Abstract
Hydroxyl radical protein footprinting (HRPF) is an MS-based technique for analyzing protein structure based on measuring the oxidation of amino acid side chains by hydroxyl radicals diffusing in solution. Spatial resolution of HRPF is limited by the smallest portion of the protein for which oxidation amounts can be accurately quantitated. Previous work has shown electron transfer dissociation (ETD) to be the most reliable method for quantifying the amount of oxidation of each amino acid side chain in a mixture of peptide oxidation isomers, but efficient ETD requires high peptide charge states which limits its applicability for HRPF. Supercharging reagents have been used to enhance peptide charge state for ETD analysis, but previous work have shown supercharging reagents to enhance charge state differently for different peptides sequences; it is currently unknown if different oxidation isomers will experience different charge enhancement effects. Here, we report the effect of m-nitrobenzyl alcohol (m-NBA) on the ETD-based quantification of peptide oxidation. The addition of m-NBA to both a defined mixture of synthetic isomeric oxidized peptides and Robo1 protein subjected to HRPF increased the abundance of higher charge state ions, improving our ability to perform efficient ETD of the mixture. No differences in the reported quantitation by ETD were noted in the presence or absence of m-NBA, indicating that all oxidation isomers were charge-enhanced to a similar extent. These results indicate the utility of m-NBA for residue-level quantification of peptide oxidation in HRPF and other applications.
Introduction
Protein tertiary and quaternary structure are fundamental to determining mechanisms of protein function. Understanding the structure and function of proteins and their interactions in macromolecular assemblies is critical to achieve an overall understanding of biological systems. Hydroxyl radical protein footprinting (HRPF) is a relatively recent covalent labeling approach coupled with mass spectrometry, and has been developed over the last decade to a powerful method for analyzing protein structure and dynamics. HRPF has several advantages that recommend it for the analysis of protein structure, particularly for difficult systems such as large, heterogeneous protein complexes, membrane proteins, and flexible protein systems [1-3]. HRPF takes advantage of the fact that the rate of oxidation of each amino acid varies directly with the solvent accessibility of that amino acid [4, 5]. This relationship allows for changes in protein structure to be monitored by monitoring the apparent rate of oxidation of a particular amino acid side chain [6, 7].
Initial uses of HRPF were limited in spatial resolution to the size of a proteolytic peptide, as the amount of oxidation of any individual amino acid within the peptide could not be accurately quantified by CID [8-10]. As sub-microsecond HRPF technologies such as Fast Photochemical Oxidation of Proteins (FPOP) [3] and pulsed electron beam radiolysis [11] began to allow for heavier oxidation of proteins, the need to quantitate isomeric peptide oxidation products became even more pronounced. Reports from Gross and coworkers have used UPLC to separate isomeric peptide products and quantify based on peak area in a selected ion chromatogram [12]; however, the only attempt to use UPLC separation coupled with peak area quantification using known oxidized peptide standards found this method to be inaccurate in some cases, while electron transfer dissociation (ETD) provided an accurate and reliable quantification of oxidation at the residue level for isomeric mixtures [13].
While ETD gave reliable results for residue-level quantification of oxidation, ETD is widely known for having poor fragmentation efficiency for doubly-charged peptides, which are commonly observed for tryptic digestion products. This poor fragmentation efficiency limits both the sensitivity of ETD-based quantification as well as the spatial resolution of HRPF information, as cleavage of each peptide bond in the peptide is required for true residue-level resolution. One approach to improve ETD fragmentation is based on addition of supercharging reagent into electrospray solution to increase the charge state of tryptic peptide ions [14, 15]. As the ability to quantify oxidation by ETD depends upon the ability of m-NBA to equally alter the charge state of each oxidation isomer of a given peptide sequence, as well as the ETD fragmentation process remaining transparent to the site of oxidation in the presence of m-NBA, the applicability of supercharging to ETD-based HRPF remains in question. In this study, we test the effect of the charge-enhancing reagent m-NBA on the ability to accurately quantify the amount of oxidation on each amino acid by ETD, as well as the ability of m-NBA to positively impact actual HRPF studies of an oxidized protein.
Experimental
m-Nitrobenzyl alcohol (m-NBA), catalase, formic acid and L-glutamine was purchased from the Sigma-Aldrich Corporation (St. Louis, MO). Hydrogen peroxide (30%) was obtained from J.T. Baker (Phillipsburg, NJ). Dithiothreitol (DTT) and HPLC-grade acetonitrile (ACN) were purchased from Fisher Scientific (Fair Lawn, NJ). Methionine amide was purchased from Bachem (Torrace, CA). Sequencing-grade modified trypsin was purchased from Promega Corporation (Madison, WI, USA). All reagents were used as provided. Purified water (18 MΩ) was obtained from an in-house Milli-Q Synthesis system (Millipore, Billerica, MA). Peptide oxidation analog standards of the peptides RPMFAIWK and MLLPSGSLFFLR were synthesized and purified as previously described [13], and detailed in Supplementary Information. Robo1 Ig1-2 protein was a gift from Prof. Kelley Moremen (University of Georgia) and expressed and purified as described in Supplementary Information.
The working stock solutions of the four synthetic peptides were prepared in 50% ACN or in 50% ACN containing 0.1% m-NBA. The working solutions with or without m-NBA were mixed in 1:1:1:1 volume ratios, respectively. The final molar concentration for each peptide in the mixture is 2 μM. HRPF and tryptic digestion of Robo1 Ig1-2 was performed using FPOP as described previously [16, 17] and summarized in Supplementary Information. Mass spectral analyses were performed as detailed in Supplementary Information.
The fragment ion intensities from ETD and/or CID are used for the calculation of oxidation rate at specific residue site using a similar approach reported previously [13, 18]. The actual fractional oxidation of a given sequence ion is defined as the ratio between the oxidized sequence ion intensity to the sum of the intensity of the corresponding oxidized and unoxidized sequence ion. This is shown in Equation 1:
| (1) |
where f (ci)actual denotes the fractional oxidation of c-ion i. I(ci) denotes the intensity of the c ion i, whether the oxidized and unoxidized form.
The relative oxidation rate for a specific residue i is calculated as the difference between the fractional oxidation of adjacent residues. This is shown in Equation 2:
| (2) |
Results and Discussion
Effect of 0.1% m-NBA on relative quantitation of synthetic peptide isomers by ETD and CID
The effect of different concentrations of m-NBA on the charge states of the oxidized peptide RPMFA*IWK (where the alanine is oxidized) is shown in Table S1. The absolute intensity of the triply-charged ion stopped increasing appreciably above a concentration of 0.1% m-NBA, which was used in the following studies. A representative ESI mass spectra of the peptide RPMFA*IWK with 0% and 0.1% m-NBA is shown in Figure S1, where it can be seen that 0.1% m-NBA did not appreciably add to the background of the spectrum. Mobile phase containing 0.1% m-NBA was not found to have a negative effect on either the chromatographic peak ion intensity or the peak shape of peptide RPMFA*IWK in LC-MS (data not shown).
In order for m-NBA to be useful for ETD-based high resolution HRPF, it is necessary that both the supercharging effect and the subsequent ETD fragmentation of the peptide be uninfluenced by the position of the site of oxidation in a mixture of isomers. Therefore, the effect of 0.1% m-NBA on the relative quantification of oxidation isomers was evaluated by direct infusion of a mixture of peptide oxidation isomers of either the peptide RPMFAIWK or MLLPSGSLFFLR, the Robo1 tryptic peptide 64-75, present at an equimolar ratio in 50% ACN solutions containing 0% and 0.1% m-NBA. The amount of oxidation at each specific residue site was determined based on the CID and ETD fragment ion intensity at different charge states and calculated by Equations 1 and 2. The measured average oxidation percentages from triplicate samples based on product ions from ETD are plotted against the theoretical percentages oxidized in the mixture (Figure 1). The percentage of oxidation in the mixture calculated based on abundance of the c-type and z-type product ion fragmented by ETD with and without m-NBA correlates very well to the theoretical values for both peptide oxidation isomer series, indicating m-NBA has no effects on ETD-based quantification of oxidation isomers with multiple oxidation sites at adjacent sites. For comparison, the same analysis for CID ion from these samples is shown in Figure S2. As reported previously [13], the precursor charge state and oxidized side chain identity plays a major role in CID quantification, with CID-based quantification remaining unreliable. UPLC analysis was previously shown to be unable to quantitate the RPMFAIWK methionine oxidation isomer [13], and was unable to separate the oxidized methionine and oxidized proline isomers in the MLLPSGSLFFLR peptide after gradient optimization, preventing quantification by UPLC (data not shown).
Figure 1.
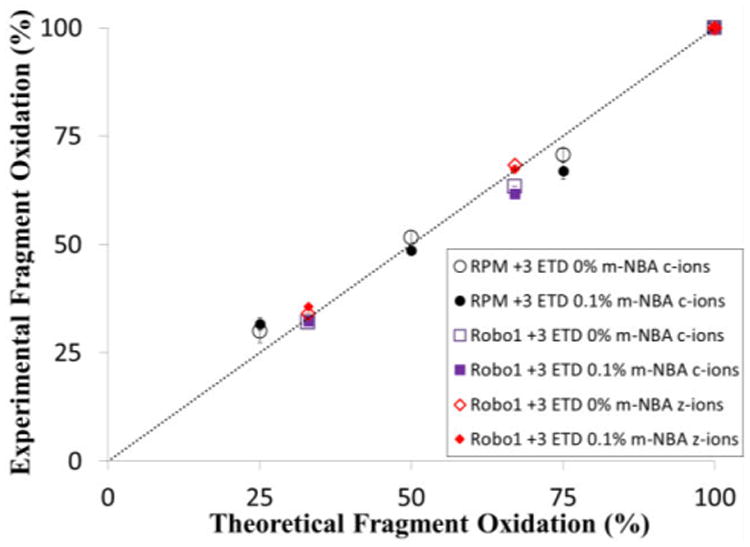
Comparison of theoretical and experimental quantification of equimolar mixtures of four oxidation isomers of the peptide RPMFAIWK based and three oxidation isomers of the peptide MLLPSGSLFFLR on product ion intensities by ETD of triply-charged ions in the presence of 0% or 0.1% m-NBA. The line represents the ideal 1:1 relationship between theoretical and measured oxidation. Error bars represent one standard deviation from a triplicate dataset.
Application of m-NBA in protein structural characterization by HRPF
The effect of m-NBA on ETD-based analysis of Robo1 subjected to HRPF was also studied. As shown in Table 1, addition of 0.1% m-NBA in mobile phase increases the relative abundance of the +3 and higher charge states for all unoxidized peptides measured (64-75 was only detected in the oxidized state after HRPF). m-NBA was found to increase the abundance of the higher charge states not only for the unoxidized peptides, but also for the oxidized products. The practical effect of the increased charge states of the oxidized peptides in the presence of 0.1%m-NBA in high resolution HRPF can be easily observed by comparing the ETD MS/MS spectrum of the highest charge state precursor detected for the oxidation product of peptide 194-205, shown in Supplementary Information Figure S3. In the absence of m-NBA, no +3 charge state precursor for the oxidized peptide was detected; the resulting ETD fragmentation of the +2 charge precursor resulted in poor ETD-based sequence coverage and lower signal-to-noise ratios. Upon addition of m-NBA, a sufficient abundance of the +3 charge state precursor ion could be detected for ETD fragmentation, and the resulting spectrum yielded many more product ions and superior signal-to-noise ratios.
Table 1. Charge State Distribution of peptides from Robo 1 trypsin digestion sample in mobile phases with or without m-NBA.
| 0% m-NBA | 0.1% m-NBA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Peptide | Charge-State Distribution | AV | Charge-State Distribution | AV | ||||||
| 1 + | 2+ | 3+ | 4+ | 1 + | 2+ | 3+ | 4+ | |||
| 14-25 | 0.00 | 96.53 | 3.47 | 0.00 | 2.03 | 0.17 | 22.52 | 77.31 | 0.00 | 2.77 |
| 35-47 | 0.00 | 52.76 | 47.24 | 0.00 | 2.47 | 0.00 | 6.05 | 92.99 | 0.96 | 2.95 |
| 64-75 (M+O) | 0.00 | 100.00 | 0.00 | 0.00 | 2.00 | 0.00 | 95.15 | 4.85 | 0.00 | 2.05 |
| 140-149 | 0.00 | 95.13 | 4.87 | 0.00 | 2.05 | 0.00 | 52.44 | 47.56 | 0.00 | 2.48 |
| 151-161 | 0.00 | 93.23 | 6.77 | 0.00 | 2.07 | 0.00 | 47.38 | 52.62 | 0.00 | 2.53 |
| 194-205 | 12.00 | 88.00 | 0.00 | 0.00 | 1.88 | 0.00 | 82.33 | 17.67 | 0.00 | 2.18 |
To ensure that the measured amount of oxidation observed from the sample analyzed in the presence of m-NBA yielded identical quantification results as the sample analyzed without m-NBA, we compared the +3 charge states of the +16 oxidation products of other peptides from Robo1. A representative MS/MS spectra of oxidized peptide 140-149 are shown in Supplementary Information, Figure S4. An obvious increase of the intensity of the triply-charged ion of this oxidized peptide was found in 0.1% m-NBA sample compared to 0% m-NBA (data not shown). In both the sample with 0% m-NBA and with 0.1% m-NBA, the ETD spectra of the oxidized peptide 140-GHPEPTISWK-149, [M+O+3H]3+ reveals no oxidation of the c3-c7 ions, and both oxidized and unoxidized c8 and c9 product ions, indicating residue S, W and K were oxidized in this mixture by HRPF. Furthermore, the quantification of oxidation extent based on ETD fragmentation intensity clearly indicates that residue S, W and K were oxidized 14%, 59%, 27% respectively in the sample with 0% m-NBA. In the sample with 0.1% m-NBA, 18% of S, 52% of W and 30% of K were oxidized, indicating that 0.1% m-NBA has no substantial effect on quantitating site-specific oxidation in HRPF samples. Similar consistency in the measured oxidation amounts were found for other oxidized peptides of Robo1 in the presence and absence of 0.1% m-NBA (data not shown).
Conclusion
In this work, we have demonstrated the ability of m-NBA in increasing charge state distribution and thus improve sequence coverage of ETD spectra without affecting the ETD-based quantification of site-specific oxidation. Both synthetic peptide mixtures and actual tryptic peptides from an HRPF experiment of Robo1 gave a robust increase in the abundance of higher charge states without negatively impacting the ETD-based quantification, indicating the oxidation isomers all have their charge states affected similarly by m-NBA. These results indicate that the use of m-NBA is a workable method for increasing sequence coverage and spatial resolution in HRPF quantification as well as quantification of protein oxidation in general by ETD-based LC-MS/MS.
Supplementary Material
Acknowledgments
This research is supported by the National Institute of General Medical Sciences-funded “Research Resource for Integrated Glycotechnology” (P41 GM103390), and in part by the National Institute of General Medical Sciences (1R01GM096049-01A1) from the National Institutes of Health. The authors would like to thank Prof. Kelley Moremen for the expression and purification of the Robo-1 Ig1-2 protein.
Footnotes
Supporting Information Available The online version of this article contains supplementary material, which is available to authorized users.
References
- 1.Sharp JS, Becker JM, Hettich RL. Protein surface mapping by chemical oxidation: Structural analysis by mass spectrometry. Anal Biochem. 2003;313(2):216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 2.Maleknia SD, Brenowitz M, Chance MR. Synchrotron radiolysis studies of peptides & proteins by mass spectrometry. Biophysical Journal. 1999;76(1):A172–A172. [Google Scholar]
- 3.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16(12):2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Xu GH, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107(8):3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 5.Sharp JS, Becker JM, Hettich RL. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal Chem. 2004;76(3):672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 6.Charvatova O, et al. Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1. J Am Soc Mass Spectrom. 2008;19(11):1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chance MR. Unfolding of apomyoglobin examined by synchrotron footprinting. Biochem Biophys Res Commun. 2001;287(3):614–621. doi: 10.1006/bbrc.2001.5628. [DOI] [PubMed] [Google Scholar]
- 8.Bridgewater JD, Srikanth R, Lim J, Vachet RW. The effect of histidine oxidation on the dissociation patterns of peptide ions. J Am Soc Mass Spectr. 2007;18(3):553–562. doi: 10.1016/j.jasms.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srikanth R, et al. Improved sequencing of oxidized cysteine and methionine containing peptides using electron transfer dissociation. J Am Soc Mass Spectrom. 2007;18(8):1499–1506. doi: 10.1016/j.jasms.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Srikanth R, Wilson J, Vachet RW. Correct identification of oxidized histidine residues using electron-transfer dissociation. Journal of Mass Spectrometry. 2009;44(5):755–762. doi: 10.1002/jms.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson C, et al. Pulsed electron beam water radiolysis for submicrosecond hydroxyl radical protein footprinting. Anal Chem. 2009;81(7):2496–2505. doi: 10.1021/ac802252y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Y, et al. Fast Photochemical Oxidation of Proteins (FPOP) Maps the Epitope of EGFR Binding to Adnectin. J Am Soc Mass Spectrom. 2014 doi: 10.1007/s13361-014-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Li Z, Xie B, Sharp JS. Improved identification and relative quantification of sites of peptide and protein oxidation for hydroxyl radical footprinting. J Am Soc Mass Spectrom. 2013;24(11):1767–1776. doi: 10.1007/s13361-013-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iavarone AT, Jurchen JC, Williams ER. Supercharged protein and peptide ions formed by electrospray ionization. Anal Chem. 2001;73(7):1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjeldsen F, Giessing AM, Ingrell CR, Jensen ON. Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Anal Chem. 2007;79(24):9243–9252. doi: 10.1021/ac701700g. [DOI] [PubMed] [Google Scholar]
- 16.Watson C, Sharp JS. Conformational analysis of therapeutic proteins by hydroxyl radical protein footprinting. AAPS J. 2012;14(2):206–217. doi: 10.1208/s12248-012-9336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem. 2009;81(16):6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumper CC, Bomgarden R, Rogers J, Etienne C, Schriemer DC. High-resolution mapping of carbene-based protein footprints. Anal Chem. 2012;84(10):4411–4418. doi: 10.1021/ac300120z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


