Abstract
Improved treatment for major depressive disorder (MDD) remains elusive due to limited understanding of its underlying biological mechanisms. Stress–induced maladaptive transcriptional regulation within limbic neural circuits likely contributes to the development of MDD, possibly through epigenetic factors that regulate chromatin structure. We establish that persistent upregulation of the ACF ATP–dependent chromatin remodeling complex, occurring in the nucleus accumbens of stress–susceptible mice and depressed humans, is necessary for stress–induced depressive–like behaviors. Altered ACF binding after chronic stress is correlated with altered nucleosome positioning, particularly around the transcription start sites of affected genes. These alterations in ACF binding and nucleosome positioning are associated with repressed expression of genes implicated in susceptibility to stress. Together, we identify the ACF chromatin remodeling complex as a critical component in the development of susceptibility to depression and in regulating stress–related behaviors.
Although major depressive disorder (MDD) is one of the most prevalent and debilitating disorders worldwide, it has been difficult to understand its pathophysiology and to develop more effective treatments1. Epidemiological studies have revealed that environmental factors, such as stressful life events, and highly complex genetic variations both act as important determinants of susceptibility and resilience to MDD2-5. Maladaptive transcriptional regulation within limbic neural circuits, including reward processing regions such as the nucleus accumbens (NAc), in response to chronic stress is thought to be a major contributor to the development of MDD4-10. Understanding this transcriptional dysregulation will be important in providing mechanistic insights into disease, as well as in identifying novel therapeutic targets.
Chromatin, histone and non–histone proteins associating with DNA, serves as an organizer of the genome by condensing the double–stranded DNA into multiple levels of higher order structures. Nucleosomes, each consisting of an octamer of core histones around which DNA is superhelically wrapped, are the basic packaging units of chromatin, and are positioned at precise locations to modulate accessibility of regulatory proteins to DNA, thus controlling eukaryotic gene regulation11. As a result, the mechanisms by which chromatin structure and nucleosome positions are specified and maintained in vivo are critical for the regulation of all DNA–dependent processes, including gene transcription. Epigenetic events—in particular, histone writers and erasers—that alter chromatin structure to regulate programs of gene expression have increasingly been associated with depression–related behavioral abnormalities in animal models and in depressed humans examined postmortem5,12-20. ATP–dependent chromatin remodeling complexes also play a key role in regulating nucleosome positioning to control gene expression, but have not yet been investigated in depression or other psychiatric disorders21-25.
Here we demonstrate that the specific and persistent upregulation of BAZ1A (also known as ACF1), a subunit of the ISWI family ACF (ATP-utilizing chromatin assembly and remodeling factor) chromatin remodeler complex, in NAc in several mouse depression models and in depressed humans, is necessary for susceptibility to stress–induced depressive–like behaviors by regulating nucleosome architecture at transcriptional start sites (TSSs) and repressing expression of a subset of genes. This identifies ATP-dependent chromatin remodeling dysregulation as a key mechanism in depression pathophysiology, and provides novel candidate targets for improved therapeutics for depression and other stress–related disorders.
Results
Regulation of ACF complex in mouse models and human depression
As ATP–dependent chromatin remodelers play a key role in regulating nucleosome positioning and transcriptional regulation, we screened subunits in 4 families of remodelers complexes (SWI/SNF, ISWI, CHD, and INO80) in NAc of an ethologically validated mouse model of depression, chronic social defeat stress (CSDS)20,26,27. Over 10 consecutive days C57BL/6J male mice were subjected to daily 10–minute aggressive encounters with CD1 mice, followed by sensory but not physical contact for the remainder of the day. Following CSDS, ~65% of test mice—termed susceptible—exhibit depression–related behavioral abnormalities including social avoidance (Fig. 1a) and reduced sucrose preference, whereas those that behave similar to control, non-stressed animals and do not exhibit these behaviors are termed resilient27. Initial profiling revealed persistent NAc mRNA expression changes in subunits from all 4 families of chromatin remodeling complexes 10 days after the last CSDS session (Supplementary Fig. 1a). Expression of the ISWI subunit Baz1a showed robust induction in NAc of susceptible mice, and thus was the focus of subsequent investigations. BAZ1A associates with the ATPase, SMARCA5 (also known as SNF2H), to form the ACF complex, which serves several cellular functions including transcriptional regulation28.
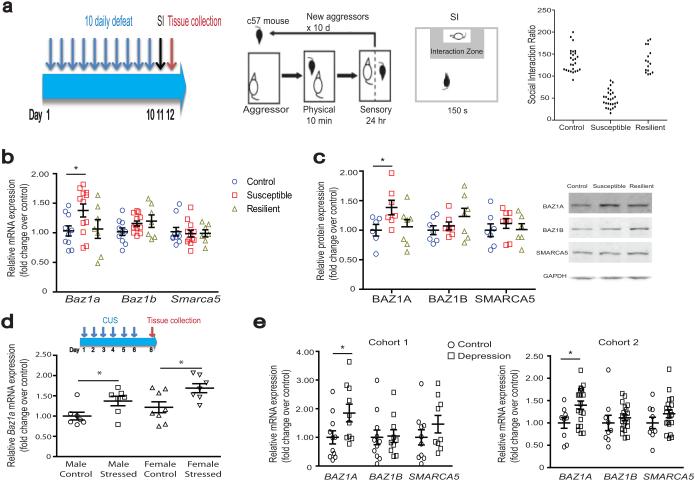
Figure 1.
Chromatin remodeling after chronic social defeat stress (CSDS) and in depressed humans. (a) Schematic of CSDS, social interaction test (including representative data for control, susceptible, and resilient mice), and time of tissue collection. d–day, hr–hour, min–minute, s–second. (b,c) NAc mRNA and protein levels 48 hours after CSDS, respectively. For b, control n = 10; susceptible n = 11; resilient n = 7. For c, control n = 6–7; susceptible n = 7–8; resilient n = 6–7. (d) NAc Baz1a mRNA levels 48 hours after 6 days of chronic unpredictable stress in male and female mice. Experimental schematic is shown in upper panel. n = 7 for control male, stressed male, stressed female, and n=8 for control female. (e) NAc mRNA levels in postmortem humans in two separate cohorts (demographic information in Supplementary Table 1). For cohort 1, control n = 10–11; depressed n = 9–10. For cohort 2, control n = 9; depressed n = 20. For b–e, post–hoc Student’s t–test * P < 0.05 compared to respective controls. See Supplementary Information for detailed statistics.
We characterized regulation of Baz1a and Smarca5, as well as the closely related ISWI accessory subunit Baz1b (also known as Wstf), over time following the last defeat session. Both BAZ1A mRNA and protein levels were upregulated in NAc of susceptible mice 48 hours after CSDS compared to control non-stressed animals (Fig. 1b,c). BAZ1A mRNA and protein levels, in contrast, were not different between resilient and control animals (Fig. 1b,c). BAZ1A upregulation in NAc of susceptible animals persisted through 10 and 28 days after the last defeat session (Supplementary Fig. 1b–e), indicating that it is a highly stable adaptation. In contrast, BAZ1B and SMARCA5 mRNA and protein levels were not altered at any of these time points (Fig. 1b,c, Supplementary Fig. 1b–e). Additionally, co–immunoprecipitation experiments revealed increased levels of ACF complexes (SMARCA5 co-precipitated BAZ1A) in NAc of susceptible mice (Supplementary Fig. 1f, P < 0.05), with no change observed for BAZ1B–SMARCA5 complexes. Persistent induction of Baz1a required CSDS as no change in gene regulation was observed 48 hours after a single defeat session (Supplementary Fig. 1g). Baz1a induction in susceptible NAc after CSDS was also region specific as no changes were observed in medial prefrontal cortex (mPFC) of susceptible mice (Supplementary Fig. 1h, i). Similar induction of Baz1a was observed in NAc of another depression model, 48 hours after 6 days of chronic unpredictable stress (CUS, a combination of tail suspension, shock, and restraint stress), an effect seen in both stressed male and female mice compared to non-stressed controls (Fig. 1d).
Next, to validate our findings from mice, we examined NAc of postmortem brains of depressed humans and matched control subjects in two separate cohorts (Supplementary Table 1). In both cohorts, we observed increased BAZ1A mRNA levels in NAc of depressed patients compared to controls, with no change seen for SMARCA5 or BAZ1B (Fig. 1e). These data together suggest that elevated levels of BAZ1A and the resulting ACF complex in NAc are associated with susceptibility to stress and depression.
Upstream regulators of BAZ1A regulation
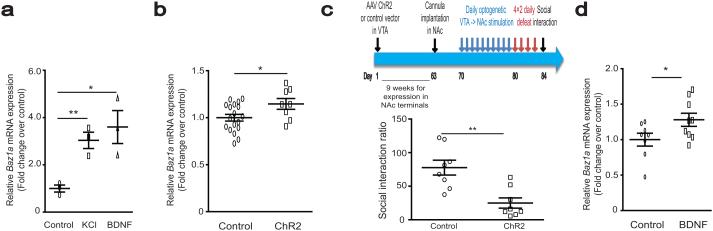
We next studied potential upstream mechanisms responsible for the persistent upregulation of Baz1a after CSDS. In cultured mouse striatal neurons, KCl–mediated depolarization (4 hours treatment) increased Baz1a mRNA levels, suggesting that its expression is activity–regulated (Fig. 2a). Previous studies have shown that increased burst firing of VTA dopamine neurons is a key mechanism driving susceptibility after CSDS27,29. Therefore we sought to investigate whether optogenetically–controlled stimulation of VTA–to–NAc projections, mimicking the increased burst firing observed in susceptible animals, similarly induces Baz1a and susceptibility to social defeat stress. AAV–CaMKIIa–ChR2 expression vectors were injected into either VTA, hippocampus, or mPFC of mice. An implanted optic fiber targeting NAc shell was used to stimulate ChR2–expressing terminals. We observed that 10 stimulation sessions (5 min per day over 10 consecutive days) of VTA neuronal projections to NAc in control animals mimicked the induction of Baz1a in NAc by CSDS (Fig. 2b). This effect was specific to the VTA–NAc pathway, as stimulation of NAc nerve terminals projecting from mPFC or hippocampus had no effect (Supplementary Fig. 2a,b). Upon subsequent behavioral analysis, these animals did not exhibit changes in basal social interaction (Supplementary Fig. 2c), but did display increased susceptibility to an accelerated social defeat stress paradigm (4 days × 2 daily defeats, equally robust in inducing susceptibility as CSDS and used here to minimize damage to optogenetic cannula) compared to non–stimulated controls (Fig. 2c). It has also previously been shown that BDNF release in NAc mediates the increased susceptibility caused by increased firing of VTA neurons30. We observed that BDNF application increased Baz1a expression both in primary striatal neurons in vitro and in NAc of control animals in vivo, suggesting that BDNF release in NAc may increase susceptibility in part via Baz1a induction (Fig. 2a,d). These results support the hypothesis that the persistent upregulation of the ACF complex in NAc, mediated at least partly via increased VTA projection firing and BDNF release in this region, is a key driver of susceptibility to stress and depression.
Figure 2.
Upstream regulators of Baz1a. (a) Baz1a mRNA levels after exposure to KCl or BDNF in primary mouse striatal neurons. n = 3 per group. (b) NAc Baz1a mRNA levels 48 hours after 10 days of 5–minute daily VTA channelrhodopsin (ChR2) optogenetic stimulations in control mice. Control n = 8 and ChR2 n=18. (c) Paradigm for the experiment is shown in top panel. Bottom panel shows that optogenetic stimulation increased susceptibility to social defeat stress. n = 8 per group. (d) NAc Baz1a mRNA levels after local BDNF infusion in mice. Control n = 8, BDNF n = 9. For a–d, Post–hoc Student’s t–test * P < 0.05, ** P < 0.01 compared to respective controls. See Supplementary Information for detailed statistics.
BAZ1A modulates depressive–like behaviors
To directly test the hypothesis that elevated BAZ1A and resulting increased ACF complex formation is a key driver of stress susceptibility, we manipulated levels of the ACF complex selectively in NAc of adult animals and then examined susceptibility to stress using a subthreshold social defeat paradigm known as “microdefeat” to reveal pro-susceptibility effects of experimental manipulation. Herpes simplex virus (HSV)–mediated gene transfer resulted in the rapid overexpression of BAZ1A or SMARCA5 in NAc (Fig. 3a, Supplementary Fig 3a,b, P < 0.05). After 3 days of virus expression, mice were subjected to microdefeat consisting of three 5 minute defeat sessions with 15 minutes rest in between within a single day, which does not induce social avoidance or reduced sucrose preference in normal mice27. Mice that received intra–NAc injections of HSV–GFP, or of HSV–BAZ1A or HSV–SMARCA5 alone, showed no behavioral effects of the stress (Fig. 3a). However, overexpression of both BAZ1A and SMARCA5 as the ACF complex increased susceptibility: these mice exhibited both social avoidance and decreased sucrose consumption (Fig. 3a). The pro–susceptibility effect ACF complex (BAZ1A + SMARCA5) overexpression was region–specific as their overexpression in mPFC did not affect social interaction or sucrose preference (Supplementary Fig. 3c).
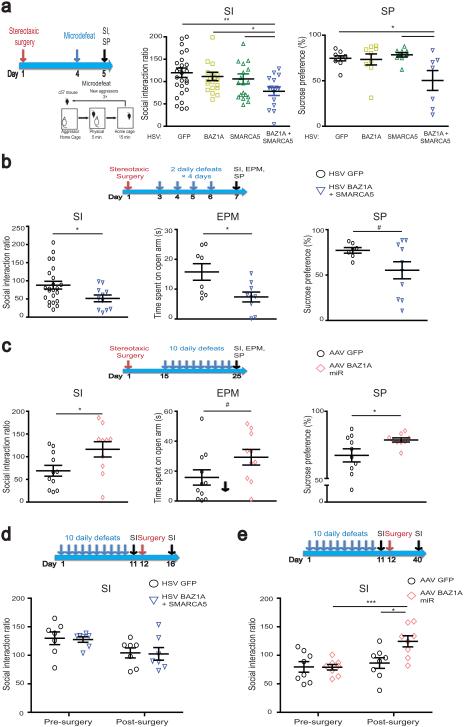
Figure 3.
ACF complex in NAc mediates stress susceptibility. Experimental schematics for each experiment are included above. (a) ACF complex (BAZ1A + SMARCA5) overexpression in NAc decreased social interaction (SI) and sucrose preference (SP) after subthreshold (micro–) defeat. GFP n = 9; BAZ1A n = 9; SMARCA5 n = 9; BAZ1A + SMARCA5 n = 7. (b) ACF complex overexpression in NAc decreased SI, time in open arm of elevated plus maze (EPM), and SP after four–day defeat paradigm. GFP n = 7; BAZ1A + SMARCA5 n = 10. (c) BAZ1A knockdown in NAc increased SI and SP after CSDS, with a trend for increased time in open arm of EPM. GFP n = 10; BAZ1A miR n = 9. (d) Overexpression of ACF complex in NAc of resilient mice after CSDS. GFP n = 7; BAZ1A + SMARCA5 n = 7. (e) ACF1 knockdown in susceptible mice after CSDS. GFP n = 8; BAZ1A miR n = 8. For a–e, post–hoc Student’s t–test # 0.05 < P <0.1; * P < 0.05; ** P < 0.01; *** P < 0.001. See Supplementary Information for detailed statistics.
To extend our finding beyond acute paradigms (i.e., microdefeat) to chronic paradigms of social defeat, we next used the accelerated social defeat protocol which induces behavioral deficits in control HSV–GFP overexpressing mice within the timeframe of maximal HSV–mediated transgene expression (Fig. 3b)27. Animals overexpressing ACF in NAc showed greater reductions in social interaction and sucrose preference. Given the high level of co-morbidities between MDD and anxiety disorders31, we examined anxiety behavior on the elevated plus maze (EPM) and found ACF overexpression increased anxiety–like behavior (decreased time spent on the open arms) compared to GFP controls after accelerated defeat (Fig. 3b). Thus, induction of the ACF complex in NAc during a course of social defeat stress increases susceptibility.
To examine whether ACF complex induction is also required for susceptibility, we virally overexpressed an miRNA that specifically reduces BAZ1A protein levels in NAc by ~50% (Supplementary Fig. 3d, P < 0.05). Mice that received AAV–BAZ1A miR in NAc exhibited increased social interaction after CSDS compared to control animals, indicative of a pro–resilience effect (Fig. 3c). AAV–BAZ1A miR animals similarly showed increased sucrose preference and a trend for decreased anxiety–like behavior (Fig. 3c). Additionally, there was a negative trending correlation between Baz1a expression levels and both social interaction (r = −0.23, P = 0.16) and sucrose preference (r = −0.39, P < 0.05) (Supplementary Fig. 3e,f), providing further support for ACF’s role in mediating stress susceptibility and depressive–like behaviors.
To test if the context of social stress is important for inducing susceptibility, we overexpressed ACF in NAc after CSDS. ACF overexpression in resilient animals after they had already undergone CSDS, or in stress–naïve mice, did not make them more socially avoidant (Fig. 3d, Supplementary Fig 4a,b). These findings suggest that ACF’s pro–susceptibility effect occurs very specifically in the context of stress exposure. Additionally, overexpression of ACF in NAc of control animals did not alter several other domains of behaviors including locomotor activity and freezing behavior in contextual fear conditioning (Supplementary Fig. 4c,d).
To determine whether persistent BAZ1A induction is necessary to maintain the susceptible phenotype, we knocked down BAZ1A in NAc of susceptible animals, by use of AAV–BAZ1A miR, after CSDS. Whereas control animals continued to display social avoidance 4 weeks after the viral infusion as we have shown previously26,27, those that received the AAV–BAZ1A miR showed reversal of this behavioral abnormality (Fig. 3e), indicating that suppression of ACF in NAc exerts antidepressant–like actions in previously stressed mice. Together, these data suggest that ACF in NAc is a necessary and causal component driving susceptibility to stress and depression.
Increased ACF binding and nucleosome remodeling after CSDS
Relatively little is known about the action of chromatin remodeling complexes in mammalian brain24,25,32,33. We used ChIP–seq to determine the location of BAZ1A and SMARCA5 binding genome–wide in NAc of susceptible and resilient mice after CSDS compared with controls (raw and genome–browser compatible files can be accessed online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=anenaoiqxzypjwz&acc=GSE54263). First, these samples all had good NSCs (normalized strand coefficients) and RSCs (relative strand coefficients), exceeding ENCODE standards, suggesting good antibody quality and good signal to noise ratios34 (Supplementary Table 2). Second, Pearson correlation analysis showed that these samples generated consistent peaks/signals across samples, suggesting that the enrichments over input are meaningful signals, not random noise (Supplementary Table 3). Third, close examination of ChIP–seq tracks identified many coincident BAZ1A and SMARCA5 peaks (Pearson correlation between BAZ1A and SMARCA5 ChIP–seq replicates: r = 0.41, adjusted P < 2.2×10−16) throughout both genic and intergenic regions, with significant enrichment over input (Supplementary Fig. 5a,b). While some regions of the genome exhibited more defined peaks of BAZ1A and SMARCA5 binding, other regions exhibited more diffuse signals across broad regions (Supplementary Fig. 5c–f). To address both types of binding, we used ChromHMM35 software to identify sites with high coincident binding of both BAZ1A and SMARCA5 (i.e., ACF complex). In agreement with our biochemical data of upregulated ACF in NAc of susceptible mice (Fig. 1, Supplementary Fig. 1) this analysis revealed more than twice the number of condition–specific ACF complex binding sites in NAc of susceptible mice (691) compared to control (329) and resilient (288) mice (Fig. 4a, for genomic loci see Supplementary Table 4). Heatmap analysis (Fig. 4b) and close examination of ChIP–seq tracks (Supplementary Fig. 5c–f) confirmed increased coincident binding of BAZ1A and SMARCA5 at identified loci in both genic (~30%) and intergenic (~70%) regions (Supplementary Fig. 6a).
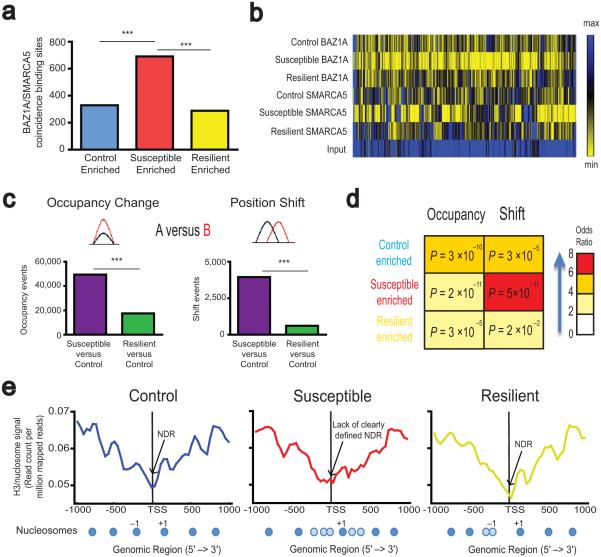
Figure 4.
Regulation of ACF complex and nucleosome positioning genome–wide in NAc by CSDS. (a) Genome–wide identification of coincident binding sites for BAZ1A and SMARCA5 enriched in NAc of control, susceptible, and resilient animals by ChromHMM. For detailed breakdown of genomic loci, see Supplementary Table 4 and Supplementary Fig. 6a. (b) Heatmap demonstrating increased BAZ1A and SMARCA5 enrichment in NAc of susceptible animals compared to control and resilient animals, as identified by ChromHMM. (c) Genome–wide identification of occupancy and shift events after CSDS. For detailed breakdown of genomic loci, see Supplementary Fig. 6b–c. (d) Overlap between ACF complex enriched sites and sites of nucleosome remodeling events in NAc after CSDS calculated with Fischer’s test. Red scale reflects odds ratios for each comparison, with numbers in the boxes reflecting adjusted probabilities for each comparison. (e) Average nucleosome profile around TSSs for genes enriched for ACF complex binding sites in NAc of susceptible animals (for gene list, see Supplementary Table 4). Representative nucleosome maps are shown below each profile. Dark blue circles represent strongly positioned nucleosomes; light blue circles represent weakly positioned nucleosomes. For overlay and quantification, see Supplementary Fig. 6d. Chi-squared tests *** P < 2.2 × 10−16.
Since ISWI complexes regulate nucleosome repositioning23,24,32, we also generated a nucleosomal map from total H3 ChIP–seq data in NAc under control and CSDS conditions. We identified over 70,000 occupancy changes (altered density of nucleosomes) and 4,000 shift changes (altered position of nucleosomes), with the majority occurring between susceptible and control animals (Fig. 4c, Supplementary Table 5), consistent with dynamic nucleosome remodeling in NAc of susceptible animals after CSDS. Approximately 2/3 of both occupancy and shift changes occurred in intergenic regions, while the rest occurred in genic regions (Supplementary Fig. 6b,c). The majority of occupancy changes represented decreased occupancy in NAc of susceptible or resilient mice compared to control, while there was an equal distribution of upstream and downstream shift change events (Supplementary Fig. 6b.c).
Sites of ACF binding across the different conditions significantly overlapped with differential nucleosome occupancy and shift changes; this overlap was particularly evident for sites with enriched ACF complex binding and altered nucleosome positioning (shift) in susceptible mice (Fig. 4d). These findings led us to speculate that genes that show increased ACF binding in NAc of susceptible animals might also show altered nucleosome architecture. We therefore plotted the promoter nucleosome distribution for this subset of genes with susceptible enriched ACF binding (Supplementary Table 4) under control, susceptible, and resilient conditions (Fig. 4e, for overlay see Supplementary Fig. 6d upper left panel), as promoter nucleosome architecture has the most well characterized role in transcription. In control animals, these genes showed a strongly positioned +1 and −1 nucleosome in addition to a well–defined nucleosome depleted region (NDR) (Fig. 4e). Susceptible animals, where ACF binding is enriched, lacked a well–defined NDR and a strongly positioned −1 nucleosome; the occupancy of the +1 nucleosome was also lower compared to control animals. These findings suggest a possible redistribution of the −1 and to a lesser extent the +1 nucleosome across TSSs in susceptibility (Fig. 4e, Supplementary Fig. 6d upper left panel). In contrast, the promoter nucleosome positioning of resilient animals after CSDS largely resembled that of control animals with a strongly defined +1 nucleosome and a well–defined NDR. The occupancy of the +1 nucleosome in resilient animals, however, was lower compared to control animals, with a −1 nucleosome that is shifted farther away from the TSS. To quantify this difference in NDR between susceptible and control/resilient animals, we calculated the “height” of the NDR (top of the +1 nucleosome − bottom at the TSS), and found that it was approximately half the magnitude in susceptible versus control and resilient animals for the subset of genes that showed increased ACF enrichment in susceptibility (Supplementary Fig. 6d lower left panel).
To further test whether the nucleosome architecture change was associated with altered ACF binding, we examined a group of approximately 500 randomly generated genes (Supplementary Table 6) that do not exhibit ACF complex enrichment in NAc of susceptible animals. Unlike the subset of genes that showed increased ACF complex enrichment in susceptible animals, the promoter nucleosome positioning for these randomly generated genes did not differ between control, susceptible, and resilient animals, with all of them exhibiting a strongly positioned +1 and −1 nucleosome as well as a well–defined NDR (Supplementary Fig. 6d upper right panel). Additionally, the NDR height did not differ between the different groups (Supplementary Fig. 6d lower right panel). However, similar to the subset of genes that showed increased ACF complex enrichment in susceptible animals, the nucleosome occupancy of these randomly generated genes appeared decreased for susceptible and resilient animals compared to control, in particular at the +1 nucleosome (Supplementary Fig. 6d). These results together suggest that certain aspects of altered nucleosome occupancy and positioning (in particular the lack of a well–defined NDR and −1 nucleosome) are associated with increased ACF enrichment in NAc of susceptible animals, whereas other aspects (in particular decreased occupancy at the +1 nucleosome) in susceptible and resilient animals seem to be more global phenomena and not specific to genes that show increased ACF enrichment.
ACF target genes mediate susceptibility to stress
Previous studies reported that ACF exerts a repressive role on gene transcription36. Here, in NAc of mouse brain, we observed that BAZ1A binding at promoters was inversely correlated with gene expression (Supplementary Fig. 6e). This suggests that increased ACF binding at gene targets, with associated altered nucleosome architecture at the respective promoters, likely mediates repression of target gene transcription in NAc of susceptible animals after CSDS.
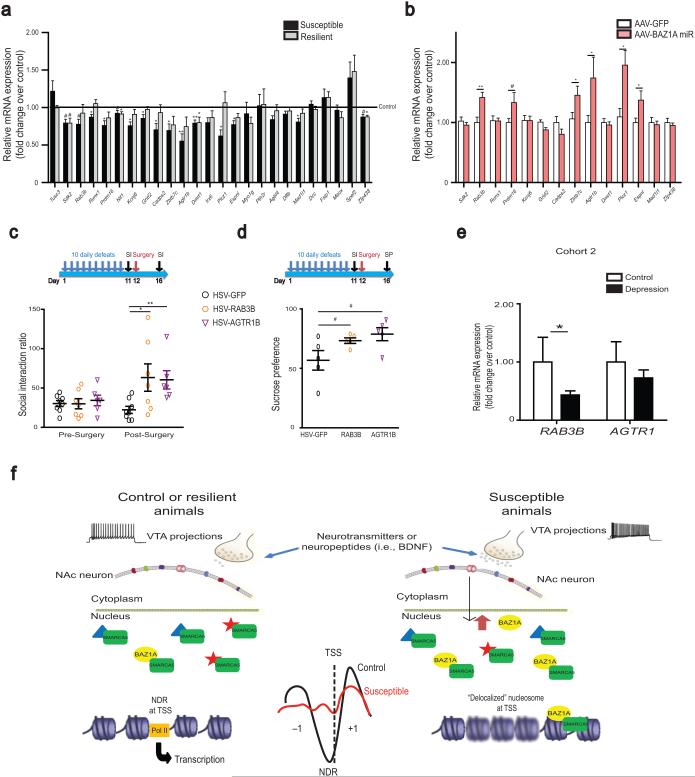
To test this hypothesis, we screened 25 randomly selected genes from the ~100 testable genes that exhibited increased ACF binding in susceptibility (Supplementary Table 4, 150 genic loci map to 145 genes, 22 are unannotated, ~30 have low to no expression in NAc) and observed that 15/25 showed significant or trending decreased mRNA expression in NAc of susceptible mice (Fig. 5a). To directly demonstrate that ACF mediates repression of these genes, we examined NAc of animals that underwent CSDS with and without BAZ1A knockdown by use of AAV–BAZ1A miR. Compared to AAV–GFP animals, BAZ1A knockdown in NAc reversed CSDS repression of 6 of 14 genes tested (Fig. 5b). Furthermore, the AAV–BAZ1A miR injected animals that showed reversal of gene repression also showed reversal of their social avoidance behavior (Fig. 3c), suggesting that the repression of genes regulated by ACF contributes to stress susceptibility.
Figure 5.
Regulation of genes enriched in ACF complex binding in NAc of susceptible animals. (a) Repression of transcription in NAc of genes with increased ACF complex binding and altered nucleosome architecture at promoter regions in susceptible animals. n = 5–8 per group. (b) Reversal of gene repression in NAc with local BAZ1A knockdown. GFP n = 11–12; BAZ1A miR n = 9–10. (c) (d) RAB3B or AGTR1B overexpression in NAc partially rescues social avoidance and sucrose preference, respectively, after CSDS. For c, GFP n = 8; RAB3B n = 7, AGTR1B n = 6. For d, n = 5 for all groups. (e) Regulation of RAB3B and AGTR1 NAc mRNA levels in postmortem humans in cohort 2 (demographic information in Supplementary Tables 1). Control n = 9; depressed n = 20. (f) Schematic of the hypothesized role of the ACF complex in NAc in mediating stress susceptibility. CSDS, via increased burst firing of VTA neurons and BDNF release, induces BAZ1A expression in NAc. The resulting upregulation of ACF complex activity, possibly through changes in TSS nucleosome positioning, represses a set of genes in NAc, the reduced expression of which contributes to susceptibility. Blurry nucleosomes in the right figure represent weakly positioned or delocalized nucleosomes at TSSs. For a–e, post–hoc Student’s t–test: # 0.05 < P < 0.1; * P < 0.05; ** P < 0.01 versus respective control. See Supplementary Information for detailed statistics.
We thus investigated directly whether decreased expression of such ACF targets contributes to the susceptible phenotype. Overexpression of two validated targets, RAB3B or AGTR1B, in NAc partially reversed the social avoidance and sucrose preference deficits in mice susceptible to CSDS (Fig. 5c,d, Supplementary Fig. 7a–c, P < 0.05), suggesting that ACF repression of these genes contributes to depressive–like abnormalities. Additionally, RAB3B expression levels were positively correlated (r = 0.6461, P < 0.01) with the extent of susceptibility reversal (Supplementary Fig. 7d). We further validated our results in human postmortem NAc, and observed a significant decrease in RAB3B mRNA levels and a trend for decreased AGTR1 (the mouse genome encodes Agtr1a and Agtr1b at distinct chromosomal loci, while the human genome encodes only AGTR1 at a single locus) mRNA levels in depressed patients compared to controls (Fig. 5e). To further demonstrate the specificity of ACF gene repression in susceptible animals, we examined 10 genes from the randomly generated gene list that had no ACF enrichment (Supplementary Table 6) nor promoter nucleosome architecture change in susceptible animals, and found 0/10 genes showed gene expression regulation in NAc by CSDS or by BAZ1A knockdown (Supplementary Fig. 7e,f). Moreover, overexpression of one of those genes, PRMT5, in NAc of susceptible animals did not rescue social avoidance behavior (Supplementary Fig. 7g).
Discussion
Here we show that the ISWI subunit BAZ1A, and the resulting ACF complex, is persistently and selectively upregulated, possibly as a result of increased VTA neuronal activity and BDNF release, in NAc of mice that are susceptible to chronic social stress, as well as in NAc of depressed humans. We further establish that ACF induction is necessary and causal for susceptibility to stress–induced depressive–like behaviors. Using ChIP–seq, we demonstrate that increased ACF binding in NAc of susceptible animals after chronic stress is correlated with altered nucleosome positioning, in particular, around the TSSs of affected genes. These alterations in ACF binding and nucleosome positioning are associated with repressed expression of a subset of genes in NAc of animals that are susceptible to chronic stress. Together, these findings establish that the ACF complex, presumably through its effects on chromatin remodeling, is a novel and key mediator of the repression of genes that contribute to stress susceptibility (Fig. 5f). Our study is in agreement with previous work demonstrating that yeast homologs of ISWI and other families of chromatin remodeling complexes repress transcription by altering nucleosome positioning, in particular around TSSs23,24,37. Several studies in other systems have further confirmed ACF’s transcriptional repressive role36,38-41.
BAZ1A and ACF upregulation alone appears insufficient for inducing depressive-like behavior without co-occurring stress, but is likely one of several key mechanisms important in mediating depression pathophysiology (for more detailed discussion, see Supplementary information). Importantly, we show that elevated BAZ1A levels in NAc are required for the induction and maintenance of depressive–like phenotypes, indicating that reversal of this elevation may have therapeutic potential (Fig. 3e). Specific inhibitors of reader domains such as bromodomains have become increasingly popular for the treatment of several cancers; specific inhibitors of BAZ1A’s bromodomain may exert antidepressant efficacy. However, as the ACF complex is also involved in other important cellular process such as DNA replication and repair, ACF may not be an ideal therapeutic target. Thus, identification of ACF target genes—by studying the location of the ACF complex and its regulation of TSS nucleosome repositioning—could reveal novel therapeutic targets that may have less off–site effects. Several bona fide targets (Rab3b, Prdm16, Zbtb7c, Agtr1b, Plcz1, and Espnl) were identified here through the following criteria: increased binding of ACF under susceptible conditions; specific suppression of gene expression under susceptible conditions, and reversal of gene suppression upon BAZ1A knockdown. The rescue of social avoidance and sucrose preference phenotypes by RAB3B or AGTR1B overexpression in NAc of susceptible mice as well as the significant decrease in RAB3B mRNA observed in NAc of depressed humans further strengthen our approach of identifying novel therapeutic targets through ChIP–seq analysis of the ACF complex.
Our analysis of chromatin remodeling factors revealed several other regulators that may contribute to stress susceptibility or resilience (Supplementary Fig. 1a), while our nucleosome maps before and after CSDS revealed many changes not accounted for by ACF. Therefore, this study is only the beginning of examining ATP–dependent nucleosome remodelers in the pathophysiology of depression. Together, these studies elucidate a novel mechanism of ATP–dependent chromatin remodeling in the gene regulation implicated in the persisting pathophysiology of depression. The studies thereby provide a new approach for the identification of novel therapeutic targets for the treatment of stress–related disorders.
Online Methods
Animals and treatments
For all experiments, 7 to 8–week–old C57BL/6J male or female mice (The Jackson Laboratory, Bar Harbor, ME, USA) were housed in a colony room set at constant temperature (23 °C) on a 12 hr light/dark cycle (lights on from 0700 to 1900 hr) with ad libitum access to food and water. All protocols involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai.
Sample/subject selection and general experimental practices
Sample sizes for all experiments were pre-determined based on extensive laboratory experience with these end points. All samples/subjects that have successfully made it through the endpoint of the experiments were included in the analysis. Values were excluded (1 maximum per group) only if they were considered outliers by the Grubbs' test. This criterion was pre-established. For all molecular and behavioral experiments, animals were randomly assigned to groups. Furthermore, for all molecular experiments, tissue collected from treated animals was randomly pooled to provide ample tissue for biochemical procedures and to minimize variance across cohorts. Additionally, the investigator was blind to treatment group until all data was collected.
NAc RNA isolation and qRT–PCR
Bilateral punches of NAc or mPFC were obtained at varying times after the last defeat session and frozen on dry ice. Samples were then homogenized in TRIzol and processed according to manufacturer’s instructions (Life technologies, NY, USA). RNA was purified with RNAeasy Micro columns (Qiagen, CA, USA) and reverse transcribed using an iScript Kit (BioRad, CA, USA). cDNA was quantified by qPCR using SYBR green. Each reaction was performed in duplicate and analyzed following the standard ΔΔCt method using glyceraldehyde–3–phosphate dehydrogenase (GAPDH) as a normalization control. See Supplementary Table 7 for complete list of primers.
Western blotting
Frozen NAc tissue was homogenized in 30 μl of RIPA buffer containing 10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X–100, 1% Sodium Deoxycholate, and protease inhibitors (Roche, Basel, Switzerland) using an ultrasonic processor (Cole Parmer, Vernon Hills, IL, USA). Protein concentrations were determined using a DC protein assay (Bio–Rad, Hercules, CA, USA), and 50 μg of protein were loaded onto 4–15% gradient Tris–HCl polyacrylamide gels for electrophoresis fractionation (Bio–Rad). Proteins were transferred to nitrocellulose membranes, blocked with Odyssey® blocking buffer (Li–Cor, NE, USA), and incubated overnight at 4°C with primary antibodies (BAZ1A: Bethyl A301–318A, 1/500; BAZ1B: Abcam ab51256, 1/500; SMARCA5: Abcam ab3749, 1/1000) in Odyssey® blocking buffer. After thorough washing with 1x Tris–Buffered Saline plus 0.1% Tween–20, membranes were incubated with IRDye® secondary antibodies (1/5000 to 1/10000; Li–Cor, Lincoln, NE, USA) dissolved in Odyssey® blocking buffer for 1 hr at room temperature. For analysis, the blots were imaged with the Odyssey® Infrared Imaging system (Li–Cor, Lincoln, NE, USA) and quantified by densitometry using ImageJ (NIH, Bethesda, Maryland, USA). The amount of protein blotted onto each lane was normalized to levels of GAPDH (Cell Signaling 2118, 1/30000).
Immunoprecipitation
Anti–SMARCA5 antibody (Bethyl A301–018A, TX, USA) was incubated overnight with anti–rabbit magnetic beads (Invitrogen) at 4°C. Frozen NAc tissue was homogenized in 100 μl of RIPA buffer as above with plastic pestles, and combined with 220 μl of immunoprecipitation buffer containing 16.7 mM Tris, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X–100, and protease inhibitors. The antibody–bead mixture was then added to the tissue lysate, and incubated overnight at 4°C. Following 3 washes with buffer containing 20 mM Tris, 150 mM NaCl, 2 mM EDTA, 1% Triton X–100, and 0.1% SDS, the pulldown was dissociated from the beads with elution buffer (50 mM Tris, 10 mM EDTA, 1% SDS) at 65°C and analyzed by Western blotting as described above.
Cell Culture
E16.5 mouse striata were dissected and dissociated in PBS and grown for 8 days in culture in neurobasal media supplemented with B27 (Gibco), glutamax (Gibco), and Pen Strep (Gibco). Neurons were stimulated for 4 hr with either BDNF (50 ng/mL, PeproTech) or KCl. Neurons were lysed and RNA was collected as described above.
Optogenetics
For circuit specific stimulations, mice were injected with AAV–CAMKIIa–ChR2 or control vectors with the following coordinates: VTA: −3.3 AP, +1.05 ML, −4.6 DV; hippocampus (HP): −3.6 AP, +3.05 ML, −4.85 DV; and medial prefrontal cortex (mPFC): +1.9 AP, +0.5 ML, −3.0 DV. After 9 weeks of recovery to allow for expression in terminals, a second stereotaxic surgery was performed to implant an optic fiber targeting NAc shell (+1.4 AP, +1.5 ML, −4 DV). After one week of recovery, mice underwent 10 days of daily 5–min stimulation sessions. Stimulation parameters were 20 Hz, 40ms, 5 spikes over 10 sec (VTA), 20 Hz, 30 pulses/burst, with 10 sec between bursts (hippocampus); or 30 Hz, 90 pulses/burst, 10 sec between burst (mPFC).
BDNF infusion
Recombinant human BDNF was infused into NAc (AP = 1.5, ML = ±1.5, and DV = −4.4; 10° angle). An infusion volume of 0.5 μL was delivered using 5 μL Hamilton syringe over the course of 5 min (at a rate of 0.1 μL/min). The infusion needle remained in place for at least 5 min after the infusion before removal to prevent backflow of the injection.
Human postmortem brain tissue
For cohort 1, NAc tissue, obtained from the Dallas Brain Collection at UT Southwestern, was analyzed for control and depressed subjects matched for age, postmortem interval, RNA integrity, and pH. Standard dissection technique was used and the tissue was snapped frozen and stored at −80°C. The UT Southwestern IRB reviewed and approved the collection of this tissue for research use and informed consent was obtained from all subjects/next of kin. .
For cohort 2, brain tissue was obtained from the Quebec Suicide Brain Bank (QSBB; Douglas Mental Health Institute, Verdun, Québec). All individuals were group–matched for age, pH, and post–mortem intervals (PMI). Inclusion criteria for both suicide completers and controls were the following: the subject had to be Caucasian and of French Canadian origin, and die suddenly without prolonged agonal state. NAc was stored at −80°C. This study was approved by the Douglas IRB and signed informed consent was obtained from next of kin. For detailed demographic information for both cohorts, please refer to Supplementary Table 1.
Generation of viral constructs
Human BAZ1A (ACF1)–pEGFP and SMARCA5 (SNF2H)–Flag cDNAs were cloned into HSV vectors. Briefly, BAZ1A–GFP was first cloned into pENTR1A by cutting it out with Asp718 (5’) and NotI (3’) from the original vector and putting it into pENTR1A cut with the same enzymes. Gateway cloning technology (Invitrogen) was then used to recombine it into HSV p1006 GW. SMARCA5–Flag was cut out of the original vector with NheI (5’) and SalI (3’). Its 5’ end was blunted with polymerase and cloned into HSV p1005+ with EcoRV (5’) and XhoI (3’). Mouse Rab3b (MC203236) and Agtr1b (MC208118) cDNA were obtained from Origene (Rockville, MD). Rab3b was cut out of the original vector with EcoRI (5’) and XhoI (3’) and cloned into HSV p1005+. Agtr1b was cut of the original vector with KpnI (5’) and XhoI (3’) and cloned into HSV p1005+. PRMT5 was excised from a human pcDNA3 plasmid using BamHI (5’) and EcoRI (3’) and cloned into HSV p1005+.
Viral–mediated gene transfer
HSV expression plasmids were packaged into high–titer viral particles as described previously42. Viral titers for these experiments were between 3–4 × 108 particles/ml. Mice were positioned in small animal stereotaxic instruments, under ketamine (100 mg/kg)/xylazine (10 mg/kg) anesthesia, and their cranial surfaces were exposed. Thirty–three gauge syringe needles were bilaterally lowered into NAc (anterior/posterior + 1.6; medial/lateral + 1.5; dorsal/ventral − 4.4 mm from Bregma, 10° angle) or PFC (anterior/posterior + 1.8; medial/lateral + 0.75; dorsal/ventral − 2.7 mm from Bregma, 15° angle) to infuse 0.5 μl of virus. Infusions occurred at a rate of 0.1 μl/min. Animals receiving HSV injections were allowed to recover for at least 24 hr following surgery.
Chronic social defeat stress (CSDS) and behavior testing
CSDS was performed as described previously26,27. Briefly, an experimental C57BL/6J mouse was placed into the home cage of a CD1 mouse for 10 min during which time it was physically defeated by the CD1 mouse. After the physical interaction, the CD1 and experimental mouse were maintained in sensory contact for 24 using a perforated plexiglass partition dividing the resident home cage in two. The experimental mice were exposed to a new CD1 mouse for 10 consecutive days. Subthreshold defeat involved exposing experimental mice to a novel CD1 male aggressor for 5 min, 3 times with 15 min intervals between each exposure. A social interaction test was performed 24 hr after the last defeat. Mice were placed in an open field which included an interaction zone and two opposing corner zones. A social target (novel CD1 mouse) was placed in a metal meshplastic box in the interaction zone, which allows sensory but not physical interaction. Ethovision XT (Noldus) tracking software was used to measure the time that the test mouse spent in the interaction zone with and without the target CD1 present (2.5 min). Subsequently, test animals were studied in a standard elevated plus maze for 5 min monitored by Ethovision XT. They were also monitored for their sucrose consumption in a sucrose–preference over 24 hr. Briefly, a solution of 1% sucrose or drinking water was filled in 50 ml tubes with stoppers fitted with ball–point sippers. The weights of solutions each mouse consumed were recorded and sucrose preference was calculated as a percentage [100 × volume of sucrose consumed /total volume consumed].
Locomotor activity
Locomotor behavior was tested in an open field box 3 days following viral infusion. Animals were placed in the box for 2.5 minutes and Ethovision XT was used to track and analyze distance travelled.
Contextual fear conditioning
Animals were put into a sound–proof, contextually distinct chamber 3 days after viral infusion for 5 minutes. Three shocks (0.7mA /1 sec for 2 seconds) were delivered 2, 3 and 4.5 minutes after the animal was placed into the chamber. 24 hours after training, the animal was put back into the same chamber and recorded for 5 minutes. Freezing behavior was scored.
ChIP, library preparation, and sequencing
3 fully independent biological replicates were obtained per mark per condition. For each ChIP–seq replicate, bilateral 14–gauge NAc punches were pooled from 5–10 mice. Tissue was lightly fixed to cross–link DNA (12 minutes with 1% formaldehyde, following by 5 minutes with glycine, and 8x cold PBS washes) with associated proteins, and the material was further sheared (for BAZ1A and SMARCA5, Bioruptor to obtain mostly 100–300 bp fragments) or MNase digested (for histone H3/nucleosome, nuclei were digested with 10 U/mL MNase at 37 °C for 10 min, reaction stopped with adding EDTA to 20 mM, >80% mononucleosome obtained) and immunoprecipitated using sheep anti–rabbit magnetic beads (Invitrogen) conjugated to an antibody that specifically recognizes BAZ1A (Bethyl), SMARCA5, or H3 (Abcam). Immunoprecipitated DNA and total (input) genomic DNA were prepared for ChIP–seq using an Illumina kit according to the manufacturer’s instructions. Each experimental condition was analyzed with independent biological triplicates. Briefly, each sample underwent end repair followed by addition of an A base to the 3′ end. Proprietary adapters were then ligated to the ends, followed by size selection on a 2% agarose gel. The range of excision was 200–300 bp. After DNA clean up, samples were amplified with 13 (H3) or 19 (BAZ1A and SMARCA5) cycles of PCR. Amplification and size selection were confirmed with a BioAnalyzer. The resulting libraries were sequenced on an Illumina HiSeq 2500 with 100 bp read length.
ChIP–seq data analysis
ChIP–seq data were aligned to the mouse genome (mm9) by CASAVA 1.8 (http://www.illumina.com/software/genome_analyzer_software.ilmn), and only unique reads were retained for analysis. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was applied for quality control, and then SAMTools (http://samtools.sourceforge.net) was used to remove potential PCR duplicates43. PhantomPeak (https://code.google.com/p/phantompeakqualtools/) was applied to estimate the quality and enrichment of the ChIP–seq dataset34. Additional ENCODE quality metrics, such as the normalized strand coefficient (NSC) and the relative strand correlation (RSC), were calculated. For all samples in our research, NSC≥1.05, RSC≥0.8. Basic filtering and quality control confirmed that these samples were of strong quality and exceeded ENCODE standards34; see Supplementary Table 3 and 4 (raw and genome–browser compatible files can be accessed online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=anenaoiqxzypjwz&acc=GSE54263). For the H3 MNase ChIP–seq libraries, 150+ million raw reads were obtained for each replicate. With ~70% reads uniquely mapped and less than 20% duplicative reads, there were ~100 million uniquely mapped, non–redundant reads per replicate. This is about 4–5x coverage of the mouse genome, and allows for sufficient analysis of genome–wide nucleosome mapping.
For visualization of the ChIP–seq data genome–wide, ngs.plot (https://code.google.com/p/ngsplot/) was applied to visualize the dataset44. All 3 replicates of the conditions were pooled, and normalized to 1 million reads. The density of BAZ1A binding 1 kb up– and downstream of TSSs of coding genes in Ensembl annotations were plotted. The Corrgram package in the R software (http://www.r–project.org/) was used to calculate and visualize the correlation between basal BAZ1A and SMARCA5. TDF files (all duplicative/redundant reads >2 removed) were applied in IGV for genome browser views of ChIP–seq tracks.
Coincident binding sites between BAZ1A and SMARCA5 were identified using ChromHMM (http://compbio.mit.edu/ChromHMM/)35. First, all BAZ1A and SMARCA5 ChIP–seq data were binarized at 200 bp intervals. The intervals were designated as enriched or “1” if the fold enrichment threshold over input was 2.0 or greater, and the Poisson tail p–value was less than or equal to 1×10−6. Otherwise, the intervals were designated as not enriched or “0”. Then the binarized marks were fed into ChromHMM to estimate the states of the 200 bp intervals, iterating the training 200 times. The initiating number of states was 4, and based on state pruning strategy, we defined two states: one state represented low to no binding of the two factors and the other state represented high binding of both factors. To analyze the effect of CSDS, sites of high binding of both factors were extracted for control, susceptible, and resilient conditions. A site is considered enriched/specific in one condition only if no other condition contained that enrichment site within 2 kb.
For analysis of nucleosome position and occupancy, DANPOS (https://code.google.com/p/danpos/) was applied in the dynamic analysis of nucleosomes45. Analysis focused on the position shift and occupancy change events. The FDR cutoff was 0.01 for both events. For shift events, the cutoff of shifting distance was between 50 and 90 bp. For occupancy events, the FDR of difference between treatment and control was >0.01. The overlap of condition–enrichment of the ACF complex and nucleosome events was calculated using Fisher’s tests, with multiple tests corrected by Benjamini–Hochberg analysis.
Statistical Analysis
Appropriate assumptions of data (e.g., normal distribution or similar variation between experimental groups) were examined before conducting statistical tests. Student’s t–tests were used whenever two groups were compared, while one–way and two–way ANOVAs were performed wherever necessary to determine significance for all other data. Significant main effects (p < 0.05) were further analyzed using post hoc tests. See individual figure legends and Supplementary Information for detailed statistics.
Supplementary Material
Acknowledgements
We thank the patients and their families for contribution to the brain banks. This work was supported by grants from the National Institute of Mental Health (P50 MH096890 and R01 MH51399) and the Hope for Depression Research Foundation.
Footnotes
Accession Codes
ChIP-seq data for BAZ1A, SMARCA5 and nucleosome positioning (total H3 MNase-seq) have been deposited with the accession code GSE54263.
Author Contributions
H.S. and E.J.N conceived the study and wrote the manuscript. H.S. performed most experiments with the help of D.D.W., K.N.S., C.D., J.R., J.W.K., E.K, R.C.B., F.H.A., M.E.C., B.L. E.M., E.A.H., H.C., S.A.G. S.J.R., P.J.K., I.M., D.M.D. N.Y.S., S.A., P.V.W and L.S conducted bioinformatics analysis. K.G., G.T., and C.T., provided post-mortem human brain samples. R.N provided the HSV overexpression vectors. C.D.A. and P.V.W provided plasmid and antibody reagents.
The authors have no competing financial interests.
References
- 1.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 3.Kessler RC. The effects of stressful life events on depression. Annu Rev Psychol. 1997;48:191–214. doi: 10.1146/annurev.psych.48.1.191. [DOI] [PubMed] [Google Scholar]
- 4.Kendler KS. Anna-Monika-Prize paper. Major depression and the environment: a psychiatric genetic perspective. Pharmacopsychiatry. 1998;31:5–9. doi: 10.1055/s-2007-979287. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology. 2013;38:124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pittenger C, Duman RS. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology. 2008;33:88–109. doi: 10.1038/sj.npp.1301574. [DOI] [PubMed] [Google Scholar]
- 7.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14:609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charney DS, Manji HK. Life stress, genes, and depression: multiple pathways lead to increased risk and new opportunities for intervention. Sci STKE. 2004;2004:re5. doi: 10.1126/stke.2252004re5. [DOI] [PubMed] [Google Scholar]
- 9.Caldji C, Hellstrom IC, Zhang TY, Diorio J, Meaney MJ. Environmental regulation of the neural epigenome. FEBS Lett. 2011;585:2049–2058. doi: 10.1016/j.febslet.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Gudsnuk K, Champagne FA. Epigenetic influence of stress and the social environment. ILAR J. 2012;53:279–288. doi: 10.1093/ilar.53.3-4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 13.Covington HE, 3rd, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobara T, et al. Altered gene expression of histone deacetylases in mood disorder patients. J Psychiatr Res. 2010;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Covington HE, 3rd, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol Med. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe N, et al. Altered sirtuin deacetylase gene expression in patients with a mood disorder. J Psychiatr Res. 2011;45:1106–1112. doi: 10.1016/j.jpsychires.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Hollis F, Duclot F, Gunjan A, Kabbaj M. Individual differences in the effect of social defeat on anhedonia and histone acetylation in the rat hippocampus. Horm Behav. 2011;59:331–337. doi: 10.1016/j.yhbeh.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 23.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 24.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel-Ciernia A, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berton O, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 27.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Eberharter A, et al. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 2001;20:3781–3788. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhury D, et al. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 2013;493:532–536. doi: 10.1038/nature11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh JJ, et al. Stress and CRF gate neural activation of BDNF in the mesolimbic reward pathway. Nat Neurosci. 2014;17:27–29. doi: 10.1038/nn.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorwood P. Generalized anxiety disorder and major depressive disorder comorbidity: an example of genetic pleiotropy? European psychiatry : the journal of the Association of European Psychiatrists. 2004;19:27–33. doi: 10.1016/j.eurpsy.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Sala A, et al. Genome-wide characterization of chromatin binding and nucleosome spacing activity of the nucleosome remodelling ATPase ISWI. EMBO J. 2011;30:1766–1777. doi: 10.1038/emboj.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 37.Yadon AN, et al. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol. 2010;30:5110–5122. doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deuring R, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 39.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YI, et al. The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets. Dev Biol. 2008;323:41–52. doi: 10.1016/j.ydbio.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ewing AK, Attner M, Chakravarti D. Novel regulatory role for human Acf1 in transcriptional repression of vitamin D3 receptor-regulated genes. Mol Endocrinol. 2007;21:1791–1806. doi: 10.1210/me.2007-0095. [DOI] [PubMed] [Google Scholar]
- 42.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, et al. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013;23:341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.