Abstract
Antibody drug conjugates (ADCs) have progressed from hypothesis to approved therapeutics in less than 30 years, and the technologies available to modify both the antibodies and the cytotoxic drugs are expanding rapidly. For reasons well reviewed previously, the field is trending strongly toward homogeneous, defined antibody conjugation. In this review we present the antibody and small molecule chemistries that are currently used and being explored to develop specific, homogenous ADCs.
Key Words: antibody drug conjugates (ADCs), homogenous conjugates, protein engineering, site-specific conjugation, targeted delivery
Introduction
Antibody drug conjugates (ADCs) are a class of biotherapeutics that combine the specificity, favorable pharmacokinetics, and biodistribution of a monoclonal antibody (mAb) with the cytotoxic potency of a small-molecule drug. This combination results in a widened therapeutic window over traditional cancer chemotherapeutics. Thus, many cytotoxic drugs that are not amenable to traditional chemotherapy can often be used in the context of an ADC (1–6).
While the concept of ADCs is in principle a simple one, their practical implementation is dependent on many factors. The conjugation method can be as important as the choice of mAb and cytotoxic drug when designing ADCs, as it potentially impacts efficacy, stability, and toxicity of the resulting product (7, 8).
Given the scarcity of orthogonally-reactive functional groups on natural antibodies, current ADCs on the market are made by conjugating small-molecule cytotoxic drugs to primary amines in lysines or thiols exposed by reduction of interchain disulfide bonds, as is the case for the recently approved ADCs Kadcyla/trastuzumab emtansine/T-DM1 (9) and Adcetris/brentuximab vedotin/SGN-35 respectively (10, 11).
Though these ADCs improve the therapeutic index of the original small-molecule drug and improve the potency over the naked antibody, ADCs made in this manner are heterogeneous mixtures, in which each species has different pharmacological properties. (12). Furthermore, development of safe and efficacious therapeutics requires their accurate characterization throughout all phases of the discovery, optimization, and development life cycle. Since ADCs made by these conventional conjugation chemistries yield heterogeneous mixtures that obscure the individual characteristics of each underlying species, they make optimization to the best possible ADC difficult. For example, much discussion has centered on the optimal number of linker warheads that can be attached to antibodies, without being detrimental to the original physicochemical characteristics. While there will be an approximation of the maximally tolerable payload of specific linkers and warheads, the numbers that have been claimed to be optimal thus far, are based on experimental data with conjugation chemistries utilizing the natural amino acids. Conjugations to available lysines and cysteines are stochastic in number and location, giving a distribution of species with DARs (drug antibody ratios) ranging between 0 and 9 for loading of warhead molecules. For example, Kadcyla, is comprised of the conjugation of an average of 3.4 linker/DM1 warheads per trastuzumab antibody. In the case of huN901-DM1, warheads are conjugated to roughly 40 of the 86 lysines within the trastuzumab antibody sequence corresponding to the accessible sites (13, 6).
The properties of individual species of ADCs in the product can vary depending on the number, position and proximity of the conjugated drugs (7, 8). Cognizant of this, the makers of Kadcyla and Adcetris, and the ADC field as a whole, have migrated toward the development of methods for site-specific attachment of drugs to antibodies, with the intention of further improving ADC therapeutics. The real but still modest increases in the therapeutic index that came about from antibody conjugation may be further increased by defined, designed, and homogenous conjugation of the cytotoxic drugs to the antibody. This review aims to give an overview of the available schemes to generate such homogeneous ADCs.
Methods to Make Homogenous ADCs
It has been proven that the site of drug conjugation modulates stability, PK and therefore efficacy of ADCs (14). The diverse methods developed to make homogenous ADC can be grouped in two overarching categories. One general strategy requires engineering of the antibody primary structure, e.g. mutation of residues to cysteine or a non-natural amino acids (nnAA), or insertion of additional amino acids or fusion tags. The second class uses native antibody sequences and instead employs novel chemistries and linker strategies to yield site-specific modification. The antibody engineering based methods offer the flexibility to mutate or insert tags at multiple defined positions, in order to directly control the DAR and determine the optimal sites for conjugation. On the other hand, methods utilizing native antibodies lack this flexibility and only allow for limited sites to be conjugated, as determined by the chosen chemistry. However, since no mutagenesis of the antibody is required, such methods offer the advantage of slotting directly into existing antibody production platforms, allowing for the rapid and efficient conjugation of any off-the-shelf or de novo antibody, without the need for proprietary expression platforms.
Homogenous ADC Requiring Antibody Engineering
nnAA Incorporation
On a conceptual level, the production of homogeneous ADCs with site-specific drug linking requires that the antibody has one or more unique features that can be exploited for attachment of the warhead. One method of incorporating such features in an antibody is the utilization of non-natural amino acids (nnAAs).
In Vivo nnAA Incorporation
In vivo systems that rely on engineered cell-lines have been utilized for incorporating nnAA into antibodies to provide bio-orthogonal conjugation handles. In vivo nnAA incorporation relies on a tRNA and synthetase (aaRS) pair that is orthogonal to all the endogenous tRNAs and synthetases in the host cell. The nnAA of choice is supplemented to the media during fermentation, making cell-permeability and stability important considerations for the nnAA. Fundamentally, three approaches based on stop codon suppression have been developed to enable in vivo incorporation of nnAAs into antibodies, with amber codon suppression being the most common (15).
One method relies on a tyrosyl aaRS/tRNA pair from Escherichia coli, which was engineered to recognize and charge para-acetyl-phenylalanine (pAcPhe). This engineered aaRS/tRNA pair was stably integrated in Chinese hamster ovary cells (16). Subsequent stable integration of light and heavy chain genes containing the amber stop codon was used to express antibodies with pAcPhe at designed sites (17). The keto group of the pAcPhe is reactive towards alkoxy-amines via oxime coupling and can be conjugated with an alkoxy-amine containing linker-drug to generate a site-specific ADC. Ambrx has been an industry leader in in vivo ADC production using this approach.
Another approach for in vivo nnAA incorporation is based on the natural amber suppressor tRNA/aaRS pair responsible for incorporation of pyrrolysine (Pyl) in Methanosarcina species (18). Here, no engineering of the nnRS/tRNA pair is required since the system is naturally occurring. Furthermore, this archea aaRS/tRNA pair is fully orthogonal to both Escherichia coli and mammalian cells (19, 20). Serendipitously, the PylRS enzyme is fairly promiscuous in its specificity towards pyrrolysine, and a variety of chemically functionalized pyrrolysine derivatives have been shown to be incorporated by the enzyme (21, 22). Much like the pAcPhe based system, the PylRS/tRNA pair can be utilized to incorporate reactive handles like ketones and azides into antibodies allowing for site-specific conjugation. This approach is currently being developed by Allozyne/MedImmune termed the “AzAbs” (23).
Selenocysteine (Sec) has also been introduced as a chemical handle in antibodies to generate conjugates via transiently transfected human embryonic kidney (HEK) 293 F cells (24). Since Sec is incorporated in naturally occurring systems, no engineering of the aaRS, tRNA or amino acid is necessary to make it suitable for ADC generation. Sec incorporation is accomplished via opal stop codon suppression that is dependent on a selenocystein insertion sequence present on the mRNA transcript. The demonstration of selecocysteine incorporation suggested the potential for site-specific ADCs via selenides.
Site-specific nnAA incorporation can also be achieved by replacing methionine using the sufficiently-promiscuous MetRS. Cells are made auxotrophic for methionine and engineered to overexpress MetRS. Supplementation of methionine analogs into the growth media is used to overcome the 500–2000-fold reduction in catalytic efficiency (Kcat/Km) of MetRS for the nnAAs. Further engineering can be carried out to recover function in the nnAA incorporated proteins (25, 26). So far this particular approach has been exemplified with E.coli expression systems, however, the applicability to ADC production has yet to be established.
In Vitro nnAA Incorporation
The approach of nnAA incorporation into antibodies has also been extended to in vitro transcription-translation platforms, although titers and scalability limitations were once a concern for cell-free protein synthesis (CFPS) systems. However, Zawada et al. (27) engineered an optimized E.coli based cell-free system for predictable high-yield protein synthesis and folding that showed scalability over multiple orders of magnitude. E. coli strains and their extracts were engineered to contain all of the necessary components for transcription, translation and energy generation from low cost substrates such as glutamate. Additional improvements upon this CFPS system led to the successful expression of antibodies and antibody fragments in the gram per liter range, in part enabled through overexpression of chaperones and disulfide isomerases in the E. coli host strain, from which the cell-free extract is derived (28, 29). In addition, nnAA incorporation strategies used in in vivo systems can also be leveraged in in vitro cell-free reactions, with the added advantage that that the open nature of CFPS systems allows direct manipulation of the chemical environment in which the protein synthesis occurs. Consequently, cell-free systems can overcome constraints of cell permeability and media stability of the nnAA present in in vivo systems.
Another factor common to most nnAA incorporation systems is the presence of competing endogenous release factors (RFs). These RFs naturally recognize certain stop codons and therefore compete for stop codon suppression of nnAA-tRNAs. Thus, full length protein production of nnAA containing antibodies can be significantly reduced in titer when compared to wild-type translation. While removal of the competing RF1 for amber suppression is therefore desirable, this is complicated by the fact that simple deletion of the gene from cells is not tolerated in either microbial or eukaryotic systems. Nonetheless, efficient non-natural amino acid incorporation was achieved by RF1 deletion in E. coli enabled by compensatory engineering of RF2 (30). In vitro transcription-translation CFPS platforms offer yet another alternative solution, which circumvents the growth rate issues that arise when cell-based systems are subjected to release factor modifications (31). Several groups have demonstrated that RF1 can be removed after extract preparation allowing E. coli to have intact RFs during growth of the biomass. Furthermore, RF1 can also be inhibited in the cell-free reaction. These examples enabled improved nnAA incorporation at many sites without reengineering the entire genome to maintain cellular viability (32–36). Hallam and co-workers (37) successfully demonstrated that it is feasible to replace wild-type RF1 with an OmpT-susceptible RF1 in strains of E.coli. During the process of extract production, RF1 degrades due to exposure to OmpT, which is localized on the outer membrane and therefore not in contact with RF1 during normal cellular growth. The result is a cell-free system that produces nnAA containing proteins at titers comparable to wild-type, as RF1 function is completely attenuated.
Thus, CFPS systems can be efficiently leveraged to produce nnAA containing mAbs for ADC generation. Recently Zimmerman et.al (38) reported production of site-specific, aglycosylated ADCs by CFPS using a new orthogonal synthetase that enables incorporation of a para-azidomethylphenylalanine (pAMF) followed by strain-promoted alkyl-azide conjugation (SPAAC) to a dibenzylcyclooctyne (DBCO) functionalized monomethyl auristatin F (MMAF) drug (Fig. 1).
Fig. 1.

Strain-promoted alkyne-azide coupling (SPAAC) between the inserted non-natural amino acid p-azidomethyl phenylalanine (pAMF) and the dibenzyloctyne (DBCO)-derivitized auristatin drug forms a triazole ring, forging a stable linkage between the small molecule and the antibody.
To arrive at a synthetase capable of charging pAMF onto its cognate tRNA, the group screened a library of 1760 Methanococcus jannaschii tyrosyl- tRNA synthetase (M.jann-TyrRS) active site variants, by cell-free expression of each synthetase and subsequent screening for its ability to suppress the amber stop codon in the presence of pAzMeF (38).
Due to the simplified expression format afforded by CFPS systems, rapid screening of many variants can be enabled in microtiter formats. By combining CFPS expression and automated high throughput protein purification techniques, screening of 100’s of site-specifically labeled ADCs was demonstrated with trastuzumab IgG1 as a base scaffold (Sutro Biopharma unpublished data). Top sites within the trastuzumab IgG1 framework were identified based on the attributes of good suppression efficiency, correct assembly of IgG, and high conjugation efficiency while maintaining properties of antigen binding, cell killing, and thermostability. This overall process allowed positional optimization of non-natural amino acids to enable discovery and production of single species ADCs with superior properties and showcases the advantages of homogenous ADC production.
In addition to the Sutro Bipharma technology, PeptiDream has developed a cell free protein expression method. Their Flexizyme system is an in vitro evolved artificial ribozyme that catalyzes the aminoacylation of any non-natural amino acid onto any tRNA, thereby functioning as an artificial aminoacyl-tRNA synthetase that efficiently introduces non-natural amino acids into the growing polypeptide sequence during a cell-free translation process (39, 40). This has been integrated with an mRNA display and screening technique called RaPID (Random non-standard Peptide Integrated Discovery), and extensive libraries of peptides have been generated and screened yielding good binding to a variety of protein targets (41). Recently this work was extended to generate a bispecific antibody Fab fragment by introducing p-acetylphenylalanine into an αCD3 Fab followed by conjugation to hydroxylamino folic acid. Simultaneous engagement of the T-cell (via CD3) and tumor cell (via the folate receptor) suppressed tumor growth in vivo at 5 mg/kg, although regrowth began after 80 days (42). At the time of this writing no ADCs have been reported from this technology.
Engineered Cysteine Conjugation
Another option for site-specific modification of antibodies is through the use of cysteines. Since all cysteines in a native antibody are paired in disulfide bonds, free cysteines can be introduced as site-specific chemical moieties through site directed mutagenesis. Due to the highly reactive nature of cysteine as a nucleophile, the free thiols can then be selectively modified through the use of maleimides or other electrophilic groups. Engineering free cysteines into antibodies for site directed conjugation has the advantage that no specialized expression system is needed, as would be the case for nnAA incorporation. However, hurdles for expression and processing have to be overcome. Cysteines are not tolerated at every position in an antibody as they may interfere with correct folding or function, disrupt correct formation of the native disulfide bonds, or form disulfide bonds with free cysteines in other proteins. Another challenge in inserting free cysteines into antibodies is that, even if the antibody is well behaved, the cysteines are often oxidized by disulfide bond formation with small molecule thiols present in the cell, such as free cysteine or glutathione. To liberate the cysteine, the antibody has to be partially reduced, which may result in reduction of some native disulfide bonds. Subsequent re-oxidation must then restore the disulfide bonds to their native configuration, while leaving the engineered cysteine in the reduced form (43). To address the issue of antibody folding, Junutula et al. at Genentech developed a Phage ELISA for Selection of Reactive Thiols (PHESELECTOR), which allows for the selection of tolerated cysteine substitutions in the context of an antibody Fab fragment (44). From the PHESELECTOR output, alanine 114 (Kabat numbering) of the heavy chain was determined to be the optimal site for cysteine modification, giving rise to Genentech’s THIOMAB platform. Furthermore, production of various full length antibodies with the A114C HC mutation showed that the cysteine mutation was well tolerated and did not adversely affect antibody function, demonstrating that the approach is applicable across different antibodies. Other sites outside the Fab domain have also been used to create free cysteine containing antibodies (14). Seattle Genetics has an anti-CD33 antibody based on their proprietary engineered cysteine platform in Phase 1 clinical trials, exemplifying the robustness of the approach (45).
Enzymatic Conjugation
Ligating enzymes that have high specificity for one substrate but relaxed specificity for the second substrate are ideal for chemoenzymatic synthesis of structurally defined protein conjugates. Bacterial enzymes, many originally identified in highly pathogenic species, have proven extraordinarily useful tools in protein engineering. Mammalian orthologs are either absent or have different substrate specificities. Moreover, genetic manipulation and screening of variants, and expression in non-pathogenic species for in situ or in vitro applications, is usually straightforward. Mammalian enzymes have also been re-engineered as biosynthetic tools. The chemoenzymatic strategies for protein-small molecule conjugation are presented below in the order of the degree of protein perturbation required, from small (3–6 residues) to medium (12–16 residues) to large (domain fusions).
A quadruple mutant of trypsin (K60E/N143H/E151H/D189K), trypsiligase, is capable of both Tyr-Arg cleavage and fusion of a ligand p-guanidinylphenyl ester (R 4-OGp) onto the newly-exposed arginine amine. Trypsiligase has been shown to efficiently label a variety of proteins modified with an N-terminal YRH tripeptide (Fig. 2). Stoichiometries of the acyl moieties are quite reasonable (3:1), reactions are complete within 1 h, and the engineered hydrolase/ligase accepts a variety of substrates (46).
Fig. 2.

An engineered trypsiligase acylates a newly-created N-terminus at arginine with a small molecule having an activated carboxylate.
Streptoverticillium mobaraense transglutaminase, EC 2.3.2.13, is a stable, Ca+-independent enzyme catalyzing the γ-acyl transfer of glutamine to the ε -amino group of lysine (47, 48). Chemists at the ETH first observed the selective acylation of amino ligands exclusively by the glutamines located at the flexible regions of an IgG heavy chain. These sites were the mutated (and therefore aglycosylated) N to Q mutation sites, which are highly exposed and available to protein-modifying enzymes. By using the bacterial transglutaminase (BTG) at these sites they synthesized homogeneous radioimmunotoxins that were tumor-uptake selective in vivo (49). Building on these results, the Rinat/Pfizer team identified a glutamine tag (Q tag) sequence LLQGA that enabled specific Gln transamination in the presence of all other protein Gln residues and introduced this pentapeptide into discrete locations along an anti-EGFR IgG1 antibody (Fig. 3). Out of 90 sites into which the sequence was introduced, 12 proved to be amendable to conjugation of amine-containing small molecules.
Fig. 3.
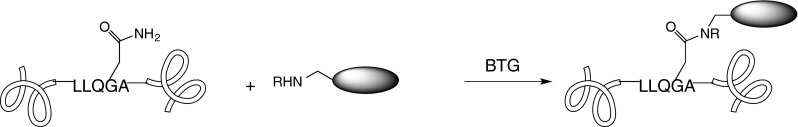
Transglutaminase inserts diverse amines into proteins at suitably exposed glutamines that are embedded in the LLQGA recongition sequence.
Engineering the Q tag into the anti-M1S1 mAb C16, and subsequent conjugation of monomethyl dolastatin 10 (MMAD) at one heavy chain (HC) and one light chain (LC) site, facilitated comparison of the two discrete DAR 2 ADCs. They observed that in the rat model the LC ADC had more favorable pharmacokinetics. Significantly, they also observed that a higher therapeutic window for both HC and LC ADCs could be achieved versus the conventional heterogenous cysteine-linked maleimidocaproyl ValCitpaAB MMAD conjugate. The emphasis in these studies was site-dependent rather than linker chemistry-dependent stability and the impact of site on overall ADC behavior in vivo (50). Subsequently, it was determined that naturally-occurring aglycosylated antibodies gave rise to 1.3% off-target conjugation at the heavy chain Q295 due to conformational effects: in the native glycosylated protein the glycosylation would sterically obstruct this transamination. This exposure effect could be overcome by the Q295N mutation to achieve a completely homogeneous conjugate (51). Thus the Q tag, when presented at solvent-exposed, enzyme-accessible sites, remains a valuable tool for defined homogeneous conjugation. Working with, rather than around, Q295, Innate Pharma recruits bacterial transglutaminase to this site in an aglyclosylated mAb, and introduces the N297S or N297Q mutation to provide homogeneous, site-specific DAR 2 or 4 aglycosylated ADCs (Innate Pharma website).
The aldehyde tag represents an entirely different chemistry for linking. It is crafted along the same principle of enzymatic recognition of a very small consensus sequences (52). Formylglycine-generating enzyme (FGE) oxidizes the cysteine in the CxPxR sequence at N- or C- termini to formyl glycine, when overexpressed in either mammalian or E. coli cells. This enables expression of antibodies with specifically-inserted aldehydes suitable for bioorthogonal reactions with alkoxylamines, hydrazines, or Pictet-Spengler-reacting tryptamines (Fig. 4) (53).
Fig. 4.

Enzymatic conversion of the cystein thiol to a formyl group enables reactions with oximino and other carbonyl-directed nucleophiles.
Site specific in vitro oximination at an internal IgG CTPSR site has recently been reported, overcoming the requirement that the aldehyde tag be restricted to the protein N- or C-terminus. Interestingly, the recombinant Mycobacterium tuberculosis enzyme, used to affect the Cys to fGly conversion in this work, was consumed stoichiometrically rather than reacting catalytically (54). Hydration of the aldehyde in the milieu of protein labeling can lower the efficiency of these reactions (55), but nonetheless Redwood Bioscience/Catalent has successfully developed aldehyde incorporation/reactivity as its SMARTagTM technology, demonstrating its application for a variety of highly controlled protein modifications (56).
Protein modification technologies that have been used for trafficking and biodynamic studies have not to date been used for generation of ADCs, but one may expect that in the future many of these will be adapted for drug bioconjugation. Several of these are included in this review as an acknowledgment of their potential.
Sortase A (SrtA) from Staphylococcus aureus, one of the many related sortases found in a variety of Gram-positive species, is a transpeptidase (57) that recognizes a C-terminal LPXTG sequence, cleaves the TG bond, and facilitates—via the thioacyl enzyme-threonine intermediate—the nucleophilic attack of the incoming protein alpha amine on the threonine. In protein or peptide ligation, the preferred attacking residue is a glycine di- or trimer, and early chemical ligation approaches attached glycines to the small molecule of interest. This was demonstrated to work for conjugation of folate, PEG, rhodamine, cholesterol, biotin, and a variety of peptides, all of which could function with a terminal (Gly)n (58, 59). Eliminating the requirement for oligoglycine, Matsumoto and coworkers (60) used purified Lactobacillus plantarum sortase (SrtLp) to site-specifically append the primary amine biotinyl-3,6-dioxaoctanediamine to LPQTx sequences that were not substrates for SrtA. There are some limitations to ‘sortagging’ in the context of ADCs: the conjugation site is restricted to the C-terminus, reaction kinetics are slow, yields are better with glycine than with general primary amines and intramolecular lysines can react to crosslink and aggregate the protein. It has also been shown that the glycine leaving group can return as a competing nucleophile, essentially reversing the reaction, unless a large (ca 10:1) stoichiometric excess of intended nucleophile is used. (61). Interest in this approach, however, has prompted creative solutions to many of these impediments, including the use of threonine esters that are recognized as substrates and release the far less nucleophilic glycolic acid, enabling conjugation with a 1:1 stoichiometry (Fig. 5) (62). Directed evolution of the enzyme using yeast-displayed sortase mutants resulted in a 140-fold increase in LPETG-conjugation efficiency (63). Moreover, the potential of protein-small molecule conjugation using sortase was demonstrated by the conjugation of an antibody Fab fragment F19 having a C-terminal LPETGG in the heavy chain to a (Gly)3-DY647 fluorophore (64). A recent adaptation of sortase demonstrated the irreversible hydrazinolysis of proteins, enabling the fusion of both protein and small molecule hydrazides at the nascent C-terminus (Fig. 6).
Fig. 5.
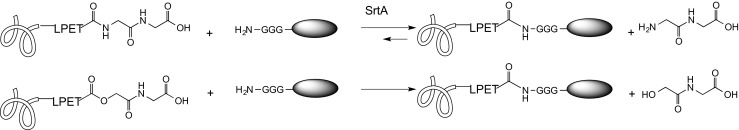
The sortase-mediated glycine splicing places the glycine-derivitized small molecule at the C-terminus of the protein; C-terminal threonine esters show improved reaction kinetcs in this strategy.
Fig. 6.

Sortagging using hydrazides in place of polyglycine on the small molecule affords an alternative linkage for the small molecule.
Sortagging has been transitioned from a tool for protein study to a tool for conjugation: with the addition of the second recognition motif of SrtB, NPQTN (65) a dual warhead ADC was designed by the NBE Therapeutics Group. Each heavy chain is C-terminally modified with one antitumor toxin while each light chain is C-terminally modified with a second toxin. Examples of pentaglycine maytansine and pentaglycin auristatins were presented at the 2014 PEGS. The use of cleavable linkers would in principle allow the (Gly)n to be universally acceptable in the linker construction since it would ultimately be separated from the released drug.
The E. coli enzyme biotin ligase (BirA), which recognizes the 15-amino acid Avi-tag GLNDIEAQKIEWHE and acylates the embedded lysine with biotin, would at first glance appear useful only in those research applications where site-specific biotinylation was important. The Ting group, however, demonstrated that BirA will accept a synthetic keto analog of biotin as a substrate, albeit at reduced reaction kinetics, and in so doing placed BirA in the same essential category as FGE, i.e. an enzyme capable of inserting a carbonyl (in this case a ketone) site-specifically into a protein (Fig. 7). Whereas FGE recognizes a small sequence, but prefers N- and C-termini, BirA requires a longer sequence, but is far less restricted in location along the substrate protein. Reaction with the installed carbonyl was demonstrated using fluorescein hydrazide (66).
Fig. 7.

BirA accepts biotin-like ketones for incorporation into the recongition sequence, enabling the carbonyl chemistry that allows oxyamine-small molecules to be inserted into the protein.
The pioneering work in the Ting lab on BirA-based ligation (BLINC) proved difficult to apply to neurons, and the group investigated the potential of E. coli lipoic acid ligase (LplA, called “LAP”). Mutating this enzyme to LplAW37A to relax the substrate specificity for the lipoic acid component enabled installation of diverse reactive moieties as long they were at the terminal end of an alkyl carboxylic acid (67–70). An acceptor sequence GFEIDKVWYDLDA was optimized to present the embedded lysine for acylation, effectively converting this lysine into a platform for presenting reactive groups including halides, azides, aldehydes (71), and trans cyclooctene (72). Currently both BLINC and LAP technologies have been used for visualization and cellular protein studies rather than for therapeutic conjugates.
Fusion Tag Technology
Another strategy for engineering site-specific, homogeneous ADCs involves fusing entire domains or proteins onto the antibody sequence and using their activity to achieve labeling. The most well know such strategy is expressed protein ligation (EPL), which relies on the activity of a self-splicing intein protein to activate the C-terminal of the target protein and therefore allowing formation of a new amide bond with a peptide, protein or small molecule. Though limited to the C-terminus of the expressed protein, the intien is self-excising and leaves only a single cysteine residue scar in the resultant ligated protein. EPL follows a mechanism of (a) C-terminal thioester formation from an intein sequence (e.g. Mycobacterium xenopi Gyr A) that spontaneous undergoes N to S-rearrangement, (b) selected thiol displacement of the intein to give an activated thioester, (c) thiol exchange of the thioester with an N-terminal cysteinyl or β-amino mercapto ligand (second protein, peptide, small molecule), followed by (d) spontaneous N-to-C rearrangement to an amide bond that forges the covalent link from the C terminus of the protein of interest to the small molecule(or ligand), with one intervening cysteine (Fig. 8).
Fig. 8.
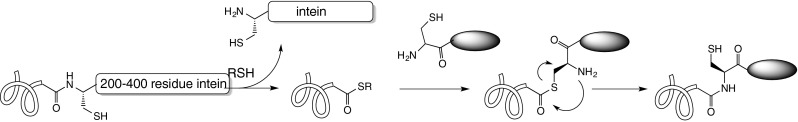
A β-mercaptoamine-derivitized small molecule (typically cysteine-modified) can be appended onto a protein via intein splicing, in which the ultimate bond is a stable amide.
Protein trans-splicing (PTS) uses an intein variant which is split into two parts, attached to the N- and C- termini of the partners to be ligated. A similar series of reactions attaches the 2 partners, while excising the intein (73). Intein splicing was demonstrated to site-specifically biotinylate the C-terminus of an antibody directed against the tumor antigen fibronectin ED-B9 (73) and to attach a peptide to an αDEC205 antibody (74). A recent modification of intein chemistry replaced the entire intein with an α-hydroxy acid, incorporated using pyrrolysyl tRNA synthetase, that could be synthetically converted to a C-terminal hydrazide, a function more generally stable than a thioester, but still a good leaving group for acylation chemistry (75). Although intein splicing has been used for site-specifically inserting quantum dots into proteins (76) and for synthesizing a selenoprotein (77), it has not been placed in the toolbox of antibody drug conjugation.
Proteins that can function with a significant insertion can exploit the O6 alkylguanine-DNA-alkyl transferase (AGT) reaction in which the functional group attached to an O6 benzyl group is covalently attacked by the cysteine on AGT and thus transferred to the protein-of-interest-AGT fusion (Fig. 9).
Fig. 9.
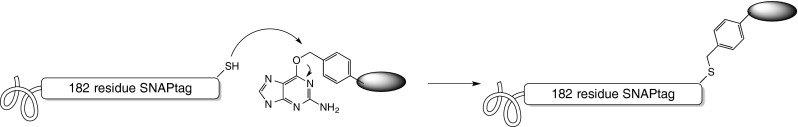
SNAP tagging uses guanine as a leaving group, forging a thioether bond to a benzyl-derivitized small molecule, but requiring a substantial additional polypeptide to be added to the protein undergoing modification.
To realize the potential of this reaction, directed evolution of human AGT was used to arrive at a mutant enzyme with kinetics comparable to wild type hAGT, while retaining the promiscuous substrate tolerance for the O-benzyl moiety (78). Referred to as the SNAP-tag, this 20 kDa fusion has found wide acceptance for site-specifically modifying proteins for chemical biology studies and therapeutics. An scFv directed against the tumor-associated epidermal growth factor receptor (EGFR) was fused to an AGT SNAP-tag and this fusion protein reacted with a benzyl derivative of the photosensitizer e-chlorin. The scFv-SNAP-e-chlorin conjugate, at a DAR 1, was used in photo-immunotherapy experiments against target and non-target cells and showed strong selectivity, good potency, and dependence on light as well as EGFR expression for cell killing (79).
Homogenous ADCs from Native Antibodies
All the methods mentioned above to make homogenous ADCs require antibody engineering. More conveniently, other types of methods don’t require protein mutagenesis for site-specific conjugation. The biggest advantage of such methods is to conjugate antibodies at a post-translational stage, which allows the rapid and efficient conjugation of any off-the-shelf or de novo antibody.
Crosslinking Two Reduced Cysteines
Instead of using engineered cysteine residues, mAbs can be partially reduced to break intermolecular disulfide bonds of the native cysteine thiols followed by bis-alkylation to introduce a three-carbon bridge to which the drug is covalently attached (Fig. 10). Depending on the degree of reduction, the numbers of free cysteines for conjugation can be 4 or 8 to generate DARs of 2 and 4 respectively. PolyTherics demonstrated that the ThioBridge ADCs are more stable than maleimide ADCs in the presence of human serum albumin (80).
Fig. 10.
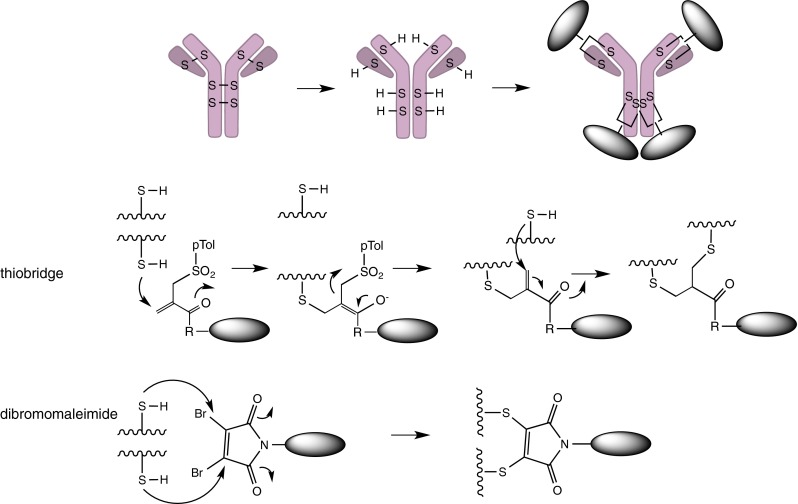
Two solutions to the disulfide re-bridging problem: The thiobridge uses two seuquential reactions and gives rise to a longer, more flexible bridge. The dibromomaleimide could in principle be simultaneously or sequentially attacked by the two thiols, giving rise to a short, rigid bridge.
Similarly, Igenica uses this approach to reduce all intermolecular disulfide bonds, exposing 8 cysteines, and then uses dibromomaleimide (DBM) to react with the free thiol groups to produce a dithiomaleimide (DTM) (81, 82). Four cytotoxic drugs with this bifunctional linker are conveniently attached to the mAbs by crosslinking the reduced cysteins in the Fab and hinge regions. To make ADCs with DARs higher than 4, additional cysteines must be engineered into the mAb.
Glycoengineering
ADCs can also be constructed by attaching the drug to chemically activated sugars, attached to the glycosylated mAb. These glyco-ADCs are constructed by engineering glycosyltranserases in order to accommodate chemically active sugars. Molecules of choice can then be conjugated to the chemical handle on the sugar moiety. Residues in the active site of the glycotransferase are mutated to allow the transfer of the chemically reactive sugar residue.
Using modified galactosyl- and sialyltransferases in vitro, terminal sialic acids were introduced into native glycans on Asn-297 of antibodies. Subsequent periodate oxidation then exposed aldehyde groups, which were used to conjugate aminooxy functionalized cytotoxic agents via oxime ligation (Fig. 11) (83). Direct introduction of a keto group was realized by NCI researchers using a mutant β1,4Gal-T1-Y289L transferase, 2-(2-oxopropyl) galactose, and a bioincorporation strategy combined with oximination of the drug-linker (84). SynAffix reports that their GlycoConnectTM technology involves two distinct steps of (a) enzymatic glycan remodeling (‘tagging’) and (b) selective chemical conjugation. The enzymatic incorporation of different sugars enables different conjugation methods: such as azide-containing sugars, thiol-modified sugars and halogenated sugars (SynAffix website).
Fig. 11.
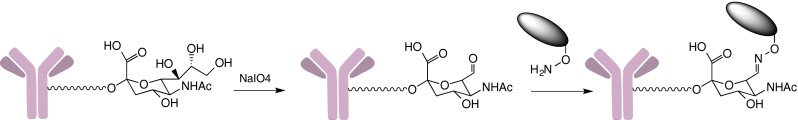
Periodate oxidation of antibody glycans is another strategy to enable carbonyl–directed chemistry.
Enzymatic Conjugation by Transglutaminase/Engineered Transglutaminase
As discussed in the section 1.3 on enzymatic conjugation, the use of bacterial transglutaminase (BTG) allows one to attach diverse compounds at multiple positions by inserting an LLQGA tag into antibodies. Deglycosylated mAb can be attached with amino containing compounds site-specifically at its Gln295 residue (49). Unfortunately with intact sugar chain, no Gln residues are available to be reactive with the BTG active site. Using rational design, Dophen Biomed engineered a BTG by enlarging the opening of the active site pocket so that a surface Gln could be docked close to catalytic Cys residue of the engineered BTGenzyme. This enables the direct site-specific conjugation of drug to an un-modified mAb’s heavy chain constant region at a surface Gln residue (85).
We summarize current methods to make homogenous ADCs and the highlights of their technologies in Table 1.
Table 1.
Regio- and Stoichiometry- Controlled ADCs have been Obtained by a Variety of Methods, Reflecting the Expanded Interest in the Field. KEY: (a) Whether the Antibody is (yes) or is not (no) Glycosylated. (b) The Expected Drug-Antibody Ratio Based on Available Sites on the Antibody. In Many Cases, the DAR may be Somewhat More Variable than the Average Values Presented Here
| Mutagenesis | Method and Conjugation chemistry | Glycosylationa | Conjugation site | DARb | Institutions | |
|---|---|---|---|---|---|---|
| Yes | Mutation introduced | |||||
| Engineered Cys | Thioether via thiol-maleimide | Yes | Engineered Cys | 2 or 4 | Genentech, Seattle Genetics | |
| Enzymatic manipulation | Transamination onto inserted glutamine | Yes | LLQGA | 2 or 4 | Rinat/Pfizer | |
| Transamination onto native Q295 (requires N297S mutation or deglycosylation) or N297Q | No | Q295 or Q297 | 2 or 4 | Innate Pharma | ||
| FGE oxidation to aldehyde (SMART tag) by insertion of FGE recognition tag cxpxr to generate formylglycine (Fg) from cysteine; several available carbonyl chemistries | Yes | FgxPxR | 2, 4, or 6 | Redwood Bioscience/Catalent | ||
| nnAA incorporation | In vivo nnAA incorporation via Met or amber suppression aldehyde-hydroxyamine reaction to oxime | Yes | nnAA | 2 or 4 | Ambrx, Allozyne/MedImmune | |
| In vitro nnAA incorporation [3 + 2]SPAAC and/or inverse electron demand Diels-Alder | No | nnAA | 2 or more | Sutro Biopharma | ||
| No | Thiol linkage | Tandem thiol reaction on bis-electrophile | Yes | Cys | 2 or more | Igenica, PolyTherics |
| Glycan enzymatic conjugation | Remodel the conserved glycan to recruit carbohydrate functional groups for conjugation to mAb; several chemistries available | Yes | Oxidized glycan carbonyl or synthetic carbohydrate functional group | 2 | Glykos, SynAffix | |
| BTG conjugation | Reaction between carboxamide group of glutamine and a primary amino group | No or yes with engineered BTG | Q295 | 2 | Dophen Biomed | |
Conjugation Chemistries on the Horizon
Biocompatible synthetic reactions are being explored to extend the toolkit for protein modification. The [3 + 2] strain-promoted azide-alkyne cycloaddition (SPAAC) shown in Fig. 1 was used by University of Georgia researchers in conjunction with glycoengineering. The azide was incorporated into the protein on an azido sugar by a mutated galactosyl transferase. The subsequent reaction with DBCO ligands, including doxorubicin, followed the same course as the Fig. 1 conjugation chemistry (86). Although none of the chemistries described below (Fig. 12) have been applied to ADCs as of the writing of this review, one may look to them for future adaptation to antibody conjugation.
Fig. 12.

Reactions once considered restricted to synthetic chemistry are being incorporated into the toolkit of bioconjugation. Solubility, stability, and stoichiometry issues are among the parameters that need to be optimized.
Sonogashira cross coupling of a terminal alkyne to an aryl iodide is in principle a promising bioorthogonal chemistry for ADCs, since either an iodoPhe or an alkynyl aminoacid could be used as the site of conjugation. Li et al. (87) took the latter route, using a homopropargyl glycine auxotrophically substituted for methionine at the C-terminus of ubiquitin. The HPG-Ub protein was converted in 30 min to a variety of aryl-substituted derivatives, in 55–93% yields, in the presence of a water-soluble (and -compatible) palladium complex developed in their group. The 50:1 stoichiometry of both ‘catalyst’ and aryl iodide to protein, however, indicate that further development of this method is necessary before using it to make cytotoxic drug conjugates on antibodies. A recent and impressive extension of Sonogashira reactions in living bacterial cells, at much lower stoichiometires and with a simple Pd(NO3)2 promoter, was reported by Li et al. (88), suggesting that these developments are attainable. Unfavorable stoichiometry has so far also impeded Suzuki-Miyaura coupling, which has been utilized for the reaction of iodophenylalanine in maltose binding protein to small molecule aryl boronates (89), from becoming a mainstream method for antibody-drug conjugation.
The facile conversion of thiols to allyl thiols, and the reactivity of cysteinyl allyl thiols in cross metathesis, promoted by water–soluble ruthenium catalysts, opened an alternative route to cysteine conjugation. Initial attempts to modify the B. lentus subtilisin protein required 10,000:1 stoichiometry of the small molecule olefin, however, due to a strong tendency toward self-metathesis. (90, 91). Binder and Raines subsequently reduced the stoichiometry to 200:1 (92). Se-allyl selenocyseine (Seac) gave somewhat improved kinetics, but nonetheless required 2000:1 small molecule reactant:protein (93). Given the current interest in thiobridging to obtain homogeneous ADCs that require no genetic manipulation, overcoming the hurdles to metathesis would seem a useful pursuit. Chemists at the ETH have been developing potassium acyl trifluroborates (KATs) for bioorthogonal reactions with suitable non-natural amino acids (94) and have recently demonstrated the formation of amide bonds between a diverse set of KATs and synthetic, but unprotected peptides bearing the O-carbamoyhydroxylamine function. (95). The ability to site-specifically incorporate O-diethylcarbamoyl-hydroxylamine into a protein would enable conjugation of small molecules to antibodies via an amide bond.
The pioneering ADCs from the last two decades have become, in turn, the substrate for further innovation. Creative small-molecule and protein chemistries, often co-mingled, are being applied to develop homogeneous, site-specific conjugates. Analytical and bio-analytical technologies are achieving higher and higher resolution to subtend this drive.
Whether derived from a modify-the-antibody side or the modify-the-linker side, the next generation of ADCs to reach the clinic is likely to have a dramatically distinct profile as compared to the current generation of ADCs in the clinic and on the market.
References
- 1.Kovtun YV, Goldmacher VS. Cell killing by antibody-drug conjugates. Cancer Lett. 2007;255(2):232–40. doi: 10.1016/j.canlet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Chari RVJ. Targeted cancer therapy: Conferring specificity to cytotoxic drugs. Acc Chem Res. 2008;41(1):98–107. doi: 10.1021/ar700108g. [DOI] [PubMed] [Google Scholar]
- 3.Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5(2):147–59. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 4.Perez HL, Cardarelli PM, Deshpande S, Gangwar S, Schroeder GM, Vite GD, et al. Antibody-drug conjugates: current status and future directions. Drug Discov Today. 2014;19(7):869–81. doi: 10.1016/j.drudis.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Chari RVJ, Miller ML, Widdison WC. Antibody-drug conjugates: An emerging concept in cancer therapy. Angew Chem Int Ed Engl. 2014;53(15):3796–827. doi: 10.1002/anie.201307628. [DOI] [PubMed] [Google Scholar]
- 6.Flygare JA, Pillow TH, Aristoff P. Antibody-drug conjugates for the treatment of cancer. Chem Biol Drug Des. 2013;81(1):113–21. doi: 10.1111/cbdd.12085. [DOI] [PubMed] [Google Scholar]
- 7.Behrens CR, Liu B. Methods for site-specific drug conjugation to antibodies. mAbs. 2014;6(1):46–53. doi: 10.4161/mabs.26632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panowski S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. mAbs. 2014;6(1):34–45. doi: 10.4161/mabs.27022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LoRusso PM, Weiss D, Guardino E, Girish S, Sliwkowski MX. Trastuzumab emtansine: A unique antibody-drug conjugate in development for human epidermal growth factor receptor 2-positive cancer. Clin Cancer Res. 2011;17(20):6437–47. doi: 10.1158/1078-0432.CCR-11-0762. [DOI] [PubMed] [Google Scholar]
- 10.Senter PD, Sievers EL. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat Biotechnol. 2012;30(7):631–7. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 11.Katz J, Janik JE, Younes A. Brentuximab Vedotin (SGN-35) Clin Cancer Res. 2011;17(20):6428–36. doi: 10.1158/1078-0432.CCR-11-0488. [DOI] [PubMed] [Google Scholar]
- 12.Hamblett KJ, Senter PD, Chace DF, Sun MMC, Lenox J, Cerveny CG, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10(20):7063–70. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Amphlett G, Blättler WA, Lambert JM, Zhang W. Structural characterization of the maytansinoid–monoclonal antibody immunoconjugate, huN901–DM1, by mass spectrometry. Protein Sci. 2005;14(9):2436–46. doi: 10.1110/ps.051478705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen B-Q, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 2012;30(2):184–9. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 15.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4(3):239–44. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 17.Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci U S A. 2012;109(40):16101–6. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296(5572):1459–62. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 19.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, et al. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431(7006):333–5. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 20.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371(4):818–22. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 21.Polycarpo CR, Herring S, Bérubé A, Wood JL, Söll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580(28–29):6695–700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fekner T, Li X, Lee MM, Chan MK. A Pyrrolysine Analogue for Protein Click Chemistry. Angew Chem. 2009;121(9):1661–3. doi: 10.1002/ange.200805420. [DOI] [PubMed] [Google Scholar]
- 23.Grabstein KH, Van BM, Marelli M. Amino acid derivatives. WO2014044873 A1, 2014
- 24.Hofer T, Skeffington LR, Chapman CM, Rader C. Molecularly defined antibody conjugation through a selenocysteine interface. Biochemistry (Mosc) 2009;48(50):12047–57. doi: 10.1021/bi901744t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiick KL, Tirrell DA. Protein Engineering by In Vivo Incorporation of Non-Natural Amino Acids: Control of Incorporation of Methionine Analogues by Methionyl-tRNA Synthetase. Tetrahedron. 2000;56(48):9487–93. doi: 10.1016/S0040-4020(00)00833-4. [DOI] [Google Scholar]
- 26.Tirrell DA, Kiick KL.Overexpression of aminoacyl-tRNA synthetases for efficient production of engineered proteins containing amino acid analogues, US20120077228 A1. 2012
- 27.Zawada JF, Yin G, Steiner AR, Yang J, Naresh A, Roy SM, et al. Microscale to manufacturing scale-up of cell-free cytokine production–a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108(7):1570–8. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin G, Garces ED, Yang J, Zhang J, Tran C, Steiner AR. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. 4. 2012(2). [DOI] [PMC free article] [PubMed]
- 29.Groff D, Armstrong S, Rivers PJ, Zhang J, Yang J, Green E. Engineering toward a bacterial “endoplasmic reticulum” for the rapid expression of immunoglobulin proteins. mAbs. 2014;6(3). [DOI] [PMC free article] [PubMed]
- 30.Johnson DBF, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7(11):779–86. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray CJ, Baliga R. Cell-free translation of peptides and proteins: From high throughput screening to clinical production. Curr Opin Chem Biol. 2013;17(3):420–6. doi: 10.1016/j.cbpa.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G. Multiple-site labeling of proteins with unnatural amino acids. Angew Chem Int Ed Engl. 2012;51(9):2243–6. doi: 10.1002/anie.201108275. [DOI] [PubMed] [Google Scholar]
- 33.Sando S, Ogawa A, Nishi T, Hayami M, Aoyama Y. In vitro selection of RNA aptamer against Escherichia coli release factor 1. Bioorg Med Chem Lett. 2007;17(5):1216–20. doi: 10.1016/j.bmcl.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Agafonov DE, Huang Y, Grote M, Sprinzl M. Efficient suppression of the amber codon in E. coli in vitro translation system. FEBS Lett. 2005;579(10):2156–60. doi: 10.1016/j.febslet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19(8):751–5. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 36.Lee KB, Kim H-C, Kim D-M, Kang TJ, Suga H. Comparative evaluation of two cell-free protein synthesis systems derived from Escherichia coli for genetic code reprogramming. J Biotechnol. 2012;164(2):330–5. doi: 10.1016/j.jbiotec.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Hallam TJ, Smider VV. Unnatural amino acids in novel antibody conjugates. Futur Med Chem. 2014;6(11):1309–24. doi: 10.4155/fmc.14.79. [DOI] [PubMed] [Google Scholar]
- 38.Zimmerman ES, Heibeck TH, Gill A, Li X, Murray CJ, Madlansacay MR, et al. Production of site-specific antibody–drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug Chem. 2014;25(2):351–61. doi: 10.1021/bc400490z. [DOI] [PubMed] [Google Scholar]
- 39.Goto Y, Suga H. Translation Initiation with Initiator tRNA Charged with Exotic Peptides. J Am Chem Soc. 2009;131(14):5040–1. doi: 10.1021/ja900597d. [DOI] [PubMed] [Google Scholar]
- 40.Niwa N, Yamagishi Y, Murakami H, Suga H. A flexizyme that selectively charges amino acids activated by a water-friendly leaving group. Bioorg Med Chem Lett. 2009;19(14):3892–4. doi: 10.1016/j.bmcl.2009.03.114. [DOI] [PubMed] [Google Scholar]
- 41.Krall N, Scheuermann J, Neri D. Small targeted cytotoxics: Current state and promises from DNA-encoded chemical libraries. Angew Chem Int Ed Engl. 2013;52(5):1384–402. doi: 10.1002/anie.201204631. [DOI] [PubMed] [Google Scholar]
- 42.Brabletz T. Abstract SY05-03: microRNAs, EMT, and cancer stem cells. Cancer Res. 2012 Apr 15;72(8 Supplement):SY05–03 – SY05–03.
- 43.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26(8):925–32. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 44.Junutula JR, Bhakta S, Raab H, Ervin KE, Eigenbrot C, Vandlen R, et al. Rapid identification of reactive cysteine residues for site-specific labeling of antibody-Fabs. J Immunol Methods. 2008;332(1–2):41–52. doi: 10.1016/j.jim.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Kung Sutherland MS, Walter RB, Jeffrey SC, Burke PJ, Yu C, Kostner H, et al. SGN-CD33A: A novel CD33-targeting antibody-drug conjugate using a pyrrolobenzodiazepine dimer is active in models of drug-resistant AML. Blood. 2013;122(8):1455–63. doi: 10.1182/blood-2013-03-491506. [DOI] [PubMed] [Google Scholar]
- 46.Liebscher S, Schöpfel M, Aumüller T, Sharkhuukhen A, Pech A, Höss E, et al. N-terminal protein modification by substrate-activated reverse proteolysis. Angew Chem Int Ed Engl. 2014;53(11):3024–8. doi: 10.1002/anie.201307736. [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama K, Nio N, Kikuchi Y. Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol. 2004;64(4):447–54. doi: 10.1007/s00253-003-1539-5. [DOI] [PubMed] [Google Scholar]
- 48.Spolaore B, Raboni S, Ramos Molina A, Satwekar A, Damiano N, Fontana A. Local unfolding is required for the site-specific protein modification by transglutaminase. Biochemistry (Mosc) 2012;51(43):8679–89. doi: 10.1021/bi301005z. [DOI] [PubMed] [Google Scholar]
- 49.Jeger S, Zimmermann K, Blanc A, Grünberg J, Honer M, Hunziker P, et al. Site-specific and stoichiometric modification of antibodies by bacterial transglutaminase. Angew Chem Int Ed Engl. 2010;49(51):9995–7. doi: 10.1002/anie.201004243. [DOI] [PubMed] [Google Scholar]
- 50.Strop P, Liu SH, Dorywalska M, Delaria K, Dushin RG, Tran TT, et al. Location matters: Site of conjugation modulates stability and pharmacokinetics of antibody drug conjugates. Chem Biol. 2013;20(2):161–7. doi: 10.1016/j.chembiol.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Farias SE, Strop P, Delaria K, Galindo Casas M, Dorywalska M, Shelton DL, et al. Mass spectrometric characterization of transglutaminase based site-specific antibody-drug conjugates. Bioconjug Chem. 2014;25(2):240–50. doi: 10.1021/bc4003794. [DOI] [PubMed] [Google Scholar]
- 52.Hudak JE, Barfield RM, de Hart GW, Grob P, Nogales E, Bertozzi CR, et al. Synthesis of heterobifunctional protein fusions using copper-free click chemistry and the aldehyde tag. Angew Chem Int Ed Engl. 2012;51(17):4161–5. doi: 10.1002/anie.201108130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agarwal P, van der Weijden J, Sletten EM, Rabuka D, Bertozzi CR. A Pictet-Spengler ligation for protein chemical modification. Proc Natl Acad Sci. 2013;110(1):46–51. doi: 10.1073/pnas.1213186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith EL, Giddens JP, Iavarone AT, Godula K, Wang L-X, Bertozzi CR. Chemoenzymatic Fc glycosylation via engineered aldehyde tags. Bioconjug Chem. 2014;25(4):788–95. doi: 10.1021/bc500061s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabuka D, Rush JS, Dehart GW, Wu P, Bertozzi CR. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat Protoc. 2012;7(6):1052–67. doi: 10.1038/nprot.2012.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rashidian M, Dozier JK, Distefano MD. Enzymatic labeling of proteins: Techniques and approaches. Bioconjug Chem. 2013;24(8):1277–94. doi: 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–3. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 58.Tsukiji S, Nagamune T. Sortase-mediated ligation: A gift from Gram-positive bacteria to protein engineering. Chem Biochem Eur J Chem Biol. 2009;10(5):787–98. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 59.Madej MP, Coia G, Williams CC, Caine JM, Pearce LA, Attwood R, et al. Engineering of an anti-epidermal growth factor receptor antibody to single chain format and labeling by Sortase A-mediated protein ligation. Biotechnol Bioeng. 2012;109(6):1461–70. doi: 10.1002/bit.24407. [DOI] [PubMed] [Google Scholar]
- 60.Matsumoto T, Takase R, Tanaka T, Fukuda H, Kondo A. Site-specific protein labeling with amine-containing molecules using Lactobacillus plantarum sortase. Biotechnol J. 2012;7(5):642–8. doi: 10.1002/biot.201100213. [DOI] [PubMed] [Google Scholar]
- 61.Ta HT, Peter K, Hagemeyer CE. Enzymatic antibody tagging: Toward a universal biocompatible targeting tool. Trends Cardiovasc Med. 2012;22(4):105–11. doi: 10.1016/j.tcm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Williamson DJ, Fascione MA, Webb ME, Turnbull WB. Efficient N-Terminal Labeling of Proteins by Use of Sortase. Angew Chem. 2012;124(37):9511–4. doi: 10.1002/ange.201204538. [DOI] [PubMed] [Google Scholar]
- 63.Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc Natl Acad Sci U S A. 2011;108(28):11399–404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Möhlmann S, Mahlert C, Greven S, Scholz P, Harrenga A. In vitro sortagging of an antibody fab fragment: Overcoming unproductive reactions of sortase with water and lysine side chains. Chem Biochem Eur J Chem Biol. 2011;12(11):1774–80. doi: 10.1002/cbic.201100002. [DOI] [PubMed] [Google Scholar]
- 65.Pucciarelli MG, Calvo E, Sabet C, Bierne H, Cossart P, García-del Portillo F. Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomic analysis. Proteomics. 2005;5(18):4808–17. doi: 10.1002/pmic.200402075. [DOI] [PubMed] [Google Scholar]
- 66.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2(2):99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 67.Fernández-Suárez M, Baruah H, Martínez-Hernández L, Xie KT, Baskin JM, Bertozzi CR, et al. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25(12):1483–7. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puthenveetil S, Liu DS, White KA, Thompson S, Ting AY. Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. J Am Chem Soc. 2009;131(45):16430–8. doi: 10.1021/ja904596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uttamapinant C, White KA, Baruah H, Thompson S, Fernández-Suárez M, Puthenveetil S, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci U S A. 2010;107(24):10914–9. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu DS, Loh KH, Lam SS, White KA, Ting AY. Imaging trans-cellular neurexin-neuroligin interactions by enzymatic probe ligation. PLoS ONE. 2013;8(2):e52823. doi: 10.1371/journal.pone.0052823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen JD, Zou P, Ting AY. Site-specific protein modification using lipoic acid ligase and bis-aryl hydrazone formation. Chem Biochem Eur J Chem Biol. 2012;13(6):888–94. doi: 10.1002/cbic.201100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. Diels–Alder Cycloaddition for Fluorophore Targeting to Specific Proteins inside Living Cells. J Am Chem Soc. 2012;134(2):792–5. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Möhlmann S, Bringmann P, Greven S, Harrenga A. Site-specific modification of ED-B-targeting antibody using intein-fusion technology. BMC Biotechnol. 2011;11:76. doi: 10.1186/1472-6750-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vila-Perello M, Liu Z, Shah NH, Willis JA, Idoyaga J, Muir TW. Streamlined expressed protein ligation using split inteins. J Am Chem Soc. 2013;135(1):286–92. doi: 10.1021/ja309126m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y-M, Yang M-Y, Huang Y-C, Li Y-T, Chen PR, Liu L. Ligation of expressed protein α-hydrazides via genetic incorporation of an α-hydroxy acid. ACS Chem Biol. 2012;7(6):1015–22. doi: 10.1021/cb300020s. [DOI] [PubMed] [Google Scholar]
- 76.Charalambous A, Andreou M, Antoniades I, Christodoulou N, Skourides PA. In vivo, site-specific, covalent conjugation of quantum dots to proteins via split-intein splicing. Methods Mol Biol Clifton NJ. 2012;906:157–69. doi: 10.1007/978-1-61779-953-2_11. [DOI] [PubMed] [Google Scholar]
- 77.Hondal RJ, Nilsson BL, Raines RT. Selenocysteine in native chemical ligation and expressed protein ligation. J Am Chem Soc. 2000;123(21):5140–1. doi: 10.1021/ja005885t. [DOI] [PubMed] [Google Scholar]
- 78.Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, et al. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol. 2003;10(4):313–7. doi: 10.1016/S1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 79.Hussain AF, Kampmeier F, von Felbert V, Merk H-F, Tur MK, Barth S. SNAP-tag technology mediates site specific conjugation of antibody fragments with a photosensitizer and improves target specific phototoxicity in tumor cells. Bioconjug Chem. 2011;22(12):2487–95. doi: 10.1021/bc200304k. [DOI] [PubMed] [Google Scholar]
- 80.Badescu G, Bryant P, Bird M, Henseleit K, Swierkosz J, Parekh V, et al. Bridging disulfides for stable and defined antibody drug conjugates. Bioconjug Chem. 2014;25(6):1124–36. doi: 10.1021/bc500148x. [DOI] [PubMed] [Google Scholar]
- 81.Robin MP, Mabire AB, Damborsky JC, Thom ES, Winzer-Serhan UH, Raymond JE, et al. New functional handle for use as a self-reporting contrast and delivery agent in nanomedicine. J Am Chem Soc. 2013;135(25):9518–24. doi: 10.1021/ja403587c. [DOI] [PubMed] [Google Scholar]
- 82.Jackson DY, Ha E. Antibody-drug conjugates and related compounds, compositions, and methods. WO2013085925 A1, 2013.
- 83.Zhou Q, Stefano JE, Manning C, Kyazike J, Chen B, Gianolio DA, et al. Site-specific antibody–drug conjugation through glycoengineering. Bioconjug Chem. 2014;25(3):510–20. doi: 10.1021/bc400505q. [DOI] [PubMed] [Google Scholar]
- 84.Zhu Z, Ramakrishnan B, Li J, Wang Y, Feng Y, Prabakaran P. Site-specific antibody-drug conjugation through an engineered glycotransferase and a chemically reactive sugar. mAbs. 2014;6(5). [DOI] [PMC free article] [PubMed]
- 85.Hu S, Yin Y. Transglutaminase for Protein Drug Modification: Pegylation and beyond. In: Tao J (Alex), Kazlauskas R, editors. Biocatalysis for Green Chemistry and Chemical Process Development [Internet]. John Wiley & Sons, Inc.; 2011 p. 151–72.
- 86.Li X, Fang T, Boons GJ. Preparation of well-defined antibody–drug conjugates through glycan remodeling and strain-promoted azide–alkyne cycloadditions. Angew Chem. 2014;126(28):7307–10. doi: 10.1002/ange.201402606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li N, Lim RKV, Edwardraja S, Lin Q. Copper-free Sonogashira cross-coupling for functionalization of alkyne-encoded proteins in aqueous medium and in bacterial cells. J Am Chem Soc. 2011;133(39):15316–9. doi: 10.1021/ja2066913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J, Lin S, Wang J, Jia S, Yang M, Hao Z, et al. Ligand-free palladium-mediated site-specific protein labeling inside gram-negative bacterial pathogens. J Am Chem Soc. 2013;135(19):7330–8. doi: 10.1021/ja402424j. [DOI] [PubMed] [Google Scholar]
- 89.Spicer CD, Davis BG. Palladium-mediated site-selective Suzuki-Miyaura protein modification at genetically encoded aryl halides. Chem Commun Camb Engl. 2011;47(6):1698–700. doi: 10.1039/c0cc04970k. [DOI] [PubMed] [Google Scholar]
- 90.Lin YA, Chalker JM, Floyd N, Bernardes GJL, Davis BG. Allyl sulfides are privileged substrates in aqueous cross-metathesis: Application to site-selective protein modification. J Am Chem Soc. 2008;130(30):9642–3. doi: 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]
- 91.Lin YA, Chalker JM, Davis BG. Olefin cross-metathesis on proteins: Investigation of allylic chalcogen effects and guiding principles in metathesis partner selection. J Am Chem Soc. 2010;132(47):16805–11. doi: 10.1021/ja104994d. [DOI] [PubMed] [Google Scholar]
- 92.Binder JB, Raines RT. Olefin metathesis for chemical biology. Curr Opin Chem Biol. 2008;12(6):767–73. doi: 10.1016/j.cbpa.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin YA, Boutureira O, Lercher L, Bhushan B, Paton RS, Davis BG. Rapid cross-metathesis for reversible protein modifications via chemical access to Se-allyl-selenocysteine in proteins. J Am Chem Soc. 2013;135(33):12156–9. doi: 10.1021/ja403191g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Molander GA, Raushel J, Ellis NM. Synthesis of an acyltrifluoroborate and its fusion with azides to form amides. J Org Chem. 2010;75(12):4304–6. doi: 10.1021/jo1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noda H, Erős G, Bode JW. Rapid ligations with equimolar reactants in water with the potassium acyltrifluoroborate (KAT) amide formation. J Am Chem Soc. 2014;136(15):5611–4. doi: 10.1021/ja5018442. [DOI] [PubMed] [Google Scholar]


