Abstract
Aging is driven by changes of the epigenetic state that are only partially understood. We performed a comprehensive epigenomic analysis of the pancreatic β cell, key player in glucose homeostasis, in adolescent and very old mice. We observe a global methylation drift resulting in an overall more leveled methylome in old β cells. Importantly, we discover targeted changes in the methylation status of β cell proliferation and function genes that go against the global methylation drift, are specific to β cells, and correlate with repression of the proliferation program and activation of metabolic regulators. These targeted alterations are associated with specific chromatin marks and transcription factor occupancy in young β cells. Strikingly, we find β cell function improved in aged mice, as predicted by the changes in methylome and transcriptome. Thus, aging of terminally differentiated cells in mammals is not always coupled to functional decline.
Introduction
DNA methylation is a major component of the mammalian epigenome, and the only one for which inheritance through multiple mitotic divisions has been demonstrated. As such, DNA methylation can provide an essential function for the stability of expression programs and the maintenance of cellular identity [for review, see (Issa, 2014)]. In mammals, methylation occurs exclusively on cytosines, most commonly in the context of CG dinucleotides, or ‘CpGs’. While most of the genome is methylated, CpG islands (CGIs) commonly near promoters remain generally unmethylated. The symmetry of the CpG palindrome explains how methylation is maintained, as the enzyme Dnmt1 (DNA methyltransferase 1) recognizes hemimethylated CpGs that are the product of DNA synthesis during S-phase as substrate for remethylation, restoring the status that existed before the onset of cell division.
The concept that DNA methylation is a constant epigenetic mark that, once established during embryonic development, is stably maintained throughout life has been challenged recently. While de novo methylation is associated with silencing of developmental regulatory genes and is perpetuated through multiple rounds of cell division [reviewed in (Bird, 2002)], recent evidence indicates that DNA methylation can be dynamic in specific contexts, including cellular differentiation (Sheaffer et al., 2014; Stadler et al., 2011), and during aging (Day et al., 2013).
The insulin-producing cells of the pancreas, or β cells, are a prime example of aging-dependent changes in essential properties such as replicative capacity. Pancreatic β cell mass grows well into adolescence to provide increased insulin secretory capacity to match the greater metabolic requirements of maturity (Ackermann and Gannon, 2007). β cell mass in postnatal humans and rodents expands mainly by replication of fully differentiated β cells (Dor et al., 2004), and in rodents can also expand in response to increased insulin requirements (Weir et al., 2001), or during pregnancy (Parsons et al., 1992; Rieck and Kaestner, 2010). The ability of mature β cells to divide has prompted speculation that restoration of β cell function through increased β cell number might someday be employed as a novel therapeutic intervention (Dor et al., 2004). However, β cell regeneration has proven to be an elusive goal, as β cell turnover declines dramatically with advanced age in mice (Rankin and Kushner, 2009). Likewise, basal replication rates in human pancreata and cultured human islets decline dramatically with donor age (Tyrberg et al., 1996). As of now, the molecular basis underlying the reduction of cell cycle entry in aged β cells has been explored only partially. Thus, epigenetic de-repression of the cyclin kinase inhibitor and senescence master gene p16Ink4a pathway by the enhancer of zeste homolog 2 (EZH2), a polycomb group protein, as well as activation of p38MAPK, contributes to this process in mice (Chen et al., 2009; Dhawan et al., 2009).
We hypothesized that age-related alterations in DNA methylation might relate to the decline in regenerative capacity of β cells. Therefore, we performed a comprehensive and integrative analysis of DNA methylation during β cell aging. We employed genome-wide base-resolution methylome analysis of highly purified β cells from pre-pubescent and post-fertile age mice, and integrated these findings with transcriptome data, histone modification profiles, and analysis of altered β cell function. In addition to defining DNA methylation changes at cell cycle regulators that may contribute to the irreversibility of β cell quiescence in old age, we find a dramatic and unexpected improvement of β cell function with aging, which coincides with specific changes in the β cell epigenome.
Results
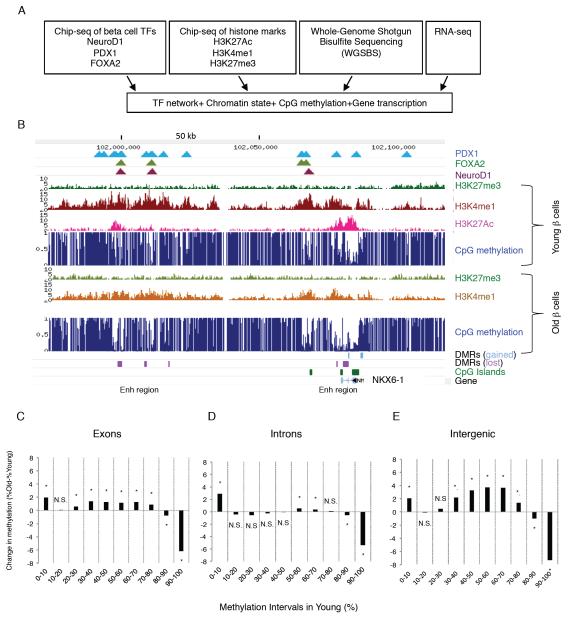
The epigenetic underpinnings of the aging-related decline in proliferative capacity of pancreatic β cells has been elucidated only partially. Therefore, we performed a comprehensive analysis of the epigenome and transcriptome of very young (pre-puberty) and very old (post fertile age) mouse β cells. Highly enriched (>98%) β cell populations were obtained by flow cytometry sorting of isolated pancreatic islets of young (4-6 weeks) and old (16-20 months) mice (Figure S1). DNA, chromatin and RNA were extracted for genome-wide, single-base resolution analysis of DNA methylation, mapping of key histone modifications, and transcriptome analysis, respectively. In addition, we integrated binding site maps of key transcription factors (TFs) essential for β-cell identity and function (NeuroD1, Pdx1 and Foxa2) from previous work (Hoffman et al., 2010; Khoo et al., 2012; Tennant et al., 2013) into our analysis (Figure 1A). This comprehensive study revealed the dynamic changes of the epigenomic landscape that occur in β cells as they age, and inform us on regulatory events in the aging process in postmitotic cells in general. Figure 1B depicts the Nkx6.1 locus, encoding an essential β cell transcription factor, as an example. Shown are two enhancers with age-related differential methylation regions (“DMRs”), one within the gene body and another about 80kb downstream of the transcriptional start site (TSS), where decreased methylation with age is correlated with chromatin marks typical of enhancers, such as acetylation of histone H3 at lysine 27 (H3K27ac) and monomethylation of histone H3 at lysine 4 (H3K4me1).
Figure 1. Integrative epigenomic analysis of pancreatic β cells aging.
(A) Outline of experimental paradigm. Our analysis integrated chromatin state marks, base-resolution methylation profiles and gene transcription in young (4-6 weeks old) and old (16-20 months old) mouse β cells with maps of key β cell transcription factors. (B) Map of the Nkx6.1 locus as an example of our integrated epigenomic map. Differentially methylated regions (DMRs) were identified between the two developmental states as described in the text. Note the multiple areas with lower DNA methylation in old β cells as indicated by the magenta boxes. (C-E) Global changes in methylation levels with aging by genomic region. Methylation states in young β cells were subdivided into bins by degree of methylation, and then the change in methylation levels of the same regions in old β cells were calculated. Note that, regardless of genomic context, regions with very high methylation in young β cells lose methylation with age, while other genomic regions gain methylation. Only CpGs pairs with a minimal read coverage of 11 in both young and old β cell methylomes were included in the analysis and combined into regions. (C) Exons, 23,048 regions, (D) introns, 21,000 regions (E) Intergenic, 21,243 regions. * p < 0.01. See also Figure S1.
A “drift” in DNA methylation levels during β cell aging
To obtain single base pair resolution of DNA methylation in old and young β cells, we employed Whole Genome Shotgun Bisulfite Sequencing (WGSBS). Genomic sequencing cumulative coverage for old and young cells was on average of 18 and 16-fold, respectively (Figure S2A). We observed little non-CpG methylation across the genome for both cell populations (Figure S2B). As described in previously published base-resolution methylomes, such as those of mouse embryonic stem and intestinal epithelial cells (Sheaffer et al., 2014; Stadler et al., 2011), most promoters are hypomethylated in β cells, while the remainder of the genome is highly methylated (Figure S2C).
When comparing global methylation levels in old and young β cells, we found that CpGs with low methylation levels in young β cells tended to gain methylation with age, with an average methylation increase of 1.5% in genic (exons and introns, Figure 1C-D) and of 3% in intergenic regions (Figure 1E), while in promoter-proximal CpG islands (CGIs) we detected an overall increase of 1% in methylation with age (4% average methylation in young versus 5% average methylation in old β-cells, p<0.0001, (Figure S2C). In contrast, CpGs that started out highly methylated (80% and higher) in young β-cells tended to lose methylation with age, with an average loss of 7% methylation in both the gene body and intergenic regions (Figure 1C-E), similar to what had been shown before for repetitive sequence elements (Bollati et al., 2009). These global, age-related changes in the level of DNA methylation support the idea of an age-related “methylation drift” process (Issa, 2014), suggesting that the maintenance of highly differential methylation patterns becomes compromised during aging, resulting in an overall somewhat more leveled methylome.
DNA methylation signatures identify likely β cell enhancers
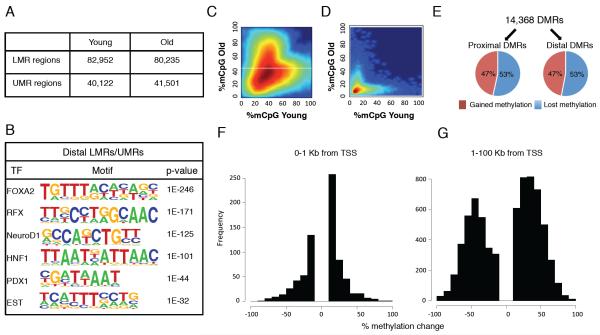
Next, we applied a Hidden Markov Model approach (Stadler et al., 2011) to segment both old and young beta cell methylomes into fully methylated regions (FMRs: >50% methylated), low methylated regions (LMRs: 30% methylated on average), and unmethylated regions (UMRs: 5.7% methylated on average), and determined into which genomic regions (promoter, repeats, etc.) these methylation classes are subdivided (Figure S3A-B). In both methylomes, the vast majority of the genome is fully methylated (about 2.5 Gbp, or > 95%), and FMRs encompass mostly repeat and intergenic sequences. As expected, UMRs were highly enriched for gene promoters, and cover about 40,000 regions in young and 41,500 regions in old β cells (Figure 2A), encompassing 23.8 and 28 megabases, respectively. Conversely, LMRs (about 83,000 regions in young, and 80,000 regions in old β cells, spanning an area of 79.5 and 70 megabases, respectively; Figure 2A) are depleted of promoters and enriched for intergenic regions. LMRs, which are formed dynamically during differentiation by cell-type-specific regulatory proteins, were previously characterized in mouse embryonic stem cells as distal regulatory regions with little or no overlap with CGIs (Stadler et al., 2011). Analyzing distal β cell LMRs and UMRs for the enrichment of transcription factor recognition sequences identified motifs specific to β cell transcription factors including Foxa2, Rfx, NeuroD1, Hnf1, and Pdx1 with high statistical significance (Figure 2B). Thus, simple segmentation of cell type-specific DNA methylation data allows for the identification of β cell-specific distal regulatory elements.
Figure 2. DNA methylation is dynamic during β cells aging.
(A) Number of genomic regions with low methylation (LMR, between 13.9% and 50% methylation) and unmethylated (UMR, between 0 and 13.9% methylation) regions in β cells isolated from prepubescent (4-6 weeks old) and post-fertile age (16-20 months old) mice. (B) Distal (more than 1 kb from the nearest transcriptional start site) LMRs and UMRs of β cells are highly enriched for the recognition motifs of key β cell transcription factors (HOMER (Heinz et al., 2010)). (C,D) Heatmap of LMRs (C) and UMRs (D) from young and old β cells. Intensity of the color reflects the number or regions, with red being high, and blue being low. Differentially methylated regions (DMRs) are the clouds that fall off the diagonal. (E) Regions with differential methylation between old and young β cells (DMRs) occur both at promoters (“proximal DMRs” - 1000 base pairs ± of TSS, 3,669 regions) and elsewhere in the genome (“distal DMRs” - >1000 base pairs from the nearest TSS, 10,699 regions), and can gain or lose methylation with aging. (F-G) The amplitude of change in DNA methylation is much larger at distal DMRs than at those close to transcriptional start sites. DMRs were subdivided by distance to transcriptional start sites as indicated. X-axis values between (−) 100 and 0 represent gain of methylation with aging and between 0-100, lose of methylation with aging. See also Figure S2 and S3.
To focus on the specific subset of hypomethylated regions that are engaged in transcriptional regulation in β cells, we overlapped LMRs and UMRs with our whole-genome map of acetylation of histone H3 at lysine 27 (H3K27ac) in young β cells, a histone modification that is enriched in promoters and active enhancers (Creyghton et al., 2010). We found that almost all genomic sites enriched for nucleosomes containing H3K27ac span LMRs and UMRs (Figure S3C-D), while conversely only a subset of LMR and UMR regions are enriched for this histone modification (9% of distal regions and 40% of proximal regions (Figure S3E-F). When we overlapped LMRs and UMRs with whole genome binding site maps of transcription factors essential for the identity and/or function of β cells for which genome-wide data are available (Pdx1, Foxa2 and NeuroD1; (Hoffman et al., 2010; Khoo et al., 2012; Tennant et al., 2013), we found that these genomic regions are frequently occupied by one or more of these proteins (Figure S3G-H). In sum, genomic regions with reduced DNA methylation levels that are distal to promoters are enriched for β cell transcription factor binding, a subset of which likely functions as enhancers.
β cell regulatory elements exhibit dynamic DNA methylation far exceeding methylation drift
Next, we compared UMRs and LMRs between old and young β cells and identified a total of 14,368 significant differentially methylated regions, or DMRs, covering 13 Mbps. Most of the DMRs were created by changes in methylation levels at LMR regions located more than one kb distal to TSSs, as previously observed for DMRs that occur during mouse ES cell differentiation towards neuronal progenitors and during development (Stadler et al., 2011)(Figure 2C) while most UMRs in promoter regions show a smaller change in methylation levels with age (Figure 2D). Whereas the same proportion of DMRs proximal or distal to promoters lose or gain methylation with aging (Figure 2E), the extent of differential methylation was quite distinct. DMRs near promoters displayed small, though statistically significant, changes in methylation levels (generally up to 10%; Figure 2F). In contrast, out of 10,699 distal DMRs, 9,373 exhibited a change grater than 15% in DNA methylation level (Figure 2G), with about a third of the distal DMRs (3,440 regions) changing methylation levels by over 50% in old β cells. Thus, putative distal regulatory elements display amplitudes of differential methylation with aging that far exceed the genome-wide trends described above as age-related DNA methylation drift.
Chromatin state and transcription factor binding precede age-related demethylation in regulatory elements
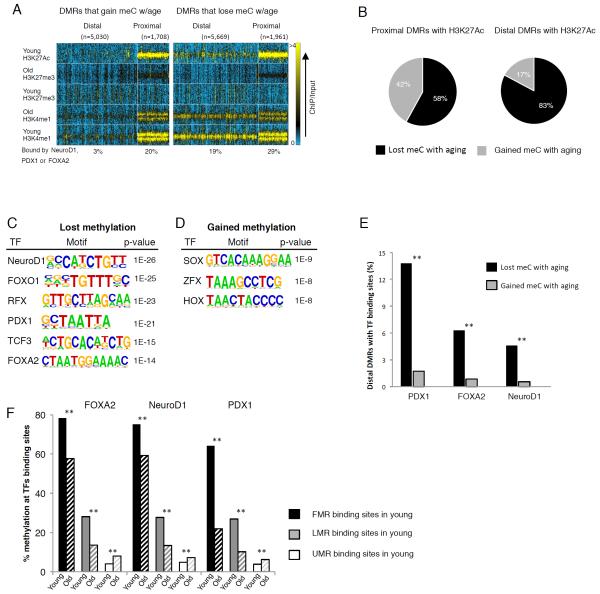
To begin to understand the molecular underpinnings of the dynamics of the β cell methylome during aging, we explored the relationship of chromatin state and DNA methylation changes. We integrated all significant DMRs with our whole genome histone modification maps from old and young β cells, including H3K4me1, which is enriched in nucleosomes that occupy potential enhancers (Heintzman et al., 2007) and H3K27me3, associated with transcriptionally silent chromatin (Schubert et al., 2006). In addition, we employed a map of H3K27ac, associated with active enhancers, obtained from young β cells. For this analysis, we divided differentially methylated regions into four groups: proximal and distal regions that gained methylation with age, and proximal and distal regions that lost methylation with age (Figure 3A). When comparing proximal promoters that gain or lose DNA methylation with aging, their profile of H3K4me1, H3K27me3 and H3K27ac is comparable, meaning that the chromatin signature at promoters of the young β cell (based on these three major histone marks), does not predict gain or loss of DNA methylation during aging. Interestingly, we observed an accumulation of the repressive H3K27me3 mark in proximal promoters with age (Figure 3A), similar to what was recently demonstrated in aged quiescence stem cells of the human muscle (Liu et al., 2013).
Figure 3. Chromatin marks at distal DMRs predict loss of DNA methylation during β cells aging.
(A) Heatmap integrating chromatin marks with four subgroups of DMR, located either proximal or distal to TSS and either gaining or losing methylation with aging. Note that greater proportion of distal DMRs that lose methylation with aging are previously marked as active enhancers by both H3K4me1 and H3K27ac in young β cells as compared with distal DMRs that gain methylation with aging. Colorbar scale is linear, representing read density with blue being low and yellow being high. (B) Integration of DMRs with the H3K27ac mark, often found at active enhancers and promoters, yielded 1,028 distal and 1,902 proximal regions that contained both properties. In both groups greater proportion of DMRs becomes demethylated with age, with a stronger effect in distal regions. Black, DMRs that lose methylation with aging, gray, DMRs that gain methylation with aging. (C) Distal DMRs that lose methylation with aging are enriched for recognition sequences for key β cell transcription factors (HOMER (Heinz et al., 2010)). (D) Distal DMRs that gain methylation with aging are enriched for binding motifs for transcription factors important in early embryonic development (HOMER (Heinz et al., 2010)). (E) Percentage of distal DMRs bound by the β cell transcription factors Foxa2, Pdx1 and NeuroD1. Black bars, distal DMRs that lose methylation with aging, gray bars, distal DMRs that gain methylation with aging. (F) DNA methylation levels at the binding sites of Foxa2, Pdx1 and NeuroD1 in young and old. Binding sites were divided into three methylation categories in young previous to analysis; fully methylated (black bars, over 50% on average), Low methylated (gray bars, between 14% - 50% on average) and Unmethylated (white bars, less than 14%). Only binding sites that had coverage depth of over 10 reads in both old and young were considered for the analysis. All methylation differences between young and old β cells were found significant by paired t-test, p<0.0001 (**). See also Figure S2 and S3.
The situation is very different, however, for distal DMRs. Here, about a third of the DMRs that lose methylation with age are previously occupied by the enhancer mark H3K4me1, and 15% are marked by the active enhancer mark H3K27ac in young β cells (Figure 3A and Table S1), while a much smaller subset of distal DMRs that gain methylation with age carry the H3K4me1 and H3K27ac marks in young β cells (10% and 3.5% respectively, Figure 3A and Table S1). Further analysis of DMRs enriched for the H3K27ac mark, often found at active enhancers and promoters, yielded 1,028 distal and 1,902 proximal regions that contained both properties. For both these subsets of proximal and distal elements, we found a greater proportion of DMRs that become demethylated with age (Figure 3B); however, this effect was much more pronounced in distal regions, where 83% of DMRs marked by H3K27ac lost methylation in old β cells. Interestingly, the distal DMRs that lose methylation with aging are enriched for recognition sequences for key β cell transcription factors such as Pdx1 and the Foxa factors (Figure 3C), while distal DMRs that gain methylation with aging instead show enrichment for binding motifs for TFs important in early embryonic development such as the Sox, Hox and Zfx gene families, expression of which is silenced in the postnatal β cell (Figure 3D). Indeed, the distal DMRs that lose methylation with aging are much more likely to be bound by the three β cell transcription factors NeuroD1, Pdx1 and Foxa2 for which genome-wide binding site maps in young mice are available (Hoffman et al., 2010; Khoo et al., 2012; Tennant et al., 2013), than those that gain methylation with age (Figure 3E). Analyzing the methylation levels at the binding sites of Foxa2, NeuroD1, and Pdx1 in young and old β cells genome-wide reveals that binding sites that started as fully or low methylated in young β cells lost methylation during aging, turning into LMRs or UMRs, while binding sites that started as unmethylated (less than 8% methylation on average) remained so while slightly increased in methylation level with age (Figure 3F). Given with our finding that the expression of Foxa2, NeuroD1, and Pdx1 is upregulated during aging (see below), the observed demethylation of distal DMRs could be at least partially explained by increased occupancy by these transcription factors with aging. Taken together, these data support the notion that chromatin state and the presence of transcription factor binding sites at distal regulatory elements in the young β cell target regulatory elements for demethylation with aging.
Demethylated enhancers are enriched near β cell function genes
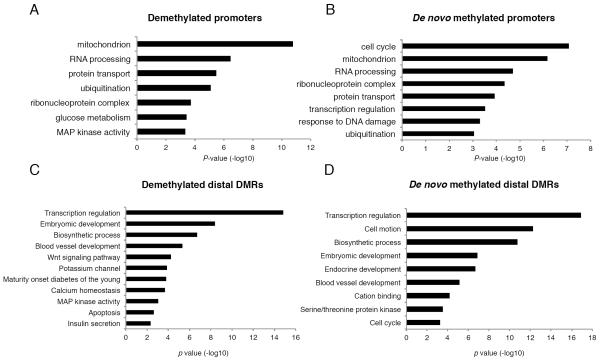
Next, we asked which biological pathways are associated with the genes nearest to distal DMRs (putative enhancers) and proximal DMRs (promoters). In general, proximal DMRs that lose or gain methylation with age are enriched for gene ontology (GO) categories essential for common cellular functions, such as mitochondrial activity and protein transport (Figure 4A,B). However, while promoter regions that are demethylated in aging β cells are additionally enriched near genes involved in the MAPK pathway and glucose metabolism (Figure 4A), the de novo methylated promoters are enriched near genes involved in cell cycle control (Figure 4B). This group of genes includes MKi67, Ccnd3, and Plk1, the promoters of which displayed increased methylation and decreased expression with aging (Figure S4). These data suggest a possible explanation for the age-dependent decline in the ability of β cells to proliferate introduced above: as the promoters of key cell cycle genes are methylated, they may become refractory to mitogenic signals.
Figure 4. Age-dependent methylation dynamics differentiate among functional gene categories.
(A-D) Gene ontology analysis (DAVID (Dennis et al., 2003)) for DMRs classified by location relative to transcriptional start site and direction of methylation change with age. (A) Demethylated proximal promoters are enriched for genes function in cellular metabolism. (B) De novo methylated promoters are frequently involved in the cell cycle. (C) In addition to biological pathways enriched in both distal DMRs that gain or lose methylation with aging, demethylated distal DMRs are enriched for β-cell function genes, while (D) de novo methylated regions are found associated with pancreas development and cell cycle genes. See also Figure S4.
When analyzing the biological pathways associated with distal DMRs we observed that both DMRs that gain or lose methylation with age are enriched near genes involved in transcription factor activity, such as the Hox proteins, and embryonic development (Figure 4C-D). However, in addition to this common enrichment of DMRs near developmental-related genes, distal DMRs that gain or lose methylation with aging are enriched for very different GO categories. Thus, DMRs that lose methylation with age are further enriched near genes essential to β cell function, such as the MODY (Maturity Onset Diabetes of the Young) genes, the ATP-dependent potassium channel genes Kcnj11 and Abcc8, and other biological pathways essential for mature β cell identity and physiology (Figure 4C). On the other hand, the distal DMRs that gain methylation with age are enriched near genes involved in endocrine development and cell cycle (Figure 4D). These findings suggested the unexpected possibility that β cell function might actually improve, not decrease, with aging, a notion we address experimentally below.
Age-related demethylation of enhancers is associated with gene activation
To study the relationship between age-dependent gene regulation and DNA methylation, we integrated only the specific subset of differentially expressed genes that changed significantly between adolescence and 16 months of age according to their steady state mRNA levels (≥1.5 FC, <10% FDR), with changes in methylation levels with age (list of significant DMRs discussed above). Overall, we found that 2,083 genes were upregulated (activated) and 4,040 were downregulated (silenced) with age (Figure S5A). For about half of these differentially expressed genes, the change in gene expression was accompanied by an altered DNA methylation state at an associated distal DMR. In contrast, the proportion of expression changes correlating with altered promoter methylation was much lower, i.e. only about 25% for the upregulated and 12% for the downregulated genes. Pathway analysis revealed that the group of genes upregulated with aging is enriched for gene ontology categories characteristic of beta cell function, such as protein production, folding and degradation, protein transport, and mitochondrial activity, while genes downregulated with aging are enriched for developmental and proliferation processes, cell to cell contact, and signaling (Figure S5B-C). Moreover, genes upregulated with aging are significantly enriched for binding sites of the β cell transcription factors Pdx1, NeuroD1 and Foxa2; 58% of the upregulated genes versus 37.8% of the remainder of the genes have at least one TF binding site in an associated distal elements and 33.5% versus 13.7% have at least one TF binding site in the promoter region. In contrast, the group of downregulated genes is depleted of such target genes; 34.4% of the downregulated genes versus 40.9% of the remainder of the genes have at least one TF binding site in an associated distal elements and 2.8% versus 18.9% in the promoter regions (p <0.01 by z test).
Next, we analyzed changes in DNA methylation separately for the genes that are activated or repressed with aging. The correlation of loss of DNA methylation with gene activation was stronger for distal DMRs (67%; p < 0.01, Figure 5A) than for those near transcriptional start sites (57%; p < 0.05, Figure 5B) (see also Tables S2A-D). The lower fraction of demethylated promoters compared with demethylated enhancers among activated genes likely reflects the fact that most promoters were already unmethylated in young β cells. However, when analyzing genes that are turned off during β cell aging, we found that they are equally likely to be de novo methylated or demethylated in their distal and proximal regulatory elements (Figure 5C-D, Tables S1E-H); Nevertheless, there was still a greater proportion of regulatory regions that became methylated with age that are associated with downregulated genes than upregulated genes (compare panel A and B to C and D in Figure 5). Importantly, the demethylated distal DMRs are enriched for β cell transcription factor binding sites as compared with distal DMRs that gained methylation with age, especially for DMRs associated with activated genes (Figure 5A, C). Specifically, within the group of genes associated with DMRs, the sub-group of upregulated genes are significantly enriched for target genes of the β cell transcription factors Pdx1, NeuroD1 and Foxa2; 19% of the upregulated genes versus 13% of the rest of the DMR-associated genes have at least one binding site in a distal DMR, and 16.6% versus 9.7% have at least one binding site in a DMR in their promoter region (p<0.01 by z test). In contrast, downregulated genes with aging associated with a change in methylation level in their regulatory elements are significantly depleted of target genes of these β cell transcription factors (9% of the downregulated genes versus 13% of the rest of the DMR-associated genes have at least one TF binding site in a distal DMR and only 1.5% versus 9.7% have at least one TF binding site in a DMR in their promoter region (p<0.01 by z test).
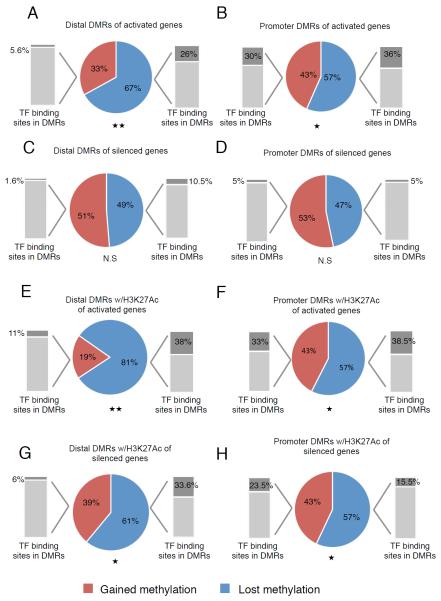
Figure 5. Age-related demethylation of enhancers is associated with gene activation.
(A-D), Expression analysis of genes associated with DMRs located within promoter regions (≤±1kb of the nearest TSS) and distal regions (≥ ±1kb from nearest TSS). DMRs that lose methylation with age (blue) correlate with activated genes compared with DMRs that gain methylation (red) in both promoters and distal regions while no such correlation is found with genes downregulated (silenced) with age. (*) p < 0.05, (**) p < 0.01 by z-test, N.S, not significant. Grey bars show the percentage of DMRs containing Pdx1, NeuroD1 and/or Foxa2 binding sites within the specific group of DMRs. (E-H), Expression analysis of only the genes associated with the subset of DMRs marked by H3K27ac. (E,F) H3K27ac enriched DMRs near activated genes show significant demethylation during aging (promoters; 57%, enhancers 81%). (G,H) H3K27ac enriched DMRs in promoters of silenced genes are demethylated with age similarly to DMR in promoters of activated genes (57%; compare F and H) but to a lesser extent than DMRs of enhancers near activated genes (61% in H versus 81% in F, p < 0.01 by t-test). See also Figure S5.
When considering only the specific subset of DMRs enriched for the H3K27ac mark of active enhancers, the association of age-dependent demethylation and gene activation is striking, with now 81% of the activated genes becoming demethylated with age in distal enhancers, while the fraction of promoters that become demethylated remains the same as for the global analysis (57%, Figure 7B,F). An association was also found between demethylation of distal regulatory elements and nearby downregulated genes (Figure 5G, 61%, p < 0.05); these elements are possibly acting as silencers. However, no such association was found for age-dependent downregulation of genes in promoter regions (Figure 5H). In the subset of DMRs enriched for the H3K27ac mark, the fraction of demethylated distal DMRs bound by TFs is even more pronounced (38%, grey bar in Figure 5E). Therefore, we conclude that age-related demethylation of distal cis-regulatory elements is associated with transcription factor binding and gene activation, whereas, in general, de novo methylation of promoters and distal elements is not a major mediator of age-related gene silencing in β cells.
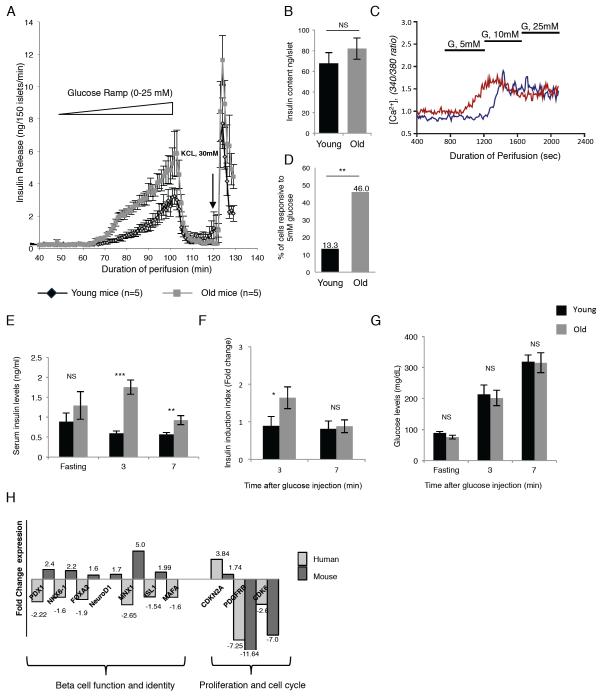
Figure 7. Insulin secretory function of β cells increase in old mice.
(A) Islet perifusion assay using a glucose ramp (0 to 25 mM) to assess glucose threshold and maximal glucose-stimulated insulin secretion in old (gray line) and young (black line) mice (n=5 each). (B) Islet insulin content in old (gray bar) and young (black bar) mice (n=5 each). (C) Assessment of old and young β cell function by glucose-stimulated calcium influx assay. Dispersed islet cells retrieved from old (16 months) and young (4 weeks old) mice were loaded with Fura-2AM, a fluorescent dye which binds to intracellular free calcium [Ca2+]i and stimulated with three concentrations of glucose. Shown are calcium traces examples of single old and young (red and blue line respectively) β cells. Note that the young β cell (blue trace) responds with elevated calcium levels only at 10 mM glucose, while the old β cell is responsive at 5 mM. (D) Quantification of glucose responsiveness as determined by calcium imaging of individual β cells obtained from old (gray bar) and young (black bar) mice (n= 30 and 37 β cells for young and old mice respectively). (E-G) In vivo insulin secretion assay. 17 months old (average weight 29 gr, gray bars) and four-week old (average weight 20 gr, black bars) male mice, n=5 each, were fasted for 16 hours before an intraperitoneal injection of glucose (2mg/gr body weight). Blood insulin levels were measured before the injection (fasting level) and 3 and 7 minutes following glucose injection. (E), average serum insulin levels (ng/ml). (F), ratios between the averages of fasting insulin levels and insulin levels 3 minutes and 7 minutes after glucose injection. (G), average blood glucose levels (mg/dL). (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; NS, not significant, by t-test. (H) Gene expression analysis of key β-cell function genes and regulators of β cell cycle activity in young (4-6 weeks) and old (16 months) old mouse and human β cells. For the human β cells, samples were from four young donors (ages 6 months, 4, 10 and 14 years old) and six older donors (ages between 28 to 64 years). Data represent the fold change between old and young β cells. Primer sets can be found in the Supplemental Material (Table S7).
β cell specific DMRs act as enhancers and silencers
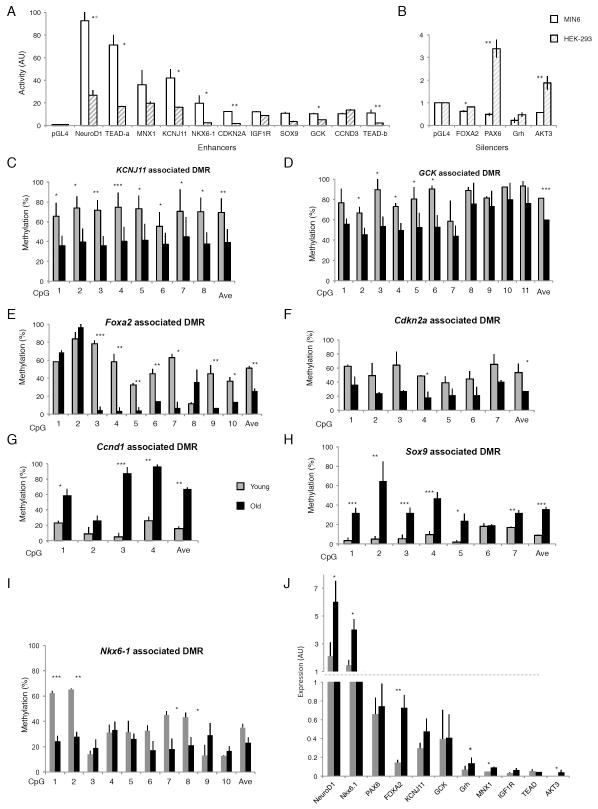
Having discovered age-related dynamics in the status of DNA methylation of distal DMRs in β cells, we next wanted to determine whether these elements indeed function as enhancers or silencers. We selected several age-dependent DMRs displaying methylation changes between 20%-90% and associated with key genes essential for β cell maturation, proliferation, and function. Some of the genes are either downregulated or activated with aging as was found by our RNAseq and qRT-PCR analyses (selected DMRs and their properties are listed in Table S3). We validated several DMRs by targeted bisulfite sequencing (Figure 6C-I) and for differential expression by qRT-PCR (Figure 6J). Both upstream (> 1 kb distal to TSS) and intronic elements, representing potential enhancers or silencers, were cloned in front of a luciferase gene expressed from a minimal promoter. We determined the potential regulatory activity of these elements in the MIN6 β cell line. Out of eighteen DMRs tested, eleven demonstrated enhancer activity in MIN6 cells while four DMRs acted as silencers and repressed the luciferase activity driven by the minimal promoter (Figure 6A and B white bars). Luciferase activity of seven enhancers and three silencers was found significantly elevated in MIN6 as compared with their activity in HEK293 cells (Figure 6A and B gray striped bars; (*) p < 0.05, (**) p<0.01, paired Student’s t-test), supporting their beta cell specificity. Out of two regions associated with cell cycle genes (Cdkn2a and Ccnd3), the regulatory region associated with Cdkn2a was beta cell specific.
Figure 6. β cell specific DMRs act as enhancers and silencers.
(A, B), DMRs activity of β cell enhancers (A) and silencers (B). DMRs associated with key genes of β cell function were cloned upstream of a luciferase gene and transiently transfected into the β cell line MIN6 (white bars) and HEK 293 cells for comparison (gray striped bars). Luciferase activity was normalized against the activity of a cotransfected Renilla construct, and mean values ± (SEM) are shown relative to the empty vector (pGL4.23). Luciferase activity of seven enhancers and three silencers was found significantly elevated in MIN6 as compared with the activity in HEK293 cells, (*) p < 0.05, (**) p<0.01, paired Student’s t-test. (C-I) Targeted bisulfite sequencing validation of age-dependent β cell DMRs identified by whole genome bisulfite sequencing. Three biological replicates of young and old β cells were used to verify methylation at the single-CpG level and each regional average for DMRs associated with Kcnj11, encoding the Kir6.2 subunit of the ATP-sensitive potassium channel (C), Gck, encoding glucokinase (D) Foxa2, encoding the β cell transcription factor FOXA2 (E), Cdkn2a, encoding the cell cycle and senescence regulator p16 (F), Ccnd1, encoding Cyclin D1 (G), Sox9, encoding the β cell transcription factor SOX9 (H) and Nkx6-1, encoding the β cell specific transcription factor NKX6-1 (I). The DMR regions associated with Kcnj11, Gck, Foxa2 and Cdkn2a decrease in their regional average methylation with age (Kcnj11: 30.4% ± 15%; Gck: 21.3% ± 10.65%, Foxa2: 25.7% ± 6.2%; Cdkn2a 26.1% ± 2.3%) while the DMR associated with Nkx6-1 contains several individual CpGs that become significantly demethylated with age. Both DMR regions associated with Ccnd1 and Sox9, increase in their regional average methylation with age significantly (Ccnd1: 51.7% ± 17.4%; Sox9 27.2% ± 7%). (*) p < 0.05; (**) p < 0.01; (***) p < 0.001 by t-test. (J), qPCR validation of differentially expressed key beta cell genes. Genes associated with the above DMRs were validated for their RNA expression levels in old and young β cells by qRT-PCR. The transcription factors NeuroD1, Nkx6-1 and FOXA2 were found significantly upregulated with age, in accordance with the RNA-seq data (*) p < 0.05; (**) p < 0.01 by t-test. See also Figure S6.
Next, we compared the DNA methylation status of two of the above distal cis-regulatory elements in other tissues, i.e. liver and kidney, to those of old and young β-cells by targeted bisulfite sequencing (Figure S6, Table S4). In both DMRs associated with the β cell genes Kcnj11 and Gck, the methylation levels of individual CpGs as well as the regional methylation average were significantly decreased with age in β cells (Figure 6C-D), but not in kidney or liver (Figure S6). Taken together, this analysis demonstrates that the cell specificity of enhancers is accompanied by low methylation levels, and moreover, that methylation dynamics are involved in the regulation of distal regulatory elements not only during development (Sheaffer et al., 2014), but also in aging. These observations provide further evidence that the age-dependent methylation changes we observed in β cells are not the result of a general, non-specific “drift” process, but part of the specific regulatory landscape of β cells.
Beta-cell function improves with age in mice
As noted above, our RNA-seq and qPCR data of β cells from adolescent and post-fertile age mice showed an unexpected increase in the expression of β cell function genes in old animals, suggestive of functional improvement, not decay, with aging (Table S5 and Figure 6J). Thus, as β cells mature, at least up to 16 months of age, there was further activation of gene expression of several transcription factors essential for β cell function, identity, and maintenance such as Pdx1, Nkx6-1, NeuroD1, Foxa2, and Mnx1. Conversely, we observed reduced expression of genes governing β cell proliferation such as the cyclin D genes and CDKs, and elevated expression of the cell cycle inhibitor p16 (encoded by Cdkn2a, Table S5), previously shown to contribute to loss of β cell replication with aging (Krishnamurthy et al., 2006).
Prompted by these surprising results regarding the activation of β cell function genes in old mice, we investigated β cell performance in old and young mouse β cells using multiple modalities. First, we employed the islet perifusion assay to assess both the glucose threshold as well as the maximal insulin secretory response to glucose, by applying a glucose ramp from 0 to 25 mM. As shown in Figure 7A, the glucose response curve was left-shifted in β cells from old mice compared to young, indicating improved responsiveness to glucose stimulation in matured β cells. In addition, the maximal insulin secretory response to glucose stimulation is about two-fold higher in old β cells, despite the similar response to KCl depolarization. This increase in functionality cannot be explained by increased insulin content per islet, as demonstrated in Figure 7B. Secondly, we performed single cell calcium imaging to assess the glucose response. A rise in intracellular calcium is the penultimate step in insulin secretion, and triggers the fusion of insulin granules with the plasma membrane. Figure 7C gives an example of single cell calcium traces from old and young β cells, where the old β cell responds to 5 mM glucose with increased calcium levels, while the young β cell becomes activated only at 10 mM glucose. When we quantified this response, we found that a much larger fraction of old β cells was responsive to low glucose levels compared with young β cells (Figure 7D), consistent with the left-shift in the glucose response of insulin secretion shown above. Thus, by two independent measures, β cells from old mice show improved function compared to those of adolescent animals. Next we measured in vivo insulin secretion in response to a small glucose stimulus in 17 months old and four week-old mice (Figure 7E-G). Basal blood insulin level was measured at fasting as well as three and seven minutes after glucose injection. As shown in Figure 7E and 7F, the mature mice responded with larger insulin excursion than the four-week old mice, consistent with the in vitro secretion data.
Finally, we asked the question if a similar shift in the gene expression profile towards functional maturation occurs in human β cells with aging. To this end, we compared mRNA levels of key genes in β cells obtained from young (ages ranging from 6 months to 14 years) to those of mature deceased organ donors (ages ranging from 28 to 64 years, Table S6-donor info). As shown in Figure 7H, the age-dependent change in expression of the cell cycle genes occurred in the same direction in humans and mice, with decreased expression of pro-proliferative genes Pdgfrb and Cdk6, and increased expression of the cell cycle inhibitor Cdkn2a in old β cells. This is consistent with the decreased proliferation rate in old β cells compared to young in both species (Meier et al., 2008; Teta et al., 2005). In striking contrast, five β cell transcription factors, key to mature β cell function, showed divergent age-dependent regulation between human and mouse. Thus, steady-state mRNA levels of PDX1, NKX6.1, FOXA2, MNX1 and ISL1 were all decreased in older human β cells, consistent with loss of β cell functionality (Table S5). This discrepancy may at least partially be the consequence of the life style and diet in the human subjects, which had elevated BMI (see Table S6 for donor info), compared to the healthier rodent chow (with less than 10% calories from fat) on which our mice were raised. We therefore propose that decreased β cell function in older mammals cannot be simply explained by aging of β cell as a predetermination.
Discussion
Type 2 diabetes is caused in part by compromised function and regeneration capacity of pancreatic β cells, and is associated with aging and unhealthy lifestyle. The proliferative capacity of β cells declines dramatically with age. Hence, we hypothesized that the epigenetic changes that occur during aging might contribute to the age-related decline in proliferative capacity as well as to the potential age-related reduction in β cell function. We employed comprehensive epigenetic profiling of β cells from young and old mice including whole genome base-resolution methylome, mapping of key histone modifications, and transcriptome analyses. We integrated all these analyses with binding site maps of key transcription factors (TFs) essential for β-cell identity and function (NeuroD1, Pdx1 and Foxa2 (Hoffman et al., 2010; Khoo et al., 2012; Tennant et al., 2013)). This allowed us to identify novel regulatory elements and study the yet uncertain role of DNA methylation in the function of distal cis-regulatory DNA elements.
While overall DNA methylation is remarkably stable with aging, reflecting the precision of methylation of hemi-methylated DNA by Dnmt1 after completion of S-phase, we observed the existence of two distinct dynamic processes affecting the DNA methylome that occur during the maturation of β cells. First, we noticed a general genomic drift in the level of methylation, pointing to a process of blunting the amplitude of differences between fully and lowly methylated areas of the genome with aging. This methylation drift exists side by side with dynamic targeted local demethylation and de novo methylation of specific regulatory elements. Thus, we found that hundreds of distal regulatory elements, identified by their low DNA methylation levels (LMRs), show dramatic changes in methylation state, while promoters remain essentially unmethylated during life. How are these distal regulatory regions targeted for demethylation or de novo methylation? Our comprehensive analysis revealed that distal regulatory elements that become demethylated with aging were previously marked with histone modifications characteristic of active enhancers, such as monomethylation of histone H3 at lysine 4 (H3K4me1) and acetylation of histone H3 at lysine 27 (H3K27ac), and are depleted of the repressive histone H3 lysine 27 trimethylation mark (H3K27me3). Moreover, these DMRs often reside near genes relevant to β cell function (Figure 4C) and are enriched for lineage-restricted transcription factors recognition motifs (Figure 3C). They are frequently bound by one or more β cell specific transcription factors, especially when associated with genes upregulated with aging (38%, Figure 5F) and exhibit enhancer activity in reporter assays (Figure 6A). Our data suggest a model of precisely orchestrated epigenetic events, where chromatin state and binding of β cell-specific transcription factors precede and possibly cause subsequent local DNA demethylation of β cell specific distal regulatory elements associated with activated gene during the aging of β cells. At present, we do not know what mechanisms drive the demethylation of these distal regulatory elements with aging. However, it is tempting to speculate that the self-reinforcing β cell transcription factor network leads to recruitment of Tet (ten-eleven translocation) enzymes to these elements, which oxidize methyl cytosines to hydroxy-methyl cytosines. This oxidation of methyl cytosine can either result in active demethylation (He et al., 2011), or in targeted passive demethylation, because hydroxy-methyl cytosines are not recognized as substrates by DNA methyltransferases. Thus, cis-regulatory elements marked by hydroxy-methylation will lose their methylation status passively after even a few cell divisions. Alternatively, the presence of TF at these elements might diminish their recognition by Dnmt1 following DNA replication. Future studies will need to address the molecular mechanism of aging-dependent demethylation in detail.
In contrast to the situation described above for demethylated elements, most distal regions that gain methylation with aging were not marked by H3K4me1 and H3K27ac by six weeks of age, and are enriched for embryonic developmental transcription factors recognition sequences (Figure 3D). Generally, they are not associated with β cell function genes (Figure 4D) and are rarely bound by β-cell specific transcription factors (i.e. 1.6% near silenced genes, Figure 5C). Furthermore, we did not find a strong correlation between de novo methylation and age-related gene silencing (Figure 5C and D). Therefore, we conclude that, unlike demethylation of distal cis-regulatory elements that correlate with transcription factor occupancy, absence of transcription factor binding is not always followed by de novo methylation. Consequently, in general, de novo methylation of promoters and enhancers may not be a major effector of age-related gene silencing. Nonetheless, increased methylation at promoters, as we observed for cell cycle genes (Figure 4B), may promote locking them in an ‘off-state’, thereby contributing to the reduced regeneration ability of the old β cell.
Unexpectedly, our analysis of old and young β cells transcriptomes revealed an upregulation of key β cell function genes such as Pdx1 and NeuroD1 with aging, and suggested that aging is not necessarily coupled with a decline in β cell function, at least in mice, contrary to current held assumptions. We employed islet perfusion assays as well as in vivo insulin secretion to confirm that β cells of mice way past their reproductive age exhibit a far better, more robust insulin secretory response than β cells of young mice (Figure 7). This is true both for the glucose threshold, and the maximal extent of the insulin secretory response. These data support the notion that declining β cell function is not an inherent property of aging, at least in rodents, and suggests that in normal, healthy aging, the decline in β cell proliferation rate could be balanced by improved function. However, an opposite scenario of gene regulation occurs in aged human β cells (Figure 7H). While the age-dependent changes in gene expression of the cell cycle regulators are similar between human and mouse β cells, reflective of the decline in proliferative capacity, β cell function genes are down-regulated in old human β cells, in contrast to the situation in the aged mouse. Potential explanations for this striking discrepancy include, but are not limited to, the different diets consumed by the average human subjects (the typical Western diet contains more than 30% calories from fat, as compared to the less than 10% in the rodent chow), and the dramatically different life spans of the two species. Thus, human β cells have to function for several decades as opposed to months in the mouse, which might contribute to the eventual decline in functional capacity. In the future, it would be interesting to perform methylome and transcriptome analysis of aged mice which are raised on a Western diet, and thus exposed to many months of insulin resistance and increased β cell workload. It is possible that under these conditions mouse β cells might show a functional decline similar to what is seen in obese or diabetic humans, as opposed to improvement with aging. Conversely, it is also possible that the specific epigenetic phenotype of the mouse β-cell proves to be protective even under these conditions. In the latter case, a comparative methylome analysis between aged human and mouse β cells might reveal critical, protective regulatory circuits. Additionally, the age-related functional decline of β cells may not be entirely cell autonomous and thus may involve deterioration of the pancreatic islet niche with aging as was recently suggested (Almaca et al., 2014).
In sum, our comprehensive epigenomic analysis of β cell aging has led to new, and sometimes unexpected insights into the biology of aging. The decline in replicative capacity of β cells coincides with increased promoter methylation and decreased expression of cell cycle regulators, possibly making old β cells refractory to mitogenic stimulation. This is exactly the opposite scenario of that often seen in cancer, where hypermethylation of tumor suppressor genes makes cancer cells independent of mitogenic stimuli (Fujiwara-Igarashi et al., 2014). Surprisingly, our functional analysis revealed improved β cell function in older mouse β cells, which correlated with and was predicted by the alterations in transcriptome and epigenome with age. Thus, functional decline is not always a predetermined outcome of aging of terminally differentiated cells.
EXPERIMENTAL PROCEDURES
Mouse pancreatic islet purification and beta cell FACS
Pancreatic islets were isolated from 4-6 weeks and 16-20 months old C57BL6 male mice as previously described (Gupta et al., 2005). Isolated islets were dissociated into single cells, stained using an anti-insulin antibody (DAKO) and sorted using FACS to obtain a 98% pure β cell population. Cells were immediately frozen in liquid nitrogen. For downstream RNA analyses, flow cytometry sorting was performed on live β cells from isolated pancreatic islets of young and old MIP-GFP mice. DNA and RNA were isolated using the AllPrep Qiagen kit (Qiagen).
DNA Methylome Analysis
WGSBS was performed as previously published (Lister et al. 2009) with some modifications; Genomic DNA isolated from pure β cell populations obtained from 10 young and 10 old mice, was fragmented into ≈300bp pieces using the S220 Focused-ultrasonicator (Covaris). Sequencing libraries were generated with ≥500ng genomic DNA using the NEBNext genomic sequencing kit (NEB) and Illumina adaptors (PE-102-1001. PE-102-1002). Libraries were bisulfite converted using Imprint DNA Modification Kit (Sigma). Size selection of the libraries to 300-600bp was performed on the Pippin Prep DNA size selection system (Sage Science). Amplification of libraries was performed in 18 rounds of PCR using Pfu Turbo Cx Hotstart DNA Polymerase (Agilent Technologies). Paired-End Sequencing was performed on the Illumina HiSeq (100bp reads). Converted DNA sequence was aligned to the mouse (mm9) genome using BS Seeker (Chen et al., 2010). Percent methylation was calculated for individual Cs and data from the C and adjacent G were combined to represent each CpG. Segmentation was performed as described previously using the R package RHmm with a three-state HMM corresponding to fully methylated (>50%), low-methylated (30% on average) and unmethylated 5.7% on average) CpGs (Stadler et al., 2011). Methylation levels were smoothed over 5 CpGs prior to HMM segmentation. Differentially methylated regions (DMRs) were determined using a Bayesian binomial model (Berninger et al., 2008). To determine DMRs, we first determined a joint set of segments, which is the union of all segments (UMRs or LMRs) identified in either old and young β cells. In case two or more segments overlapped, the segments were fused. We then determined for each segment (in the joint set) its methylated and total counts in each methylome by summing the counts over all reads mapping to the segment. The potential DMRs had a median of seven and a mean of 16 CpGs. To assess whether methylation levels were significantly different between the methylomes, we used a Bayesian binomial model that considers two hypotheses, the hypothesis that the average methylation levels of the segments are the same (H1) versus the hypothesis that the average methylation levels in the two methylomes can take on any value and are independent of each other (Berninger et al., 2008). We used a conservative Bonferroni correction of 148,856 to the posterior probability for H2 and accepted regions as DMRs when the corrected posterior probability of H1 was less than 0.1. This type of correction is judged appropriate for large numbers of independent tests when the null hypothesis is the favored hypothesis, i.e., that most regions are not DMRs (Westfall). These significant regions had a median of 10 and a mean of 32 CpGs and thus have a strong statistical foundation.
Supplementary Material
Highlights.
DNA methylation of regulatory elements is dynamic with aging of pancreatic β-cells
Promoters of pro-proliferation genes become de novo methylated with β-cell aging
Enhancers near β-cell function genes become demethylated with aging
Insulin secretory function increases with β-cell maturation
eTORC Blurb.
Aging is driven by epigenetic modifications only partially understood. Avrahami et al. show that targeted methylation changes in β cell genes correlate with repression of the proliferation program and activation of metabolic regulators. Strikingly, β cell function is improved in aged mice, as predicted by methylome and transcriptome changes.
ACKNOWLEDGEMENTS
We thank Drs. Yuval Dor, Marisa Bartolomei, Karyn Sheaffer and Mitch Lazar for critical comments on the manuscript, Olga Smirnova, Haleigh Zillges, Christina Theodouru, and Tia Bernard for excellent technical support. This study was supported by the NIDDK (UC4DK104119 to KHK and BG). DA was supported by NIDDK 5T32DK007314. We thank the University of Pennsylvania Diabetes Research Center (DRC) for the use of the Functional Genomics, Radioimmunoassay, and Islet Cell Biology Cores (P30-DK19525). D.A. and K.H.K. designed the experiments and wrote the manuscript. D.A. performed the majority of experiments, and CL and JZ performed some. JS, RA, SR, MS, LB, and DS performed computational analyses. BG edited the manuscript. K.H.K. directed the whole study.
Footnotes
The authors declare no competing financial interests.
ACCESSION NUMBER
Genomic data have been deposited in Gene Expression Omnibus: GSE68618.
Supplemental Information including supplemental methods, 7 Tables and 6 Figures can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann AM, Gannon M. Molecular regulation of pancreatic beta-cell mass development, maintenance, and expansion. Journal of molecular endocrinology. 2007;38:193–206. doi: 10.1677/JME-06-0053. [DOI] [PubMed] [Google Scholar]
- Almaca J, Molina J, Arrojo EDR, Abdulreda MH, Jeon WB, Berggren PO, Caicedo A, Nam HG. Young capillary vessels rejuvenate aged pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17612–17617. doi: 10.1073/pnas.1414053111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mechanisms of ageing and development. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes & development. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, Myers RM, Absher D. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome biology. 2013;14:R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes & development. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Fujiwara-Igarashi A, Goto-Koshino Y, Mochizuki H, Sato M, Fujino Y, Ohno K, Tsujimoto H. Inhibition of p16 tumor suppressor gene expression via promoter hypermethylation in canine lymphoid tumor cells. Research in veterinary science. 2014;97:60–63. doi: 10.1016/j.rvsc.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Vatamaniuk MZ, Lee CS, Flaschen RC, Fulmer JT, Matschinsky FM, Duncan SA, Kaestner KH. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. The Journal of clinical investigation. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome research. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. Aging and epigenetic drift: a vicious cycle. The Journal of clinical investigation. 2014;124:24–29. doi: 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo C, Yang J, Weinrott SA, Kaestner KH, Naji A, Schug J, Stoffers DA. Research resource: the pdx1 cistrome of pancreatic islets. Molecular endocrinology. 2012;26:521–533. doi: 10.1210/me.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell reports. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JJ, Butler AE, Saisho Y, Monchamp T, Galasso R, Bhushan A, Rizza RA, Butler PC. Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes. 2008;57:1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JA, Brelje TC, Sorenson RL. Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology. 1992;130:1459–1466. doi: 10.1210/endo.130.3.1537300. [DOI] [PubMed] [Google Scholar]
- Rankin MM, Kushner JA. Adaptive beta-cell proliferation is severely restricted with advanced age. Diabetes. 2009;58:1365–1372. doi: 10.2337/db08-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieck S, Kaestner KH. Expansion of beta-cell mass in response to pregnancy. Trends in endocrinology and metabolism: TEM. 2010;21:151–158. doi: 10.1016/j.tem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. The EMBO journal. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schubeler D, Kaestner KH. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes & development. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Tennant BR, Robertson AG, Kramer M, Li L, Zhang X, Beach M, Thiessen N, Chiu R, Mungall K, Whiting CJ, et al. Identification and analysis of murine pancreatic islet enhancers. Diabetologia. 2013;56:542–552. doi: 10.1007/s00125-012-2797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- Tyrberg B, Eizirik DL, Hellerstrom C, Pipeleers DG, Andersson A. Human pancreatic beta-cell deoxyribonucleic acid-synthesis in islet grafts decreases with increasing organ donor age but increases in response to glucose stimulation in vitro. Endocrinology. 1996;137:5694–5699. doi: 10.1210/endo.137.12.8940401. [DOI] [PubMed] [Google Scholar]
- Weir GC, Laybutt DR, Kaneto H, Bonner-Weir S, Sharma A. Beta-cell adaptation and decompensation during the progression of diabetes. Diabetes. 2001;50(Suppl 1):S154–159. doi: 10.2337/diabetes.50.2007.s154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.