presented are locomotion parameters in UAS-RNAi controls descended from cross of CSBDSC with VDRC RNAi line for a gene indicated (CS, second columns) and in flies derived by crossing CSBDSC with host strain #60100 (CS, first, hatched column). Mean values with standard errors are shown. N = 40 for each data point. Significant difference from a corresponding control is indicated by filling (2-sided randomization test, 10,000 iterations, P < 0.05). Comparisons excluded from consideration (see text) are marked with asterisk. For details of genotypes see Methods and Table 2.
Figure 2.
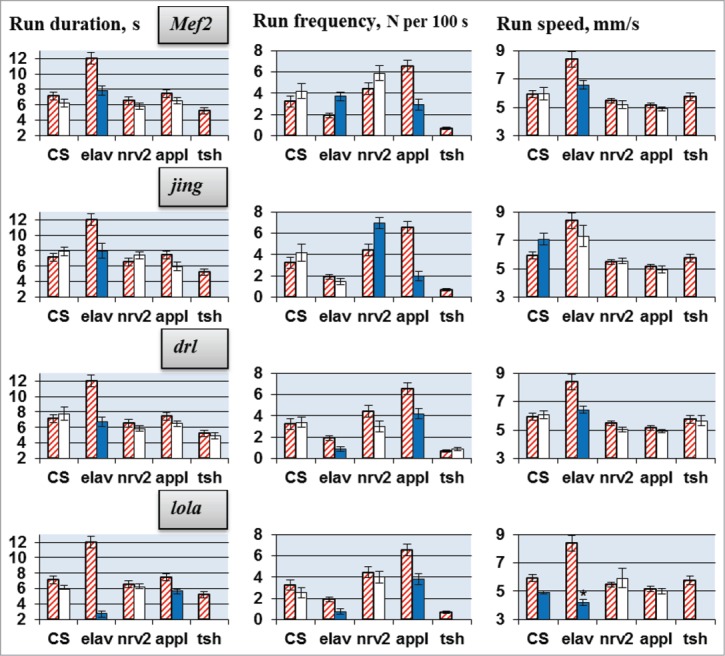
Locomotion parameters in flies with tissue-specific knockdown of the candidate genes (TRiP RNAi lines). Flies with spatially restricted knockdown derived by crossing elav/nrv2/appl/tsh-GAL4 drivers with TRiP RNAi lines. The host strain #36303 was used for all control crosses. Absence of a value indicates lethality or gross morphological effects of the knockdown. Other explanations, see legend for Figure 1.
RNAi of each candidate gene led to deviations in one or more locomotor parameters (Figs. 1, 2). The results depended greatly on the gene being suppressed and the CNS-specific driver used. Run duration and run frequency were especially variable. Under different drivers, RNAi of a target gene often caused opposite changes in these parameters, while RNAi under some drivers produced no effect. Run speed was more consistent. For all candidate genes, RNAi led to either no changes or to a decrease in run speed. For CG6746, 3 different drivers produced a decrease in run speed, while for all other genes this effect was only observed for one of the 4 drivers.
In general, we can conclude that all selected candidate genes are involved in the neural processes responsible for locomotion. However, their roles are determined by unique spatiotemporal patterns of gene expression in various neurons. This explains why the RNAi effects are strongly dependent on the GAL4 driver, which specifies the location, stage and intensity of gene suppression.
Courtship song parameters in flies with CNS-specific knockdown of candidate genes
Parameters of courtship song were assessed by CNS-specific RNAi knockdown of the candidate genes, Sps2, CG15630, CG6746, CG34460 and Mef2, which showed significant deviations in locomotor behavior and, according to the literature and bio-informational data, had more perspectives to be considered as a molecular players in CPG functioning. For all genes no own effect of UAS-RNAi transgene insertions was observed (Fig. 3). As for locomotion, deviations in courtship song parameters varied according to different ‘driver - target gene’ combinations. Similar to locomotion parameters, opposite changes occurred in pulse train duration and IPI according to the different GAL4 drivers used for RNAi. We paid special attention to IPI because this parameter directly reflects CPG functioning (motor pattern of wing vibration). Knockdown of all candidate genes with at least one GAL4 driver led to IPI deviations. The most prominent changes were seen in flies with RNAi of Sps2 or CG15630. Suppression of either gene resulted in IPI reduction.
Figure 3.
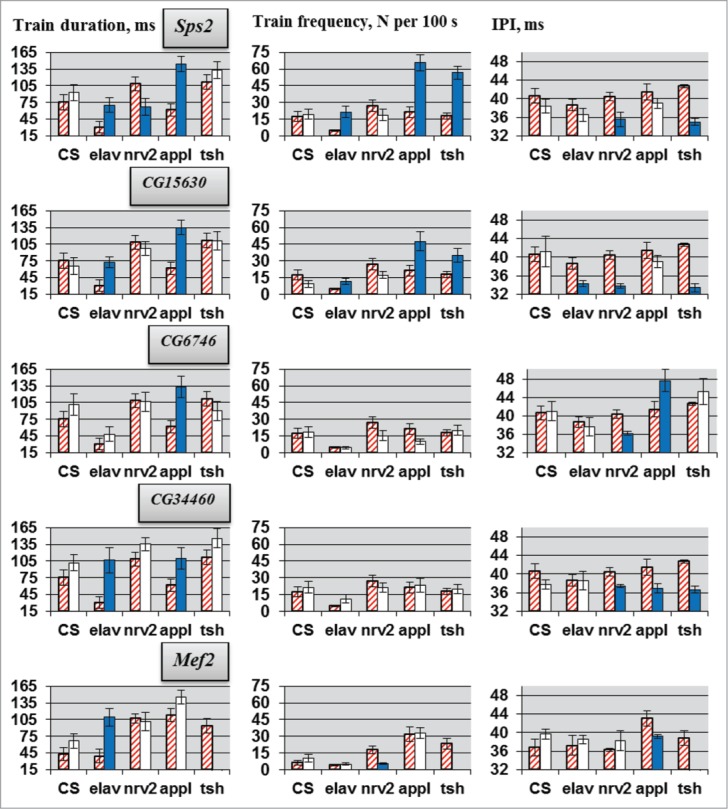
Courtship song parameters in flies with tissue-specific knockdown of the candidate genes. Flies with tissue-specific knockdown derived by crossing elav/nrv2/appl/tsh-GAL4 with VDRC/TRiP RNAi lines. N = 20 for each data point. Explanations are presented in legends for Figures 1 and 2.
Sps2, CG15630 and Mef2 expression levels in flies with tissue-specific knockdown
In many cases, behavioral deviations had opposite directions under control of different GAL4 drivers (Figs. 1–3). We chose 3 genes, most perspective for further investigations. The quantitative assessment of Sps2, CG15630 and Mef2 expression levels at the imago stage showed a reduction in the expression of each target gene (Fig. 4). However, the magnitude of suppression varied according to different ‘driver - target gene’ combinations. The differences in suppression levels are determined by the extent of concordance between endogenous expression patterns of candidate genes and drivers. For example, RNAi of CG15630 under elav-GAL4 leads to more pronounced suppression compared with Sps2 and Mef2. Indeed, according to the high-throughput data of Chintapalli et al.26 (cited in FlyAtlas27), the expression pattern of CG15630 in different adult tissues is more similar to the elav expression pattern than that of Sps2.
Figure 4.
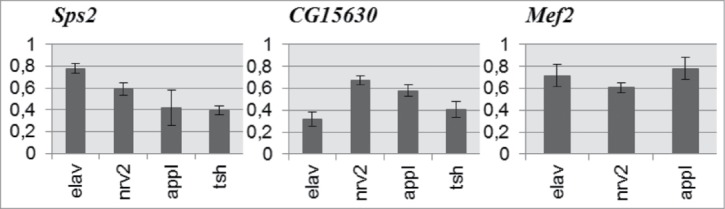
Expression level of Sps2, CG15630 and Mef2 genes in RNAi-mediated knockdowns relative to control. Mean expression ratios (knockdown/control) with standard errors are shown. Expression levels of all candidate genes are reduced in all experimental samples (2-sided randomization test, p < 0.05, REST 2009 software).
We also analyzed GFP fluorescence under the control of elav/appl/nrv2/tsh-GAL4 drivers at embryo stage 13–16 to confirm tissue-specificity of expression patterns. All drivers showed expression patterns consistent with previously published results.28-31 elav- and appl-GAL4 produced GFP fluorescence in the brain and ventral ganglion, while tsh-GAL4 expression was mainly in the ventral ganglion (data not shown).
Locomotor and courtship song parameters in flies with glia-specific Sps2 or CG15630 knockdown
For each candidate gene, the effect of RNAi on motor activity was strongly dependent on the CNS-specific GAL4 driver used (see the run and pulse train durations in Sps2 knockdowns, Figs. 1, 3). elav and nrv2 are both reported to be expressed in neurons and glia.32,33 Conversely, the appl driver was used for selective expression in neuronal structures but not in glia.34 Therefore, we tested flies with RNAi under control of the glia-specific repo-GAL4 driver to determine the consequences of glial suppression of the candidate genes. We revealed that knockdown of Sps2 or CG15630 in glia has no significant effect on either locomotor and courtship song parameters, except reduction in run frequency for CG15630 knockdown (Fig. 5). Importantly, it was not observed for neurospecific drivers but was revealed as nonspecific alteration in flies from cross of CSBDSC with RNAi line for CG15630 (Fig. 1). Thus, the effect of CG15630 RNAi on motor functions seems to be related to underexpression of CG15630 in neurons rather than in glia.
Figure 5.
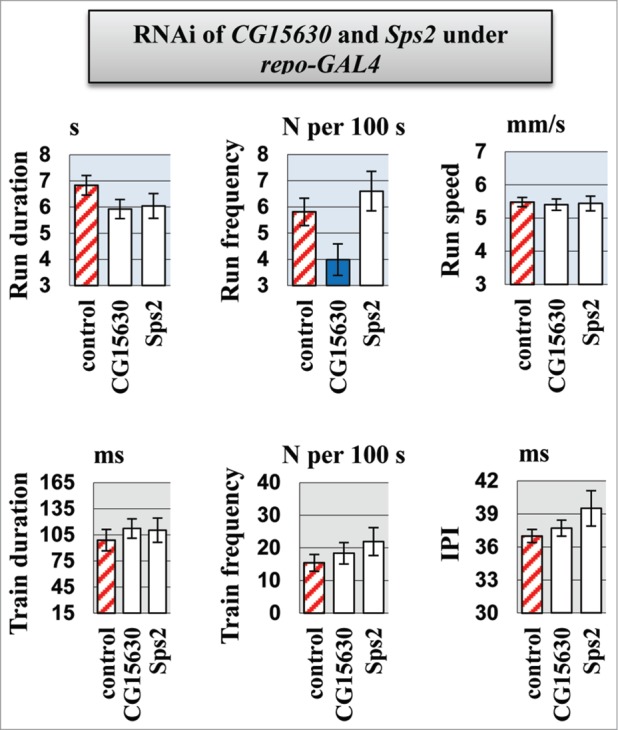
Motor parameters in flies with knockdown of CG15630 under the glia-specific repo-Gal4 driver. Glia-specific knockdown flies derived by crossing repo-GAL4 driver with VDRC RNAi lines #107797 for CG15630 and #105268 for Sps2. Control flies derived by crossing repo-GAL4 driver with host line #60100. Mean values and standard errors are shown. N = 40 for locomotor parameters, N = 20 for courtship song parameters. Significant difference from a corresponding control is indicated by filled columns (2-sided randomization test, 10,000 iterations, p < 0.05).
Tissue-specificity of repo-GAL4 driver expression was assessed by observing UAS-GFP expression in stage 13–16 embryos (data not shown). The repo-GAL4 expression pattern was consistent with previously described results.35
Locomotor and courtship song parameters in flies with estrogen-induced knockdown of CG15630 in adults
CNS-specific RNAi of CG15630 caused significant deviations in locomotor (Fig. 1) and courtship song (Fig. 3) parameters, the most prominent being for IPI. This may result from RNAi effects at either developmental or imago stages. Therefore, using the estrogen-induced GAL4 driver (GAL4.ER), we tested motor activity in flies with CG15630 RNAi turned on at the imago stage.
Knockdown of CG15630 in adult flies resulted in deviations of both locomotor and courtship song parameters: increasing the run duration and decreasing the run frequency and IPI (Fig. 6). Similar effects were observed for RNAi under some of the constitutive drivers (Figs. 1, 3). Neither estrogen-induced nor constitutive RNAi of CG15630 changed the run speed (Figs. 1, 6).
Figure 6.
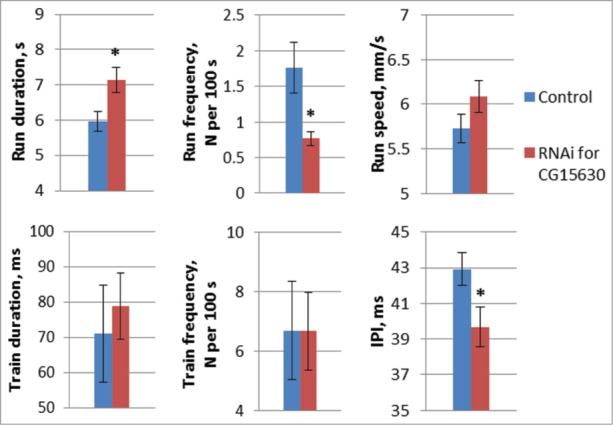
Motor parameters in flies with estrogen-induced knockdown of CG15630 in adults. Estrogen-induced knockdown flies derived by crossing GAL4.ER driver with VDRC RNAi line #107797 for CG15630. Control flies derived by crossing GAL4.ER driver with host line #60100. Both experimental and control flies were kept on estrogen-containing medium from the moment of imago eclosion. Mean values and standard errors are shown. N = 40 for locomotor parameters, N = 20 for courtship song parameters. Significant differences between RNAi and control data are marked with asterisks (2-sided randomization test, 10,000 iterations, P < 0.05).
Given that CG15630 is expressed predominantly in the CNS26,27 and that GAL4.ER drives expression in the CNS,36 and taking into account the conclusions in the previous section, we can assert that neuronal expression of CG15630 at the imago stage is required for the neural mechanisms that determine run duration and the mechanisms responsible for formation of pulse song rhythmicity (IPI). It is very probable that CG15630 RNAi influences pulse song patterning through participation of the gene product in the molecular mechanism of song CPG.
Estrogen-induced knockdown of CG15630 did not lead to changes in train duration or frequency (Fig. 6), changes that were produced by some of the non-induced CNS-specific knockdowns (Fig. 3). Therefore, it is likely that, in the latter case, deviations in these parameters were caused by developmental abnormalities.
CG15630 suppression in flies with estrogen-induced knockdown was confirmed by quantitative assessment of gene expression in adults, mean expression ratio (knockdown/control) was 0.649, standard error ±0.044.
Discussion
The genes identified as candidates for involvement in motor functions (Table 1) encode 8 transcription or translation factors, 4 cell surface receptor components, 2 regulatory RNAs, a microtubule binding protein and various metabolic enzymes. To determine CNS-specific effects of selected genes on motor functions, we assessed locomotor (10 genes) and courtship song (5 genes) parameters in flies with tissue-specific knockdowns.
Comparison of cross between the wild-type strain CSBDSC and host strain (60100 or 36303) with crosses between CSBDSC and RNAi lines (Fig. 1, 2, the first pair of columns on each graph) revealed several differences in locomotion parameters. The descendants of 2 TRiP RNAi lines demonstrated deviations in run speed, while descendants of VDRC RNAi lines showed decreased run frequency or increased run duration. These deviations could be caused either by own effect of UAS-RNAi transgenes or by difference in genetic background between the host strains and RNAi lines. Recently Green et al.37 reported that transgenic RNAi-inducing lines from the VDRC libraries are subject to dominant phenotypic effects. Specifically, on crossing 39 randomly selected KK lines to the panneuronal driver elav-GAL4c155, 9 of them produced F1 progeny unable to properly inflate their wings. They found that the main integration site for pKC26 vector (carrying the shRNA sequences) in the VDRC KK library is the non-annotated pKC43 target (occupied in all 39 lines tested), whereas only the 9 lines displaying the elav-GAL4c155-dependent non-inflating wing phenotype were found to have a pKC26 integration into the annotated pKC43 insertion. In our experiments we did not observe any gross morphological or wing defects in experimental flies generated by crossing GAL4 and VDRC RNAi lines.
RNAi of lola and drl, under the control of elav/appl-GAL4 drivers, repressed locomotion (run duration and frequency, Fig. 2). lola and drl, code for a transcription factor and a receptor tyrosine kinase, respectively, and are important determinants of motor activity. Feminization of cells expressing lola produced a ‘no song’ phenotype,25 and drl was identified to affect locomotor reactivity in a quantitative trait loci study.21 At the same time, mutations in these genes led to structural defects in the adult brain.38-40 Therefore, the probable explanation of locomotion reduction in lola and drl RNAi knockdowns is the disturbance of regulatory inputs from the brain to the locomotor CPG, but not a malfunction of the latter.
Knockdown of 2 other candidate genes encoding transcription factors, Mef2 and jing, also reduced run duration, but only under elav-GAL4 control; different drivers produced opposite deviations in run frequency. Contra-directional deviations, depending on the driver used, were also found for run duration in flies with RNAi of CG15630 and Dgp-1 (Fig. 1, increase in run duration in Sps2 knockdown under nrv2-GAL4 is not taken into consideration due to coincidence of its effect with own effect of UAS-RNAi transgene insertion). Contrary effects of elav-GAL4 and appl-GAL4 on physiological parameters were reported earlier. Simonsen et al.41 noted that enhanced expression of Atg8a (autophagy-related gene) under appl-GAL4 control produced a dramatic extension of adult longevity, but under elav-GAL4 control the lifespan was, conversely, reduced. The authors suggested that this difference may be related to a difference in age specificity of gene expression under these drivers, suggesting that the timing of Atg8a expression in the aging CNS is critical for its ability to extend longevity.
The designers of nrv2-GAL4 31 revealed a few notable differences between patterns of nrv2-GAL4- and elav-GAL4-directed GFP fluorescence that could explain contrary effects of the drivers. Fluorescence was not seen in many peripheral sensory neurons in the nrv2-GAL4/GFP flies in late embryo and larval stages, in developing photoreceptor cell neurons and in the third-instar larval eye disks, fluorescence that was observed in the elav-GAL4/GFP flies. Moreover, Dumstrei et al.42 expressed a target gene (DE-cadex) specifically in cortical glia of the larval brain using the nrv2-Gal4 driver and specifically in cortical neurons using the elav-GAL4 driver.
Unlike for run duration and frequency, RNAi knockdowns caused only decreases in run speed. Almost all mutants with structural defects of the central complex walked more slowly than wild-type flies.43 Strauss et al.44 reported that reduced walking speed in nob flies, which have a medially lesioned protocerebral bridge, was caused by their inability to increase step length concomitant with an increase in stepping frequency. In contrast, nob swing phases were of normal duration and frequency throughout the whole frequency range, because timing is most probably a function of the thoracic ganglia. Accordingly, we did not observe (with the exception of CG6746) speed alterations in RNAi flies under the control of tsh-Gal4, which is expressed in the thoracic segments,45 predominantly in the CNS26,27 where the insect locomotor CPG is most likely situated.46,47
Compared with locomotion, deviations in the courtship song parameters were more uniform under the different GAL4 drivers (Fig. 3). Contra-directional alterations in pulse train duration were observed only in males with Sps2 RNAi, while IPI was oppositely changed only in males with CG6746 RNAi. It should be noted that CG6746 suppression under tsh-Gal4 control did not significantly influence IPI. Therefore, it seems likely that the product of this gene (a predicted 3-hydroxyacyl-CoA dehydratase) does not participate in the functioning of the song CPG,48 while products of the genes, Sps2 (a predicted monoselenphosphate synthetase), CG15630 (see below), CG34460 (product unknown) and, probably, Mef2 (a predicted transcription factor) do. The conclusion for Mef2 is uncertain because the tsh-Gal4/Mef2 combination is lethal. The most prominent IPI changes were observed for CG15630, a CNS-specific gene26,27 that encodes a protein-binding protein, with immunoglobulin-like and fibronectin type III domains (accession number Q9VR25).49 In contrast to other drivers, CG15630 RNAi under appl-Gal4 control did not produce a significant IPI reduction. Because appl is not expressed in glia, it is possible that IPI reduction in CG15630 RNAi flies is associated with abnormality of glial cells but not neurons. However, CG15630 RNAi under glia-specific repo-Gal4 control does not affect courtship song parameters, indicating neuronal effects of the knockdowns on impulse song.
In principle, changes in courtship song parameters may result from both developmental effects of RNAi and malfunction of the neural structures determining song CPG activity in adults. Conditional estrogen-induced CG15630 RNAi in adults causes IPI reduction (Fig. 6), suggesting direct involvement of CG15630 in the functioning of the song CPG. Absence of alterations in train frequency and duration following RNAi under GAL4.ER control indicates that deviations of these parameters in flies with gene suppression under other drivers (Fig. 3) are possibly associated with developmental defects. However, it cannot be excluded that the difference between consequences of induced and persistent CG15630 RNAi is determined by the distinctive expression pattern of GAL4.ER.
The effects of CG15630 RNAi driven by GAL4.ER on courtship song production are similar to those driven by tsh- and nrv2-Gal4 (Fig. 3). tsh is predominantly expressed in the thoracic-abdominal ganglion, so tsh-Gal4-driven RNAi can directly affect CPG functioning but not brain regulatory mechanisms. The expression of the nrv2-Gal4 driver is controlled by transcriptional regulatory elements present in the flanking DNA of the Drosophila Na+,K+-ATPase β-subunit gene, Nervana2.31 In imagoes, nrv2-Gal4-driven expression is more pronounced than that driven by appl- and elav-Gal4.50,51 Together, these data indicate that CG15630 is an important genetic determinant whose expression in adults is necessary for proper functioning of the courtship song CPG. Von Philipsborn et al.11 discovered a class of fru neurons (vPR6) in Drosophila mesothoracic ganglia whose activity was both necessary and sufficient for robust song production. In addition, they reported that the level of vPR6 activity had tight inverse coupling with IPI. Therefore, IPI reduction in flies with CG15630 RNAi under tsh-Gal4, nrv2-Gal4 and GAL4.ER control may be related to reduced activity of vPR6. Future studies will ascertain the role of the CG15630 gene product in the functioning of vPR6 and other fru neurons, which comprise the courtship song CPG.48
Conclusions
The product of the CNS-specific gene, CG15630 (a predicted cell surface receptor), is likely to be directly involved in the functioning of the CPG generating the pulse song pattern. Future studies should ascertain its functional role in the neurons that produce the song CPG. Other genes (Sps2, CG34460), whose CNS-specific knockdown resulted in IPI reduction, are also worthy of detailed examination.
Materials and Methods
Drosophila stocks
The 22 lines used for gene identification were isolated from a collection previously created by the authors. This consisted of 1064 lines carrying a single random P element insertion (PdL transposon) in one of the autosomes. The 22 lines displayed extreme deviations in locomotor and/or courtship song parameters compared with the whole collection (the results of behavioral screening will be published elsewhere).
Tissue-specific suppression of the candidate genes was achieved by synthesis of interfering RNA using the GAL4/UAS system.52 The GAL4/UAS flies were produced by crossing females of GAL4 lines carrying various GAL4 transgenes under CNS specific promotors (drivers) and males of RNAi lines carrying interfering RNA transgenes under UAS (upstream activation sequence) control (Table 2). All GAL4 lines were obtained from the Bloomington Drosophila Stock Center (BDSC), including lines with various CNS-specific drivers: elav-GAL4, nrv2-GAL4, appl-GAL4, tsh-GAL4, GAL4.ER and the glia-specific driver, repo-GAL4. phiC31-based RNAi lines with site specific insertion53 of shRNA sequences within the vector were obtained from the Vienna Drosophila RNAi Center (VDRC KK lines) and the Transgenic RNAi Project (TRiP lines). The GAL4 controls, specific for each driver, were produced by crossing the corresponding GAL4 line and the line used for creating RNAi lines, without an interfering RNA transgene (Table 2, host strains for VDRC KK and TRiP RNAi lines, #60100 and #36303 respectively). Additionally, we tested flies from crosses between wild-type strain Canton-S (CSBDSC) and RNAi line for each gene (Table 2), as well as hybrids between CSBDSC and host controls for RNAi lines, 60100 and 36303. In GFP fluorescence experiments, UAS-GFP stock from BDSC was used (Table 2).
Females of the outbred wild-type strain Canton-S (CSPIF) have been maintained as mass-bred stock for several decades in the Pavlov Institute of Physiology in Koltushi (St. Petersburg) and were used as a standard opponent for male courtship in experiments with courtship song registration.
Experimental conditions
Flies were raised and kept on standard yeast-raisin medium at 25°C and under a 12-h light/dark cycle. For the locomotion test, experimental males were collected using CO2 anesthesia soon after eclosion and kept in groups of 30 flies in culture vials with yeast-raisin medium for 3 d until the experiment was performed. For courtship song registration, males were collected without anesthesia and kept individually. Females (CSPIF) were collected as virgins (10–12 flies per vial). A day before the experiment, 10–12 3-day-old females were brought together with 15–18 3-day-old males (CSPIF) in one vial and left to mate for 18–22 h. Behavioral assays were performed at 25°C between 9 a.m. and 6 p.m.
Hormone treatment
β-estradiol (Sigma-Aldrich International GmbH, cat. # Е8875) was dissolved in dimethyl sulfoxide (Sigma-Aldrich International GmbH, cat. # D5879) at a concentration of 260 mg/ml and mixed 1:3 (vol/vol) with wet yeast paste. From the moment of eclosion, males were maintained in culture vials with yeast-raisin medium and a drop of yeast (with or without hormone), refreshed daily.
Locomotion and courtship song assays and analysis
Individual males were freely moving in Perspex experimental chambers (15 mm diam., 5 mm high), which had transparent Perspex covers and a lateral entry (3 mm diam.) with stopper. Before locomotion recording, flies were agitated by chamber shaking. The coordinates of 20 individual male flies were simultaneously recorded for 1 h at a frequency of 10 Hz using 2 web cameras and Drosophila tracks software (© N.G. Kamyshev). Data processing was performed by the analysis module of Drosophila tracks. The percentage of time spent in locomotion (LA index), run frequency (number of runs per 100 s), run duration (s), run speed (mm/s) and other parameters of locomotor behavior were calculated for each fly. The flies were tested in duplicate (2 × 20 males).
Courtship song was recorded in a soundproof room for 5 min from 4 males simultaneously, each male being placed together with a fertilized CS female in a Perspex chamber with a latticed bottom on top of a microphone in a foam box, which functioned as an anechoic chamber. The sound signal was filtered, cutting off frequencies below 100 and above 800 Hz, processed with an analog-to-digital converter at 11025 Hz and saved as a sound file. The software Drosophila courtship song analysis (DCSA, © N.G. Kamyshev) autodetects the pulse song in sound recordings and allows manual editing of the results for automatic recognition. The DCSA analysis module performed primary data processing and created data tables for experimental groups. The calculated parameters involved percentage of time spent in pulse song (pulse song index), the frequency of pulse trains (trains per 100 s), number of pulses in a train, train duration (ms), intertrain interval (ms) and interpulse interval within a train (IPI, ms).
Statistical comparisons were made using the randomization test at significance level α = 0.05.54 We did not use the Bonferroni method to adjust statistical significance for the number of tests that have been performed in the study because we share the opinion that it creates more problems than it solves.55
Mapping of the P element insertion site by inverse PCR and DNA sequencing
Genomic DNA was prepared from 20 adult flies as described in Kim et al.56 Genomic DNA equivalent to 2 adult flies was digested with TaqI (SibEnzyme Ltd., cat. #E133) or BstKTI (SibEnzyme Ltd., cat. #E119) and ligated with T4 DNA ligase (SibEnzyme Ltd., cat. #E319). The ligation mixture was used as a template for PCR. HSP (5′CTGCAGATTGTTTAGCTTGTTC3′) and IRS (5′CGGGACCCACCTTATGTTAT3′) served as PCR primers (Beagle). The PCR products were run on agarose gels, eluted, and sequenced with the HSP primer using an Applied Biosystems 3130 Genetic Analyzer (USA).
Real-time reverse transcription PCR analysis
Target gene expression levels were assessed using real-time reverse transcription PCR. Total RNA was isolated from 30 adult males using TRIzol,57 treated with DNaseI and reverse transcribed by M-MuLV (SibEnzyme Ltd., cat #E317) reverse transcriptase and random hexamer primer. Quantitative real-time PCR was performed on a StepOnePlus (Applied Biosystems, Inc., USA) using EvaGreen® fluorescent dye (Biotium, Inc., BT-31000). Baseline and cycle threshold values were determined by automated threshold analysis with StepOne software v.2.0 (Applied Biosystems, USA). Both non-template and non-reverse transcribed RNA samples were used as negative controls for each sample. Expression levels of the RpL32 transcript were used as an internal control, and relative mRNA levels were calculated using the comparative ΔCt method. Primers used (Beagle) were CG15630-RA (5′-ATTCGTTGAGATTCTCGCAATGCG-3′ / 5′-CGGCGATTTCCAATGGAGCT-3′), Sps2-RA (5′-TGAGAAGGAACGCGACGTTGTG-3′ / 5′-GCTGCGCGTTTGACGGTAGTATTAT-3′), Mef2-RA (5′;-TGTACCAGTACGCCAGCACCGA-3′ / 5′-TTGTACTTGGCCTCCGTTCGC-3′) and RpL32-RA (5′-TATGCTAAGCTGTCGCACAAATGGC-3′ / 5′-GTTCTGCATGAGCAGGACCTCCA-3′).
Data were statistically analyzed using the randomization test at a significance level of α = 0.05 using the free software REST 2009.58
Wide field fluorescence microscopy
GAL4 driver expression patterns were assayed in GFP fluorescence experiments (at 25°C). Tissue-specific GAL4 lines were crossed with the UAS-GFP line. Embryos from the cross were dechorionated with 50% bleach and mounted in a 1:1 mixture of glycerol/PBS in a small viewing chamber constructed of a filter paper support on a standard coverslip. Samples were observed with a Mikmed 2 fluorescence microscope (LOMO, Russia) under Hg illumination using standard FITC fluorescence filters. Digital images were recorded using a CХ05 Baumer Optronic camera (Switzerland). Image files were processed using PHOTOSHOP (Adobe Systems).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. John Tower (University of Southern California, Los Angeles, USA) for providing us with PdL(X)A and other Drosophila stocks from his laboratory, the Bloomington Drosophila Stock Center (NIH P40OD018537) at Indiana University (USA), the Vienna Drosophila RNAi Center at Campus Science Support Facilities (Austria) and TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing us with transgenic GAL4 and RNAi fly stocks. We are grateful to Dr. Konstantin G. Iliadi (The Hospital for Sick Children, Toronto, ON, Canada) for propagation, maintenance and transcontinental shipment of these stocks to our laboratory.
Funding
The study was supported by a grant from the Ministry of Education and Science of the Russian Federation (contract P316), the programs of the Presidium of the Russian Academy of Sciences #7 and #30, and grants #13–04–12030 and #13–04–02153 from the Russian Foundation for Basic Research.
References
- 1. Briggman KL, Kristan WB. Multifunctional pattern-generating circuits. Annu Rev Neurosci 2008; 31:271-94;PMID:18558856; http://dx.doi.org/ 10.1146/annurev.neuro.31.060407.125552 [DOI] [PubMed] [Google Scholar]
- 2. Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol 2007; 69:291-316; PMID:17009928; http://dx.doi.org/ 10.1146/annurev.physiol.69.031905.161516 [DOI] [PubMed] [Google Scholar]
- 3. Kiehn O. Development and functional organization of spinal locomotor circuits. Curr Opin Neurobiol 2011; 21:100-9; PMID:20889331; http://dx.doi.org/ 10.1016/j.conb.2010.09.004 [DOI] [PubMed] [Google Scholar]
- 4. Lu TZ, Feng Z-P. NALCN: A regulator of pacemaker activity. Mol Neurobiol 2012; 45:415-23; PMID:22476981; http://dx.doi.org/ 10.1007/s12035-012-8260-2 [DOI] [PubMed] [Google Scholar]
- 5. Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol 2002; 12:2152-8; PMID:12498692; http://dx.doi.org/ 10.1016/S0960-9822(02)01358-1 [DOI] [PubMed] [Google Scholar]
- 6. Banerjee S, Lee J, Venkatesh K., Wu C-F, Hasan G. Loss of flight and associated neuronal rhythmicity in inositol 1,4,5-trisphosphate receptor mutants of Drosophila. J Neurosci 2004; 24:7869-78; PMID:15356199; http://dx.doi.org/ 10.1523/JNEUROSCI.0656-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burnet B, Burnet L, Connolly K, Williamson N. A genetic analysis of locomotor activity in Drosophila melanogaster. Heredity 1988; 61:111-9; http://dx.doi.org/ 10.1038/hdy.1988.96 [DOI] [Google Scholar]
- 8. Krishnan KS, Nash HA. A genetic study of the anesthetic response: Mutants of Drosophila melanogaster altered in sensitivity to halothane. Proc Nati Acad Sci USA 1990; 87:8632-6; PMID:2122464; http://dx.doi.org/ 10.1073/pnas.87.21.8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mendes C, Bartos I, Akay T, Marka S, Mann RS. Quantification of gait parameters in freely walking wild type and sensory deprived Drosophila melanogaster. eLife 2013; 2:e00231; PMID:23326642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ewing AW, Bennet-Clark HC. The courtship songs of Drosophila. Behaviour 1968; 31:288-301; http://dx.doi.org/ 10.1163/156853968X00298 [DOI] [Google Scholar]
- 11. Von Philipsborn AC, Liu T, Yu JY, Masser C, Bidaye SS, Dickson BJ. Neuronal control of Drosophila courtship song. Neuron 2011; 69:509-22; PMID:21315261; http://dx.doi.org/ 10.1016/j.neuron.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 12. Ewing AW. Attempts to select for spontaneous activity in Drosophila melanogaster. Anim Behav 1963; 11:369-78; http://dx.doi.org/ 10.1016/S0003-3472(63)80128-1 [DOI] [Google Scholar]
- 13. Ewing AW. Genetics and activity in D. melanogaster. Experientia 1967; 23:330-2; PMID:6065753; http://dx.doi.org/ 10.1007/BF02144495 [DOI] [PubMed] [Google Scholar]
- 14. Connolly K. Locomotor activity in Drosophila. II. Selection for active and inactive strains. Anim Behav 1966; 14:444-9; PMID:5972802; http://dx.doi.org/ 10.1016/S0003-3472(66)80043-X [DOI] [PubMed] [Google Scholar]
- 15. Connolly K. Locomotor activity in Drosophila III. A distinction between activity and reactivity. Anim Behav 1967; 15:149-52; PMID:6031102; http://dx.doi.org/ 10.1016/S0003-3472(67)80026-5 [DOI] [PubMed] [Google Scholar]
- 16. Luchnikova EM. The nature of genetic determination of general locomotor activity in Drosophila melanogaster. Genetica (Russ.) 1966; 6: 36-46; [in Russian] [Google Scholar]
- 17. van Dijken FR, Scharloo W. Divergent selection on locomotor activity in Drosophila melanogaster. I. Selection response. Behav Genet 1979a; 9:543-53; PMID:122270; http://dx.doi.org/ 10.1007/BF01067350 [DOI] [PubMed] [Google Scholar]
- 18. Luchnikova EM, Borisov GV. Identification of the linkage group responsible for interstrain differences in the level of locomotor activity in Drosophila. Bulletin of Leningrad State University 1971; 4:67-75 [Google Scholar]
- 19. van Dijken FR, van Sambeek MP, Scharloo W. Divergent selection on locomotor activity in Drosophila melanogaster. III. Genetic analysis. Behav Genet 1979b; 9:563-70; PMID:122272; http://dx.doi.org/ 10.1007/BF01067352 [DOI] [PubMed] [Google Scholar]
- 20. Costa R, Zonta LA, Osti M, Jayakar SD. Genetic analysis of adult locomotor activity in Drosophila melanogaster. Heredity 1989; 63:107-17; http://dx.doi.org/ 10.1038/hdy.1989.81 [DOI] [Google Scholar]
- 21. Jordan KW, Morgan TJ, Mackay TF. Quantitative trait loci for locomotor behavior in Drosophila melanogaster. Genetics 2006; 174:271-84; PMID:16783013; http://dx.doi.org/ 10.1534/genetics.106.058099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jordan KW, Carbone MA, Yamamoto A, Morgan TJ, Mackay TF. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol 2007; 8:R172; PMID:17708775; http://dx.doi.org/ 10.1186/gb-2007-8-8-r172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strauss R. A screen for EMS-induced X-linked locomotor mutants in Drosophila melanogaster. J Neurogenet 1995; 10:53-4 [Google Scholar]
- 24. Tauber E, Eberl DF. Acoustic communication in Drosophila. Behavioural Processes 2003; 64:197-210; http://dx.doi.org/ 10.1016/S0376-6357(03)00135-9 [DOI] [Google Scholar]
- 25. Moran CN, Kyriacou CP. Functional neurogenomics of the courtship song of male Drosophila melanogaster. Cortex 2009; 45:18-34; PMID:19054505; http://dx.doi.org/ 10.1016/j.cortex.2008.05.010 [DOI] [PubMed] [Google Scholar]
- 26. Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 2007; 39:715-20; PMID:17534367; http://dx.doi.org/ 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- 27. FlyAtlas The Drosophila gene expression atlas. ; [ http://flyatlas.org] [Google Scholar]
- 28. Luo LQ, Martin-Morris LE, White K. Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci 1990; 10:3849-61; PMID:2125311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fasano L, Röder L, Coré N, Alexandre E, Vola C, Jacq B, Kerridge S. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell 1991; 64:63-79; PMID:1846092; http://dx.doi.org/ 10.1016/0092-8674(91)90209-H [DOI] [PubMed] [Google Scholar]
- 30. Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev 1994; 8:1787-802; PMID:7958857; http://dx.doi.org/ 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- 31. Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci USA 1999; 96:10438-43; PMID:10468627; http://dx.doi.org/ 10.1073/pnas.96.18.10438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun B, Xu P, Wang W, Salvaterra PM. In vivo modification of Na(+),K(+)-ATPase activity in Drosophila. Comp Biochem Physiol B, Biochem Mol Biol 2001; 130:521-36; PMID:11691629; http://dx.doi.org/ 10.1016/S1096-4959(01)00470-5 [DOI] [PubMed] [Google Scholar]
- 33. Berger C, Renner S, Lüer K, Technau GM. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn 2007; 236:3562-8; PMID:17994541; http://dx.doi.org/ 10.1002/dvdy.21372 [DOI] [PubMed] [Google Scholar]
- 34. Fang Y, Soares L, Bonini NM. Design and implementation of in vivo imaging of neural injury responses in the adult Drosophila wing. Nat Protoc 2013; 8:810-9; PMID:23589940; http://dx.doi.org/ 10.1038/nprot.2013.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reifegerste R, Schreiber J, Gülland S, Lüdemann A, Wegner M. mGCMa is a murine transcription factor that overrides cell fate decisions in Drosophila. Mech Dev 1999; 82:141-50; PMID:10354478; http://dx.doi.org/ 10.1016/S0925-4773(99)00027-1 [DOI] [PubMed] [Google Scholar]
- 36. Han DD, Stein D, Stevens LM. Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development 2000; 127:573-83; PMID:10631178 [DOI] [PubMed] [Google Scholar]
- 37. Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nature methods 2014; 11: 222-3; PMID:24577271; http://dx.doi.org/ 10.1038/nmeth.2856 [DOI] [PubMed] [Google Scholar]
- 38. Yamamoto A, Zwarts L, Callaerts P, Norga K, Mackay TF, Anholt RR. Neurogenetic networks for startle-induced locomotion in Drosophila melanogaster. Proc Natl Acad Sci USA 2008; 105:12393-8; PMID:18713854; http://dx.doi.org/ 10.1073/pnas.0804889105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moreau-Fauvarque C, Taillebourg E, Boissoneau E, Mesnard J, Dura JM. The receptor tyrosine kinase gene linotte is required for neuronal pathway selection in the Drosophila mushroom bodies. Mech Dev 1998; 78:47-61; PMID:9858681; http://dx.doi.org/ 10.1016/S0925-4773(98)00147-6 [DOI] [PubMed] [Google Scholar]
- 40. Yao Y, Wu Y, Yin C, Ozawa R, Aigaki T, Wouda RR, Noordermeer JN, Fradkin LG, Hing H. Antagonistic roles of Wnt5 and the Drl receptor in patterning the Drosophila antennal lobe. Nat Neurosci 2007; 10:1423-32; PMID:17934456; http://dx.doi.org/ 10.1038/nn1993 [DOI] [PubMed] [Google Scholar]
- 41. Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 2008; 4:176-84; PMID:18059160; http://dx.doi.org/ 10.4161/auto.5269 [DOI] [PubMed] [Google Scholar]
- 42. Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci 2003; 23:3325-35; PMID:12716940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J Neurosci 1993; 13:1852-61; PMID:8478679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strauss R, Hanesch U, Kinkelin M, Wolf R, Heisenberg M. No-bridge of Drosophila melanogaster: portrait of a structural brain mutant of the central complex. J Neurogenet 1992; 8:125-55; PMID:1460532; http://dx.doi.org/ 10.3109/01677069209083444 [DOI] [PubMed] [Google Scholar]
- 45. Roder L, Vola C, Kerridge S. The role of the teashirt gene in trunk segmental identity in Drosophila. Development 1992; 115:1017-33; PMID:1360402 [DOI] [PubMed] [Google Scholar]
- 46. Büschges A, Akay T, Gabriel JP, Schmidt J. Organizing network action for locomotion: insights from studying insect walking. Brain Res Rev 2008; 57:162-71; PMID:17888515; http://dx.doi.org/ 10.1016/j.brainresrev.2007.06.028 [DOI] [PubMed] [Google Scholar]
- 47. Yellman C, Tao H, He B, Hirsh J. Conserved and sexually dimorphic behavioral responses to biogenic amines in decapitated Drosophila. Proc Natl Acad Sci USA 1997; 94: 4131-6; PMID:9108117; http://dx.doi.org/ 10.1073/pnas.94.8.4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Clyne JD, Miesenböck G. Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell 2008; 133:354-63; PMID:18423205; http://dx.doi.org/ 10.1016/j.cell.2008.01.050 [DOI] [PubMed] [Google Scholar]
- 49. InterPro Protein sequence analysis & classification ; [http://www.ebi.ac.uk/interpro/] [Google Scholar]
- 50. Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, et al. The developmental transcriptome of Drosophila melanogaster. Nature 2010; 471:473-9; PMID:21179090; http://dx.doi.org/ 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. FlyBase A Database of Drosophila Genes & Genomes ; [http://flybase.org/] [Google Scholar]
- 52. Duffy JB. GAL4 system in Drosophila: A fly geneticist's Swiss army knife. Genesis 2002; 34:1-15; PMID:12324939; http://dx.doi.org/ 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- 53. Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage φC31. Genetics 2004; 166:1775-82; PMID:15126397; http://dx.doi.org/ 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Edgington ES. Randomization tests. New York: Marcel Dekker; 1995 [Google Scholar]
- 55. Perneger TV. What's wrong with Bonferroni adjustments. BMJ 1998; 316: 1236-8; PMID:9553006; http://dx.doi.org/ 10.1136/bmj.316.7139.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim J, Lee S, Hwang M, Ko S, Min C, Kim-Ha J. Bunched specifically regulates α/β mushroom body neuronal cell proliferation during metamorphosis. Neuroscience 2009; 161:46-52; PMID:19328837; http://dx.doi.org/ 10.1016/j.neuroscience.2009.03.054 [DOI] [PubMed] [Google Scholar]
- 57. Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat Protoc 2006; 1:581-5; PMID:17406285; http://dx.doi.org/ 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- 58. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res 2002; 30:e36-e36; PMID:11972351; http://dx.doi.org/ 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]


