Abstract
Meiotic recombination initiates with DNA double-strand breaks (DSBs) made by Spo11. In Saccharomyces cerevisiae, many DSBs occur in “hotspots” coinciding with nucleosome-depleted gene promoters. Transcription factors (TFs) stimulate DSB formation in some hotspots, but TF roles are complex and variable between locations. Until now, available data for TF effects on global DSB patterns were of low spatial resolution and confined to a single TF. Here, we examine at high resolution the contributions of two TFs to genome-wide DSB distributions: Bas1, which was known to regulate DSB activity at some loci, and Ino4, for which some binding sites were known to be within strong DSB hotspots. We examined fine-scale DSB distributions in TF mutant strains by deep sequencing oligonucleotides that remain covalently bound to Spo11 as a byproduct of DSB formation, mapped Bas1 and Ino4 binding sites in meiotic cells, evaluated chromatin structure around DSB hotspots, and measured changes in global messenger RNA levels. Our findings show that binding of these TFs has essentially no predictive power for DSB hotspot activity and definitively support the hypothesis that TF control of DSB numbers is context dependent and frequently indirect. TFs often affected the fine-scale distributions of DSBs within hotspots, and when seen, these effects paralleled effects on local chromatin structure. In contrast, changes in DSB frequencies in hotspots did not correlate with quantitative measures of chromatin accessibility, histone H3 lysine 4 trimethylation, or transcript levels. We also ruled out hotspot competition as a major source of indirect TF effects on DSB distributions. Thus, counter to prevailing models, roles of these TFs on DSB hotspot strength cannot be simply explained via chromatin “openness,” histone modification, or compensatory interactions between adjacent hotspots.
Keywords: Spo11, meiotic recombination, double-strand break, transcription factor, chromatin
MEIOSIS is a specialized cell division in which one round of DNA replication is followed by two successive rounds of chromosome segregation to produce haploid gametes from diploid cells. In meiotic prophase, most sexually reproducing organisms use homologous recombination to form physical connections between homologous chromosomes that are essential for accurate chromosome segregation (Petronczki et al. 2003). Recombination also disrupts linkage of sequence polymorphisms on the same chromosome and thus promotes genome diversity and evolution (Kauppi et al. 2004).
Recombination is initiated by the programmed formation of DNA double-strand breaks (DSBs). Approximately 150–200 DSBs are formed per meiosis in Saccharomyces cerevisiae (Buhler et al. 2007; Chen et al. 2008; Mancera et al. 2008; Pan et al. 2011), generated by a dimer of the topoisomerase-like protein Spo11 (Bergerat et al. 1997; Keeney et al. 1997). After break formation, Spo11 remains covalently bound to the 5′ DNA strand termini (de Massy et al. 1995; Keeney and Kleckner 1995; Liu et al. 1995). Single-stranded nicks nearby then release Spo11 still bound to a short oligonucleotide (Spo11-oligo complexes; Figure 1A) (Neale et al. 2005) and permit further 5′-to-3′ resection to generate 3′ single-stranded tails, which are substrates for strand exchange proteins (Sun et al. 1991; Schwacha and Kleckner 1994; Goldfarb and Lichten 2010; Zakharyevich et al. 2010).
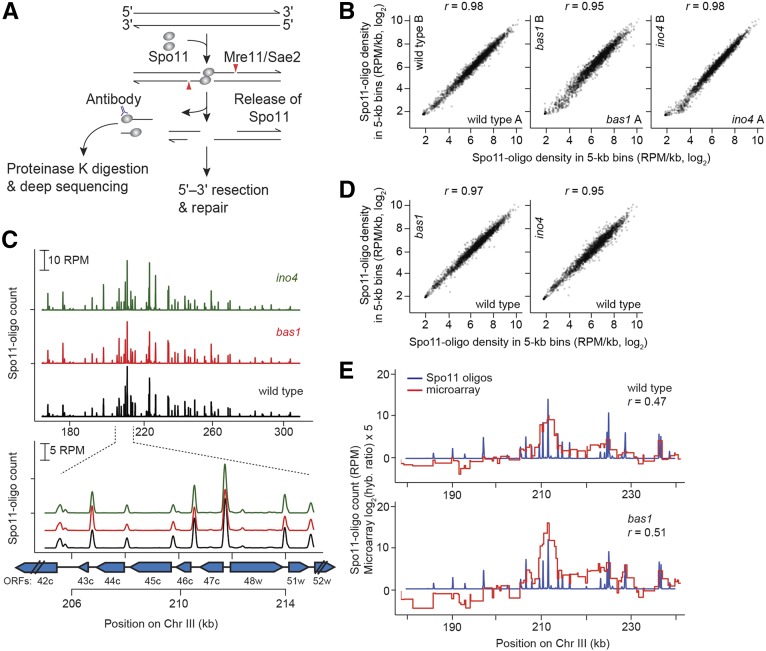
Figure 1.
Spo11-oligo mapping in bas1 and ino4 mutants. (A) Formation and processing of meiotic DSBs (Spo11-oligo mapping method on left). (B) Quantitative reproducibility of Spo11-oligo maps. Comparisons are shown for individual wild-type, bas1, or ino4 datasets. Spo11 oligos were summed in nonoverlaping 5-kb bins and expressed as RPM per kilobase (plotted on a log scale). Correlation coefficients (Pearson’s r) are shown on the top. (C) Top, bas1 and ino4 mutations do not cause widespread change in the large-scale patterns of the DSB landscape. Bottom, DSBs form at the same hotspots in bas1 and ino4 as in wild type. Spo11-oligo counts here and in E are the per-base pair frequency, smoothed with a 201-bp Hann window. Positions of open reading frames are indicated by blue arrows. (D) Comparison between wild-type and TF mutant datasets. For each genotype, Spo11-oligo maps were averaged from biological replicates. (E) The Spo11-oligo map agrees with previous rad50S Spo11 ChIP microarray analysis (Mieczkowski et al. 2006) but provides higher resolution. A representative genomic region is shown. The value for each microarry probe is plotted along the length of sequence covered by the probe. The correlation coefficient was calculated between the normalized microarray signal and the sum of Spo11-oligo counts within 2 kb on either side of each microarray probe.
The distribution of meiotic DSBs across the S. cerevisiae genome is highly nonrandom (Baudat and Nicolas 1997; Gerton et al. 2000; Borde et al. 2004; Blitzblau et al. 2007; Buhler et al. 2007; Pan et al. 2011). There are large DSB-hot and -cold domains (tens of kilobases), within which are short regions called hotspots (typically several hundred base pairs wide), where DSBs preferentially form. Most hotspots in S. cerevisiae coincide with the nucleosome-depleted regions at gene promoters (Wu and Lichten 1994; Baudat and Nicolas 1997; Pan et al. 2011). The global DSB landscape is shaped by a hierarchical combination of factors, including whole chromosome variation, large subchromosomal domains, presence of cohesins and other chromosome structure proteins, local chromatin structure, and histone modification (Lichten and Goldman 1995; Petes 2001; Kauppi et al. 2004; Lichten and de Massy 2011; Pan et al. 2011; Acquaviva et al. 2013; de Massy 2013; Sommermeyer et al. 2013).
In some cases, DSB hotspot activity has been shown to depend on the binding of sequence-specific transcription factors (TFs), e.g., at the HIS4 locus in S. cerevisiae and the ade6-M26 allele in Schizosaccharomyces pombe (Schuchert et al. 1991; White et al. 1991; Kon et al. 1997; Steiner et al. 2002). However, the role of TFs in shaping genome-wide DSB patterns is complex and poorly defined. In certain S. cerevisiae strains, the HIS4 promoter displays strong hotspot activity that requires the binding of TFs Bas1, Bas2, and Rap1 (White et al. 1991). A bas1 null mutation reduces DSB activity at a number of chromosomal sites, but it also causes increased DSB frequency at many other sites (Mieczkowski et al. 2006). Thus, the effect of Bas1 on DSB formation is context dependent, and local DSB stimulation by this TF at HIS4 cannot be simply extrapolated as a general feature to all genomic loci. Moreover, loci whose DSB activity changes in bas1 mutants are poorly correlated with Bas1 binding sites defined by chromatin immunoprecipitation (ChIP), suggesting Bas1 has both direct and indirect roles in DSB regulation (Mieczkowski et al. 2006).
Even in those instances where DSB hotspot activity is clearly linked to TF binding, the nature of this link has remained elusive. Like most yeast promoters, the HIS4 promoter exhibits hypersensitivity to DNase I digestion of chromatin, but it becomes nuclease resistant when it loses DSB hotspot activity in bas1bas2 double mutants (Fan and Petes 1996). Another example is the PHO5 promoter, which is bound by the TF Pho4 (Wu and Lichten 1994). When PHO5 transcription is inhibited by deleting Pho4, its promoter is occupied by a well-positioned nucleosome array and meiotic DSBs are detected as a narrow band by Southern blot. However, in the transcriptionally induced state, the nucleosomes are absent and the DSB band becomes dramatically wider across the whole nucleosome-depleted promoter. Thus, TF binding can promote DSB formation nearby, possibly via establishing an open chromatin structure in the surrounding area. However, the available data are anecdotal: HIS4 and PHO5 are the only TF-dependent hotspots for which the effect of TFs on meiotic DNA accessibility and DSBs has been examined in budding yeast. It is unknown whether TF binding can cause DNA accessibility change at DSB sites in all TF-affected hotspots.
Thus far, Bas1 is the only TF that has been studied genome-wide for its influence on meiotic DSB distribution. Bas1 is a Myb-related TF involved in the expression of genes acting in the adenine and histidine biosynthetic pathways (Arndt et al. 1987; Daignan-Fornier and Fink 1992; Denis et al. 1998). Bas1 binds to a site containing a GAGTCA motif found in the ADE2 and ADE5,7 promoters (Hovring et al. 1994). This motif is required for regulation of ADE2 by Bas1 and another TF Bas2/Pho2 (Daignan-Fornier and Fink 1992). It has been proposed that formation of a complex between Bas1 and Bas2 unmasks an activator function of Bas1, but there may be other partners that can bind Bas1 as well (Denis and Daignan-Fornier 1998). Bas1 and Bas2 also regulate several genes involved in one-carbon metabolism, such as GLN1, SHM2, and MTD1 (Denis and Daignan-Fornier 1998; Gelling et al. 2004).
Meiotic DSBs were previously mapped genome-wide in a bas1 mutant by microarray hybridization of Spo11-attached DNA from a rad50S mutant (Mieczkowski et al. 2006). In the rad50S background, Spo11 is not released from DSB ends and thus repair is blocked and DSBs accumulate (Alani et al. 1990; Keeney et al. 1997). A total of 153 open reading frames (ORFs) were identified as regions with different DSB frequency between wild type and bas1, but the spatial resolution (usually several kilobases) was not high enough to precisely locate DSB hotspots in these regions (Mieczkowski et al. 2006). Besides bas1, there are no genome-wide DSB maps of other TF mutants.
In this study, we wished to more precisely define the contributions of specific TFs to global DSB patterns. We examined a bas1 mutant because, while it is known to affect DSBs, nucleotide-resolution information about changes in DSB patterns was lacking. To determine whether another TF affects break formation similarly to Bas1, we also examined an ino4 mutant. Ino4 motifs were previously found to be enriched in strong DSB hotspots (Pan et al. 2011), but it was unknown if Ino4 stimulates DSB activity in those hotspots. Ino4 controls genes involved in phospholipid synthesis and is required for derepression of inositol/choline-regulated genes such as INO1, CHO1, CHO2, and OPI3 (Santiago and Mamoun 2003). Ino4 forms a complex with Ino2 and binds the inositol-choline-responsive element through a basic helix-loop-helix domain (Schwank et al. 1995).
Here, we ask the following questions: Is the context dependence for DSB frequency modulation seen for Bas1 a unique situation, or does it also hold for another TF? Are TF effects on DSB frequency mediated principally via opening or closing of local chromatin structure, as has been hypothesized (Wu and Lichten 1994; Mieczkowski et al. 2006; Lichten 2008)? Are indirect effects of TFs mediated by local competition between hotspots? And, separate from effects on DSB numbers, do TFs influence fine-scale DSB patterns around their binding sites, and if so, how?
Materials and Methods
Yeast strains and culture methods
Strains used in this study are of the SK1 background and are listed in Supporting Information, Table S1. The bas1 and ino4 deletions were made by replacing the coding sequence with the hygromycin B phosphotransferase gene (hphMX4). The sae2 deletion was made by replacing the coding sequence with the ClonNAT resistance gene (natMX). Gene disruption was verified by PCR and Southern blotting. The SPO11-FLAG construct was provided by Kunihiro Ohta (Kugou et al. 2009). All mutants analyzed were moved into the desired tester strain backgrounds by crossing and tetrad dissection. Synchronous meiotic cultures were prepared as described (Alani et al. 1990; Padmore et al. 1991). Briefly, cells were grown in YPA (1% yeast extract, 2% Bacto Peptone, 1% potassium acetate) for 13.5–14 hr at 30° (starting OD600 = 0.2, ending OD600 = 2∼4), harvested, resuspended in 2% potassium acetate (OD600 = 4.5∼6), and sporulated at 30°. Null bas1 and ino4 mutants show normal division kinetics and spore viability (data not shown).
Spo11-oligo mapping
Spo11-oligo mapping was performed with 3000–4000 OD600 units of cells harvested at 4 hr in meiosis from cultures prepared as described above, except with supplementation of YPA with 0.001% antifoam 204. Cell lysates were prepared by grinding frozen cell paste in a bead mill, similar to methods described (Thacker et al. 2014). The lysate was then diluted with an equal volume of 2× immunoprecipitation (IP) buffer (2% Triton X-100, 30 mM Tris-HCl, pH 8.0, 300 mM NaCl, 2 mM EDTA) and incubated with protein G agarose beads (Roche) for mock IP (4 hr at 4° mixing end over end, 400 µl beads per 50 ml extract). Supernatant was removed into fresh tubes and the mock IP beads were stored on ice. The supernatant was incubated with 80 µl anti-FLAG antibody (Sigma) and 400 µl protein G agarose beads per 50 ml of extract for 4 hr at 4° mixing end over end, and then beads were recovered. Mock and IP beads were washed three times with cold 1× IP buffer. Protein was eluted from mock or IP beads with 400 µl 2× NuPAGE LDS buffer (Invitrogen) by boiling for 5 min, followed by a second elution with 400 µl 0.5× NuPAGE LDS buffer. The eluates were combined and diluted with 800 µl of 2× IP buffer, then incubated with 125 µl fresh protein G agarose beads (mock) or 25 µl anti-FLAG antibody plus 125 µl protein G agarose beads (IP), 4° overnight with end-over-end rotation.
The beads were recovered and resuspended in 400 µl proteinase K buffer (100 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.5% SDS, 1 mM CaCl2) and 100 µg purified proteinase K, and incubated overnight at 50° with end-over-end rotation. The supernatant was collected using a SPIN-X tube (Corning) and ethanol precipitated with 0.3 volume of 9 M ammonium acetate, 10 μg of DNA-free glycogen, and 2.5 volumes of 100% ethanol. Spo11 oligos were then quantified and used for library preparation as described (Thacker et al. 2014). Sequencing was performed using Illumina HiSeq in the Memorial Sloan Kettering Cancer Center (MSKCC) Integrated Genomics Operation core facility.
Statistical analyses were performed using R version 3.0.1 (http://www.r-project.org/). Clipping of library adapters and mapping of reads to the sacCer2 genome assembly was performed using a custom pipeline as described (Pan et al. 2011; Thacker et al. 2014). Sequence read totals and mapping statistics are described in Table S2. After mapping, the reads were separated into unique and multiple mapping sets, but only uniquely mapping reads were analyzed in this study (multiple mapping reads constituted a small minority of the total) (Table S2). Each map was normalized to the total number of uniquely mapped reads (reads per million, RPM; excluding reads mapping to mitochondrial DNA or the 2μ plasmid), then maps for biological replicates were averaged. Hotspot calling was performed as described (Pan et al. 2011) on a merged map made by combining the averages of the wild-type, bas1, and ino4 maps. Hotspot positions and their Spo11-oligo counts are compiled in Table S3. In analyses evaluating the fold change in TF mutants, we assumed genome-wide numbers of DSBs are approximately the same in wild type and in the TF mutants, which agreed with direct DSB measurement by Southern blotting (Figure 2, G, H, and K–N). The bas1 B, bas1 C, bas1 D, ino4 A, and ino4 B datasets contained a small number of spurious reads (<0.3% of total in bas1dataset B, <0.04% of total in the other datasets) at positions 244,583 bp and 244,584 bp on chromosome V as well as 152,223 bp on chromosome III, likely from contamination of the Spo11-oligo sequencing libraries with PCR primers from those loci. These reads were deleted from the maps. Sequence reads and compiled Spo11-oligo maps have been submitted to the Gene Expression Omnibus (GEO) repository.
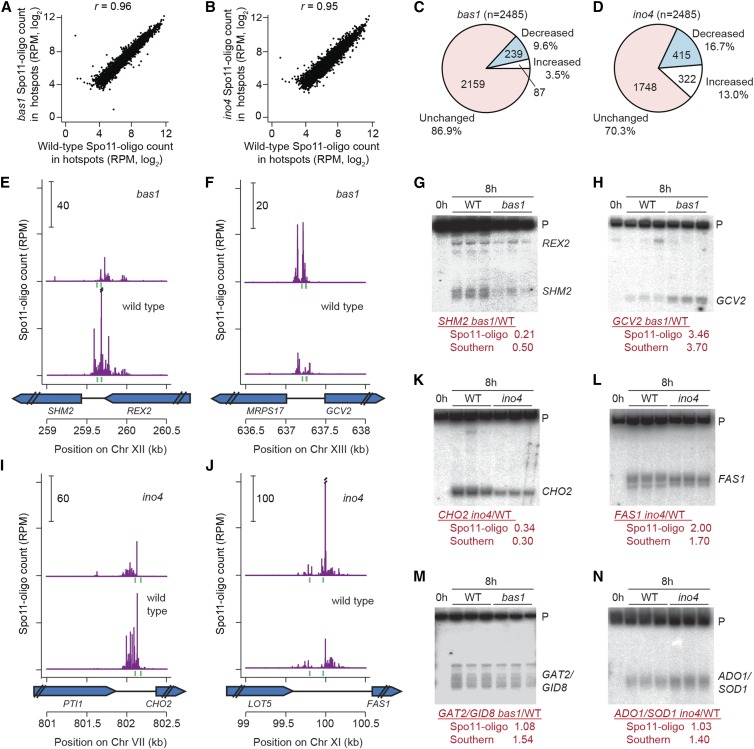
Figure 2.
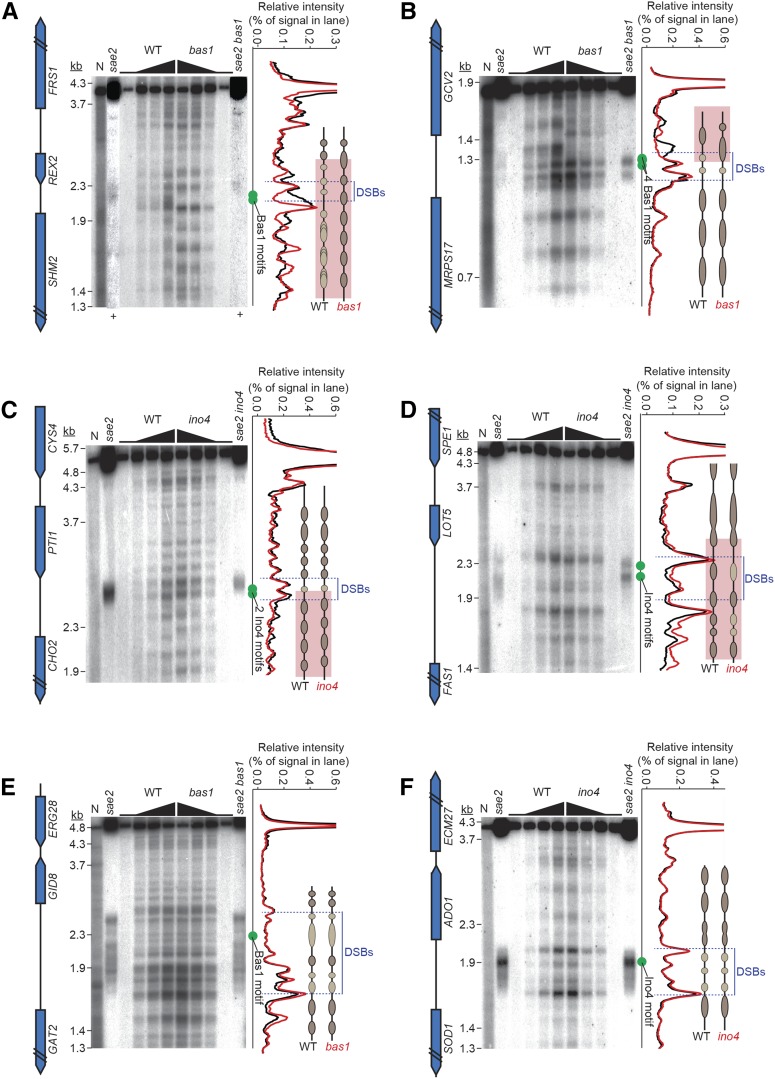
Effects of bas1 and ino4 mutations on DSB activity at individual hotspots. (A and B) Comparisons between wild type and TF mutants for all hotspots. (C and D) Proportion of hotspots that displayed at least a 20% change in Spo11-oligo counts relative to wild type. (E, F, I, and J) Spo11 oligos in hotspots at the promoters of SHM2 (E), GCV2 (F), CHO2 (I), and FAS1 (J). Double slashes indicate where Spo11-oligo counts are off scale. Green vertical ticks indicate sequence matches to Bas1 (E and F) or Ino4 (I and J) binding motifs. (G, H, and K–N) Southern blot results confirming the Spo11-oligo data for hotspots at the promoters of SHM2 (G), GCV2 (H), CHO2 (K), FAS1 (L), GAT2–GID8 (M), and ADO1–SOD1 (N). Genomic DNA was purified from three independent 8-hr sae2Δ cultures for each genotype and analyzed by Southern blotting and indirect end labeling. Fold change (mutant/wild type) for each hotspot is shown below the blots. P, parental.
Detection of meiotic DSBs by Southern blot
Genomic DNA was prepared in agarose plugs as described (Borde et al. 2000; Murakami et al. 2009). DSB analyses at individual hotspots were performed as described (Murakami et al. 2009; Pan et al. 2011). Restriction enzymes and primer sequences for amplification of probes were as follows: GCV2–MRPS17 hotspot, StuI, MRPS17 probe (5′-TCCATCATGACGTGCTTTCT, 5′-AGCCCAAAAGGCAAAAGAAT); GAT2–GID8 hotspot, XbaI, PSO2 probe (5′-GCTTTCGTCCCACATGTCAT, 5′-TCGAAGCAACGCCAAAATGA); SHM2 hotspot, NcoI-HF, SHM2 probe (5′-GAGAATCGGTACCGTTGGAA, 5′-CAGGTGTCATCCCATCTCCT); CHO2 hotspot, XbaI, CYS4 probe (5′-CTGGTGGGACTATTAGCGGT, 5′-AAGAGTCAAAACGGGCCAAC); ADO1–SOD1 hotspot, XbaI, YPS25 probe (5′-AATAGAGCAAGCTTTCGGGC, 5′-ACCCGTCAACCCAATTCTTT); and FAS1 hotspot, XbaI, ASH1 probe (5′-CGCTTAGAGGAGTAGAGGCC, 5′-GCTTGGAGTGTATGCCTTGG). Blots were quantified by phosphoimager.
ChIP-seq to define Bas1 and Ino4 binding sites
Bas1 and Ino4 were C-terminally tagged with eight copies of the Myc epitope (BAS1-Myc and INO4-Myc). ChIP was performed as described (Murakami and Keeney 2014). ChIP efficiencies (percent of input) were measured by qPCR. DNA from ChIP and input samples collected from the tagged strains were sequenced (50-bp paired-end reads) on the HiSeq platform (Illumina). Reads were mapped to the S. cerevisiae sacCer2 genome assembly using BWA mem (version 0.7.5a-r405) to generate coverage maps for each dataset. To remove regions with spurious mapping, the maps from the input samples of the BAS1-myc or INO4-Myc strains were used to define ad hoc “mask regions” where the coverage was out of a fixed range (>5 SD from mean coverage for Bas1 maps, and >4 SD from mean coverage for Ino4 maps; these values were chosen to achieve similar extents of mask coverage for the two sets of samples, accommodating fluctuation of input signals between samples, presumably due to experimental variation in crosslinking efficiency, sonication, and chromatin solubilization). Masks covered 7.2% and 6.6% of the genome for Bas1 and Ino4 maps, respectively; 74.2% of the Bas1 mask regions overlapped Ino4 mask regions (i.e., 81.6% of the Ino4 mask regions were accounted for). These regions were censored from further analysis. Coverage was normalized to the mean coverage calculated from the masked map and made into ChIP/input ratio. The ratio maps were smoothed using a 501-bp Parzen (triangular) sliding window. Using the smoothed maps, peaks were called using as a threshold 2.0 × genome average. De novo motif discovery on sequences corresponding to windowed peaks was performed using MEME (Machanick and Bailey 2011). Matches to position weight matrices for each motif were called using MEME default settings.
Micrococcal nuclease digestion of chromatin
Isolation of nuclei, micrococcal nuclease (MNase) digestion of chromatin, and proteinase K treatment were performed as described (Keeney and Kleckner 1996; Sasaki et al. 2013). After proteinase K treatment, reactions were precipitated with an equal volume of isopropanol and dissolved in 50 µl TE buffer. SHM2 and GCV2 hotspots were analyzed using the same restriction enzymes and probes as in direct DSB measurement above. Additional restriction enzymes and primer sequences for amplification of probes were as follows: GAT2–GID8 hotspot, XhoI, GAT2 probe (5′-CGCGCCTCTTCAAAAGTTAC, 5′-TGGTCCCTTTCTCCATTCTG); CHO2 hotspot, BmgBI (isoschizomer of BtrI), CHO2 probe (5′-TTGATGAAAGCAAGAACTCCAA, 5′-CTCCAATGACCACCTGATCC); ADO1–SOD1 hotspot, HpaI, URA8 probe (5′-ACCACGTTCCCTTATTGCTG, 5′-TCCACCAGGAACCAAAATTC); and FAS1 hotspot, SalI-HF, FAS1 probe (5′-TGGTGCGAAGAATCTAGTCG, 5′-CTCGGAAATGGAACCTGAAA).
ChIP-seq of histone H3 lysine 4 trimethylation
Approximately 250 OD units of cells were collected for each sample. Cells were crosslinked with 1% formaldehyde for 15 min and quenched with 125 mM glycine for 5 min at room temperature. Cells were collected and disrupted by bead beating in 200 µl lysis buffer (50 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate). Chromatin was prepared and washed with MNase reaction buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mercaptoethanol, 0.1% Igepal CA-630 (Sigma-Aldrich) and digested to predominantly mono-nucleosomes with 2 units MNase at 37° for 30 min. ChIP-seq was then performed on nucleosomes with anti-H3 (Abcam, ab1791) or anti-H3 lysine 4 trimethylation (H3K4me3) (Abcam, ab8580) as described (Zhang et al. 2011b; Murakami and Keeney 2014). Reads were mapped to the sacCer2 genome assembly using BWA mem (version 0.7.5a-r405). The midpoint position of each paired-end read was extracted. The coordinate of the midpoints was shifted 73 bp toward both the 5′ and 3′ ends to generate nucleosome coverage maps. Each H3 or H3K4me3 map was normalized to the mean coverage. H3K4me3 enrichment was measured by taking the ratio of H3K4me3 ChIP signal to H3 ChIP signal.
RNA-seq
Deep sequencing of RNA (RNA-seq) was performed on two biological replicates each for wild type, bas1, and ino4. For each sample, 15 OD600 units of cells were collected at 4 hr in meiosis. Cells were resuspended in 500 μl Complete Buffer A (50 mM sodium acetate pH 5.2, 10 mM EDTA, 1% SDS) and 750 μl of phenol equilibrated with Complete Buffer A was added. Cells were disrupted by incubating at 65° for 5 min with vortexing for 10 s every 1.5 min, and then placed on ice for 3 min. After spinning in a mini centrifuge at 13,000 rpm for 6 min, the bottom layer was removed and fresh phenol was added to the top layer. Incubation and vortexing were repeated, except that the top layer was then transferred to a new 1.5-ml tube. RNA was purified by two rounds of phenol/chloroform extraction, one round of chloroform extraction, and ethanol precipitation. Precipitates were dissolved in 100 μl TE (10 mM Tris-HCl pH 6.8, 1 mM EDTA). Messenger RNA (mRNA) was enriched by poly-A selection and sequenced using Illumina HiSeq in the Genomics Resources Core Facility of Weill Cornell Medical College. Reads were mapped to the S. cerevisiae sacCer2 genome assembly using the Spliced Transcripts Alignment to a Reference (STAR) software (Dobin et al. 2013). Transcriptome assembly and differential expression analysis were performed using the Cufflinks software (Trapnell et al. 2010) and R Cummerbund package (http://compbio.mit.edu/cummeRbund/).
Data availability
Strains are available upon request. Next-generation sequencing data were deposited at the Gene Expression Omnibus (GEO) under accession nos. GSE67910 (Spo11-oligo mapping), GSE67912 (Bas1 and Ino4 ChIP-seq), GSE67907 (H3K4me3 ChIP), and GSE70911 (RNA-seq).
Results
High-resolution maps of meiotic DSBs in bas1 and ino4 mutants
Each covalently bound Spo11 oligo is a unique tag from a DSB site, so sequencing Spo11 oligos generates quantitative, high-resolution DSB maps (Pan et al. 2011). Spo11-oligo maps from biological replicate cultures of wild type and bas1 and ino4 mutants agreed well with each other (Pearson’s r = 0.92–0.98; Figure 1B) and thus were averaged (after normalization to millions of reads mapped, RPM) into consensus maps for further analysis.
The DSB landscape is shaped by a combination of many factors that operate at different scales. At large scales (tens of kilobases), bas1 and ino4 Spo11-oligo maps agreed with the wild-type map (Figure 1C, top). At small scales (<1 kb), DSB hotspots are usually located in nucleosome-depleted promoters. This pattern was retained in bas1 and ino4 mutants, in that DSBs formed in the same hotspots (Figure 1C, bottom). The correlation of Spo11-oligo density in 5-kb bins was high across the genome (Pearson’s r = 0.95–0.97) (Figure 1D). Thus, these TF mutants do not grossly alter the global DSB landscape.
To compile a list of hotspots to facilitate direct comparison of each mutant to wild type, we pooled the averaged wild-type map, the averaged bas1 map, and the averaged ino4 map, then called hotspots (clusters of Spo11 oligos) on this combined map using a previously described algorithm (Pan et al. 2011). We identified 3994 hotspots (Table S3). Of these, 3524 (88.2%) overlapped the 3600 hotspots previously identified in wild type (Pan et al. 2011) (i.e., 97.9% of the previous hotspots were accounted for). The extra hotspots in the current study were weak hotspots (≤250 RPM, equivalent to a DSB frequency of ≤0.45% of DNA, as estimated by comparing Southern blot data to Spo11-oligo counts) (Pan et al. 2011), probably emerging from the greatly increased sequencing depth in the current study.
The wild-type and bas1 Spo11-oligo maps correlated moderately with the published microarray maps (Mieczkowski et al. 2006) (Pearson’s r = 0.47–0.51) but had much higher spatial and quantitative resolution (Figure 1E). The high-resolution maps allowed us to analyze DSB activity at the individual hotspot level, which was not possible with the earlier datasets. First, we examined Spo11-oligo counts in mutants compared with wild type. Most hotspots were affected little if at all in the mutants (Figure 2, A and B), but a small number were increased or decreased so we sought to identify all changed hotspots. To minimize impact of experimental variation on calculated fold change of weak hotspots, only hotspots with >100 RPM in at least one map were analyzed (100 RPM is equivalent to a DSB frequency of ∼0.16% of total DNA). After this filtering of weak hotspots, 2485 hotspots remained. We defined altered hotspots as having >20% change in Spo11-oligo counts with a P-value <0.1 (Student’s t-test across all biological replicates, without adjustment for multiple correction).
Even with these very nonstringent criteria, only a small fraction of hotspots were detectably altered by the bas1 mutation: 239 (9.6%) hotspots were decreased, and 87 (3.5%) were increased (Figure 2C). Of these, only 18 hotspots had changed by more than 2-fold (10 down and 8 up) (Table S3). The most decreased hotspot was in the ADE17 promoter (17% of wild type) and the most increased was a relatively weak hotspot inside the ADE4 open reading frame (6.1-fold higher than wild type; a stronger hotspot in the ADE4 promoter was decreased 2.0-fold). Only 7 of the altered hotspots are in regions where the prior microarray study by Mieczkowski et al. (2006) also detected changes in bas1 (at HIS4, SHM2, GCV2, ADE1, AIM9, PLP2 and YER091C-A). Regions that did not show agreement between the studies are likely attributable to strain differences (including use of the rad50S mutation in prior work) and/or the low sensitivity and spatial resolution of the prior study, which did not allow precise determination of hotspot locations.
The ino4 mutation affected more hotspots: 415 (16.7%) were decreased by >20% and 322 (13.0%) were increased (Figure 2D). Of these, 79 had changed by more than two-fold (47 down and 32 up) (Table S3). The range of hotspot activity changes in ino4 relative to wild type was 0.23 (OPI3–MOG1 hotspot) to 4.4 (INO1 hotspot). Thus, deletion of these TFs is capable of changing hotspot activity, but affects only a small portion of total hotspots.
To validate results from Spo11-oligo mapping, we analyzed meiotic DNA formation directly by standard Southern blot analysis for hotspots at the promoters of SHM2, GAT2–GID8, GCV2, CHO2, ADO1–SOD1, and FAS1 in the rad50S-like (DSB repair deficient) mutant sae2. Consistent with Spo11-oligo maps, DSB activity was significantly reduced for SHM2 and elevated for GCV2 in the bas1 mutant (Figure 2, E–H), in agreement with previous results in a different strain background (Mieczkowski et al. 2006). In ino4, CHO2 showed decreased DSB formation and FAS1 showed increased DSB levels as well as altered spatial pattern (Figure 2, I–L). GAT2–GID8 and ADO1–SOD1 were unchanged in TF-mutant Spo11-oligo maps, but both hotspots were slightly increased by Southern blot analysis (Figure 2, M and N). It is important to note that sae2 mutants suppress late-forming DSBs (Borde et al. 2000). The discrepancies may arise from the use of a rad50S-like mutant background for the Southern blot determinations or may trace to measurement imprecision in either Spo11-oligo maps or Southern blot determinations. Prior studies in S. cerevisiae and S. pombe have established excellent overall quantitative agreement between Spo11 (Rec12) oligo counts and DSB frequencies measured by Southern blotting, but small differences between the methods have been noted in the past for some hotspots (Pan et al. 2011; Sasaki et al. 2013; Fowler et al. 2014), comparable to the differences seen here. Thus, as expected, the Spo11-oligo maps in general showed excellent quantitative agreement with direct assays of DSBs and, equally important, provided excellent spatial accuracy in defining DSB positions (Pan et al. 2011; Fowler et al. 2014).
Identification of Bas1 and Ino4 binding sites in meiotic cells
To evaluate whether hotspots altered in bas1 or ino4 mutants might be directly influenced by the TFs, we mapped Bas1 and Ino4 binding sites during meiosis by ChIP of Myc-tagged Bas1 and Ino4 followed by high-throughput sequencing. ChIP peaks were called as described (Murakami and Keeney 2014) (Materials and Methods), identifying 36 Bas1 binding peaks and 197 Ino4 binding peaks (Figure 3, A and B; Table S4 and Table S5). Most of these peaks (30 of the 36 Bas1 binding peaks and 176 of the 197 Ino4 binding peaks) were located in or near the promoters of genes. Specific enrichment of Bas1 at the promoters of GCV2 and GCV3 and of Ino4 at the FAS2 promoter was also observed by ChIP followed by qPCR, validating results from ChIP-seq (Figure 3C).
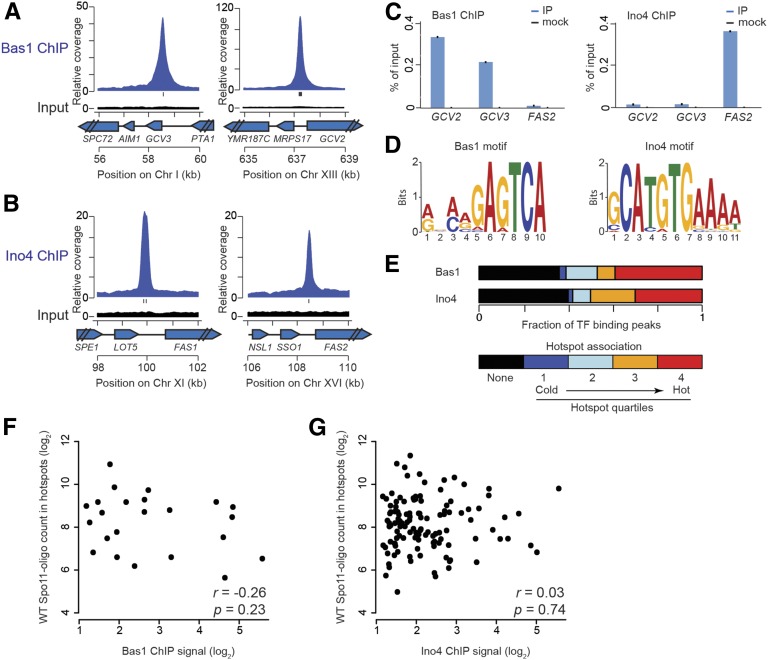
Figure 3.
TF binding sites and their association with DSB hotspots in meiosis. (A and B) Examples of Bas1 (A) or Ino4 (B) ChIP-seq data. Plots show the sequence coverage relative to genome average for ChIP and input samples. Each input is plotted on the same scale as the respective ChIP data. Tick marks under the ChIP plots indicate matches to Bas1 or Ino4 motifs. (C) ChIP-qPCR at GCV2, GCV3, and FAS2 promoters. (D) Consensus Bas1 and Ino4 motifs identified in meiotic ChIP-seq datasets. (E) Association of TF binding peaks with hotspots. Bars depict the fraction of ChIP-seq peaks not in DSB hotspots (black) and divide the remainder according to hotspot quartile. (F and G) For hotspots overlapping TF binding sites, there is no correlation between DSB hotspot activity in wild type (WT) and TF ChIP-seq signal (summed in a 401-bp window around the ChIP-seq midpoint).
We searched for likely TF recognition motifs in the 500 bp encompassing each ChIP-seq peak (Bailey et al. 2009). This analysis identified a motif containing the core sequence 5′-GAGTCA for Bas1 binding peaks (Figure 3D), in agreement with previous studies (Daignan-Fornier and Fink 1992; Mieczkowski et al. 2006; Zhu et al. 2009). More than half of the Bas1 ChIP-seq peak regions (21 of 36) contained one or more match to this motif. Peaks without a match to the motif may contain degenerate Bas1 binding sequences, may be targeted by Bas1 via interactions with other proteins, or may be false positives from the ChIP-seq. The Bas1 ChIP-seq peaks accounted for most (27 of 37; 70% within 500 bp) of a previously annotated set of Bas1 binding sites (MacIsaac et al. 2006).
Mieczkowski et al. (2006) identified 56 promoter intergenic regions with ChIP evidence for binding by Bas1 in meiotic cells and with a perfect match to the Bas1 recognition motif. Twenty-one of these regions overlapped with our Bas1 ChIP-seq peaks (Table S4). These overlapping Bas1 binding sites are in the promoters of many known Bas1-regulated genes involved in the adenine and histidine biosynthetic pathways (ADE1, ADE2, ADE4, ADE6, ADE8, ADE17, and HIS1) and in one-carbon metabolism (GCV1, GCV2, GCV3, MDT1, and SHM2) (Denis et al. 1998; Denis and Daignan-Fornier 1998; Gelling et al. 2004). However, HIS4 and a few other known Bas1 targets were not identified in our dataset. For HIS4, a very weak ChIP-seq signal was present below the peak-calling threshold (Figure S1), presumably reflecting a quantitative difference in Bas1 occupancy of the HIS4 promoter between SK1 (where HIS4 is only a modest DSB hotspot (Table S3) and the strain background used by Petes and colleagues (Stapleton and Petes 1991; Mieczkowski et al. 2006).
For Ino4, MEME identified the motif 5′-GCATGTGAAAA (Figure 3D), which matches well the upstream activation sequence recognized by Ino2/4 (UASino, C/AATGTGAAAT) (Bachhawat et al. 1995). Most (162 of 197) Ino4 ChIP-seq peaks contained at least one match to the identified motif. Furthermore, Ino4 ChIP-seq peaks accounted for all but one of the small number of previously annotated Ino4 motifs (21 of 22; 95.5%) (MacIsaac et al. 2006).
Hotspots containing Bas1 and Ino4 binding sites
In a prior study using a compilation of TF binding sites by MacIsaac et al. (2006), we observed that 32 of 37 annotated Bas1 sites were contained within 18 hotspots, three-fourths of which were among the hottest 50% of all hotspots (Pan et al. 2011). For Ino4, all but 1 of the 22 annotated binding sites were inside 17 hotspots, nearly all of which were among the hottest 50% of all hotspots (Pan et al. 2011). These findings suggested that presence of a potential binding site(s) for Bas1 or especially Ino4 was a good predictor of strong local DSB activity. Superficially at least, these findings also seemed to support the hypothesis that simple sequence motifs targeted by these TFs are a dominant factor shaping local DSB patterns (Wahls and Davidson 2010). However, the available TF binding site data were incomplete and did not represent DNA binding in meiotic cells. Specifically, the sites compiled by MacIsaac et al. (2006) marked only a minority of the ChIP-seq peaks (38.9% of Bas1 peaks; 8.5% for Ino4). We therefore revisited comparisons between TF binding and DSBs with the new TF binding site data in hand.
We divided hotspots into quartiles according to Spo11-oligo counts and calculated the percentage of TF ChIP-seq peaks that overlapped hotspots in each quartile. Unexpectedly, ∼40% of each TF’s ChIP-seq peaks were in loci that did not score as hotspots, even with our nonstringent hotspot definition (Figure 3E). Thus, binding of neither Bas1 nor Ino4 is sufficient for high local DSB activity. Interestingly, however, those Ino4 sites that were in hotspots tended to be in relatively hot ones, similar to our prior findings (Figure 3E). TFs bind to their target sites with different fractional occupancies in vivo, and the range of ChIP-seq peak intensities was large (Table S4 and Table S5), so we asked whether DSB hotspot activity correlates with TF binding strength (ChIP signal). No significant correlation was observed (Figure 3, F and G). Therefore, counter to expectation from our prior study, the binding of these TFs has essentially no predictive power for DSB activity.
Despite this lack of predictive power overall, we asked whether Bas1 or Ino4 influences the activity of the subset of hotspots that is directly bound by these TFs during meiosis. We used the same nonstringent criteria for defining hotspot changes as above (P < 0.1; >20% change from wild type; only hotspots with >100 RPM in at least one dataset). In bas1 mutants, about half of the 23 hotspots overlapping Bas1 ChIP-seq peaks had altered Spo11-oligo counts: 9 were decreased, 3 were increased (Figure 4A). Although only a fraction, this number (12/23) is significantly higher than expected by chance (P < 9.9 × 10−6, Fisher’s exact test comparing to the fraction (∼13%) of total hotspots changed in the bas1 mutant) (Figure 2C). In ino4 mutants, only one-third of the 119 hotspots overlapping Ino4 peaks were changed: 23 had decreased Spo11-oligo counts and 15 were increased (Figure 4B; compare to Figure 2D). This number (38/119) is no different than expectation from random (29.7% of hotspots were changed in the ino4 mutant) (P = 0.61).
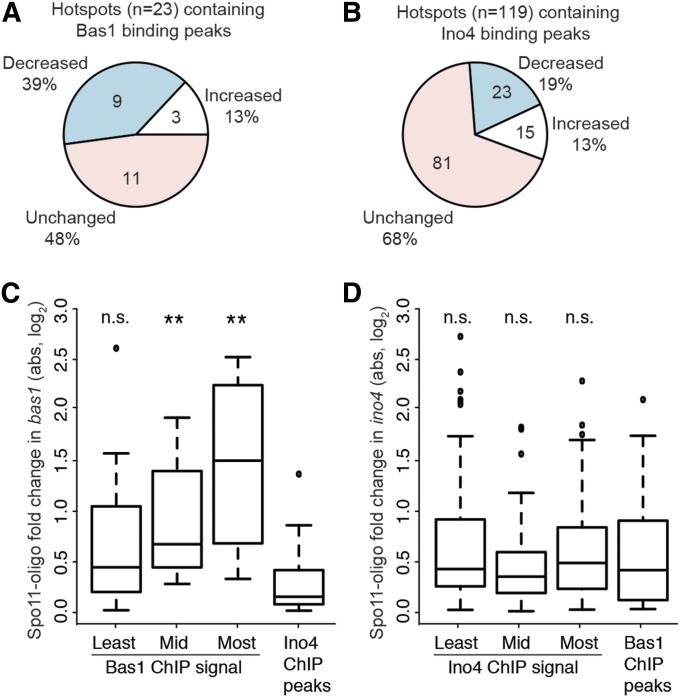
Figure 4.
Context-dependent effects of bas1 and ino4 mutations around TF binding sites. (A and B) Proportion of hotspots overlapping TF ChIP-seq peaks that displayed at least 20% change in Spo11-oligo counts relative to wild type. (C and D) Changes in Spo11-oligo counts around all TF binding sites in the TF mutants. For each TF, all detected binding sites (regardless of whether the site was scored as a hotspot) were divided into three groups according to ChIP-seq signal, and Spo11-oligo counts were summed in 401-bp windows surrounding the peaks. Absolute log fold changes relative to wild type are plotted. Boxes indicate median and interquartile range; whiskers indicate the most extreme data points, which are ≤1.5 times the interquartile range from the box; individual points are outliers. For the bas1 mutant (C), 12 Ino4 ChIP-seq peaks were randomly chosen to serve as a negative control set. For the ino4 mutant (D), all Bas1 ChIP-seq peaks were used as the negative control. Results are shown for Wilcoxon rank-sum tests comparing each TF binding site group to the respective negative control group (n.s., not significant; **P < 0.01).
We further evaluated the effects of the TF mutations on local DSB activity by examining DSB changes in the TF mutants around all Ino4 and Bas1 binding peaks, regardless of whether these loci scored as DSB hotspots. We divided TF binding peaks into three groups according to their ChIP-seq signals, and then measured the absolute fold change in local Spo11-oligo counts in the TF mutants (i.e., considering all changes equivalently, whether increasing or decreasing). Relative to control loci, Bas1 binding peaks tended to show greater change in local Spo11-oligo density in the bas1 mutant, and this trend was statistically significant for the two-thirds of binding peaks with the highest ChIP-seq signal (Figure 4C). No such trend was observed for Ino4 (Figure 4D).
These findings agree with and substantially extend prior results implying that Bas1 often influences DSB formation nearby, but that the magnitude and direction (increasing or decreasing) of influence is context dependent (Mieczkowski et al. 2006). In interesting contrast to Bas1, even though ino4 mutation has a larger net effect on DSB distribution genome-wide, Ino4 modulates DSB formation near its binding sites only rarely and thus appears to exert more of its influence indirectly. Thus, the previously apparent correlation of Ino4 binding sites with strong DSB hotspots (Pan et al. 2011) was not predictive of a widespread functional relationship.
Chromatin structure in and near hotspots in bas1 and ino4 mutants
Open chromatin structure provides a window of opportunity for Spo11 (Ohta et al. 1994; Wu and Lichten 1994; Fan and Petes 1996; Keeney and Kleckner 1996; Berchowitz et al. 2009; Pan et al. 2011). Given precedents of HIS4 and PHO5 (Introduction), a simple prediction might be that hotspots whose DSB activity is affected in TF mutants would display correlated changes in chromatin structure: increased accessibility if TF removal increases DSBs, and decreased accessibility in hotspots with decreased DSBs (Mieczkowski et al. 2006). We tested this prediction by examining chromatin structure in the hotspots validated by Southern blotting (Figure 2). As described below, the relationship between chromatin and DSB changes is context specific and more complex than the simplest model would predict.
Intact nuclei were prepared from meiotic cultures of wild-type and mutant cells and partially digested with MNase. DNA was then purified and analyzed by Southern blotting (Figure 5). MNase-digested naked DNA verified that banding patterns were chromatin dependent. Genomic DNA from meiotic cultures of sae2 mutants was run in parallel to mark the positions of meiotic DSBs.
Figure 5.
Chromatin structure in and around hotspots in wild type and TF mutants. MNase digestion patterns are shown for SHM2 (A), GCV2 (B), CHO2 (C), FAS1 (D), GAT2–GID8 (E), and ADO1–SOD1 (F). Intact meiotic nuclei were treated with zero or increasing amounts of MNase, and then DNA was purified, digested with restriction enzymes, and analyzed by Southern blotting and indirect end labeling. DSB positions are shown using DNA purified from sae2 meiotic cultures. N, naked DNA (purified genomic DNA from vegetative cells, treated with MNase). Approximate positions of Bas1 or Ino4 motifs are indicated with green dots. The hybridization signals from the lanes with highest MNase concentration are plotted on the right of each panel, with interpretive cartoons illustrating regions of MNase resistance: darker brown ovals for regions that are relatively more MNase-resistant, lighter tan ovals for relatively less resistant, black lines for MNase-hypersensitive, and pink highlighting for regions with changed MNase digestion patterns. Note that the ovals do not necessarily correspond to positioned nucleosomes; some are smaller than nucleosomes and may reflect footprints of transcription factors and/or subnucleosomal histone particles. + signs in A indicate lanes for which contrast was separately enhanced to show the DSB postions better.
In wild-type cells, the SHM2 promoter (where bas1 mutation reduces DSB formation) (Figure 2, E and G) displayed a broad zone of MNase hypersensitivity encompassing the Bas1 binding motifs, and DSBs formed across part of this zone (Figure 5A). The DSB distribution did not match the fine-scale MNase pattern, analogous to observations at HIS4 (Fan et al. 1995). In the bas1 mutant, the Bas1 motifs became strongly protected from MNase, indicating presence of a nucleosome. The remaining DSBs were now restricted to a smaller region corresponding to an MNase-hypersensitive site flanking the new positioned nucleosome (Figure 5A). Narrowing of the DSB zone was also seen in the Spo11-oligo map (Figure 2E). Previous studies showed that mutating these Bas1 motifs diminishes DSB formation in strains expressing Bas1 (Mieczkowski et al. 2006), so the chromatin changes we observed are presumably direct effects of Bas1.
In broad strokes, the parallel changes in chromatin and DSB formation near the Bas1 binding sites appeared similar to those at HIS4 and PHO5 and seemed consistent with the interpretation that reducing DNA accessibility via placement of a nucleosome may be sufficient by itself to reduce and spatially constrain DSB formation. However, the bas1 mutant displayed substantial alteration in chromatin structure beyond the immediate vicinity of the Bas1 motifs, corresponding to reduced transcription of SHM2 (based on RNA-seq results, discussed further below). The SHM2 transcription unit showed a nucleosomal ladder that was broad and shallow in wild type, indicating variable nucleosome positioning in the population. This ladder was more pronounced in bas1, indicating a more regularly positioned array (Figure 5A). The neighboring gene (REX2) also had a more regular nucleosome array at its 3′ end when Bas1 was absent. Furthermore, the bas1 mutant retained just as much MNase hypersensitivity as wild type right where the residual DSB formation occurred (Figure 5A), indicating that most cells still present a usable window of opportunity for Spo11. Because the bas1 mutant still has accessible DNA that can be targeted for DSB formation, and because absence of Bas1 triggers chromatin restructuring several kilobases around this locus, it is possible that the change in DSB frequency is caused by other factors besides a change in nucleosome occupancy at the positions where Spo11 cleaves (addressed further in Discussion).
Substantial chromatin changes were also observed in the GCV2–MRPS17 intergenic region, which experiences more DSBs in bas1. Wild-type cells displayed a broad MNase-hypersensitive site near the 5′ end of GCV2 (Figure 5B). This site was flanked on the GCV2-proximal side by a zone of protection, presumably from a well-positioned, high-occupancy nucleosome overlapping the 5′ end of GCV2. On the GCV2-distal side, two more strong MNase-hypersensitive sites were interdigitated with two areas of modest protection smaller than nucleosomes, presumably from subnucleosomal histone particles or other DNA binding proteins. One of the strong MNase sites overlapped four closely spaced Bas1 motifs. A nucleosome ladder extended into the adjacent MRPS17 gene, which is expressed during sporulation and is not regulated by Bas1 (see below). DSBs occurred diffusely throughout a region encompassing the MRPS17-proximal MNase-hypersensitive sites and trailing weakly into the hypersensitive site that overlapped most of the Bas1 motifs. As at SHM2, DSBs correlated little with the fine-scale MNase pattern (Figure 5B).
In bas1, the broad MNase-hypersensitive site nearest to GCV2 disappeared; apparently the nucleosome that should overlap the 5′ end of GCV2 was now positioned over this region (Figure 5B). The nucleosome array over the GCV2 transcription unit was also repositioned and less distinct. In contrast, the nucleosome ladder in MRPS17 and the MNase-hypersensitive sites closer to MRPS17—including the one overlapping the Bas1 motifs—were unaffected. Meiotic DSBs not only became more frequent, they were now more spatially restricted and more closely followed the pattern of MNase cleavage, including prominent cleavage overlapping the Bas1 motifs (Figure 2F and Figure 5B).
A straightforward interpretation is that DSBs in wild type occur in portions of the adjacent MRPS17 and GCV2 promoters. Absent Bas1, part of the GCV2 promoter becomes occluded by a nucleosome that partially blocks Spo11 access. Nonetheless, hotspot activity increases, with Spo11 now more constrained to cleave within a pair of MNase-hypersensitive sites closer to the MRPS17 promoter. Note that, while this intergenic region became more MNase resistant overall, accessibility of the MRPS17-proximal MNase-hypersensitive sites themselves did not detectably change. Thus, the fine-scale DSB distribution within the hotspot reflects DNA accessibility changes, but quantitative measures of MNase accessibility are a poor predictor of total DSB levels.
The CHO2 promoter contains two Ino4 motifs and experiences a decrease in DSBs in the ino4 mutant (Figure 2, I and K). In wild type, a pair of strong MNase-hypersensitive sites bracketed a subnucleosomal-sized zone of modest protection overlapping the motifs (Figure 5C). Positioned nucleosomes were present across the CHO2 transcription unit and the 3′ end of the adjacent gene (PTI1). DSBs were concentrated across the MNase-hypersensitive sites around the Ino4 motifs, including the motifs themselves and their small MNase-protected zone (Figure 2I and Figure 5C). The ino4 mutant had a small decrease in MNase cleavage in the more CHO2 proximal of the pair of MNase-hypersensitive sites near the motifs, but this segment overall remained strongly MNase hypersensitive (Figure 5C). There was a pronounced change in the spacing of nucleosomes across the CHO2 transcription unit, but no changes were seen upstream of the Ino4 motifs across PTI1. DSBs had similar spatial patterning in the TF mutant as in wild type, agreeing with the Spo11-oligo maps (Figure 2I and Figure 5C). Thus, the essentially unchanged chromatin accessibility right at the DSB hotspot paralleled the relatively unchanged DSB distribution, whereas the number of DSBs and the chromatin structure in the adjacent transcription unit changed substantially.
The FAS1 promoter also contains two Ino4 motifs, but this region experiences an increase in DSB formation in ino4 (Figure 2, J and L). In wild type, two strong MNase-hypersensitive sites were located on either side of a more modest doublet of MNase sensitivity, with the Ino4 motifs in a more MNase-resistant portion of this segment (Figure 5D). DSBs formed a pair of smears across the region containing the Ino4 motifs and bore little similarity to the fine-scale MNase pattern. In the ino4 mutant, much of the intergenic region became modestly more sensitive to digestion, but the spatial array of bands was not detectably altered and the strong MNase-hypersensitive sites were largely unchanged (Figure 5D). DSBs were spread across a similar total area as in wild type, but redistributed so that segments containing the Ino4 motifs received a higher fraction of total DSBs. In this case, increased DSB frequency correlated with increased overall MNase hypersensitivity.
Finally, as controls we examined the GAT2–GID8 and SOD1–ADO1 intergenic regions, which contain single Bas1 or Ino4 motifs, respectively, and whose DSB levels were unchanged in the TF mutants (Figure 2, M and N). GAT2–GID8 displayed a complex MNase pattern, with numerous MNase-hypersensitive sites upstream of the 5′ end of GAT2 and a smaller hypersensitive zone near the 5′ end of GID8 (Figure 5E). DSBs were distributed across much of the intergenic region, and again the DSB spatial pattern correlated poorly with the fine-scale MNase pattern. Absence of Bas1 caused little or no change in the MNase pattern, even around the Bas1 motif, and there was likewise little change in the DSB pattern (Figure 5E). Similarly, SOD1–ADO1 displayed several MNase hypersensitive sites spanning the region where DSBs appeared as a diffuse smear, with no clear correlation of DSBs with MNase cleavage (Figure 5F). We observed no change in either chromatin structure or DSB distribution in the ino4 mutant.
Taken together, these examples show that the relationships between TF binding, local chromatin structure, and DSB patterns are complex and context specific. In three cases where removing the TF affected chromatin structure right where DSBs would normally form, we observed corresponding changes in the fine-scale DSB distribution (Figure 5, A, B, and D). Conversely, fine-scale DSB patterns were not altered at three loci with unchanged MNase patterns around the wild-type DSB positions (Figure 5, C, E, and F). Importantly, however, there was no clear correlation between quantitative changes in MNase sensitivity and changes in overall DSB frequencies, ruling out effects on chromatin accessibility alone as a universal explanation for how TFs modulate DSB hotspot activity.
Changes in DSB frequency do not correlate with changes in histone H3 lysine 4 trimethylation around hotspots
In meiosis, sister chromatids form a linear protein axis (the axial element), with chromatin emanating out in loops; DSBs are more likely to occur in DNA sequences that are in loops, but many of the proteins needed for DSB formation are enriched on the axis (Blat et al. 2002; Kumar et al. 2010; Panizza et al. 2011). This and other observations have led to a “tethered loop–axis complex” model in which DSBs are formed in loop segments captured by axis-associated DSB machinery (Blat et al. 2002; Panizza et al. 2011) (Figure 6A). Recently, it was discovered that the DSB-promoting protein Mer2 interacts with Spp1, a protein containing a PHD (Plant Homeo Domain) finger that binds H3K4me3 marks and may thereby tether the chromatin loop to the axis, promoting DSB formation in nucleosome-depleted regions in loops (Acquaviva et al. 2013; Sommermeyer et al. 2013). Thus, one hypothesis to account for altered DSB frequencies could be that deletion of TFs alters the H3K4me3 modification status around certain promoters and thereby affects Spp1-dependent targeting efficiency.
Figure 6.
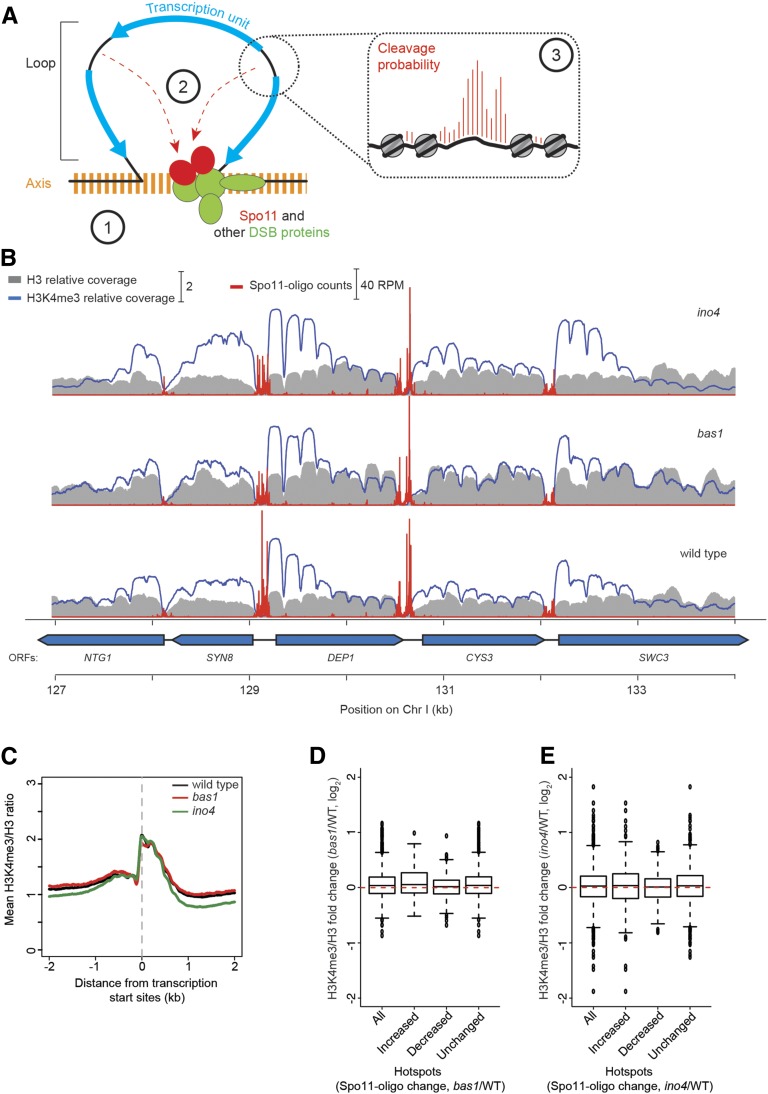
Tethered loop–axis model for DSB formation, and H3K4me3 levels in bas1 and ino4 mutants. (A) Tethered loop–axis model for DSB formation (based on Blat et al. 2002; Panizza et al. 2011; Acquaviva et al. 2013; Sommermeyer et al. 2013). Encircled numerals highlight potentially TF-dependent levels of the hierarchical combination of factors that shape the DSB distribution. (1) DSB-forming potential over regions of ∼5–20 kb is established by the loop–axis structure of meiotic chromosomes and the axis-association of DSB-promoting proteins. (2) Within a loop, gene promoters are preferentially targeted for DSB formation, in part by interactions between the DSB machinery and promoter-proximal H3K4me3. (3) Within a hotspot, the fine-scale distribution of DSBs is shaped in part by accessibility of the DNA to Spo11, in competition with nucleosomes and other DNA binding proteins. (B) Snapshot of anti-H3K4me3 and anti-H3 ChIP-seq data from wild-type, bas1, and ino4 datasets. Sequence coverage values are normalized to genome average. Spo11-oligo counts are superimposed for comparison. (C) Average profiles of H3K4me3/H3 ratios relative to transcription start sites. (D and E) Log fold change of H3K4me3/H3 ratios in groups of hotspots that responded as indicated to the bas1 (D) or ino4 (E) mutations. Hotspot groups are as indicated in Figure 2, C and D. Because the hotspots themselves are highly nucleosome depleted, we summed H3K4me3/H3 ratios in 500-bp windows offset by 200 bp away to the left and right of each hotspot’s midpoints.
To test this hypothesis, we performed ChIP-seq to measure H3K4me3 levels in wild type and TF mutants. Cells from 4-hr meiotic cultures were crosslinked with formaldehyde and disrupted. Chromatin was digested into mononucleosomes with MNase, immunoprecipitated with antihistone H3 and anti-H3K4me3 antibodies, and deep sequenced. Resulting maps showed the expected pattern of preferential H3K4me3 enrichment for nucleosomes near the 5′ ends of most genes (Figure 6, B and C) (Pokholok et al. 2005; Borde et al. 2009; Zhang et al. 2011b). However, counter to the hypothesis motivating this analysis, we observed no significant difference between the increased, decreased, and unchanged DSB hotspot groups when we examined the change in H3K4me3-to-H3 ratios caused by the bas1 or ino4 mutation (Figure 6, D and E). Thus, changes in the level of this histone modification in TF mutants provide no predictive power for changes in DSB frequency. This finding fits with the observation that, even though H3K4me3 promotes Spo11 activity in most promoter regions (84% of DSB sites exhibited a severely reduced DSB frequency in the absence of H3K4me3) (Borde et al. 2009; Acquaviva et al. 2013; Sommermeyer et al. 2013), quantitative measures of H3K4me3 correlate poorly if at all with DSB hotspot strength (Tischfield and Keeney 2012).
Changes in DSB frequencies do not correlate with changes in gene expression
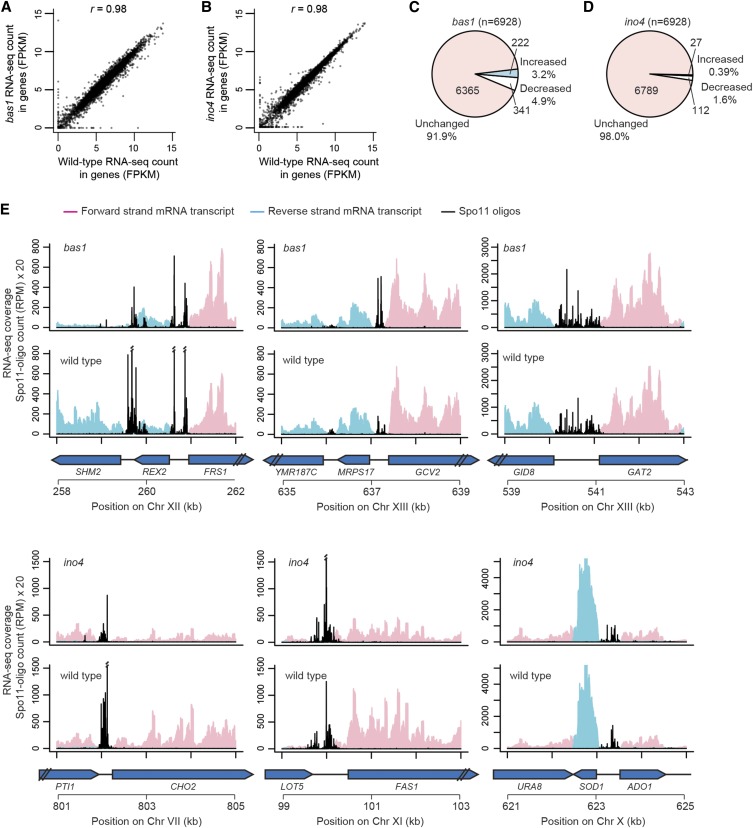
Mutation of the TATA box at HIS4 greatly reduces transcription with little effect on DSB frequency, so transcription is not required for hotspot activity of a promoter (White et al. 1992). Furthermore, genome-wide correlations between transcription activity and DSB frequencies in promoters are weak to nonexistent (Tischfield and Keeney 2012). Nonetheless, to address the possibility that changes in hotspot activity in TF mutants might reflect alterations in transcription, RNA-seq was carried out during meiosis in wild-type, bas1, and ino4 strains (Figure 7, A and B). We identified 222 significantly up-regulated and 341 down-regulated genes in bas1 mutants, as well as 27 up-regulated and 112 down-regulated genes in ino4 mutants (false discovery rate q < 0.05) (Figure 7, C and D; Table S6 and Table S7). Bas1-regulated genes were enriched for Gene Ontology (GO) terms purine nucleotide metabolic process (31 of 563 genes, 5.5%), phosphorus metabolic process (93 genes, 16.5%), carboxylic acid metabolic process (56 genes, 9.9%), and meiotic cell cycle process (68 genes, 12.1%). Ino4-regulated genes were enriched for GO terms lipid biosynthetic process (13 of 139 genes, 9.4%) and meiotic cell cycle process (25 genes, 18.0%). Thus, in addition to known Bas1- or Ino4-regulated pathways, some genes involved in meiosis were also differentially expressed in bas1 and ino4. This may reflect direct or indirect effects of these TF mutations on meiotic entry or progression or genotype-independent differences from culture-to-culture variation in timing.
Figure 7.
mRNA levels in bas1 and ino4 mutants. (A and B) Comparisons of RNA-seq read counts (in fragments per kilobase per million mapped reads, FPKM, log2-transformed) between wild type and TF mutants for all annotated genes. (C and D) Proportion of genes that were significantly changed relative to wild type (false discovery rate q < 0.05). (E) Snapshots of RNA-seq data around hotspots at the promoters of SHM2, GCV2, GAT2–GID8, CHO2, FAS1, and ADO1–SOD1. Per-base pair RNA-seq coverage is plotted separately for forward and reverse reads after normalization to set the total coverage as equal in all samples. The profile for each genotype is the average of the normalized coverage maps from two biological replicates. Double slashes indicate where Spo11-oligo counts are off scale.
Next, we examined the gene expression patterns around the six hotspots characterized above. Each of these hotspots contained a Bas1 or Ino4 ChIP-seq peak, but TF deletion had different effects on gene expression. As expected (Denis and Daignan-Fornier 1998), SHM2 mRNA level was substantially reduced (8.9-fold) after deleting Bas1 (Figure 7E; Table S6). In contrast, mRNA levels of GCV2 and MRPS17 were unchanged in bas1, even though the chromatin structure at GCV2 was substantially altered (Figure 5B) and DSB frequency in this divergent intergenic region was increased. For Ino4-bound hotspots, both CHO2 and FAS1 showed similarly reduced mRNA levels in ino4 (3.3- and 2.5-fold, respectively) (Figure 7E; Table S7), but had DSB levels that moved in opposite directions. For the genes flanking the control (unchanged) hotspots GID8–GAT2 and ADO1–SOD1, mRNA levels were unchanged in the TF mutants. Thus, there is no simple correlation between changes in Spo11-oligo counts and mRNA levels.
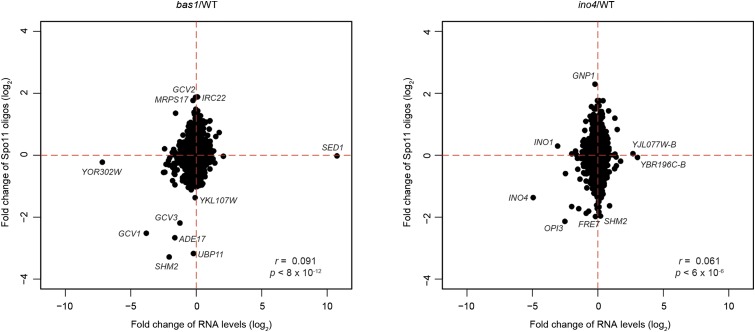
To extend this analysis to the whole genome, we plotted the fold change of mRNA level for each nondubious gene against the fold change of Spo11-oligo counts in its promoter (Figure 8). Both mutants showed significant but very weak correlations between these changes, such that variation in transcript levels explained ≤0.8% of the variation in promoter DSB frequency. For several Bas1 and Ino4 targets (SHM2, GCV1, ADE17, and GCV3 for Bas1 and OPI3 for Ino4), both gene expression and hotspot activities were decreased in the respective TF mutant. However, there were numerous exceptions to this trend. For example, GCV2 and IRC22 showed increased promoter DSB frequency but unchanged RNA levels in bas1; GNP1 behaved similarly in the ino4 mutant. Meanwhile, expression of INO1 in the ino4 mutant and YOR302W in the bas1 mutant was substantially reduced with little accompanying change in promoter DSB activity. Thus, changes in mRNA level in TF mutants provide little or no predictive power for changes in DSB frequency. These results strengthen and extend the conclusion from previous work (White et al. 1992) that DSB hotspot activity of promoters is not tied to transcriptional activity per se.
Figure 8.
Comparing changes in mRNA levels with changes in promoter DSBs in bas1 and ino4 mutants. Spo11 oligos were summed in 300-bp windows upstream from each gene’s start codon. Points corresponding to exemplar genes are labeled.
Discussion
DNA sequence-dependent targeting of meiotic recombination was first discovered more than two decades ago in two highly diverged organisms, fission yeast and budding yeast (Ponticelli et al. 1988; Schuchert et al. 1991; White et al. 1991). Additional regulatory DNA sites were identified subsequently in both yeasts (White et al. 1993; Fan et al. 1995; Steiner et al. 2009), and more recently it was found that mouse and human hotspots are defined by the histone methyltransferase PRDM9 binding to specific DNA sequences (Baudat et al. 2010; Myers et al. 2010; Grey et al. 2011; Brick et al. 2012; Pratto et al. 2014). These and other findings have focused attention on the roles of sequence-specific DNA binding proteins in shaping the DSB landscape and have even led to proposals that one or a few such proteins and their binding sites are the principal architects of DSB hotspot location and activity across all of both yeasts’ genomes (e.g., Wahls and Davidson 2010).
Sequence-specific DNA binding by PRDM9 is clearly a primal determinant of hotspots in most mammalian species (Prdm9 knockout mice still have many hotspots, but at other places) (Brick et al. 2012). However, the situation has been less clear for TFs in yeasts. Even those sequence motifs and their binding factors that are clearly capable of specifying strong DSB hotspot activity in some genomic contexts work poorly or not at all in other contexts (Ponticelli and Smith 1992; Mieczkowski et al. 2006; this work). Moreover, presence of a sequence motif with DSB-targeting potential is by itself a poor predictor of local DSB frequency (Mieczkowski et al. 2006; Fowler et al. 2014).
An alternative to a DNA sequence-centric view is a model in which DSB distributions are shaped by combinatorial action of many chromosomal factors that work in a hierarchical and scale-dependent manner (Petes 2001; Mieczkowski et al. 2006; Lichten 2008; Lichten and de Massy 2011; Pan et al. 2011; de Massy 2013). In this view, individual sequence-specific DNA binding proteins may contribute significantly to the DSB landscape, but in a circumscribed and highly context-dependent manner. Our findings in this study, in agreement with earlier work by Mieczkowski et al. (2006), support the latter model.
Because the earlier genome-wide study of Bas1 used a low-resolution microarray and focused on information for probes corresponding to entire individual ORFs (Mieczkowski et al. 2006), it was difficult to assign the hybridization signal to individual DSB hotspots. Therefore, we wished to revisit this question with the higher resolution and greater dynamic range afforded by Spo11-oligo mapping. Mieczkowski et al. (2006) scored 153 ORFs as having statistically significantly different rad50S ChIP-chip signal in the bas1 mutant (68 with decreased signal in bas1 and 85 with increased signal). We resolved more total DSB hotspots by Spo11-oligo mapping, at least in part because we were able to more precisely localize changes in DSB formation, including fine-scale changes of local DSB distribution within hotspots that were previously impossible to discern. Importantly, however, both studies support the same general conclusion: Bas1 effects on DSB formation are highly context dependent. Our results with Ino4 were similar in this regard, suggesting that context dependence is likely to be true for many if not all TFs that impinge on Spo11 activity.
Deletion of Bas1 affected DSB activity in about half of hotspots containing Bas1 binding sites, similar to frequencies observed previously (Mieczkowski et al. 2006). Deletion of Ino4 affected less than one-third of the hotspots to which Ino4 binds directly during meiosis. Moreover, only a minority fraction of the hotspots that changed in the TF mutants were detectably bound by Bas1 or Ino4 protein during meiosis. This modest correspondence between TF binding and DSB activity is analogous to the relation of TFs to transcriptional regulation: numerous studies have documented that only a small fraction of genes whose promoters are bound by a given TF show expression changes when the TF is deleted, and only a small fraction of the genes that change expression are direct binding targets (reviewed in MacQuarrie et al. 2011; Hughes and De Boer 2013). Although it is possible that the ChIP-seq failed to detect direct Bas1 or Ino4 binding that does in fact occur, the results strongly indicate that a substantial portion of the DSB-modulating activity of these TFs is indirect. One possibility is that each TF mutant causes changes in cell physiology that, by crosstalk between transcriptional regulation networks, alter DSB activity at numerous gene promoters genome-wide via changes in local chromatin structure or loop–axis architecture. In this context, it is noteworthy that recombination distributions can be altered by auxotrophies for certain amino acids or nucleobases, including auxotrophies caused by mutation of several of the genes whose expression is known to be Bas1 dependent, such as HIS4 and ADE1 (Abdullah and Borts 2001; Cotton et al. 2009). We found no correlation between DSB changes and gene expression changes in bas1 and ino4 mutants, but it remains possible that chromatin and higher order chromosome structure in and around the indirectly changed hotspots is affected.
In principle, indirect effects could also arise if hotspots that are direct TF targets influence neighboring hotspots via “hotspot competition.” Insertion of very strong artificial hotspots can suppress the activity of nearby hotspots over distances up to tens of kilobases (Wu and Lichten 1995; Xu and Kleckner 1995; Fan et al. 1997; Ohta et al. 1999; Jessop et al. 2005). If a DSB hotspot becomes substantially hotter or colder in bas1 or ino4 mutants, competition could cause compensatory changes in DSB frequency in nearby hotspots. Such an effect predicts that change of a DSB hotspot in the TF mutants should be inversely correlated with changes of neighboring hotspots. However, we did not detect such a pattern in our data whether considering all hotspots relative to their neighbors or considering only the subset of direct TF targets that were most altered in the TF mutants (Figure S1, C and D). It is possible that hotspot competition is a property of artificial hotspots but not natural hotspots, but we favor instead the interpretation that the hotspots affected by bas1 or ino4 mutation are generally not strong enough to reveal hotspot competition in a cell population. The hottest hotspot that was directly bound by Bas1 or Ino4 and also showed at least a twofold change in activity had a Spo11-oligo count of only 1134 RPM (Table S3), equivalent to a DSB frequency of <2.5% of DNA according to the regression relationship in Pan et al. (2011), and most such hotspots had an inferred DSB frequency of <1%. For a hotspot with a DSB frequency of 2.5% of DNA, DSBs occur at this locus in ≤10% of the cell population (assuming one DSB per four chromatids in most cells) (Zhang et al. 2011a). Any potential suppression by this hotspot of its neighbors would likely be masked by the behavior of the majority of unaffected cells. These considerations indicate that hotspot competition is unlikely to be a significant source of the indirect effects of TF mutations on hotspot activity.
The context dependence for Bas1 effects on DSBs has previously been framed as reflecting locus-specific differences in the balance between chromatin “loosening” and “tightening” activities (Mieczkowski et al. 2006). However, for those sites we examined directly, there was no obvious correlation between changes in DSB frequency and changes in overall MNase hypersensitivity, suggesting that chromatin accessibility is not the principal link between TFs and DSB frequency at most hotspots. In contrast, if chromatin structure immediately within a DSB hotspot was altered by TF mutation, then the fine-scale DSB pattern was also generally altered. In particular, a shift of nucleosome position or appearance of a new nucleosome at SHM2 and GCV2 appeared to result in local occlusion of Spo11 access to the DNA, consistent with inferences from genome-wide studies of DSBs relative to well-positioned nucleosomes (Pan et al. 2011). We interpret these results to mean that open chromatin provides windows of opportunity where Spo11 can cut, but it is not generally sufficient to determine overall break frequencies.
Interpreting our findings in the context of the tethered loop–axis complex model, we envision that regional factors (working on ∼5- to 20-kb scales) shape loop–axis architecture and determine the DSB potential of a particular loop region (level 1 in Figure 6A). Within a loop, interactions between axis-bound DSB proteins and loop chromatin promotes targeting of Spo11 activity to particular locations (i.e., hotspots). These interactions determine the relative likelihood that particular hotspots will fulfill the DSB-forming potential of the loop they reside in (level 2 in Figure 6A). Within a hotspot, fine-scale DSB patterns are shaped by geometry of the targeting mechanism plus competition of Spo11 with nucleosomes and other DNA-binding proteins (level 3 in Figure 6A). TFs can affect DSB formation at any (or all) of the levels shown; the context dependence presumably reflects differences in how a given TF influences each of these levels at various genomic regions. Thus, while there are common themes and general patterns, each hotspot tells its own story and TFs can play many and varied roles.
Supplementary Material
Acknowledgments
We thank Agnes Viale and the Memorial Sloan Kettering Cancer Center (MSKCC) Integrated Genomics Operation for DNA sequencing; Nick Socci and the MSKCC Bioinformatics Core Facility for assistance with data analysis; Stewart Shuman (MSKCC) for gifts of T4 RNA ligase; Kunihiro Ohta (Tokyo University) and Mariko Sasaki (present address: Tokyo University) for strains; Beate Schwer (Weill Cornell Medical College) for assistance with RNA-seq experiments; Cong Huang, Hajime Murakami, Jing Pan, and Jodi-Ann Sampson for experimental assistance; and members of the S.K. laboratory, especially Corentin Claeys Bouuaert, Sarah Kim, Isabel Lam, Neeman Mohibullah, Hajime Murakami, Sam Tischfield, and Shintaro Yamada, for discussion and comments on the manuscripts. This work was supported by National Institutes of Health grant R01 GM058673. S.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: N. Hunter
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178293/-/DC1.
Literature Cited
- Abdullah M. F., Borts R. H., 2001. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaviva L., Szekvolgyi L., Dichtl B., Dichtl B. S., de La Roche Saint Andre C., et al. , 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. [DOI] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N., 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436. [DOI] [PubMed] [Google Scholar]
- Arndt K. T., Styles C., Fink G. R., 1987. Multiple global regulators control HIS4 transcription in yeast. Science 237: 874–880. [DOI] [PubMed] [Google Scholar]
- Bachhawat N., Ouyang Q., Henry S. A., 1995. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 270: 25087–25095. [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Nicolas A., 1997. Clustering of meiotic double-strand breaks on yeast Chromosome III. Proc. Natl. Acad. Sci. USA 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Hanlon S. E., Lieb J. D., Copenhaver G. P., 2009. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 19: 2245–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., de Massy B., Gadelle D., Varoutas P. C., Nicolas A., et al. , 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R. U., Hunter N., Kleckner N., 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Blitzblau H. G., Bell G. W., Rodriguez J., Bell S. P., Hochwagen A., 2007. Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr. Biol. 17: 2003–2012. [DOI] [PubMed] [Google Scholar]
- Borde V., Goldman A. S., Lichten M., 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. [DOI] [PubMed] [Google Scholar]
- Borde V., Lin W., Novikov E., Petrini J. H., Lichten M., et al. , 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell 13: 389–401. [DOI] [PubMed] [Google Scholar]
- Borde V., Robine N., Lin W., Bonfils S., Geli V., et al. , 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C., Borde V., Lichten M., 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisae. PLoS Biol. 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Y., Tsubouchi T., Rockmill B., Sandler J. S., Richards D. R., et al. , 2008. Global analysis of the meiotic crossover landscape. Dev. Cell 15: 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton V. E., Hoffmann E. R., Abdullah M. F., Borts R. H., 2009. Interaction of genetic and environmental factors in Saccharomyces cerevisiae meiosis: the devil is in the details. Methods Mol. Biol. 557: 3–20. [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B., Fink G. R., 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89: 6746–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., 2013. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47: 563–599. [DOI] [PubMed] [Google Scholar]
- de Massy B., Rocco V., Nicolas A., 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 14: 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis V., Daignan-Fornier B., 1998. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 259: 246–255. [DOI] [PubMed] [Google Scholar]
- Denis V., Boucherie H., Monribot C., Daignan-Fornier B., 1998. Role of the myb-like protein bas1p in Saccharomyces cerevisiae: a proteome analysis. Mol. Microbiol. 30: 557–566. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Xu F., Petes T. D., 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15: 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q. Q., Petes T. D., 1996. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q. Q., Xu F., White M. A., Petes T. D., 1997. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics 145: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler K. R., Sasaki M., Milman N., Keeney S., Smith G. R., 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 24: 1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelling C. L., Piper M. D., Hong S. P., Kornfeld G. D., Dawes I. W., 2004. Identification of a novel one-carbon metabolism regulon in Saccharomyces cerevisiae. J. Biol. Chem. 279: 7072–7081. [DOI] [PubMed] [Google Scholar]
- Gerton J. L., DeRisi J., Shroff R., Lichten M., Brown P. O., et al. , 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T., Lichten M., 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C., Barthes P., Chauveau-Le Friec G., Langa F., Baudat F., et al. , 2011. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 9: e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovring I., Bostad A., Ording E., Myrset A. H., Gabrielsen O. S., 1994. DNA-binding domain and recognition sequence of the yeast BAS1 protein, a divergent member of the Myb family of transcription factors. J. Biol. Chem. 269: 17663–17669. [PubMed] [Google Scholar]
- Hughes T. R., de Boer C. G., 2013. Mapping yeast transcriptional networks. Genetics 195: 9–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Allers T., Lichten M., 2005. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L., Jeffreys A. J., Keeney S., 2004. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 5: 413–424. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Keeney S., Kleckner N., 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA 92: 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Kleckner N., 1996. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells 1: 475–489. [DOI] [PubMed] [Google Scholar]
- Kon N., Krawchuk M. D., Warren B. G., Smith G. R., Wahls W. P., 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94: 13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugou K., Fukuda T., Yamada S., Ito M., Sasanuma H., et al. , 2009. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol. Biol. Cell 20: 3064–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Bourbon H. M., de Massy B., 2010. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24: 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten, M., 2008 Meiotic chromatin: The substrate for recombination initiation, pp. 165–193 in Recombination and Meiosis, edited by R. Egel and D. H. Lankenau. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- Lichten M., de Massy B., 2011. The impressionistic landscape of meiotic recombination. Cell 147: 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Goldman A. S. H., 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29: 423–444. [DOI] [PubMed] [Google Scholar]
- Liu J., Wu T. C., Lichten M., 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 14: 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T. L., 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac K. D., Wang T., Gordon D. B., Gifford D. K., Stormo G. D., et al. , 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuarrie K. L., Fong A. P., Morse R. H., Tapscott S. J., 2011. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 27: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Dominska M., Buck M. J., Gerton J. L., Lieb J. D., et al. , 2006. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H., Borde V., Nicolas A., Keeney S., 2009. Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions. Methods Mol. Biol. 557: 117–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H., Keeney S., 2014. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell 158: 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C., et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M. J., Pan J., Keeney S., 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Shibata T., Nicolas A., 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 13: 5754–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Wu T. C., Lichten M., Shibata T., 1999. Competitive inactivation of a double-strand DNA break site involves parallel suppression of meiosis-induced changes in chromatin configuration. Nucleic Acids Res. 27: 2175–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N., 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S., Mendoza M. A., Berlinger M., Huang L., Nicolas A., et al. , 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Petes T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2: 360–369. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Siomos M. F., Nasmyth K., 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., et al. , 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Smith G. R., 1992. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc. Natl. Acad. Sci. USA 89: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Sena E. P., Smith G. R., 1988. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics 119: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F., Brick K., Khil P., Smagulova F., Petukhova G. V., et al. , 2014. DNA recombination. Recombination initiation maps of individual human genomes. Science 346: 1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago T. C., Mamoun C. B., 2003. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278: 38723–38730. [DOI] [PubMed] [Google Scholar]
- Sasaki M., Tischfield S. E., van Overbeek M., Keeney S., 2013. Meiotic recombination initiation in and around retrotransposable elements in Saccharomyces cerevisiae. PLoS Genet. 9: e1003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P., Langsford M., Kaslin E., Kohli J., 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 10: 2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Schwank S., Ebbert R., Rautenstrauss K., Schweizer E., Schuller H. J., 1995. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 23: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V., Beneut C., Chaplais E., Serrentino M. E., Borde V., 2013. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell 49: 43–54. [DOI] [PubMed] [Google Scholar]
- Stapleton A., Petes T. D., 1991. The Tn3 beta-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics 127: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W. W., Schreckhise R. W., Smith G. R., 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9: 847–855. [DOI] [PubMed] [Google Scholar]
- Steiner W. W., Steiner E. M., Girvin A. R., Plewik L. E., 2009. Novel nucleotide sequence motifs that produce hotspots of meiotic recombination in Schizosaccharomyces pombe. Genetics 182: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W., 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Thacker D., Mohibullah N., Zhu X., Keeney S., 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield S. E., Keeney S., 2012. Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 11: 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls W. P., Davidson M. K., 2010. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 26: 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Wierdl M., Detloff P., Petes T. D., 1991. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc. Natl. Acad. Sci. USA 88: 9755–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Detloff P., Strand M., Petes T. D., 1992. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr. Genet. 21: 109–116. [DOI] [PubMed] [Google Scholar]
- White M. A., Dominska M., Petes T. D., 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90: 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.-C., Lichten M., 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518. [DOI] [PubMed] [Google Scholar]
- Wu T. C., Lichten M., 1995. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 140: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Kleckner N., 1995. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 14: 5115–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Ma Y., Tang S., Hwang P. Y.-H., Boiteux S., et al. , 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40: 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kim K. P., Kleckner N. E., Storlazzi A., 2011a Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. USA 108: 20036–20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ma H., Pugh B. F., 2011b Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 21: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Byers K. J., McCord R. P., Shi Z., Berger M. F., et al. , 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. Next-generation sequencing data were deposited at the Gene Expression Omnibus (GEO) under accession nos. GSE67910 (Spo11-oligo mapping), GSE67912 (Bas1 and Ino4 ChIP-seq), GSE67907 (H3K4me3 ChIP), and GSE70911 (RNA-seq).