Abstract
Genetic variation in an infectious disease pathogen can be driven by ecological niche dissimilarities arising from different host species and different geographical locations. Whole genome sequencing was used to compare E. coli O157 isolates from host reservoirs (cattle and sheep) from Scotland and to compare genetic variation of isolates (human, animal, environmental/food) obtained from Scotland, New Zealand, Netherlands, Canada and the USA. Nei’s genetic distance calculated from core genome single nucleotide polymorphisms (SNPs) demonstrated that the animal isolates were from the same population. Investigation of the Shiga toxin bacteriophage and their insertion sites (SBI typing) revealed that cattle and sheep isolates had statistically indistinguishable rarefaction profiles, diversity and genotypes. In contrast, isolates from different countries exhibited significant differences in Nei’s genetic distance and SBI typing. Hence, after successful international transmission, which has occurred on multiple occasions, local genetic variation occurs, resulting in a global patchwork of continental and trans-continental phylogeographic clades. These findings are important for three reasons: first, understanding transmission and evolution of infectious diseases associated with multiple host reservoirs and multi-geographic locations; second, highlighting the relevance of the sheep reservoir when considering farm based interventions; and third, improving our understanding of why human disease incidence varies across the world.
Most (61%) emerging infectious diseases are of zoonotic origin and spill over from animal reservoirs to humans, followed to a lesser or greater extent by cycling in the human population1. Drivers of genetic variation play a major role in the emergence and evolution of these pathogens2. These drivers, acting through selection, include factors that are associated with the relationship between pathogen and host leading to host association as well as geographical separation3.
E. coli O157 is a relatively rare human pathogen, but sequelae can be severe. The incidence of human E. coli O157 infection is 0.51, 0.98, 3.3 and 4.1 cases per 100,000 population in the Netherlands4, USA5, New Zealand6 and Scotland7 respectively. Human disease symptoms include bloody diarrhoea in ~90% of cases which can result in haemorrhagic colitis (HC) with 10–15% of cases progressing to haemolytic uraemic syndrome (HUS) and occasionally death8.
Cattle and sheep are regarded as the natural asymptomatic reservoirs of E. coli O1579 which can also be found intermittently in other animals including goats, deer, pigs, wild birds and invertebrates10. The scientific literature provides twelve times as many references on E. coli O157 and cattle (n = 7,729) compared with sheep (n = 649) (Source: Web of Knowledge, 2014). However, sheep shed similar loads of E. coli O157 into the environment as cattle11 and are also linked with outbreaks of human infection (e.g. in Scotland for every 3 outbreaks associated with cattle one was associated with sheep12). There have been no published studies that have investigated the genetic variation of E. coli O157 obtained from cattle and sheep populations at a genomic level and as such the extent of host association is unclear. There is also a need to determine the relative importance of infectious disease reservoirs in order to inform the selection of interventions to reduce infection13.
The E. coli O157 pathogen was first recognised in 198214 and has since been reported in all continents except Antarctica15. Transmission between countries has been hypothesised to be due to animal movements (e.g. transport of cattle and sheep or bird migration) and/or transport of contaminated feed16. It has also been found that cattle and human clinical isolates are considerably different between the USA and The Netherlands17 whilst Locus Specific Polymorphism Analysis (LSPA6) genotyping and analysis of Shiga toxin bacteriophage insertion (SBI) sites differentiated between isolates from the USA, Argentina and Australia18. However, it is unclear whether genetic differences caused by geographic separation of E. coli O157 are more pronounced than those caused by host specificity in its primary reservoirs (i.e. cattle and sheep).
Recent developments in next generation sequencing now make it possible to routinely whole genome sequence (WGS) bacterial pathogens and utilise single nucleotide polymorphisms (SNPs) to characterise isolates19,20. This has proven useful to identify clades of differing virulence and to differentiate between isolates from cattle and clinically ill people21,22.
Here, WGS is used to determine the extent of host association between cattle and sheep isolates originating from NE Scotland and compares this with clinical isolates obtained from the human population and from food and the environment. These Scottish isolates are then compared with a group of international isolates to determine whether geographic separation is more important than host association as a driver for population differentiation.
Results
Analysis of Scottish Isolates
All 145 isolates provided positive in silico PCR results for rfb (O-antigen-encoding region of O157), eaeA (intimin) and hlyA (enterohemolysin) confirming they were E. coli O157 and had these putative virulence factors present (Table 1).
Table 1. Abundance of genotype/phenotype factors by source (Bovine, Ovine, Food/Environment and Clinical) for the Scottish E. coli O157 isolates.
| Factor | Sub-type | No. (%) of strains with factora |
||||
|---|---|---|---|---|---|---|
| Bovine(n = 26) | Ovine(n = 25) | Food/Env.(n = 20) | Clinical(n = 74) | Statistically significantpairwise comparisonsb | ||
| O encoding antigen | rfbO157 | 26 (100.0) | 25 (100.0) | 20 (100.0) | 74 (100.0) | NA |
| Intimin | eae | 26 (100.0) | 25 (100.0) | 20 (100.0) | 74 (100.0) | NA |
| Enterohemolysin | hlyA | 26 (100.0) | 25 (100.0) | 20 (100.0) | 74 (100.0) | NA |
| Shiga toxin | stx1a/stx2a | 0 (0.0) | 0 (0.0) | 1 (5.0) | 3 (4.1) | none |
| stx1a/stx2c | 6 (23.1) | 4 (16.0) | 0 (0.0) | 12 (16.2) | BF | |
| stx2a | 0 (0.0) | 0 (0.0) | 3 (15.0) | 5 (6.8) | none | |
| stx2a/stx2c | 18 (69.2) | 14 (56.0) | 13 (65.0) | 45 (60.8) | none | |
| stx2c | 2 (7.7) | 6 (24.0) | 0 (0.0) | 5 (6.8) | CO, FO | |
| −ve | 0 (0.0) | 1 (4.0) | 3 (15.0) | 4 (5.4) | none | |
| Phage Type | PT2 | 1 (3.8) | 1 (4.3) | 2 (22.2) | 3 (5.8) | none |
| PT21/28 | 16 (61.5) | 11 (47.8) | 4 (44.4) | 27 (51.9) | none | |
| PT32 | 2 (7.7) | 3 (13.0) | 2 (22.2) | 8 (15.4) | none | |
| PT8 | 5 (19.2) | 4 (17.4) | 0 (0.0) | 5 (9.6) | none | |
| Otherc | 2 (7.7) | 4 (17.4) | 1 (11.1) | 9 (17.3) | none | |
| SBI | Sub-type | |||||
| ASY2a2c | 9 (34.6) | 11 (44.0) | 4 (20.0) | 27 (36.5) | none | |
| SY1a2c | 5 (19.2) | 2 (8.0) | 0 (0.0) | 12 (16.2) | none | |
| SY2a2c | 8 (30.8) | 2 (8.0) | 9 (45.0) | 14 (8.9) | CF, CO, FO | |
| SY2c | 1 (3.8) | 2 (8.0) | 0 (0.0) | 3 (4.1) | CF, FO | |
| YN | 0 (0.0) | 1 (4.0) | 1 (5.0) | 3 (4.1) | none | |
| Otherd | 3 (11.5) | 7 (28.0) | 6 (30.0) | 15 (20.3) | none | |
aNo of strains phage typed: bovine (n = 26), ovine (n = 23), food/env (n = 9) and clinical (n = 52).
bListed are statistically different pairwise comparisons where B = bovine, O = ovine, F = Food/Environmental and C = clinical. Statistical significance determined by Fisher’s exact test.
cOther defines all Phage Types present in <5 Isolates.
dOther defines all SBI genotypes present in <5 isolates.
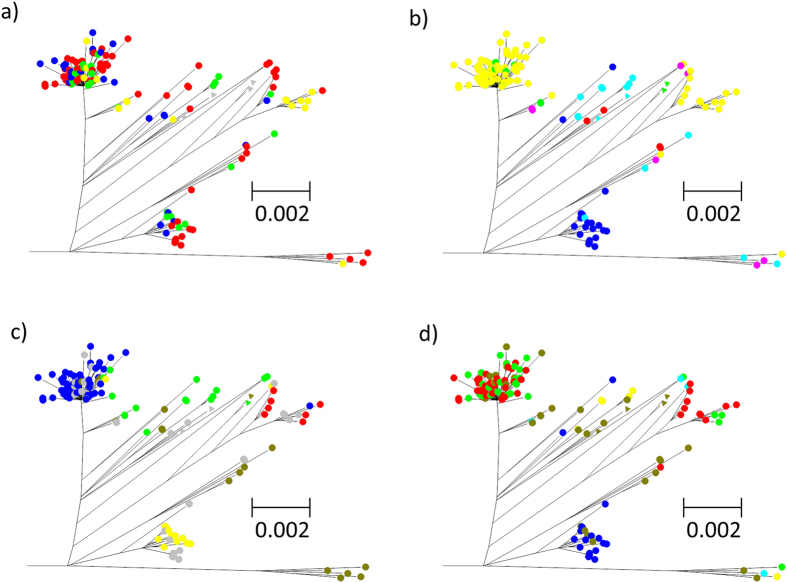
A phylogenetic tree of the E. coli O157 isolates was generated utilising the 8559 SNPs, of which 871 were phylogentically informative, from the core genome (1.223 Mb) generated by PanSeq (Fig. 1, Fig. S1 and Table S2). The tree visualises the radiation of E. coli O157:H7 and is parsimonious with the theory of the evolution of E. coli O157:H7 from O55:H7. Figure 2(a) shows the distribution of the sources (clinical, cattle, sheep, food/environment) of the isolates across the E.coli O157:H7 portion of the phylogeny and it can readily be observed that a number of these sources are present on the same branches of the tree. However, Nei’s genetic distance (Table S6), which provides a measure of the relative distribution of sources across the tree, is significantly different (P < 0.05) between each pair of sources except for cattle and sheep (P > 0.05).
Figure 1. Phylogenetic tree of 145 Scottish E. coli O157:H7 isolates.
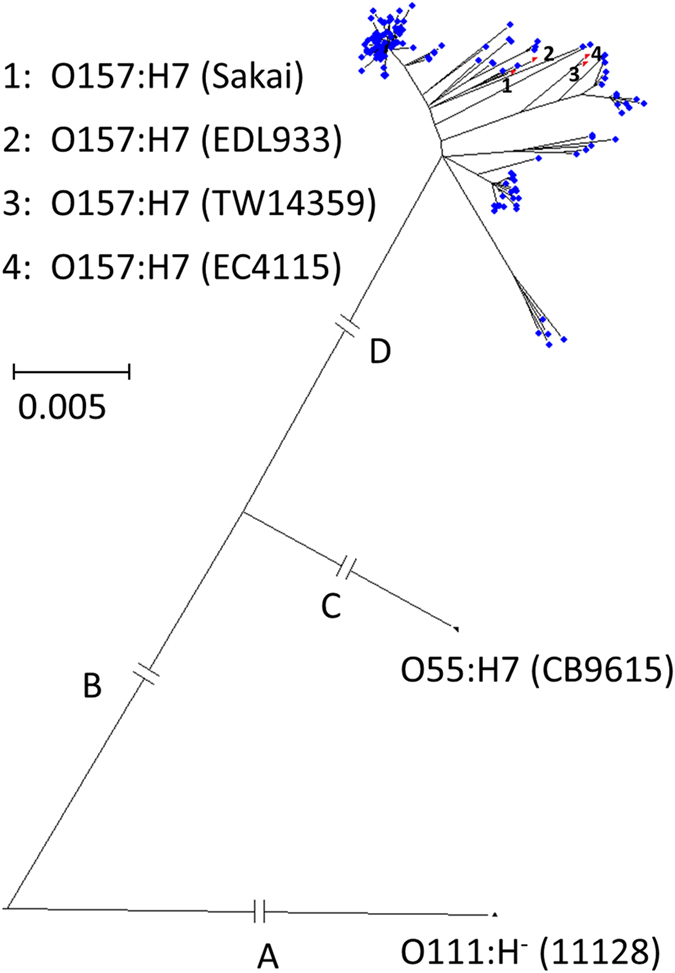
( ), 4 reference USA E. coli O157 isolates (
), 4 reference USA E. coli O157 isolates ( ) and E. coli O55:H7 (CB9615) and E. coli O111:H- (11128). The branches A & B are scaled down by a factor of 5 and C & D by a factor of 2.
) and E. coli O55:H7 (CB9615) and E. coli O111:H- (11128). The branches A & B are scaled down by a factor of 5 and C & D by a factor of 2.
Figure 2. Phylogenetic trees of 145 Scottish E. coli O157:H7 isolates and 4 reference USA isolates.
Illustrating (a) Sources: Clinical ( ); Cattle (
); Cattle ( ); Sheep (
); Sheep ( ); Food/Environment (
); Food/Environment ( ); Unknown (
); Unknown ( ); International (▵), (b) Shiga toxins: stx1a/stx2a (
); International (▵), (b) Shiga toxins: stx1a/stx2a ( ); stx1a/stx2c (
); stx1a/stx2c ( ); stx2a (
); stx2a ( ); stx2a/stx2c (
); stx2a/stx2c ( ); stx2c (
); stx2c ( ); negative (
); negative ( ); Unknown (
); Unknown ( ); International (▵), (c) Phage types: PT2 (
); International (▵), (c) Phage types: PT2 ( ); PT21/28 (
); PT21/28 ( ); PT32 (
); PT32 ( ); PT8 (
); PT8 ( ); Other PTs (
); Other PTs ( ); Unknown (
); Unknown ( ); International (▵) and (d) SBI genotypes: ASY2a2c (
); International (▵) and (d) SBI genotypes: ASY2a2c ( ); SY1a2c (
); SY1a2c ( ); SY2a2c (
); SY2a2c ( ); SY2c (
); SY2c ( ); YN (
); YN ( ); Other SBI types (
); Other SBI types ( ); and International (▵).
); and International (▵).
The most common Shiga toxin subtypes in the Scottish isolates (Table 1) were stx2a/stx2c (62%) and stx1a/stx2c (15%). The distribution of the Shiga toxin subtypes did show some association with source (χ2 = 26.4, df = 15, P = 0.03). Fisher’s exact test indicated a preponderance of the stx1a/stx2c subtype in bovine isolates compared to those from food and the environment and also that the stx2c subtype was more prevalent in ovine than clinical and food/environmental isolates. Figure 2(b) shows clusters of Shiga toxin subtypes in the E. coli O157 phylogenetic tree confirmed by Nei’s genetic distance (Table S6).
The majority (53%) of isolates from all sources were PT21/28 (Table 1). The distribution of phage types by source was found to be indistinguishable (χ2 = 10.0, df = 12, P > 0.05). Phage types appeared to cluster on the phylogenetic tree (Fig. 2(c)) and significant genetic distances were found between all pairwise combinations of phage types (Table S6).
The LSPA6 lineage I/II subtype 211111 was present in 94% of the Scottish isolates and was also found in >90% in each source (Table S5), whilst only 5 Scottish isolates (4 clinical and 1 Food/Environment) were tir 255 allele A. In total 14 Scottish isolates were Manning clade 8, 2 were clade 3 and the remaining 129 clades 4/5/6/7/9 (Table S5).
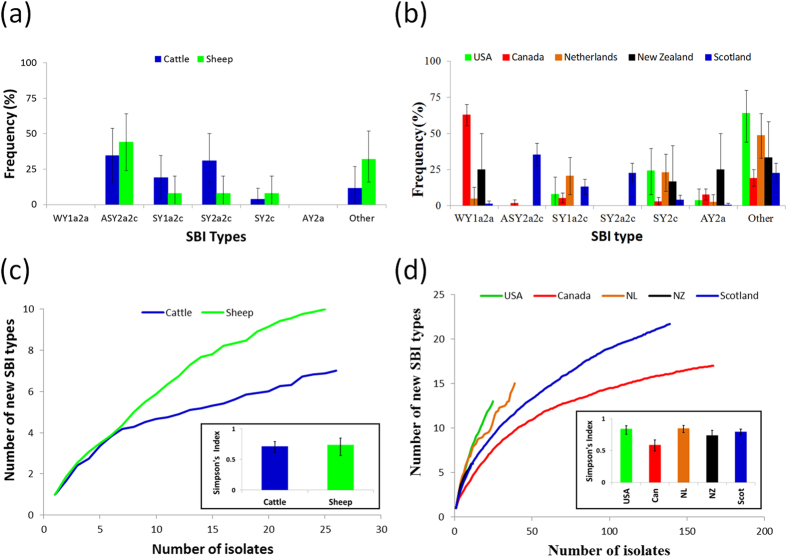
The three most common SBI genotypes ASY2a2c (35.2%), SY1a2c (13.1%) and SY2a2c (22.8%) comprised approximately 60% of all isolates (Table 1). However, the remaining isolates exhibited a considerable diversity of SBI types (n = 24). Overall there were no significant differences in the distribution of SBI genotypes by source (χ2 = 19.6, df = 15, P = 0.19). Fisher’s exact test utilising pairwise comparisons from ovine and bovine isolates also showed that the main SBI genotypes were in similar proportions (Table 1 and Fig. 3(a)). When SBI genotype was plotted on the phylogenetic tree (Fig. 2(d)) it appears that there is clustering between all the main genotypes which was confirmed by Nei’s genetic distance (P < 0.05, Table S6).
Figure 3. Variation of Shiga toxin Bacteriophage Insertion (SBI) types between ruminant hosts and between countries.
Frequency of predominant SBI types (a,b), rarefaction and Simpson’s diversity index (c,d).
Although rarefaction appears to show sheep have a greater diversity of SBI types than cattle (Fig. 3(c)) this was not significant (Table S7). Simpson’s index (Fig. 3(c)) also found no difference in diversity between Scottish cattle and sheep sources (Table S7). Isolates from food were found to have the most significant differences in SBI genotypes when compared pairwise with other sources (6 of the 8 significant differences in Table 1).
Analysis of International isolates
Of the 429 genomes 6, 9 and 20 gave negative in silico PCR results for rfb (O-antigen-encoding region of O157:H7), eaeA (intimin) and hlyA (enterohemolysin) respectively (Table S1). Of the 6 that were negative for rfbO157, 2 were positive for a single primer and all 6 were positive for a SNP in uidA which is an indicative marker for E. coli O15723. The Shiga toxin types, LSPA6 genotypes, SBI types, tir 255 A/T variants and Manning clade allocation are given in Table S1.
There were significant differences (χ2 = 127.0, df = 24, P < 0.0001) in the distribution of SBI types by country (Fig. 3(b)) with an excess of type WY1a2a from Canada and ASY2a2c and SY2a2c of Scottish origin. Although rarefaction (Fig. 3(d)) appears to show a lower diversity of Canadian SBI types compared with other countries this was not found to be statistically significant (Table S7). Whereas Simpson’s diversity index (Fig. 3(d) and Table S7) showed statistically significant differences in diversity amongst a number of countries but with Canada having the lowest diversity of all.
In total, 813 SNPs were obtained from PanSeq from a core genome of 827 kb (Table S3). The resulting bootstrapped concensus phylogeny was broken into clades (Fig. 4, Figs S3 & S4). Clade A is ancestral from which the root to E. coli O55:H7 is located (Fig. 4). This clade comprises isolates from USA, the Netherlands and other European countries. Clade B comprises isolates from the Netherlands and America. Clades C-G all branch from the same point with sub-clade structure being apparent in clades E, F and G. Clades C and E(ii) are European, E(i) being predominantly North American and Dutch and D comprising New Zealand and Scottish isolates. Clade F is formed from a number of different countries with a lack of Scottish isolates being apparent in F(ii). Clades G(i) – G(iv) are primarily European with G(ii) Scottish and G(iii) Scottish, Dutch and New Zealand isolates. Clades G(v) and G(vi) are mainly North American, with some Dutch and Asian isolates. Nei’s genetic distance utilising the 813 SNPs demonstrated significant inter-country differences (Table S9) which is in agreement with the geographical structuring of isolates on the phylogenetic tree. This phylogeny also appears to be parsimonious with previous typing approaches (Fig. S4).
Figure 4. Phylogenetic tree of international isolates.
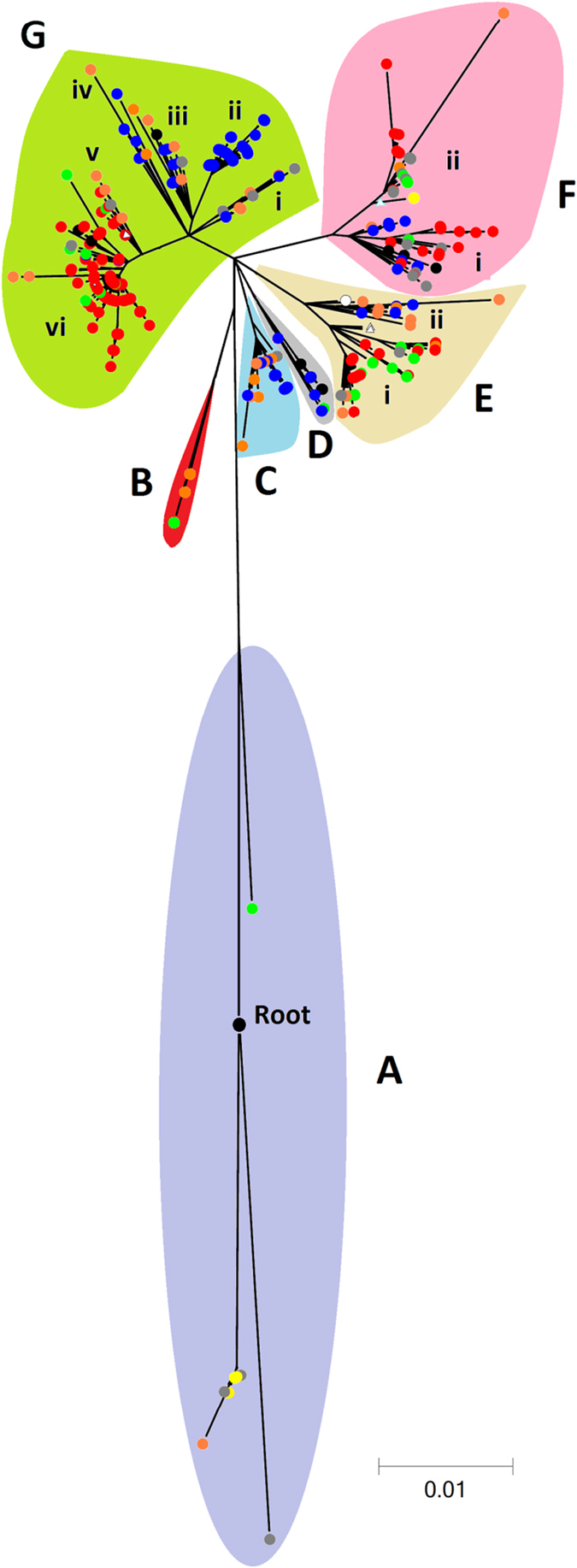
Scotand ( ); Canada (
); Canada ( ); USA (
); USA ( ); The Netherlands (
); The Netherlands ( ); New Zealand (
); New Zealand ( ); Europe (
); Europe ( ); Egypt (○); Asia (▵); South America (
); Egypt (○); Asia (▵); South America ( ); Unknown (
); Unknown ( ). Letters A–G denote the phylogenetic clades identified.
). Letters A–G denote the phylogenetic clades identified.
Discussion
The finding that an indistinguishable population of E. coli O157 is circulating through both cattle and sheep agrees with results from Sweden that used MLVA typing24. This is of importance to the many countries (e.g. UK, New Zealand etc.) that have significant populations of both cattle and sheep as there is opportunity for transmission between species when they are located on the same farm, co-grazing or at livestock markets. On farm intervention strategies (e.g. feeding probiotics, providing clean bedding etc.25) targeted at cattle may be hampered by sheep reseeding E. coli O157 into the agricultural environment. Hence, there is a need to develop and apply multi-species transmission network models for this pathogen26. The carriage of E. coli O157 by both cattle and sheep appears not to increase the genetic diversity of this pathogen perhaps because the ecological niche is similar within these hosts.
Although it appears that E. coli O157 genotypes exhibit a generalist behaviour within ruminants it is unknown whether all of these genotypes have sufficient phenotypic plasticity to be found in a range of other animal hosts9. Other gastrointestinal pathogens (e.g. Campylobacter and Salmonella) have a number of specialist subtypes associated with particular animal host species27,28, but it is yet to be established whether this is also the case for E. coli O157.
The international phylogeographic tree of E. coli O157 (Fig. 4) shows geographical structuring. There appears to be greater isolation exhibited by the Scottish and North American isolates. Whereas, both the Dutch and New Zealand isolates cluster on occasions most closely with those from Scotland and on other occasions with those from North America. This is suggestive of multiple transmission events between New Zealand, Europe and North America followed by genetic radiation of isolates within a country. This geographic structuring agrees with previous PFGE and sequence based studies and sequence based analysis6,16,18.
The question arises as to how and when these transmission events may have occurred. There is documented evidence of intermittent trade comprising relatively low numbers of live cattle and sheep, predominantly for breeding purposes, between the UK, USA, Canada, New Zealand and the Netherlands for the past 20 years (www.uktradeinfo.com). Prior to this international transport of live animals certainly occurred29 and may have taken place via intermediate countries (e.g. UK to Australia to New Zealand). E. coli O157 prevalence in cattle/sheep is approximately 5%30, and shedding within a cattle herd can last >5 months31. This time is sufficient for the organism to survive the sea crossing (50 days) between the UK and New Zealand. Transmission via contaminated feed/food exported between countries is also plausible because of the complex transportation networks32. Carriage of E. coli O157 by humans between countries can occur33, but secondary transmission between humans is low34 and transmission to cattle or sheep is also likely to be low because of the general effectiveness of sewerage systems. It seems unlikely that direct transmission by wild animals and migrating birds between continents takes place because of the intermittent nature of carriage in these hosts but certainly over shorter distances (e.g. between adjacent regions) transmission is plausible35.
From a public health perspective there is considerable interest in elucidating why there is heterogeneity in the E. coli O157 clinical disease burden between countries. Exposure is of importance with foodborne routes being dependent on local food preferences and patterns of food distribution10 and environmental routes being dependent on exposure to cattle/sheep faeces and contaminated water. Susceptibility is also of importance with young children being at particular risk36. However, establishment of E. coli O157 in the ruminant population within a country is initially dependent on inward transmission and subsequent reservoir competence. Further, since humans are effectively an evolutionary dead end for E. coli O15710, its pathogenicity in humans is an accidental consequence of its ecology within the extant agricultural system. Hence, the reason why human disease incidence varies between continents, countries and even states/regions is likely to be multifactorial. NE Scotland has a high disease burden, and a large ruminant population which offers the opportunity of environmental and foodborne exposure. Characteristics of the pathogen are also important with PT21/28 isolates (clade G(ii)) dominating, which is known to be excreted at high levels by cattle37 and carries the most potent Shiga toxin gene stx2a38.
Comparisons between cattle sources and humans within countries (Scotland, USA, Netherlands and Canada) also showed significant differences (Table S9). This was not found to be the case for New Zealand here, possibly because sample size was small, but previous studies observed differences when investigating the distribution of SBI types both in the USA39 and New Zealand6. These results indicate that there is a different population structure of E. coli O157 between cattle and humans in each country. It is plausible that some isolates from cattle are more likely than others to cause infection in humans as reported elsewhere17. It can be concluded here that human cases arise from all of the clades and sub-clades across the tree and that in Clade F most isolates have the potent stx2a toxin (this agrees with the hypothesis of Manning clade 8 being virulent21) whilst Clades C, D and E have a very low occurrence of stx2a which suggests that human disease arising from these clades is likely to be less severe.
The aim of this work was not to identify the origin of E. coli O157 from its E. coli O55:H7 progenitor but the Clade A isolates closest to the E. coli O55:H7 root of the international tree (Fig. 4) originate from North America and Europe suggesting that inter-continental spread of this organism occurred fairly early in its evolutionary origins. This is corroborated by a survey of isolates deposited in culture collections prior to 1982 which noted 7 from North America and 1 from the UK40.
This work has also been able to illustrate how part of the dynamic accessory genome (e.g. Shiga toxin encoding phage) relate to the evolutionary core genome of this pathogen. There remains the vast majority of the accessory genome to explore which may contain genes for virulence and/or adaptive advantage in specific niches19. The comparisons of the international SNP based phylogeny with existing typing techniques (e.g. LSPA6 typing, tir A/T, Manning clades etc see Fig. S4 and Table S5 & S8) shows the backward compatibility of the SNP based methodology with existing methods.
Conclusion
E. coli O157 continues to be a pathogen that causes significant levels of disease and morbidity across the world. It transmits freely between its primary reservoirs of cattle and sheep and there is a lack of evidence of genetic differences between the populations of isolates originating from these hosts. It is therefore important to consider the sheep reservoir when investigating interventions to reduce the presence of this pathogen, particularly at the farm level. The pathogen appears to have been transmitted a number of times between countries and continents but the rate of transmission is low enough that a strong phylogeographic signal remains. Achieving a global understanding of this pathogen through whole genome sequencing is important in terms of understanding the potential origins of outbreaks at national and continental spatial scales, evolution of this pathogen and the relative importance of its animal reservoirs and subsequent pathogenicity to humans.
Materials and Methods
Isolates
A total of 145 Scottish isolates (Table S1), comprising clinical isolates (74) originating from patients resident in NE Scotland, cattle (26), sheep (25) and food/environmental (20) isolates from previous studies. Isolates were selected to be independent of each other (i.e. not to be obtained from the same farm at the same time) and to be obtained over a broad time period. Isolates were phage typed as described previously41.
Sequenced isolates
Sequenced international E. coli O157 isolates from Canada (164), Netherlands (39), New Zealand (12), Scotland (145), USA (26) and other countries (45) were obtained along with varying levels of metadata (Table S1). The Canadian and Dutch sequenced isolated were selected using the same principles as the Scottish isolates, the New Zealand isolates were obtained from two calendar years but variation was ensured by selection across their PFGE profiles. The genomic sequence data of these isolates will be uploaded to an open access database on acceptance of this paper for publication (Table S1).The isolates from USA and the rest of the world were downloaded from the NCBI and PATRIC databases.
Whole genome sequencing of Scottish isolates
Overnight cultures of E. coli O157 were grown on Harlequin SMAC-BCIG agar plates (Hal 6, Lab M, Topley House, Lancashire) at 37 °C. A single, well isolated sorbitol negative colony was selected and tested with E. coli O157 latex (code DR0620M, Oxoid, Basingstoke) and the latex plated onto Columbia Agar at 37 °C for 24 hours (Code CM0031, Oxoid, Basingstoke). DNA was extracted with the Wizard Genomic DNA Purification Kit (Promega UK Ltd, Southampton) as per the manufacturers instructions with an additional Proteinase K step (25 μl, lyophilized Proteinase K reconstituted at a concentration of 6 mg/260 μl Proteinase buffer) post treatment with nucleic acid lysis solution. The concentration of DNA was determined by Picogreen assay and then submitted to the Wellcome Trust Centre for Human Genetics (WTCHG), Oxford for WGS. This was conducted using an Illumina HiSeq sequencer with 100 base paired-end sequencing. The FASTQ paired-end reads were assembled using Velvet42, coverage was typically 30x and assembled genome size approximately 5.5 Mb.
Analysis of genomes
In silico PCR and probe based assays (Table S4) were carried out both for backwards compatibility with previous studies and identification of known virulence markers. SNP discovery methods were also implemented for phylogenetic analysis.
Detection of E. coli O157 antigen encoding, intimin and enterohemolysin genes: in silico PCRs (Table S4), performed using a Perl script were conducted for a portion of the rfb (O-antigen-encoding) region of E. coli serotype O157 and for the putative accessory virulence factors intimin (eaeA) and the plasmid-encoded enterohemolysin (hlyA). The Perl script blast searched for the primers in the genome sequences with tolerance of 90%. If the pair of primers were found, the size of the PCR product was determined and if matching the predicted size the PCR was assigned as positive.
Shiga toxin subtyping: an in silico combined BLAST and SNP based approach was employed to detect the presence of Shiga toxin genes (Table S4). This allows for the homology between the different Shiga toxin types and compares the FASTQ reads and/or genome assemblies with reference Shiga toxin genes43. Odds ratios (OR) were calculated to determine whether particular Shiga toxin types were more or less frequent in particular sources (cattle, sheep, clinical and food/environment). P values were obtained from a two-tailed Fisher’s exact test.
LSPA6 sub-typing, tir 255T > A polymorphism analysis and Clade Identification (Manning): was performed in silico (Table S4).
SBI sub-typing: in silico PCRs were performed to determine prophage occupancy of E. coli O157 loci argW, sbcB, wrbA, yehV and SBI results were concatenated using the terminology described previously44 (see Table S4).
Rarefaction: was used to compare the diversity and distribution of SBI types between animal host species and between countries and testing for significance was carried out using a randomization test45 implemented with Poptools (http://www.cse.csiro.au/poptools/; accessed 2014 June 23).
Simpson’s Index: was used as a measure of the taxonomic richness of SBI types from different host species or different countries46. An index value of 0 indicates homogenous types and a value of 1 indicates a totally heterogeneous population. Testing for significance was carried out using a randomization test as above.
Pan-genomic SNP analysis: two analyses were performed. The first comprised the 145 Scottish genomes together with four reference E. coli O157:H7 published genomic sequences (TW14359 and EC4115 from the USA spinach outbreak47, EDL933 and Sakai), along with an E. coli O55:H7 (CB9615) and an E. coli O111:H- (11128). The second comprised the Scottish and all of the international E. coli O157 genomes in the study. PanSeq was used to construct a non-redundant pan-genome for each of the two analyses20. The pan-genome was constructed by using a seed genome and identifying regions of ≥1000 bp not found in the seed but present in any other genome at a 99 percent sequence identity cutoff. The pan-genome was subsequently fragmented into 1000 bp segments, and the presence/absence of each locus in every genome determined at a 99 percent sequence identity threshold. Loci present in all genomes underwent multiple sequence alignment using Muscle48, and were concatenated together. This aligned pan-genome was used to identify SNPs in the core genome of all isolates. A phylogeny of E. coli O157 isolates’ genomes was rooted using the E. coli O55:H7 strain CB9615 as a proximal outgroup (O55:H7 is considered to be the immediate ancestor of E. coli O157), and E. coli O111:H- strain 11128 as a more distal outgroup. A neighbour joining tree was generated and visualised in MEGA49. Bootstrapping with 500 iterations was also performed and concensus trees generated.
Genetic distance: Nei’s standardized genetic distance d1, was calculated for each pairwise combination of sources (e.g. cattle and sheep), phage types, Shiga toxin types, LSPA6 lineages, SBI genotypes and countries50 utilising the pan-genomic SNP data detailed above. The genetic distance ranges from 1 (where there are no SNPs in common between the two groups) and 0 (where the two groups have an identical distribution of SNPs).
Additional Information
How to cite this article: Strachan, N. J. C. et al. Whole Genome Sequencing demonstrates that Geographic Variation of Escherichia coli O157 Genotypes Dominates Host Association. Sci. Rep. 5, 14145; doi: 10.1038/srep14145 (2015).
Supplementary Material
Acknowledgments
The authors acknowledge the support of the Food Standards Agency, Scotland (FS102029) and the University of Aberdeen for funding sequencing of the Scottish E. coli O157 genomes, to Chris Low at the Scottish Agricultural College, Edinburgh for supplying a number of the Scottish sheep isolates, Iain Ogden for commenting on the manuscript and Patricia Jaros (Massey University) for preparing the New Zealand isolate DNA for sequencing.
Footnotes
Author Contributions All listed authors contributed to the idea generation, design, and completion of this work. Genome experimental work was carried out by B.L., M.H., L.A. (Scotland), C.L. and V.G. (Canada), E.F. and A.v.H. (Netherlands), N.F., T.G. and P.B. (New Zealand). Analysis of was performed by O.R., N.S., K.F., P.A., T.D. and C.L. The manuscript was drafted by N.S. and K.F. and all authors reviewed the manuscript.
References
- Wolfe N. D., Dunavan C. P. & Diamond J. Origins of major human infectious diseases. Nature 447, 279–283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S. S. Factors in the emergence of infectious-diseases. Emerging Infectious Diseases 1, 7–15 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg E. P. & Brooks D. R. A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host-parasite systems. J. Biogeogr. 35, 1533–1550 (2008). [Google Scholar]
- Friesema I. H. M. et al. Surveillance van Shigatoxineproducerende Escherichia coli (STEC) in Nederland, 2012. Infectieziekte Bulletin 24, 285–290 (2013). [Google Scholar]
- CDC. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2011 (Final Report). 2011, 1–52 (2012).
- Jaros P. et al. Geographic divergence of bovine and human shiga toxin-producing Escherichia coli O157:H7 genotypes, New Zealand. Emerging Infectious Diseases 20, 1980–1989 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locking M., Browning L., Smith-Palmer A. & Brownlie S. Gastro-intestinal and foodborne infections: Incidence of E. coli O157, Salmonella and Campylobacter reported to HPS: 2012. Health Protection Scotland Weekly Report 47, 44–45 (2013). [Google Scholar]
- Tarr P. I., Gordon C. A. & Chandler W. L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086 (2005). [DOI] [PubMed] [Google Scholar]
- Ferens W. A. & Hovde C. J. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog. Dis. 8, 465–487 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington H. Escherichia coli O157. Lancet 376, 1428–1435 (2010). [DOI] [PubMed] [Google Scholar]
- Strachan N. J., Macrae M. & Ogden I. D. Quantification of the Escherichia coli O157 reservoir in Grampian, Scotland. Vet. Rec. 156, 282–283 (2005). [DOI] [PubMed] [Google Scholar]
- Fraser E. Determining the main source of human Escherichia coli O157 infection and the effects of brassicas on the shedding of this pathogen in sheep. University of Aberdeen, PhD thesis. 269 (2014). [Google Scholar]
- Pires S. M. et al. Attributing the human disease burden of foodborne infections to specific sources. Foodborne Pathog. Dis. 6, 417–424 (2009). [DOI] [PubMed] [Google Scholar]
- Riley L. et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685 (1983). [DOI] [PubMed] [Google Scholar]
- Chase-Topping M., Gally D., Low C., Matthews L. & Woolhouse M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6, 904–912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. et al. Correlation between geographic distance and genetic similarity in an international collection of bovine faecal Escherichia coli O157 : H7 isolates. Epidemiol. Infect. 131, 923–930 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E. et al. Genetic features differentiating bovine, food, and human isolates of shiga toxin-producing Escherichia coli O157 in the Netherlands. J. Clin. Microbiol. 50, 772–780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor G. E. et al. Geographically distinct Escherichia coli O157 differ by lineage, Shiga toxin genotype and total Shiga toxin production. Journal of Clinical Microbiology 53, 579–586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E. et al. Exploiting the explosion of information associated with whole genome sequencing to tackle Shiga toxin-producing Escherichia coli (STEC) in global food production systems. International Journal of Food Microbiology 187, 57–72 (2014). [DOI] [PubMed] [Google Scholar]
- Laing C. et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11, 461-2105-11-461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning S. D. et al. Variation in virulence among clades of Escherichia coli O157 : H7, associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 105, 4868–4873 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. K. et al. Lineage and genogroup-defining single nucleotide polymorphisms of Escherichia coli O157:H7. Appl. Environ. Microbiol. 79, 7036–7041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinneman K. C., Yoshitomi K. J. & Weagant S. D. Multiplex real-time PCR method to identify Shiga toxin genes stx1 and stx2 and Escherichia coli O157 : H7/H- serotype. Appl. Environ. Microbiol. 69, 6327–6333 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund R. et al. Molecular typing of Escherichia coli O157:H7 isolates from swedish cattle and human cases: population dynamics and virulence. J. Clin. Microbiol. 52, 3906–3912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon J. M., Chadd S. A. & Baines R. N. Escherichia coli O157:H7 in beef cattle: on farm contamination and pre-slaughter control methods. Animal Health Research Reviews 12, 197–211 (2011). [DOI] [PubMed] [Google Scholar]
- Zhuang L., Cressie N., Pomeroy L. & Janies D. Multi-species SIR models from a dynamical Bayesian perspective. Theor. Ecol. 6, 457–473 (2013). [Google Scholar]
- Griekspoor P. et al. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol. Ecol. 22, 1463–1472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires S. M., Vieira A. R., Hald T. & Cole D. Source attribution of human salmonellosis: an overview of methods and estimates. Foodborne Pathog. Dis. 11, 667–676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binney B. M., Biggs P. J., Carter P. E., Holland B. M. & French N. P. Quantification of historical livestock importation into New Zealand 1860-1979. N. Z. Vet. J. 62, 309–314 (2014). [DOI] [PubMed] [Google Scholar]
- Rhoades J. R., Duffy G. & Koutsoumanis K. Prevalence and concentration of verocytotoxigenic Escherichia coli, Salmonella enterica and Listeria monocytogenes in the beef production chain: A review. Food Microbiol. 26, 357–376 (2009). [DOI] [PubMed] [Google Scholar]
- Williams K. J., Ward M. P., Dhungyel O. P., Hall E. J. S. & Van Breda L. A longitudinal study of the prevalence and super-shedding of Escherichia coli O157 in dairy heifers. Vet. Microbiol. 173, 101–109 (2014). [DOI] [PubMed] [Google Scholar]
- Ercsey-Ravasz M., Toroczkai Z., Lakner Z. & Baranyi J. Complexity of the international agro-food trade network and its impact on food safety. Plos One 7, e37810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locking M., Allison L., Rae L., Pollock K. & Hanson M. VTEC in Scotland 2004: enhanced surveillance and reference laboratory data. Health Protection Scotland Weekly Report 39, (51–52), 290–295 (2006). [Google Scholar]
- Snedeker K. G., Shaw D. J., Locking M. E. & Prescott R. J. Primary and secondary cases in Escherichia coli O157 outbreaks: a statistical analysis. BMC Infect. Dis. 9, 144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway T. R., Edrington T. S. & Nisbet D. J. Isolation of Escherichia coli O157:H7 and Salmonella from migratory brown-headed cowbirds (Molothrus ater), common grackles (Quiscalus quiscula), and cattle egrets (Bubulcus ibis). Foodborne Pathog. Dis. 11, 791–794 (2014). [DOI] [PubMed] [Google Scholar]
- Strachan N. J., Dunn G. M., Locking M. E., Reid T. M. & Ogden I. D. Escherichia coli O157: burger bug or environmental pathogen? Int. J. Food Microbiol. 112, 129–137 (2006). [DOI] [PubMed] [Google Scholar]
- Matthews L. et al. Predicting the public health benefit of vaccinating cattle against Escherichia coli O157. Proc. Natl. Acad. Sci. USA 110, 16265–16270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller C. A., Pellino C. A., Flagler M. J., Strasser J. E. & Weiss A. A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79, 1329–1337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono J. L. et al. Association of Escherichia coli O157 : H7 tir polymorphisms with human infection. BMC Infect. Dis. 7, 98 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin P. & Tauxe R. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic Escherichia coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13, 60–98 (1991). [DOI] [PubMed] [Google Scholar]
- Khakhria R., Duck D. & Lior H. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol. Infect. 105, 511–520 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton P. et al. Insight into Shiga toxin genes encoded by Escherichia coli O157 from whole genome sequencing. PeerJ PrePrints 2:e652v1, 1–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor G. E. et al. Multilocus genotype analysis of Escherichia coli O157 isolates from australia and the United States provides evidence of geographic divergence. Appl. Environ. Microbiol. 79, 5050–5058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly B. F. J. in Randomization, Bootsrap and Monte Carlo Methods in Biology (Chapman & Hall/CRC, Boca Raton, Florida, USA, 2007). [Google Scholar]
- Simpson E. H. Measurement of diversity. Nature 163, 688–688 (1949). [Google Scholar]
- Kulasekara B. R. et al. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77, 3713–3721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J. & Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9, 299–306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. in Molecular Population Genetics and Evolution (North-Holland Publishing Company, Amsterdam, the Netherlands, 1975). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.