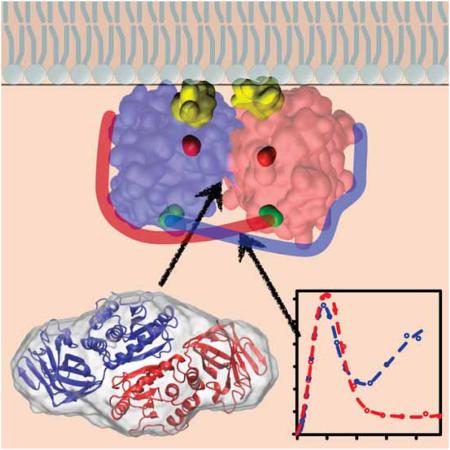
Summary
As the phosphoinositol-3-kinase antagonist in the PI3K pathway, the PTEN tumor suppressor exerts phosphatase activity on diacylphosphatidylinositoltriphosphate in the plasma membrane. Even partial loss of this activity enhances tumorigenesis, but a mechanistic basis for this aspect of PTEN physiology has not yet been established. It was recently proposed that PTEN mutations have dominant-negative effects in cancer via PTEN dimers. We show that PTEN forms homodimers in vitro and determine a structural model of the complex from SAXS and Rosetta docking studies. Our findings shed new light on the cellular control mechanism of PTEN activity. Phosphorylation of PTEN’s unstructured C-terminal tail reduces PTEN activity, and this result was interpreted as a blockage of PTEN’s membrane-binding interface through this tail. The results presented here instead suggest that the C-terminal tail functions in stabilizing the homodimer, and that tail phosphorylation interferes with this stabilization.
Keywords: PI3K/Akt pathway, PTEN phosphatase, dimer structure, SAXS, protein docking
Introduction
The diacylphosphatidylinositol-3,4,5-triphosphate (PI(3,4,5)P3)-specific lipid phosphatase PTEN (Li et al., 1997; Steck et al., 1997) is frequently mutated in human cancers (Simpson and Parsons, 2001; Stiles, 2009) and suppresses cell proliferation by limiting AKT phosphorylation in the PI3K signaling pathway. Even partial loss of PTEN activity (haploinsufficiency) enhances tumorigenesis (Berger et al., 2011). Genetic loss of Pten and mutations that affect functionality of the expressed protein are not equivalent, as patients with missense mutations develop lesions at a higher frequency than patients with gene deletion or drastic truncations (Marsh et al., 1998), so that missense mutations are, paradoxically, worse than nothing (Leslie and den Hertog, 2014). These observations can be rationalized by postulating that PTEN dimerizes in its active form, and indeed, a recent study presented evidence for PTEN dimerization in vivo and inferred that dimers are more active phosphatases than monomers (Papa et al., 2014). Here, we study structural aspects of PTEN dimerization in vitro. We find that the dimer state of bacterially expressed PTEN is favored over the monomeric form and derive a structural model of the PTEN dimer complex from small-angle x-ray scattering (SAXS) and docking studies that is consistent with earlier NR and MD results (Shenoy et al., 2012a). The PTEN monomer includes multiple disordered segments, the largest of which is its C-terminal tail (Lee et al., 1999). Whilst the monomer is partially unstructured as shown by the SAXS results, the dimer is well-folded and forms a compact particle, suggesting that the C-terminal tail plays a role in dimer stabilization. Phosphorylation of the tail was shown to inhibit PTEN membrane phosphatase activity (Rahdar et al., 2009). In addition, it affects the efficiency of dimerization (Papa et al., 2014). In combination with our structural results reported here, this suggests a novel control mechanism in which phosphorylation weakens the association of the two C-terminal tails with the protein domains, thereby destabilizing the dimer — while dimerization is presumably required for the phosphatase to reach its full enzymatic activity.
PTEN is a 403 amino acid (AA) protein with an N-terminal, dual-specificity phosphatase domain and a C-terminal, non-canonical C2 domain that binds anionic lipids independent of Ca2+ (Lee et al., 1999). In addition, PTEN includes a short (13 AA) N-terminus and the 51 AA C-terminus, both of which are unstructured. While the tumor suppressor function of PTEN depends on the interaction of the phosphatase with the plasma membrane (PM), the vast majority of the protein resides in the cytosol and interacts with the PM only sporadically (Redfern et al., 2010; Vazquez et al., 2006). Cellular control of this dynamic interaction has been debated (Ross and Gericke, 2009); in particular, phosphorylation of the C-terminal tail affects PTEN membrane localization (Rahdar et al., 2009). While other post-translational modifiers may impact PTEN membrane binding (Huang et al., 2012), we showed that bacterially expressed PTEN binds lipid membranes in vitro with high affinity and a strong dependence on lipid composition (Shenoy et al., 2012b).
Results
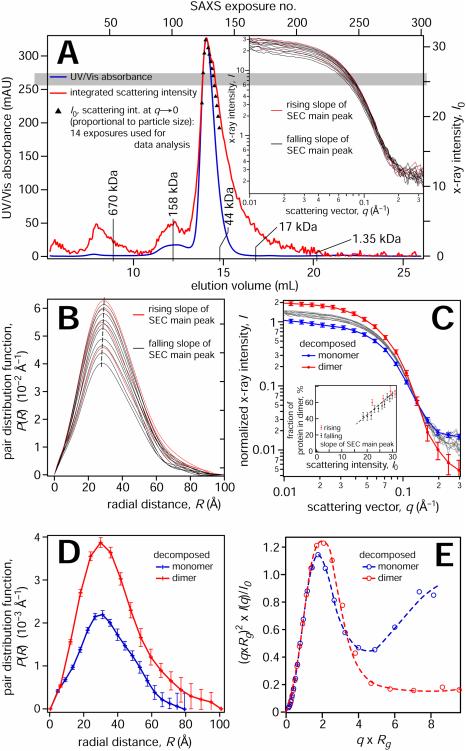
As a test for PTEN homodimerization in vitro, a GST pull-down assay with purified GST-PTEN and PTEN-His6 on a glutathione column showed His-tagged protein after elution. This signal was confirmed by Western blotting using a His-tag specific antibody (Supplementary Fig. 1). Next, we used SAXS to characterize the structure of bacterially expressed PTEN. In distinction from the protein used in the pull-down assay, this PTEN was tag-free. The protein was eluted from a size-exclusion column (SEC) and tracked by UV absorbance (Figure 1A). This trace is overlaid with the total x-ray scattering intensity, collected in > 250 individual exposures of the eluted protein as it passes through the x-ray beam, ca. 1 min. after passing the UV detector. In addition, the extrapolated (q → 0), background-corrected x-ray intensities, I0, are shown for 14 exposures across the elution peak.
Figure 1.
SAXS measurements and data analysis. For a biochemical characterization of PTEN dimer formation, see Fig. S1. (A) Protein concentration determined by UV absorbance (blue) and by the x-ray scattering intensity of a sample of PTEN eluted from an SEC. Molecular weight markers were derived from a calibration run with standards from Bio-RAD between 1.35 kDa (vitamin B12) and 670 kDa (thyroglobulin). The integrated x-ray scattering intensity on the detector is shown in red; black data points show I0, the background-corrected radial averages of the forward scattered x-ray intensities extrapolated to q = 0. I0 values are only shown for exposures that were further used for data analysis, close to the maximum of the monomer elution peak at ≈ 14.1 mL. Because the exact length of a ≈ 1 min delay between passage of the protein solution through the UV detector and the x-ray beam is not precisely known, signals were horizontally shifted to coincide at the peak positions. The time lag resulting from the travel of the sample in the capillary results in a broadening of the protein concentration-dependent x-ray intensities in comparison to the concentration-dependent UV absorbance. The difference between integrated x-ray intensity and I0 at elution volumes > 14.5 mL is likely due to protein adsorption on the cuvette walls. The inset shows the reduced SAXS data associated with these 14 exposures. (B) Pair distribution functions of the scattering centers derived from the scattering curves in the inset in (A) show a systematic dependence of their maximum positions and the protein concentration in the beam (proportional to their integrated areas). As a guide for the eye, the dashed line indicates these maxima of P(R). (C) Normalized SAXS intensities (thin lines) and their decomposition into two basis vectors from a simultaneous fit to all 14 data sets. The component coefficients that represent the fraction of protein in the dimer are shown in the inset. (D) PDFs corresponding to the basis vector scattering curves in (C). (E) Normalized Kratky plots of the two basis vectors in panel (D) for the monomer and the dimer. Dashed lines are guides to the eye. Error bars indicate 68% confidence intervals. For Guinier plots of the SAXS data, see Fig. S2.
We selected 14 SAXS exposures of protein from the major elution peak, indicated by their I0 values in Fig. 1A and shown in the inset, for a detailed evaluation. While we expected to observe scattering from homogenous PTEN fraction, a detailed analysis raised doubts about this interpretation. The maxima of the pair distribution functions (PDFs), P(R), shifted to higher R values with increasing I0 (proportional to the protein concentration in the beam), as shown in Fig. 1B. Similarly, we noticed differences in the slopes of the Guinier plots, i.e., the radii of gyration of the scattering particles (Supplementary Fig. S2). While small, these differences showed a systematic dependence on protein concentration. In view of these concentration-dependent variations, we decomposed the 14 SAXS exposures into linear combinations of two basis vectors and found that this two-state model fitted all experimental data simultaneously within experimental errors. The basis vectors (colored lines in Fig. 1C) and their weights in each SAXS curve correspond to the scattering of two distinct species and their relative concentrations in each exposure. These relative concentrations depend systematically on total protein concentration in the sample (inset in Fig. 1C). Figure 1D shows the corresponding P(R) profiles. The two PTEN species identified in the decomposition have radii of gyrations, Rg = 2.49 nm and 2.93 nm. Their Porod volumes were VP = 55 ± 10 nm3 and 98 ± 2 nm3, suggesting that the particle with the lower Rg is a PTEN monomer, and that with the larger Rg is a homodimer. The same conclusion was derived from a more elaborate analysis of the masses of the scattering particles based on scaling relations (Rambo and Tainer, 2013; Watson and Curtis, 2014), as shown in detail in the Supplemental Materials, Fig. S3 and Table S1. Normalized Kratky plots for the monomer and dimer differ significantly at high values of (q×Rg), see Fig. 1E. Whereas the dimer nearly returns to zero baseline, the monomer doesn’t show convergence. In line with current interpretation of SAXS protein signatures (Rambo and Tainer, 2011), we conclude that the monomer is partially disordered, while the dimer is well folded in its entirety, including the C-terminal tails.
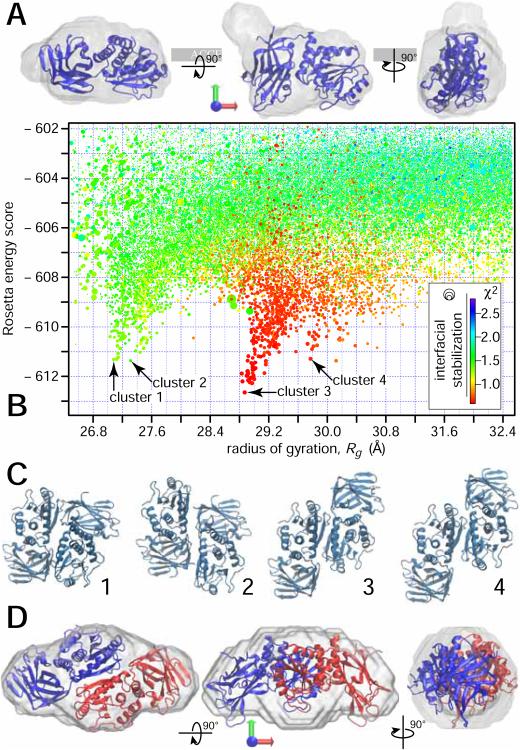
GASBOR (Svergun et al., 2001) was used for a protein shape reconstruction based on the monomer and dimer vectors. For the monomer, the corresponding envelope was found to fit the PTEN crystal structure well (Figure 2A). This structure was determined for a truncated protein that lacks ≈ 18% of the mass of full-length PTEN (Lee et al., 1999), and the visible underfilling of the protein volume defined by the SAXS results is therefore expected. This approach to data modeling appears reasonable, as all-atom MD simulations of PTEN suggested that there are only subtle differences between the crystal structure of the folded PTEN domains and its solution structure (Shenoy et al., 2012a). As expected from the dimer vector, the envelope computed for the PTEN dimer showed about twice the volume of the monomer envelope. Due to the lack of atomic-scale structural information on the unstructured protein segments, our search for trial dimer structures using Rosetta (Lyskov et al., 2013) was performed with the truncated crystal structure (Lee et al., 1999). Independent runs with and without constraints to C2 symmetry yielded similar low-energy results. Eventually, an unconstrained run yielded the structure funnel that led to the result with the overall lowest energy score. By sorting the entire set of Rosetta results (≈ 105 trial structures) according to their Rg values (Fig. 2B), four clusters were identified. The configurations associated with all four score funnels are structurally related in that dimerization is driven by interactions between the phosphatase domains while they differ by a translational offset along the protein binding interface (Fig. 2C). In addition, the membrane binding interfaces of both monomers face in the same direction in all these models. However, among these clusters there is only one that fits the experimental data well and is compatible with the Rg value, ≈ 2.9 nm, determined for the decomposed PDF vector. The configurations with the lowest Rosetta energy score within this funnel are almost identical in their structure and filled the dimer envelope particularly well, as shown in Fig. 2D for the hit with the overall lowest energy score.
Figure 2.
Structural modeling the decomposed SAXS results. (A) Reconstructed envelope of scattering density for the PTEN monomer in solution with a ribbon model of the truncated crystal structure (Lee et al., 1999) superimposed. When docked to the membrane, the direction of the membrane normal coincides with the green arrow. two orthogonal rotations transform the view shown in the center into the views to the right and the left. (B) Rosetta score vs. Rg for trial configurations from global and local Rosetta runs that yielded a Rosetta energy score ≤ −602, and a radius of gyration 26.6 Å ≤ Rg ≤ 32.6 Å. The symbol color encodes the fit quality (χ2) between the SAXS curves calculated from the configurations and the experimental SAXS curve obtained for the putative dimer from the decomposition. Because the error bars are slightly overestimated by the data reduction software provided by the facility, the best models show χ2 values below unity. The symbol size represents the interfacial energy score Isc from Rosetta. All symbols larger than the minimum size have interfacial energies that are considered ‘good’ with values 5 ≤ Isc ≤ 10. The score funnel at Rg = 29 Å yields the globally lowest Rosetta scores and the best fit to the data. (C) Graphical representation of configurations with the lowest Rosetta scores for the four score funnels indicated in panel (B). (D) Reconstructed envelope of scattering density for the large particle obtained from the decomposition, as in panel (A). The PTEN dimer corresponding to configuration 3 in panel (B) is superimposed as a ribbon structure.
Discussion
Although the formation of PTEN homodimers is well supported by genetic evidence (Papa et al., 2014) and provides an intriguing hypothesis relevant to cancer formation following mutation of a single PTEN allele (Berger et al., 2011), the structure and function of such dimers are poorly understood. Using pulldown experiments, we detected dimer formation in bacterially expressed PTEN. With tag-free PTEN protein, this result was verified by SAXS on selected SEC eluent fractions in which we identified PTEN monomer and dimer as a function of protein concentration. By decomposition of the SAXS data into independent contributions, we determined electron density envelopes of two distinct particles that fit the crystal structure of a truncated PTEN (Lee et al., 1999) monomer and dimer well. Supported by Rosetta docking simulations, this suggests a candidate structure for the PTEN homodimer. In recent MD simulations of PTEN monomers in solution we observed that the regulatory C-terminal tail shows some flexibility and associates with the surface of the PTEN domains in multiple, similar conformations (Shenoy et al., 2012a). Whilst SAXS cannot locate the tail in the candidate dimer structure, it shows conclusively that PTEN has a more compact conformation in the dimer than the monomer configuration, suggesting that the tail is stably associated with the protein domain surfaces. If this association occurs across the dimer, analogous to three-dimensional domain swapping common for other proteins (Liu and Eisenberg, 2002; Rousseau et al., 2003), this can provide a novel mechanism to stabilize the PTEN homodimer.
While SAXS provides only low-resolution structural information, our refinement of the scattering results with Rosetta leads to an attractive model that shows features consistent with previous biochemical characterizations of the PTEN dimer (Papa et al., 2014). Furthermore, the importance of the C-terminal tail for dimerization explains why the truncated PTEN protein used for x-ray crystallography did not show a dimer (Lee et al., 1999). This model also motivates predictions that can be tested in future work. (1) Without imposing constraints, all low-energy Rosetta models show approximate C2 symmetry and arrange the monomers such that their membrane-binding interfaces are coplanar. This is consistent with the fact that no higher order oligomers are experimentally observed and suggests that the membrane affinity of the dimer is considerably higher than that of the monomer. (2) In our structural model, the two phosphatase domains form the dimer interface whereas the C2 domains are not involved in this interaction. This agrees with results by Papa and coworkers which showed that an N-terminal fragment of PTEN that contained the phosphatase domain was more effective in binding to full-length PTEN in a pull-down assay than the C-terminal portion of the protein (Fig. 2B in (Papa et al., 2014)), suggesting that the phosphatase domain is indeed critical for dimerization. While the two C2 domains thus act independently of each other in membrane binding, the phosphatase domains might mutually affect each other in the tightly bound dimer state to optimize the efficiency of their catalytic sites. (3) Rosetta predicts that major contacts within the dimer occur between the two phosphatase domains and implicates the pα2 helix and pβ4 sheet in dimer stabilization. These predictions can be directly tested in future mutation studies aimed at controlling the monomer-dimer equilibrium. (4) Finally, we suggest that the C-terminal tails stabilize the dimer by crossing between its monomeric constituents in a domain-swapping exchange. If this is confirmed it will be interesting to test if inhibition of PTEN’s tumor suppressor function in cancer-associated mutations results from a reduction of dimer stability, protein misfolding, or both. In our model, we speculate that cellular control of PTEN activity results from dimer destabilization upon phosphorylation of the C-terminal tail. This hypothesis is consistent with previous results by Papa et al. These investigators showed that PTEN with a non-phosphorylatable version of the C-terminal tail (PTEN4A), which is functionally more active than wt PTEN (Vazquez et al., 2000), has increased dimeric fractions in gel filtration assays (Fig. 2I in (Papa et al., 2014)). Moreover, MD simulations of soluble PTEN monomer (Shenoy et al., 2012a) suggest that the C-terminal tail has a tendency to fold against the PTEN domains and is sufficiently long to obstruct the membrane-binding interface, which may interfere with dephosphorylation of the membrane-bound lipid substrate.
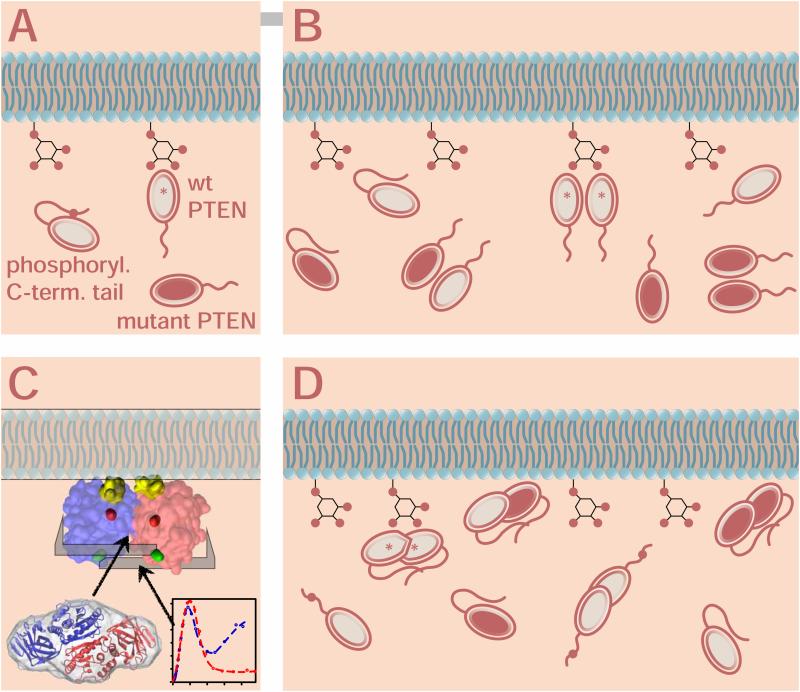
The results of this study lead to significant refinements of our understanding of the mechanism for PI(3,4,5)P3 dephosphorylation by PTEN and its cellular control. The evolution of the underlying models is schematically summarized in Fig. 3, starting with the hypothesis that phosphorylation of the C-terminal tail interferes with PM binding of the PTEN monomer in Fig. 3A (Rahdar et al., 2009; Ross and Gericke, 2009). Biochemical and genetic evidence recently implied a PTEN homodimer in maintaining PI(4,5)P2/PI(3,4,5)P3 homeostasis in healthy cells, as shown in Fig. 3B (Leslie and den Hertog, 2014; Papa et al., 2014). Here, we refine this model by providing a structural basis to the PTEN dimer hypothesis (Fig. 3D), based upon experimental observations in vitro and computational modeling using the truncated PTEN x-ray structure (Fig. 3C). Consistent with this model, it was recently shown that the binding of the phosphoinositide diC6PI(4,5)P2 to PTEN’s N-terminal sequence was associated with PTEN dimer formation in solution (Wei et al., 2015). Thus, high concentrations of PI(4,5)P2 in lipid rafts may further promote PTEN accumulation and dimerization in vivo, in agreement our in vitro experiments carried out with high PTEN concentrations. Of note, refolding of domain swap dimers may occur as a function of protein concentration (Rousseau et al., 2004).
Figure 3.
Evolution of PTEN membrane interaction models, redrawn after Fig. 1 in (Leslie and den Hertog, 2014). The enzymatically productive PTEN species in each model are marked with asterisks. (A) Cellular control of PTEN membrane interaction through phosphorylation (red dots) of the unstructured C-terminal tail (Rahdar et al., 2009). In this model, the phosphorylated tail blocks the membrane binding interface of wild-type PTEN, interfering with its enzymatic processing of PI(3,4,5)P3 in the plasma membrane (Ross and Gericke, 2009). Mutant PTEN (red filled) may interact with the membrane but is enzymatically inactive. (B) The PTEN dimer hypothesis (Papa et al., 2014) explains the dominant-negative behavior of PTEN mutants. Independent of tail phosphorylation, only homodimers of wild-type PTEN are enzymatically active while monomers or dimers that involve mutant PTEN are inactive or reduced in their activity. (C, D) Refinement of the dimer model through this work. The SAXS results provide a structural basis for the PTEN dimer hypothesis and lead to a reinterpretation of the role of tail phosphorylation. The structural model, Fig. 2D, predicts that the PTEN homodimer is formed through interactions between the two phosphatase domains, in agreement with results from the pull-down assays conducted by Papa and coworkers. Furthermore, the two membrane binding interfaces in the dimer are oriented in the same direction and form a flat, partially hydrophobic plane with exposed cationic residues for association with the plasma membrane (panel C). Within the outline of the folded protein domain in this schematic view, yellow residues mark the two CBR3 loops, the catalytic cores (C124 residues) are shown in red, and the location where the C-terminal tails emerge from the folded domains (E352 residues) are shown in green. As indicated by the Kratky analysis, the C-terminal tails are firmly bound against the folded PTEN domains, which suggests that the tails form ‘brackets’ that stabilize the dimer. These observations lead to the following refinement of the dimer model (panel D). Wild-type PTEN homodimer formation may be required to activate the phosphatase through structural adjustments around the substrate binding pocket, making the dimer more productive than the wild-type PTEN monomer. Alternatively, the increased productivity of the PTEN dimer could also result just from a higher affinity to the anionic inner plasma membrane than that of a wild-type PTEN monomer. However, this would not explain why the wild-type/mutant PTEN heterodimer has a strongly reduced enzymatic activity. In distinction to the scenario in panel (A), phosphorylation of PTEN’s C-terminal tail may decrease the interaction of the tail with the folded PTEN domains, thereby reducing the stability of the dimer.
In conclusion, we show that the bacterially expressed PTEN phosphatase dimerizes efficiently in vitro at micromolar concentrations and provide a candidate structure of the homodimer with critical interactions between the two phosphatase domains. Dimer formation may thereby result in cooperativity in PTEN membrane binding through the paired C2 domains, leading to increased enzyme affinity for the PM. In addition, the presumed tight binding of the juxtaposed phosphatase domains to each other could lead to conformational changes around the catalytic site that enhance the efficiency of lipid dephosphorylation. Clearly, the conjectures derived from our results need to be tested in mutation studies of the proposed dimer binding interface and manipulations of the phosphorylation state of PTEN’s C-terminal tail.
Experimental Procedures
Protein expression and GST pull-down assay
PTEN protein was expressed and purified as described (Redfern et al., 2008). Human PTEN with a C-terminal His-tag was expressed in E. coli BL21 (DE3). For the SAXS experiments, the His-tag was cleaved off using enterokinase. The GST pull-down assay was carried out using a batch method (results, see Supplementary Figure 1). Purified GST-PTEN and PTEN-His6 were mixed in an equimolar ratio and allowed to incubate a bed of pre-equilibrated glutathione Sepharose 4B (GE Healthcare Life Sciences) for an hour at 4 °C on a rocker. As a negative control, GST protein and PTEN-His6 were mixed using the same protocol. The resin was washed with buffers containing 0.5% triton X-100, 0.1% triton X-100, and finally detergent-free wash buffer. The remaining protein was eluted using 10 mmol/L reduced glutathione in Tris at pH 8.0. The eluted fractions were analyzed by SDS-PAGE. To confirm the presence of PTEN-His6, a Western blot was carried out using a His-tag specific antibody.
Small-angle x-ray scattering
Bacterially expressed, tag-free PTEN protein dissolved in 10 mM HEPES, 250 mM NaCl, 1 mM DTT at pH 7.4 was investigated in SAXS experiments at room temperature. Measurements were carried out at the APS BioCAT beamline (sector 18) of Argonne National Laboratory, as described earlier (Mathew et al., 2004). The 12 keV x-ray beam (λ = 1.03 Å) was focused on a 1.5 mm quartz capillary sample cell. The scattering, in the momentum transfer range, q = 0.0065 – 0.3 Å−1, was collected on a Mar165 CCD detector ca. 2.5 m downstream from the sample position. The protein solution was fed into the x-ray beam after passing through a Superdex-200 10/300 GL gel size-exclusion column onto which ≈ 500 μL were loaded at 4 mg/mL. A capillary fed the eluent first through a UV detector and then to the SAXS sample cell. The delay between protein emerging from the SEC and its arrival at the beam position was about 1 min. SAXS exposures with a length of 1 s were collected every 5 s during the gel-filtration chromatography run. Exposures before and after sample elution were averaged and used as buffer background. Exposures during elution that coincided with the UV peak on the chromatogram were treated as sample (protein + buffer) SAXS curves. Pair distribution functions, P(R), of the scattering centers were computed from the scattering curves using GNOM (Svergun, 1992). To analyze the systematic shift of scattering curves with sample concentration, we decomposed these into two basis functions by global fitting of all 14 SAXS spectra simultaneously with a Monte Carlo Markov Chain (MCMC), similar to a procedure previously described for the evaluation of neutron reflection data (Kirby et al., 2012). Full details are provided in part (B) of the Supplemental Material, Figs. S21 and S3 and Table S1.
Rosetta protein docking
Prior to the docking simulations, the truncated x-ray structure of the PTEN monomer (Lee et al., 1999) was supplemented with hydrogen atoms using MolProbity4 (Chen et al., 2010), and pre-packed using the Rosetta 3.5 Prepack Protocol (Gray et al., 2003). Unconstrained global docking simulations using the Rosetta 3 Protein Docking Protocol (Gray et al., 2003) were performed using two copies of the pre-packed structure as input. The orientations of both docking partners were randomized and default options for adding extra side-chain rotamers were applied (Wang et al., 2005). Local docking simulations without symmetry constraint did not randomize the orientations of the docking partners but, instead, allowed for a random perturbation of the input structures using a Gaussian for translation and rotation with standard deviations of 8 Å and 8°, respectively. The Rosetta 3.5 Symmetric Docking Protocol (André et al., 2007) was used for the docking simulation with C2 symmetry constraint. Default options for adding side-chain rotamers were applied. All docking simulations were performed with the low and the high resolution part of the protocol. ATSAS Crysol (Svergun et al., 1995) was used with default parameters to calculate the radius of gyration for every configuration, and to fit the theoretical SAXS curve to the experimental data. The option of a constant subtraction was enabled during the fit.
Supplementary Material
HIGHLIGHTS.
The PTEN tumor suppressor forms a homodimer in solution
SAXS/Rosetta determine a unique dimer structure with planar membrane binding surface
PTEN’s disordered regulatory C-terminal tail is well folded on the dimer
A structural basis emerges for the PTEN dimer hypothesis and cellular control of PTEN
Acknowledgments
We thank Weifeng Shang and Thomas Irving for assistance with data reduction. This research was supported by the Department of Commerce (MSE 70NANB11H8139 and 70NANB13H009), NIGMS (R01 GM101647), NINDS (R01 NS021716) and the NSF (CHEM 1216827) and used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated under contract no. DE-AC02-06CH11357. The BioCAT facility is supported by the NIGMS (P41 GM103622).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supplemental Information
The supplemental information provides a biochemical characterization of the PTEN dimer in part (A) and details of the SAXS data evaluation in part (B): Data decomposition, Guinier analysis, scaling estimates of scattering particle masses, and shape reconstruction of the scattering particles. It includes three figures and one table.
Author Contributions
F. H. & M. L. designed the research and planned the experiments. R. K. H. with the help of A. H. R. purified the protein and performed its biochemical characterization. F. H. and S. C. conducted the experiments. F. H. conceived and conducted the data evaluation. F. H. and H. N. performed the modeling studies. F. H., A. H. R., A. G. and M. L. wrote the manuscript. All authors discussed the results and commented on the manuscript at all stages.
References
- André I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. USA. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- Huang J, Yan J, Zhang J, Zhu S, Wang Y, Shi T, Zhu C, Chen C, Liu X, Cheng J, et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 2012;3:911–922. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- Kirby BJ, Kienzle PA, Maranville BB, Berk NF, Krycka J, Heinrich F, Majkrzak CF. Phase-sensitive specular neutron reflectometry for imaging the nanometer scale composition depth profile of thin-film materials. Curr. Opin. Colloid Interf. Sci. 2012;17:44–53. [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi PP, Pavletich NP. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Leslie NR, den Hertog J. Mutant PTEN in Cancer: Worse Than Nothing. Cell. 2014;157:527–529. doi: 10.1016/j.cell.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Chou F-C, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, et al. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE) PloS ONE. 2013;8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, Liaw D, Caron S, Duboué B, Lin AY, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- Mathew E, Mirza A, Menhart N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchrotron Radiat. 2004;11:314–318. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]
- Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl. Acad. Sci. USA. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern RE, Daou M, Li L, Munson M, Gericke A, Ross AH. A mutant form of PTEN linked to autism. Protein Sci. 2010;19:1948–1956. doi: 10.1002/pro.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern RE, Redfern DA, Furgason ML, Munson M, Ross AH, Gericke A. PTEN phosphatase selectively binds phosphoinositides and undergoes structural changes. Biochemistry. 2008;47:2162–2171. doi: 10.1021/bi702114w. [DOI] [PubMed] [Google Scholar]
- Ross AH, Gericke A. Phosphorylation keeps PTEN phosphatase closed for business. Proc. Natl. Acad. Sci. USA. 2009;106:1297–1298. doi: 10.1073/pnas.0812473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Schymkowitz JWH, Itzhaki LS. The unfolding story of three-dimensional domain swapping. Structure. 2003;11:243–251. doi: 10.1016/s0969-2126(03)00029-7. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Schymkowitz JWH, Wilkinson HR, Itzhaki LS. Intermediates control domain swapping during folding of p13suc1. J. Biol. Chem. 2004;279:8368–8377. doi: 10.1074/jbc.M310640200. [DOI] [PubMed] [Google Scholar]
- Shenoy S, Nanda H, Lösche M. Membrane association of the PTEN tumor suppressor: Electrostatic interaction with phosphatidylserine-containing bilayers and regulatory role of the C-terminal tail. J. Struct. Biol. 2012a;180:394–408. doi: 10.1016/j.jsb.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S, Shekhar P, Heinrich F, Daou M-C, Gericke A, Ross AH, Lösche M. Membrane association of the PTEN tumor suppressor: Molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS ONE. 2012b;7:e32591. doi: 10.1371/journal.pone.0032591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Parsons R. PTEN: Life as a tumor suppressor. Exp. Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Stiles BL. Phosphatase and tensin homologue deleted on chromosome 10: Extending its PTENtacles. Int. J. Biochem. Cell Biol. 2009;41:757–761. doi: 10.1016/j.biocel.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch M. CRYSOL— a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- Svergun DI, Petoukhov M, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Matsuoka S, Sellers WR, Yanagida T, Ueda M, Devreotes PN. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers W. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein Sci. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson MC, Curtis JE. Probing the average local structure of biomolecules using small-angle scattering and scaling laws. Biophys. J. 2014;106:2474–2482. doi: 10.1016/j.bpj.2014.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Stec B, Redfield AG, Weerapana E, Roberts MF. Phospholipid-binding sites of phosphatase and tensin homolog (PTEN): Exploring the mechanism of phosphatidylinositol 4,5-biphosphate activation. J. Biol. Chem. 2015;290:1592–1606. doi: 10.1074/jbc.M114.588590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.