Abstract
The spirochete Treponema pallidum subsp. pallidum is the causative agent of syphilis, a chronic, sexually transmitted infection characterized by multiple symptomatic and asymptomatic stages. Although several other species in the genus are able to cause or contribute to disease, T. pallidum differs in that it is able to rapidly disseminate via the bloodstream to tissue sites distant from the site of initial infection. It is also the only Treponema species able to cross both the blood-brain and placental barriers. Previously, the T. pallidum proteins, Tp0750 and Tp0751 (also called pallilysin), were shown to degrade host proteins central to blood coagulation and basement membrane integrity, suggesting a role for these proteins in T. pallidum dissemination and tissue invasion. In the present study, we characterized Tp0750 and Tp0751 sequence variation in a diversity of pathogenic and nonpathogenic treponemes. We also determined the proteolytic potential of the orthologs from the less invasive species Treponema denticola and Treponema phagedenis. These analyses showed high levels of sequence similarity among Tp0750 orthologs from pathogenic species. For pallilysin, lower levels of sequence conservation were observed between this protein and orthologs from other treponemes, except for the ortholog from the highly invasive rabbit venereal syphilis-causing Treponema paraluiscuniculi. In vitro host component binding and degradation assays demonstrated that pallilysin and Tp0750 orthologs from the less invasive treponemes tested were not capable of binding or degrading host proteins. The results show that pallilysin and Tp0750 host protein binding and degradative capability is positively correlated with treponemal invasiveness.
INTRODUCTION
Pathogenic Treponema species include Treponema pallidum subspecies pallidum (syphilis), T. paraluiscuniculi (rabbit venereal syphilis), T. denticola (one of the species contributing to the polymicrobial infection periodontitis), and T. phagedenis (a constituent of the polymicrobial infection bovine digital dermatitis). Of the Treponema species, T. pallidum subsp. pallidum (here referred to as T. pallidum) is the most medically important, causing a sexually transmitted, multistage disease in more than 11 million people each year (1) and increasing the risk for HIV acquisition and transmission (2). The public health importance of this disease underscores the need to identify T. pallidum proteins that are directly involved in pathogenesis. However, the fact that the pathogen is inherently fragile due to its unusual ultrastructure (3, 4), combined with its inability to be cultured continuously in vitro, has hindered identification of T. pallidum virulence factors. Analysis of the genome sequence has not identified any virulence factors found in other microbial pathogens, such as toxins or adhesins, that could shed light on the infection process for this important human pathogen (5).
Treponema pallidum subsp. pallidum is the most invasive of the treponemal species and one of the most invasive pathogens identified to date. Infection with T. pallidum typically manifests as a painless chancre at the site of infection approximately 3 weeks after pathogen exposure (6). Multiple studies have demonstrated that the pathogen is capable of invading tissue barriers and undergoing rapid widespread dissemination via the circulatory system (7–10). The highly invasive nature of T. pallidum is further highlighted by its ability to traverse the placental barrier to cause congenital syphilis and the blood-brain barrier to invade the central nervous system (9) and by the diverse and widespread clinical manifestations associated with secondary and tertiary syphilis (6). Numerous mechanisms have been suggested to contribute to the invasiveness and persistence of T. pallidum, including its corkscrew motility (4, 11), chemotactic ability (6), antigenic variation capability (6, 12–17), low outer membrane protein content (18, 19), host component adhesion capabilities (20–24), and its ability to cause host damage by inducing host-mediated inflammatory and immune responses (6).
Our laboratory previously identified and characterized two proteins, pallilysin (Tp0751) and Tp0750, that bind and degrade host components. Pallilysin exists as a proprotease that, upon activation through cleavage of the prodomain, binds and degrades laminin (basement membrane lining blood vessels), fibrin, and fibrinogen (a component of the host's coagulation cascade that functions to entrap bacteria and prevent dissemination), while Tp0750 binds fibrinogen and is also capable of degrading fibrinogen and fibronectin, but not fibrin (21, 23–25). The genes encoding Tp0750 and pallilysin are cotranscribed and the two proteins coassociate to form a heterodimeric complex in vitro, suggesting shared function (24). The results from opsonophagocytosis assays are in support of pallilysin residing on the treponemal surface, suggesting pallilysin can directly mediate host-pathogen interactions (25). These findings implicate T. pallidum-mediated host component degradation as an additional pathogenic mechanism promoting treponemal invasion and dissemination during infection.
In the present study, we identified and compared orthologs of Tp0750 and pallilysin in a diversity of Treponema species. We also performed in vitro assays to determine the host component binding and degradation potential of selected pallilysin and Tp0750 orthologs. We show that Tp0750 is well conserved in pathogenic treponemes, whereas pallilysin has limited overall sequence conservation. Further, we show that Tp0750 and pallilysin from T. pallidum, but not the orthologous proteins from two less invasive treponemes, are capable of binding and degrading host components. These findings suggest that the pallilysin and Tp0750 proteins may contribute to the difference in invasiveness observed among the Treponema species.
MATERIALS AND METHODS
Identification of Tp0750 and pallilysin orthologs in 19 Treponema species.
As a first step, we identified orthologs of Tp0750 and pallilysin using BLASTp, T. pallidum query sequences (Tp0750 [NP_219187] and Tp0751/pallilysin [NP_219188]) and the National Center for Biotechnology Information (NCBI) nr database. All identified orthologs had an obvious E value cutoff, above which E values rose to greater than zero. This value was 2 × 10−5 for Tp0750 and 2 × 10−6 for pallilysin. In the second step, amino acid sequences of the orthologs were aligned using CLUSTAL W2 (for multiple alignments, www.ebi.ac.uk/tools/msa/clustalw2), EMBOSS Needle (for pairwise alignments, www.ebi.ac.uk/tools/psa/emboss_needle), and by hand within the BioEdit sequence alignment editor (26). The accession numbers used to perform the alignments are listed in Table S1 in the supplemental material. Amino acid sequence homology analysis was also performed using the NCBI conserved domain database search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (27). The SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) (28) and the LipoP 1.0 server (http://www.cbs.dtu.dk/services/LipoP/) (29) were used to predict the presence and location of potential signal peptide cleavage sites and lipoprotein signal peptides, respectively. The number of amino acids, the predicted molecular masses, and the theoretical isoelectric points (pIs) of each Tp0750 and pallilysin ortholog were computed using ProtParam (http://web.expasy.org/protparam/).
To display amino acid sequence similarity and dissimilarity we generated a “WebLogo” for the region of pallilysin between amino acid residues L126 and Y224 (http://weblogo.berkeley.edu/) (30). Sequence logos are graphical representations of a multiple amino acid sequence alignment comprised of stacks of amino acids at each position in the aligned sequence. Increasing stack height (bit value) indicates increasing amino acid conservation at the corresponding position. Increasing amino acid residue height within each stack indicates a higher amino acid frequency at the corresponding position (30).
Phylogenetic analysis of pallilysin orthologs.
A phylogenetic tree of Treponema pallilysin ortholog sequences corresponding to the C-terminal region (L126-Y224) was inferred using the neighbor-joining method (31). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) was calculated (32). The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method (33) and were in the units of the number of amino acid substitutions per site. All ambiguous positions were removed for each sequence pair. There were a total of 100 amino acid positions in the final data set. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 5 software (34). The tree was rooted in a way that was consistent with taxonomy, which was based upon species-level relationships determined from 16S rRNA sequences (35).
Ethics statement.
All animal studies were approved by the local institutional review board at the University of Victoria and were conducted in strict accordance with standard accepted principles as set forth by the Canadian Council on Animal Care and the National Institutes of Health in a facility accredited by the American Association for the Accreditation of Laboratory Animal Care.
Bacterial growth and genomic DNA purification.
T. pallidum subspecies pallidum (Nichols strain) was propagated in, and extracted from, New Zealand White rabbits as described elsewhere (36). Treponema denticola (strain ATCC 35405) was kindly provided by Sheila Lukehart (University of Washington) and was grown at 37°C in prereduced, anaerobic TYGVS medium (37) supplemented with 10% heat-inactivated normal rabbit serum (Sigma-Aldrich Canada, Ltd., Oakville, Ontario, Canada) in the presence of 10 μg of rifampin ml−1. Treponema phagedenis (strain 4A) was isolated from lesions on Iowa dairy cattle as previously described (38). Treponema phagedenis was grown in prereduced, anaerobic oral treponeme isolation broth (39) containing 10% heat-inactivated fetal bovine serum (Sigma-Aldrich Canada, Ltd.) and 25 μg of rifampin ml−1. All cultures were grown in Oxoid anaerobic jars (Fisher Scientific, Ottawa, Ontario, Canada) under anaerobic conditions (<1.0% oxygen, 9 to 13% carbon dioxide) using Oxoid AnaeroGen packs (Fisher Scientific). Genomic DNA was purified from treponemes using the Qiagen DNeasy tissue kit (Qiagen, Toronto, Ontario, Canada). All DNA purifications were conducted according to the manufacturer's instructions for Gram-negative bacteria, with the following modification: the recommended amount of proteinase K (20 μl per purification) was increased 2-fold during the cell lysis step to ensure maximal genomic DNA recovery.
Construct cloning.
DNA fragments encoding S29-E491 of Tde0840 (Tp0751 ortholog) and G23-P220 of Tde0841 (Tp0750 ortholog) were PCR amplified from T. denticola (strain ATCC 35405) genomic DNA using the primers listed in Table 1 and cloned into the expression vector pDEST-17 (Invitrogen, Carlsbad, CA). DNA fragments encoding M12-H219 of DD750 (Tp0750 ortholog) and L22-D497 of DD751 (Tp0751 ortholog) were PCR amplified from T. phagedenis (strain 4A) (38) genomic DNA using the primers listed in Table 1 and cloned into the NdeI/XhoI sites of pET28a (VWR International, Mississauga, Ontario, Canada) and pDEST17 expression vectors, respectively. All cloning procedures were conducted according to the manufacturer's instructions for each expression vector. Constructs encoding wild-type pallilysin (encompasses C24 to P237), inactive pallilysin (E199A; encompasses C24 to P237), Tp0750 (encompasses G23 to D223), and the control protein Tp0327 have been previously described (23–25). All constructs were confirmed as correct by DNA sequencing.
TABLE 1.
Primers used to amplify DNA for recombinant protein expression
| Open reading frame | Primer |
|
|---|---|---|
| Orientation | Sequence (5′-3′)a | |
| Tde0840 (S29-E491) | Sense | GGGGACAAGTTTGTACAAAAAAGCAGGCTCTAATACAAAAGGTGAAGAAAAAATTG |
| Antisense | GGGGACCACTTTGTACAAGAAAGCTGGGTTTTATTCTCTTTCAAAGACAATGGG | |
| Tde0841 (G23-P220) | Sense | GGGGACAAGTTTGTACAAAAAAGCAGGCTCAGGCGAAAGAACAATGCCTG |
| Antisense | GGGGACCACTTTGTACAAGAAAGCTGGGTTTTACGGATTATCTTTTTTGATTAAG | |
| DD750 (M12-H219) | Sense | CAGTGCATATGCTGCTAACAATTCTTACAAAC |
| Antisense | GTCAGCTCGAGTTAATGCTCATCACGTATCAAG | |
| DD751 (L22-D497) | Sense | GGGGACATTGTACAAAAAAGCAGGCTTAAATCGAATAAAAATTGAAAGTAC |
| Antisense | GGGGACCACTTTGTACAAGAAAGCTGGGTTAATCTGTTTGTTTTGATTTGGG | |
Restriction sites are indicated in boldface.
Recombinant protein expression and purification.
Recombinant Tp0750, wild-type pallilysin, inactive pallilysin (E199A), and the control protein Tp0327 were expressed and purified as previously described (24, 25). Recombinant orthologs of Tp0750 (Tde0841 and DD750) and pallilysin (Tde0840 and DD751) were expressed and purified under identical conditions used for the corresponding T. pallidum proteins to ensure consistent levels of protein purity between each ortholog.
Host proteins.
Plasminogen-depleted human fibrinogen and bovine casein (both from Calbiochem) were purchased from VWR International (Mississauga, Ontario, Canada). Laminin isolated from Engelbreth-Holm-Swarm murine sarcoma basement membrane and fibronectin isolated from human plasma were purchased from Sigma-Aldrich Canada, Ltd.
Host protein binding and degradation assays.
To compare the host protein-binding capabilities of pallilysin, Tp0750, Tde0840, Tde0841, DD751, and DD750, plate-based in vitro host protein binding assays were performed as previously described (21) with the modifications that wells were blocked with 0.001% casein prepared in Tris-buffered saline and the level of host component background binding to the control protein Tp0327 was subtracted from the experimental wells.
To compare the host protein-degrading capabilities of Tp0750, Tde0841, and DD750, SDS-PAGE-based in vitro host protein degradation assays were performed as previously described (23) with the following modifications. The three recombinant proteins (40 μg of each) were independently incubated with 60 μg of plasminogen-free human fibrinogen, 30 μg of laminin, or 30 μg of fibronectin, in protease reaction buffer (20 mM HEPES [pH 7.5], 25 mM CaCl2). An inactive form of pallilysin (E199A) that previously failed to degrade host proteins (25) was included in the assays as the negative-control recombinant protein (40 μg per assay). The reaction mixtures were incubated at 37°C for 24 h. Samples were removed at 0, 4, and 24 h postincubation and analyzed for degradation of the three fibrinogen chains (α, β, and γ), two fibronectin chains (α and β), or three laminin chains (A, B1, and B2) by SDS-PAGE and Coomassie brilliant blue staining. Unlike Tp0750, pallilysin is capable of degrading fluorescein isothiocyanate (FITC)-labeled substrates. Therefore, a fluorescence-based degradation assay was performed to compare the host protein-degrading capabilities of pallilysin, Tde0840, DD751, and inactive pallilysin (E199A) using our previously described methodology (23) with the following modifications. Recombinant proteins were diluted in protease reaction buffer (20 mM HEPES [pH 7.0], 25 mM CaCl2) and preincubated at 37°C for 24 h. This preincubation step was performed since it has been demonstrated that attainment of full pallilysin proteolytic activity is dependent upon autocatalytic cleavage of an N-terminal prodomain, a process that can take up to 24 h in vitro (25). The physiological pH of the buffer used for these studies (pH 7.0) was selected to be representative of the pH of the local environment encountered by the treponemes within the host. Recombinant proteins (1 μg per well) were added in triplicate to sterile Corning Costar 96-well plates (Fisher Scientific) and incubated with FITC-labeled fibrinogen, fibronectin, or laminin (10 μg per well) as previously described (23).
RESULTS
Tp0750 orthologs are highly conserved in selected pathogenic Treponema species.
It has been demonstrated that Tp0750 binds and degrades host proteins involved in blood coagulation and fibrinolysis and may therefore contribute to the highly invasive nature of T. pallidum (24). In order to determine whether Tp0750 orthologs are present in other treponemes, we used BLASTp and full-length Tp0750 (M1-D223) as a query to survey the completed genomes included in the NCBI database. We found Tp0750 orthologs in 10/19 sequenced Treponema species, including T. paraluiscuniculi, T. phagedenis (bovine isolate strain 4A), T. phagedenis (human isolate strain F0421), T. medium, T. vincentii, T. pedis, T. denticola, T. putidum, T. azotonutricium, and T. bryantii. The biophysical properties of these 13 orthologs are summarized in Table 2. BLASTp analysis failed to identify Tp0750 orthologs in the pathogenic treponemes T. socranskii, T. brennaborense, T. maltophilum, T. lecithinolyticum and in the nonpathogenic treponemes T. saccharophilum, T. primitia, T. succinifaciens, and T. caldaria. Interestingly, Tp0750 orthologs were also found outside Treponema in three Spirochaeta (smaragdinae, bajacaliforniensis, and cellobiosiphila) and one Leptonema (illini) species.
TABLE 2.
Biophysical properties of treponemal Tp0750 orthologs
| Tp0750 treponemal orthologa | No. of amino acids (full length/signal sequence truncated) | Molecular size (kDa) (full length/signal sequence truncated) | pI (full length/signal sequence truncated) | % Identity (M1-Y194/M1-D223)b | Signal sequence prediction (SpI or SpII)c |
|---|---|---|---|---|---|
| T. pallidum (p) | 223/201 | 25.9/23.5 | 9.1/9.1 | 100/100 | SpI (A22-G23) |
| T. paraluiscuniculi (p) | 223/201 | 26.0/23.5 | 9.1/9.1 | 99/98 | SpI (A22-G23) |
| T. phagedenis (strain 4A) (p) | 219 | 25.6 | 5.1 | 63/55 | NS |
| T. phagedenis (strain F0421) (p) | 219 | 25.6 | 5.1 | 63/55 | NS |
| T. medium (p) | 220 | 25.6 | 6.0 | 60/54 | NS |
| T. vincentii (p) | 224/202 | 26.0/23.5 | 6.0/5.5 | 58/49 | SpI (A22-G23) |
| T. putidum (p) | 220 | 25.4 | 5.7 | 55/50 | NS |
| T. denticola (p) | 220 | 25.4 | 6.2 | 55/49 | NS |
| T. pedis (p) | 220 | 25.2 | 8.8 | 54/45 | NS |
| T. azotonutricium (np) | 172 | 18.3 | 8.9 | 26/22 | NS |
| T. bryantii (np) | 495/471 | 56.0/53.3 | 8.9/8.6 | 21/9 | SpI (A24-Q25) |
p, pathogen; np, nonpathogen.
M1-Y194, C-terminal-truncated Tp0750; M1-D223, full-length Tp0750.
Cleavage sites are indicated in parentheses. NS, not secreted.
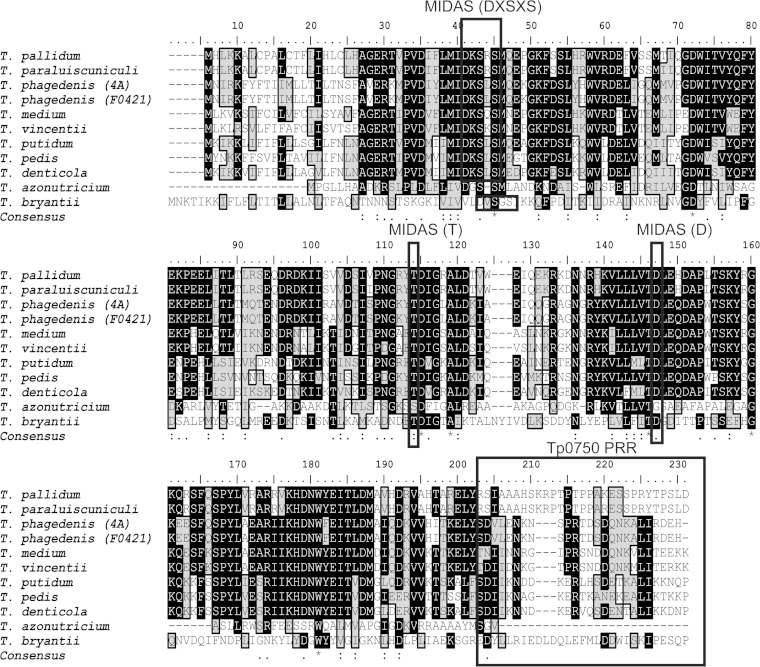
For the treponemal species in which a Tp0750 ortholog was found, the overall degree of sequence similarity was assessed by aligning the full-length sequences of the orthologs (Table 2). This analysis identified two subsets of Tp0750 orthologs: (i) orthologs from pathogenic treponemes that exhibit high amino acid identity to Tp0750 from T. pallidum and (ii) orthologs from nonpathogenic treponemes that have a lower sequence identity with Tp0750. The primary region of sequence divergence among the orthologs occurred in the C-terminal region (Fig. 1). In particular, a unique C-terminal proline-rich region (PRR; encompassing the sequence R195-D223) is found in the Tp0750 sequences from T. pallidum and T. paraluiscuniculi. Pairwise amino acid alignments were performed to compare residues M1-Y194 of Tp0750 with the corresponding region from the orthologs. This analysis showed that Tp0750 exhibits a range of 54 to 99% sequence identity with orthologs from pathogenic treponemes, while Tp0750 and the orthologs found in the nonpathogenic treponemes demonstrated lower sequence identity ranges of 21 and 26% (Table 2). Thus, similar to the full-length alignments, these analyses confirmed the highest conservation of Tp0750 orthologs occurs among pathogenic treponemal species.
FIG 1.
Amino acid alignment of Tp0750 treponemal orthologs. Full-length Tp0750 (M1-D223) was aligned with treponemal orthologs using CLUSTAL W2 and the BioEdit sequence alignment editor. Residues I232 to K495 of the T. bryantii ortholog are not included in the alignment. Identical residues (black background) and similar residues (gray background) are highlighted. The NCBI conserved domain database search was used to identify potential MIDAS motifs (indicated by rectangles). Sequences were searched manually for the presence or absence of PRRs (rectangle). At each position of the consensus sequence, an asterisk indicates full conservation among the 11 treponemes, a colon indicates strong conservation, and a dot indicates lower conservation.
Previous bioinformatic analyses of Tp0750 predicted the presence of a metal ion-dependent adhesion site (MIDAS)-containing von Willebrand factor type A (vWFA) domain comprising residues V29 to D143 (24). Similar to Tp0750, NCBI CDD analysis predicted the presence of a vWFA domain within all orthologs, with conservation of the MIDAS motif (DXSXS…T…D) in all orthologs except for the T. azotonutricium ortholog (Fig. 1). Tertiary structure prediction analyses using Phyre2 structure homology modeling server (40) predicted the structural similarity of 8/11 treponemal Tp0750 orthologs with the vWFA domain. This analysis was unable to generate high confidence structural models for the two low-identity orthologs from T. bryantii and T. azotonutricium and, surprisingly, the high sequence identity T. putidum ortholog. Further, the vWFA domain present within the protein likely was not acquired from the host in which the treponeme has a propensity to infect, since bioinformatic analyses did not detect homologs in vWFA-domain- containing proteins from humans, pigs, cows, or termites. Taken together, these findings demonstrate that approximately half of the treponemal species sequenced to date contain Tp0750 orthologs and that the sequence is highly conserved among pathogenic, but not nonpathogenic, treponemes.
Pallilysin orthologs are widespread within the genus Treponema.
We repeated the BLASTp survey using T. pallidum pallilysin as a query and found orthologs in all 19 Treponema species whose genomes have been sequenced. For the current analyses, we focused on 13 representative pallilysin orthologs from genetically and phenotypically characterized treponemes identified by BLASTp searches, including pathogenic species (T. pallidum, T. paraluiscuniculi, T. phagedenis [strain 4A], T. phagedenis [strain F0421], T. medium, T. vincentii, T. pedis, T. denticola, T. socranskii, and T. lecithinolyticum) and nonpathogenic species (T. primitia, T. saccharophilum, and T. succinifaciens). The biophysical properties of these 13 orthologs are summarized in Table 3.
TABLE 3.
Biophysical properties of treponemal Tp0751 orthologs
| Tp0751 treponemal orthologa | No. of amino acids (full length/signal sequence truncated) | Molecular size (kDa) (full length/signal sequence truncated) | pI (full length/signal sequence truncated) | % Identity (L126-Y224/M1-P237)b | Signal sequence prediction (SpI or SpII)c |
|---|---|---|---|---|---|
| T. paraluiscuniculi (p) | 237/214 | 25.8/23.3 | 7.3/7.0 | 100/99 | SpII (S23-C24) |
| T. phagedenis (strain 4A) (p) | 497 | 56.3 | 5.5 | 47/13 | NS |
| T. phagedenis (strain F0421) (p) | 497 | 56.3 | 5.5 | 47/13 | NS |
| T. vincentii (p) | 477 | 54.5 | 5.0 | 40/12 | NS |
| T. medium (p) | 477 | 54.4 | 5.3 | 41/13 | NS |
| T. pedis (p) | 493 | 56.3 | 6.1 | 32/11 | NS |
| T. denticola (p) | 491 | 56.8 | 5.3 | 34/11 | NS |
| T. lecithinolyticum (p) | 508/494 | 57.0/55.5 | 5.5/5.3 | 30/11 | SpII (A14-C15) |
| T. socranskii (p) | 528 | 59.6 | 5.3 | 26/9 | NS |
| T. saccharophilum (np) | 792 | 89.0 | 4.9 | 26/8 | NS |
| T. succinifaciens (np) | 495 | 55.6 | 5.4 | 30/14 | NS |
| T. primitia (np) | 501 | 56.5 | 5.0 | 33/12 | NS |
| T. paraluiscuniculi (p) | 237/214 | 25.8/23.3 | 7.3/7.0 | 100/99 | SpII (S23-C24) |
p, pathogen; np, nonpathogen.
L126-Y224, conserved region of Tp0751; M1-P237, full-length Tp0751.
Cleavage sites are indicated in parentheses. NS, not secreted.
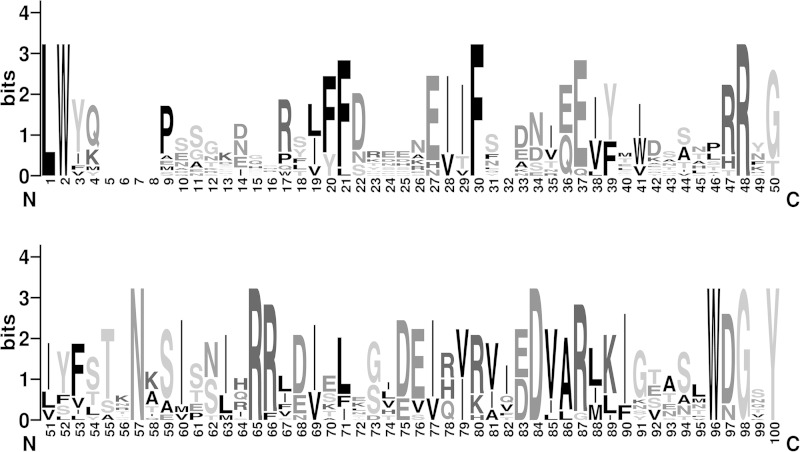
Pallilysin from the highly invasive treponemes T. pallidum and T. paraluiscuniculi was smaller (predicted molecular masses of 25.8 kDa) than pallilysin orthologs from the 11 less invasive treponemes (Table 3). With the exception of the T. saccharophilum ortholog (89.0 kDa), the predicted molecular masses of the remaining orthologs range from 54.4 to 59.4 kDa. Our amino acid alignment showed that difference is primarily due to extended N-terminal regions (∼100 to 120 residues) and C-terminal regions (∼120 to 130 residues or ∼410 residues [T. saccharophilum ortholog]) that are absent from the T. pallidum and T. paraluiscuniculi orthologs (Fig. 2). Analyses using NCBI conserved domain database searches did not uncover conserved domains within these N- and C-terminal regions and BLASTp analyses indicated that the terminal regions of the larger pallilysin orthologs are unique to these treponemal proteins. These analyses identified the central region of the pallilysin sequence, corresponding to amino acid residues L126-Y224, as being the most conserved among pallilysin orthologs. Pairwise amino acid alignment of this region of T. pallidum pallilysin with the ortholog from T. paraluiscuniculi revealed 100% amino acid identity, while alignment with the other 11 orthologs from less invasive treponemes revealed lower levels of pallilysin sequence conservation (26 to 47%; Table 3). This is well illustrated by WebLogo analysis of the 13 pallilysin orthologous sequences corresponding to residues L126 to Y224 (Fig. 3). Interestingly, the HEXXH motif that has been previously shown in T. pallidum pallilysin to be important for host-component degradation (25) was observed solely in the ortholog from T. paraluiscuniculi.
FIG 2.
Amino acid alignment of full-length treponemal pallilysin orthologs. Amino acid sequences of 13 pallilysin orthologs were aligned using CLUSTAL W2 and the BioEdit sequence alignment editor. Identical residues (black background) and similar residues (gray background) are highlighted. At each position of the consensus sequence, an asterisk indicates full conservation among the 13 orthologs, a colon indicates strong conservation, and a dot indicates lower conservation.
FIG 3.
Homology analyses of Treponema pallilysin orthologs. WebLogo graphical representation of the amino acid sequence alignment corresponding to the C-terminal domain of T. pallidum pallilysin and 12 treponemal orthologs. Amino acids L126 to Y224 of pallilysin were aligned with 12 orthologs identified from the BLASTp search and analyzed using WebLogo to generate a 100-amino-acid sequence logo for the 13 sequences.
T. pallidum pallilysin is phylogenetically distinct from orthologs of less and noninvasive treponemes.
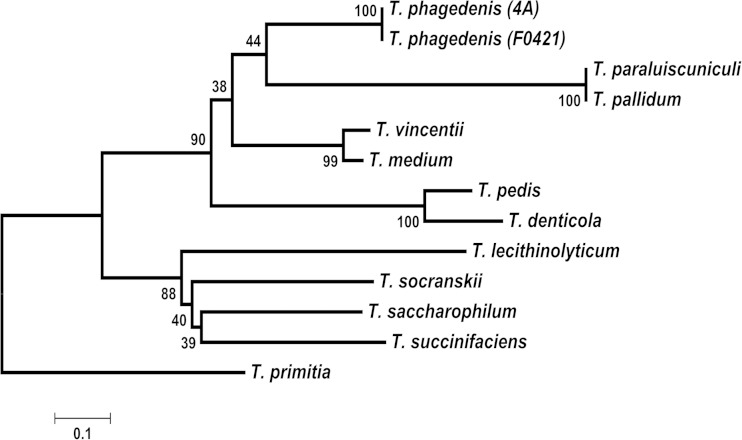
To assess the evolutionary conservation of T. pallidum pallilysin among the treponemal orthologs, a phylogenetic tree as inferred from sequences corresponding to the most conserved region of T. pallidum pallilysin (L126-Y224) was constructed using the neighbor-joining method (Fig. 4). In accordance with the homology-based results described above, this analysis revealed that the pallilysin sequence from T. pallidum is most closely related to the T. paraluiscuniculi orthologous sequence. Both sequences are most closely related to the homologs from the human and bovine T. phagedenis isolates. However, the phylogenetic tree inferred that this four-protein cluster descended from a common ancestor with only moderate reliability (bootstrap percentage, 44%). This analysis also placed T. pallidum pallilysin with 90% confidence in a well-defined major cluster containing more distantly related orthologs from T. vincentii, T. medium, T. denticola, and T. pedis. The T. pallidum pallilysin sequence was estimated to be less related to orthologous sequences from a well-defined cluster comprising T. lecithinolyticum, T. socranskii, T. saccharophilum, and T. succinifaciens. Together, these findings indicate that the pallilysin sequence has maintained more conservation within pathogens compared to nonpathogens during the course of treponemal evolution. However, the long branch lengths and separation of pallilysin orthologous sequences into well-defined clades on the phylogenetic tree demonstrates that, with the exception of the rabbit venereal syphilis-causing T. paraluiscuniculi ortholog, the most conserved region of T. pallidum pallilysin (L126-Y224) has diverged significantly from its most closely related orthologs in less invasive treponemes throughout the course of evolution.
FIG 4.
Phylogenetic analysis of Treponema pallilysin orthologs. Phylogenetic analysis of the treponemal pallilysin orthologs corresponding to the C terminus of pallilysin (L126-Y224). The optimal phylogenetic tree is drawn to scale, with the percentage of replicate trees in which the associated taxa clustered together shown next to the branches. Branch lengths are in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA5. The “root” position is based upon species-level relationships determined from 16S rRNA sequences.
Unlike T. pallidum Tp0750 and pallilysin, orthologs from less invasive treponemes do not bind or degrade host proteins.
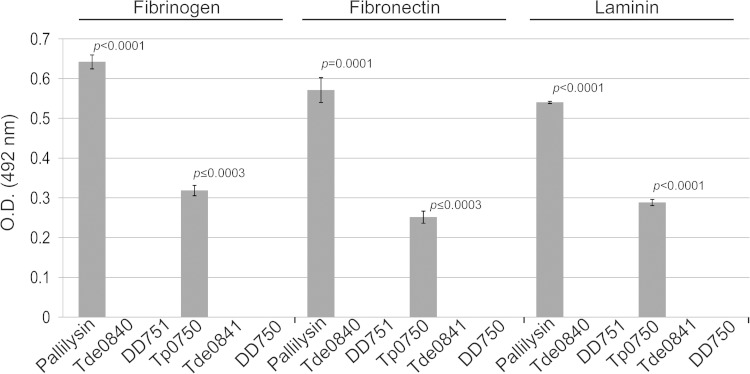
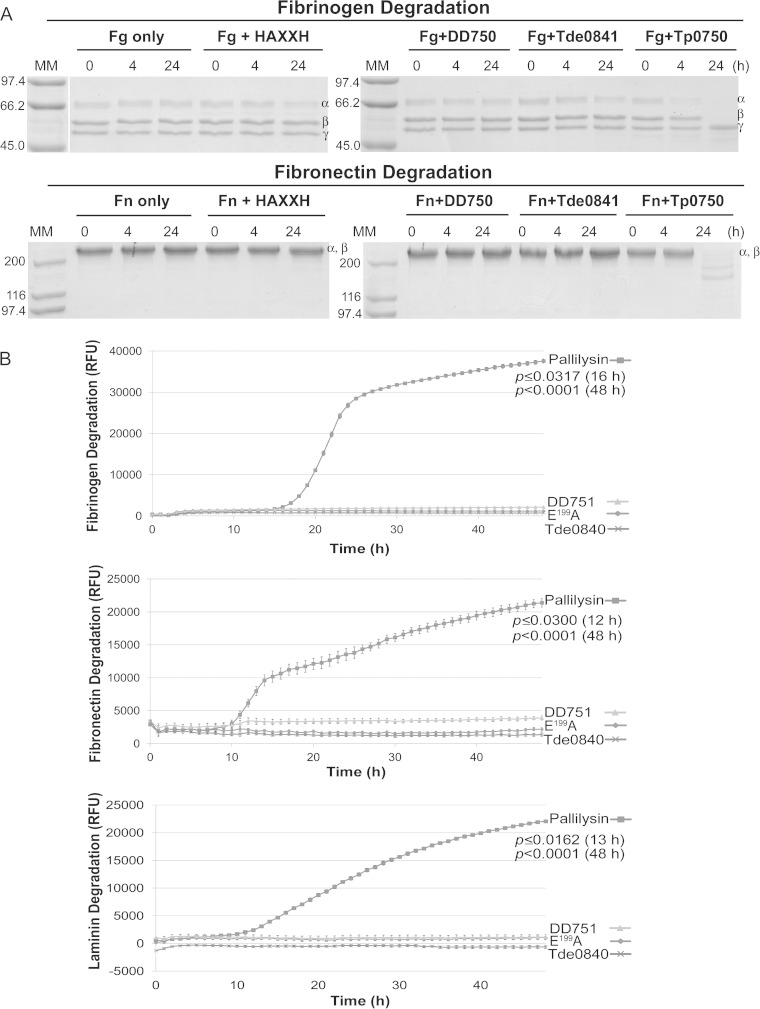
It has been demonstrated that Tp0750 and pallilysin bind and degrade host proteins and therefore are predicted to be important in promoting T. pallidum tissue invasion and dissemination during infection (23, 24). In order to experimentally compare the binding and proteolytic capabilities of Tp0750 and pallilysin with orthologs from less invasive treponemes, the orthologs from T. phagedenis (strain 4A; DD750 and DD751) and T. denticola (Tde0841 and Tde0840) were selected. Binding studies performed with Tp0750, pallilysin, and their orthologs showed a lack of binding of the orthologs to the host components fibrinogen, fibronectin, and laminin (Fig. 5).
FIG 5.
Tp0750 and pallilysin, but not orthologs from T. denticola and T. phagedenis strain 4A, bind host proteins. Binding assays were performed to compare attachment of 2 μg of each of recombinant Tp0750, pallilysin, Tde0841, DD750, Tde0840, and DD751 to 0.5 μg of each of immobilized fibrinogen, fibronectin, and laminin. Average readings of triplicate wells are presented with bars indicating standard error, and the results are representative of three independent experiments. For statistical analyses, attachment to each host protein by Tp0750 and pallilysin was compared to attachment by the corresponding orthologs using a Student two-tailed t test. Tp0750 and pallilysin exhibited statistically significant levels of binding to all three host proteins (P ≤ 0.0003) compared to the level of binding by the T. denticola and T. phagedenis orthologs.
Similarly, degradation assays showed a clear difference in degradation potential exists between the T. pallidum proteins and the corresponding orthologs from the less invasive treponemes. For Tp0750 and orthologs, purified recombinant proteins were incubated with the host proteins fibrinogen, fibronectin, or laminin for 24 h at 37°C and degradation was analyzed by SDS-PAGE (Fig. 6A). Tp0750 initiated degradation of the fibrinogen α-chain within 4 h postincubation, with complete degradation of the fibrinogen α- and β-chains by 24 h postincubation. Tp0750 also degraded the 220-kDa α- and β-chains of fibronectin (which migrate together in SDS-PAGE) between 4 and 24 h postincubation. DD750, Tde0841, and a negative-control protein (inactive pallilysin E199A) failed to degrade either host protein throughout the course of the 24 h experiments. Both fibrinogen and fibronectin were shown to be stable over 24 h when incubated at 37°C in the absence of recombinant proteins. None of the four recombinant proteins degraded laminin (data not shown).
FIG 6.
Tp0750 and pallilysin, but not orthologs from T. denticola and T. phagedenis strain 4A, degrade host proteins. (A) In vitro SDS-PAGE-based assays were performed to compare the host component-degrading capabilities of Tp0750, Tde0841, and DD750. The three recombinant proteins (40 μg each) and the negative-control protein (40 μg), were each incubated with fibrinogen (Fg; 60 μg) or fibronectin (Fn; 30 μg) at 37°C for 24 h. Samples were removed at 0, 4, and 24 h postincubation and analyzed for degradation of the three fibrinogen chains (α, β, and γ) or two comigrating fibronectin chains (α and β) by SDS-PAGE and Coomassie brilliant blue staining (6 μg loaded per lane in the absence of degradation). Numbers to the left of the lanes indicate the size (kDa) of the corresponding molecular mass (MM) markers. All recombinant proteins failed to degrade laminin (data not shown). (B) Fluorescence-based 96-well plate degradation assays were performed to compare the host component-degrading capabilities of pallilysin, Tde0840, and DD751. Recombinant proteins (1 μg per well) were added in triplicate to sterile 96-well plates and incubated with FITC-labeled fibrinogen, fibronectin, or laminin (10 μg per well) at 37 °C for 48 h in the dark. The degree of host component degradation was determined by measuring the increase in relative fluorescence units (RFU) every hour over 48 h using standard fluorescein excitation/emission filters. Average fluorescence intensity readings from triplicate measurements are presented with bars indicating the standard error (SE), and the results are representative of three independent experiments. For statistical analyses, host component degradation by wild-type pallilysin was compared to the highest fluorescence reading from either Tde0840, DD751, or Tp0751 E199A (negative control) using a Student two-tailed t test. Wild-type pallilysin exhibited a statistically significant level of fibrinogen (16 h postincubation; P ≤ 0.0317), fibronectin (12 h postincubation; P ≤ 0.0300), and laminin (13 h postincubation; P ≤ 0.0162) degradation compared to the levels exhibited by Tde0840, DD751, and the negative-control protein.
To compare the host protein-degrading capabilities of pallilysin with orthologs from T. phagedenis (DD751) and T. denticola (Tde0840), fluorescence-based proteolytic assays were performed. Purified recombinant proteins were preincubated at 37°C to allow for proteolytic activation. Recombinant proteins were then incubated with the FITC-labeled host proteins fibrinogen, fibronectin, or laminin for 48 h at 37°C in the dark. Nondigested FITC-labeled protein substrates remain quenched due to the close proximity of the FITC labels in the intact substrates. Conversely, digestion of FITC-labeled protein substrates results in dequenching and increased fluorescence which was measured over the course of the experiments to determine whether the pallilysin orthologs were capable of degrading the three host proteins. As shown in Fig. 6B, pallilysin exhibited a statistically significant level of cleavage of fibrinogen (16 h postincubation, P ≤ 0.0317; 48 h postincubation, P < 0.0001), fibronectin (12 h postincubation, P = 0.0300; 48 h postincubation, P < 0.0001), and laminin (13 h postincubation, P ≤ 0.0162; 48 h postincubation, P < 0.0001), compared to the degradation levels exhibited by DD751, Tde0840, and the negative-control recombinant protein (inactive pallilysin [E199A]). These results demonstrate that Tp0750 and pallilysin from T. pallidum, but not orthologs from two less invasive treponemes, are capable of degrading host proteins under the conditions used in our in vitro host component degradation assays.
DISCUSSION
In the present study, we demonstrated that Tp0750 is almost identical to the ortholog from the causative agent of rabbit venereal spirochetosis, T. paraluiscuniculi. Tp0750 was found to be less conserved, but still highly similar, among orthologs from less invasive pathogenic treponemes, including treponemes contributing to the polymicrobial infections human periodontitis (T. medium, T. vincentii, T. putidum, and T. denticola) and bovine digital dermatitis (T. phagedenis and T. pedis). Tp0750 orthologs were also identified in two nonpathogenic treponemes, namely, the termite hindgut and bovine rumen commensals, T. azotonutricium and T. bryantii, respectively. However, the Tp0750 orthologs from these two noninvasive symbionts were found to exhibit low amino acid identities compared to orthologs from pathogenic treponemes. We also showed that almost 50% of treponemes whose genomes have been sequenced do not possess a Tp0750 ortholog. These Treponema species included (i) pathogens associated with periodontal infections (T. socranskii, T. maltophilum, and T. lecithinolyticum), (ii) a pathogen associated with bovine digital dermatitis (T. brennaborense), (iii) symbionts from bovine rumen (T. saccharophilum), swine intestine (T. succinifaciens), and termite hindgut (T. primitia), and (iv) a free-living thermophile (T. caldaria). Therefore, ∼70% (9/13) of pathogenic treponemes were found to contain a highly conserved Tp0750 ortholog. Conversely, only ∼33% (2/6) of nonpathogenic treponemes were found to possess a Tp0750 ortholog with low amino acid identity. These findings infer a positive correlation between Tp0750 relatedness and treponemal invasive capacity.
Interestingly, we also found low identity Tp0750 orthologs in four nonpathogenic, non-Treponema species, namely, Spirochaeta smaragdinae, Spirochaeta bajacaliforniensis, Spirochaeta cellobiosiphila, and Leptonema illini. These findings, together with the high conservation of Tp0750 orthologs in the majority of pathogenic treponemes and low conservation or absence in nonpathogenic treponemes, suggest Tp0750 orthologs are either in the process of being lost or have been lost in the nonpathogenic treponemes, as opposed to having been gained in the pathogenic treponemes, during the course of spirochetal evolution.
All Tp0750 orthologs from pathogenic treponemes were also predicted to possess MIDAS-containing vWFA domains. These domains are often involved in mediating protein-protein interactions and are frequently found in cell adhesion proteins (such as integrins), plasma proteins, and extracellular matrix-binding proteins (41). The MIDAS motif coordinates a divalent metal cation that stabilizes protein-protein interactions (41, 42). Consistent with MIDAS-containing vWFA domains, it was previously demonstrated that Tp0750 coordinates calcium (24). The calcium-coordinating MIDAS-containing vWFA domain may be involved in mediating interactions of Tp0750 with host components, including the previously observed interactions of Tp0750 with the coagulation protein fibrinogen and the fibrinolytic protein annexin A2 (24). The high level of amino acid conservation and the presence of potential protein interaction domains suggested that Tp0750 orthologs in treponemes that primarily cause localized infections may similarly function as adhesins. Although binding assays demonstrated a lack of binding capability to the substrates tested (fibronectin, fibrinogen, and laminin), the possibility exists that the Tp0750 orthologs selectively bind to alternative substrates that were not tested in the present study. MIDAS-containing and MIDAS-free vWFA domains were also predicted to be present in the low amino acid identity orthologs from the bovine rumen symbiont, T. bryantii, and the termite hindgut commensal, T. azotonutricium, respectively. These findings suggest the T. bryantii ortholog, but not the T. azotonutricium ortholog, may also have a protein-protein interaction function.
The N and C termini of Tp0750 share low amino acid conservation with the treponemal orthologs. Unlike Tp0750 and orthologs from the pathogens T. paraluiscuniculi and T. vincentii, and the nonpathogen T. bryantii, which are predicted to contain SpI cleavage sites, bioinformatic analysis in the present study failed to predict signal peptides in the N termini of the remaining orthologs. It is well established that subcellular localization is an essential aspect of protein function and that protein localization and function are codependent (43). The potential differential subcellular localizations that we predicted here may differentiate ortholog function by regulating access to their respective colocalized molecular interaction partners during infection. In addition, we identified a proline-rich C-terminal region in Tp0750 and in the T. paraluiscuniculi ortholog (R195-D223) that was absent in all other orthologs. The function of this region remains to be defined; however, the unique biophysical properties of proline residues contained within proline-rich regions (PRRs) have been demonstrated to be important in mediating protein-protein interactions in a wide array of biological systems (44, 45). In addition to the potential vWFA-mediated protein interaction function common to all Tp0750 orthologs, it is possible that the unique C-terminal PRR of Tp0750 in T. pallidum and T. paraluiscuniculi provides these two systemic pathogens with additional host-interacting adhesive capabilities that are absent in less invasive pathogens and nonpathogens.
Unlike Tp0750, bioinformatic analysis in the present study indicated that pallilysin orthologs are found in all treponemes whose genomes have been sequenced and, with the exception of the T. paraluiscuniculi ortholog, amino acid conservation of the full-length protein among treponemes is very low. Using a phylogenetic approach, we assessed the evolutionary relationship among treponemal pallilysin orthologs. This analysis demonstrated that the T. pallidum pallilysin sequence used to generate the tree is identical to the T. paraluiscuniculi orthologous sequence. This result is consistent with a comparative genomics study that demonstrated identical gene orders in the genomes of T. pallidum and T. paraluiscuniculi and a genetic identity of >99% in conserved regions (46). Similar to T. pallidum, infection with T. paraluiscuniculi results in tissue invasion and systemic disease, with treponemes capable of disseminating from the initial site of infection (genitals) to distal sites, including eyelids, nose, lips, paws, and lymph nodes (47, 48).
Phylogenetic analysis inferred that orthologous and identical sequences from T. phagedenis isolates are the next most closely related orthologs to T. pallidum. Although T. phagedenis was originally isolated from a human lesion, it is now considered to be a commensal of the urogenital tract of humans (49, 50). In contrast, T. phagedenis isolates from cattle are considered pathogenic and are associated with digital dermatitis, an ulcerative disease of cattle that primarily affects the skin on the bulbs of the heel but can also result in lesions within the interdigital cleft of the hooves (38, 51). Studies have shown that T. phagedenis migrates intercellularly deep within the epidermis and, if left untreated, digital dermatitis lesions eventually affect the whole hoof (52, 53). Recent biochemical, molecular, and physiological characterizations, together with genome-wide comparisons of T. phagedenis strain F0421 and bovine T. phagedenis isolates (including strain 4A), indicated that the two spirochetes are almost identical in terms of biochemistry and genetics, leading to the conclusion that these bacteria belong to the same species and that the current description of T. phagedenis should be broadened to include both commensal strains of humans and pathogenic strains in cattle (54).
Treponema pallidum pallilysin was estimated to be more distantly related to orthologs from the human oral pathogens T. vincentii, T. medium, T. denticola, T. lecithinolyticum, and T. socranskii, the bovine and porcine pathogen T. pedis, the termite hindgut symbiont T. primitia, the bovine commensal T. saccharophilum, and the swine intestinal commensal T. succinifaciens. The human oral pathogenic treponemes are usually associated with localized infections, primarily affecting the periodontium (55). However, treponemal species are also present, albeit in low numbers, in the oral cavity of healthy individuals and thus represent members of the normal oral microbiota (55). Thus, intriguingly, increased pallilysin evolutionary conservation positively correlates with enhanced treponemal dissemination capacity. It should also be noted that in our scaled phylogenetic tree, branch lengths are proportional to the evolutionary changes that are estimated to have occurred between nodes. The extended length of the T. pallidum/T. paraluiscuniculi branch in the current phylogenetic analysis demonstrates that, even within this phylogenetically distinct genus, the T. pallidum/T. paraluiscuniculi pallilysin sequence has diverged significantly from less invasive treponemal orthologs.
The expression of host-interacting adhesins/proteases by pathogenic bacteria is an important virulence mechanism that promotes host attachment, host protein degradation, tissue destruction, pathogen invasion, and dissemination of infection (56–58). To date, the only host component binding/degrading adhesins that have been identified in T. pallidum are Tp0750 and pallilysin (23–25). In the present study, we compared the adhesive/proteolytic capability of these two proteins with orthologs from the less invasive treponemes, T. denticola and T. phagedenis strain 4A, using in vitro host substrate binding and degradation assays. Tp0750 bound to fibronectin, fibrinogen, and laminin but degraded only fibrinogen and fibronectin, while pallilysin similarly bound to all three host components and exhibited proteolytic activity against all three. In contrast, binding to and degradation of laminin, fibrinogen, and fibronectin by the orthologs from the less invasive treponemes was not detected under the physical conditions utilized in our in vitro assays.
We also showed here that the predicted molecular masses of pallilysin orthologs from treponemes that are not strictly host associated to be at least 2-fold greater than pallilysin from T. pallidum and T. paraluiscuniculi. Bioinformatic analyses in the current study failed to provide insight into the potential function(s) of the extra N- and C-terminal regions in the larger pallilysin orthologs. It is possible that these extra protein regions perform ancestral functions in the less invasive treponemes. Due to the complex and rich intercellular host environments encountered by the strictly host-associated and highly invasive pathogenic treponemes during infection, these additional domains may have been rendered functionally redundant or unnecessary, resulting in loss during the course of treponemal evolution. Obligate pathogenic bacteria, such as T. pallidum, frequently undergo genomic reduction (59, 60) and compensate for this genetic loss by encoding “multitasking” proteins, thereby increasing protein functional complexity compared to orthologous proteins from bacteria that are not strictly host-dependent (59, 61). The gain of an adhesion/protease-mediated function in the T. pallidum Tp0750 and pallilysin orthologs is consistent with increased protein functional complexity resulting from evolutionary genomic reduction and an obligate pathogenic lifestyle. In support of this, bioinformatic analyses predict that only pallilysin and the T. paraluiscuniculi and T. lecithinolyticum orthologs possess a predicted SpII cleavable signal sequence, indicating the potential for surface exposure of these proteins.
Isoelectric point analyses indicated that the isoelectric points of Tp0750 (9.1) and pallilysin (7.0) from T. pallidum and T. paraluiscuniculi differ greatly from most of the corresponding treponemal orthologs (6/11 Tp0750 ortholog pIs range from 5.1 to 6.2; 11/13 pallilysin ortholog pIs range from 4.9 to 6.1). Differences in the abundance of acidic and basic amino acids between the T. pallidum/T. paraluiscuniculi proteins and orthologs from less invasive treponemes may have arisen during adaptation to different subcellular localizations and/or habitats. This finding provides further support to the concept that the orthologs from the two highly invasive pathogens, T. pallidum and T. paraluiscuniculi, have diverged significantly from the less invasive treponemal orthologs during the course of evolution.
Fibrinogen is a key structural protein from the coagulation cascade and the primary component of fibrin clots. Fibronectin is a multifunctional extracellular matrix glycoprotein that plays a role in clot stabilization. It has been well established that clot formation is an important host defense process which facilitates bacterial containment (62–67). Laminin is an abundant glycoprotein component of both the blood-brain barrier and the basement membranes underlying endothelial cells, barriers which T. pallidum must penetrate during infection. After infection, T. pallidum invades the tissue barrier and rapidly disseminates to distant sites via the bloodstream. The ability of the T. pallidum Tp0750 and pallilysin orthologs to degrade host components may contribute to the ability of the bacterium to escape host-mediated containment and disseminate via the bloodstream and may partially account for the difference in the invasiveness of the treponemal species.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sheila Lukehart, University of Washington, for supplying T. denticola strain ATCC 35405 and the U.S. Department of Agriculture National Animal Disease Center for supplying the T. phagedenis strain 4A.
This study was funded by Public Health Service grant AI-051334 from the National Institutes of Health, and partial support was provided by awards from the Canada Foundation for Innovation, the Michael Smith Foundation for Health Research, the British Columbia Knowledge Development Fund, and Swedish Research Council Formas grant 211-2012-1612. C.E.C. is a Canada Research Chair in Molecular Pathogenesis and a Michael Smith Foundation for Health Research Scholar.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00643-15.
REFERENCES
- 1.World Health Organization. 2011. Prevalence and incidence of selected sexually transmitted infections Chlamydia trachomatis, Neisseria gonorrhoeae, syphilis, and Trichomonas vaginalis: methods and results used by WHO to generate 2005 estimates. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Nusbaum MR, Wallace RR, Slatt LM, Kondrad EC. 2004. Sexually transmitted infections and increased risk of co-infection with human immunodeficiency virus. J Am Osteopath Assoc 104:527–535. [PubMed] [Google Scholar]
- 3.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La VC, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol 403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Watthey L, Weidman J, Smith HO, Venter JC. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 6.LaFond RE, Lukehart SA. 2006. Biological basis for syphilis. Clin Microbiol Rev 19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collart P, Franceschini P, Durel P. 1971. Experimental rabbit syphilis. Br J Vener Dis 47:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cumberland MC, Turner TB. 1949. Rate of multiplication of Treponema pallidum in normal and immune rabbits. Am J Syph Gonorrhea Vener Dis 33:201–212. [PubMed] [Google Scholar]
- 9.Lukehart SA, Hook EW, Baker-Zander SA, Collier AC, Critchlow CW, Handsfield HH. 1988. Invasion of the central nervous system by Treponema pallidum: implications for diagnosis and treatment. Ann Intern Med 109:855–862. doi: 10.7326/0003-4819-109-11-855. [DOI] [PubMed] [Google Scholar]
- 10.Raiziss GW, Severac M. 1937. Rapidity with which Spirochaeta pallida invades the bloodstream. Arch Dermatol Syphilol 35:1101–1109. doi: 10.1001/archderm.1937.01470240093008. [DOI] [Google Scholar]
- 11.Norris SJ. 1993. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema pallidum Polypeptide Research Group. Microbiol Rev 57:750–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centurion-Lara A, Giacani L, Godornes C, Molini BJ, Brinck RT, Lukehart SA. 2013. Fine analysis of genetic diversity of the tpr gene family among treponemal species, subspecies and strains. PLoS Negl Trop Dis 7:e2222. doi: 10.1371/journal.pntd.0002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacani L, Brandt SL, Puray-Chavez M, Reid TB, Godornes C, Molini BJ, Benzler M, Hartig JS, Lukehart SA, Centurion-Lara A. 2012. Comparative investigation of the genomic regions involved in antigenic variation of the TprK antigen among treponemal species, subspecies, and strains. J Bacteriol 194:4208–4225. doi: 10.1128/JB.00863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giacani L, Molini B, Godornes C, Barrett L, Van VW, Centurion-Lara A, Lukehart SA. 2007. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect Immun 75:104–112. doi: 10.1128/IAI.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giacani L, Molini BJ, Kim EY, Godornes BC, Leader BT, Tantalo LC, Centurion-Lara A, Lukehart SA. 2010. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J Immunol 184:3822–3829. doi: 10.4049/jimmunol.0902788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaFond RE, Centurion-Lara A, Godornes C, Van Voorhis WC, Lukehart SA. 2006. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain Infect Immun 74:1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaFond RE, Molini BJ, Van Voorhis WC, Lukehart SA. 2006. Antigenic variation of TprK V regions abrogates specific antibody binding in syphilis. Infect Immun 74:6244–6251. doi: 10.1128/IAI.00827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron CE. 2005. T. pallidum outer membrane and outer membrane proteins, p 237–266. In Radolf JD, Lukehart SA (ed), Pathogenic Treponema: molecular and cellular biology. Caister Academic Press, Norfolk, England. [Google Scholar]
- 19.Radolf JD. 1995. Treponema pallidum and the quest for outer membrane proteins. Mol Microbiol 16:1067–1073. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- 20.Brinkman MB, McGill MA, Pettersson J, Rogers A, Matejkova P, Smajs D, Weinstock GM, Norris SJ, Palzkill T. 2008. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect Immun 76:1848–1857. doi: 10.1128/IAI.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron CE. 2003. Identification of a Treponema pallidum laminin-binding protein. Infect Immun 71:2525–2533. doi: 10.1128/IAI.71.5.2525-2533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron CE, Brown EL, Kuroiwa JMY, Schnapp LM, Brouwer NL. 2004. Treponema pallidum fibronectin-binding proteins. J Bacteriol 186:7019–7022. doi: 10.1128/JB.186.20.7019-7022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houston S, Hof R, Francescutti T, Hawkes A, Boulanger MJ, Cameron CE. 2011. Bifunctional role of the Treponema pallidum extracellular matrix binding adhesin Tp0751. Infect Immun 79:1386–1398. doi: 10.1128/IAI.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houston S, Russell S, Hof R, Roberts AK, Cullen P, Irvine K, Smith DS, Borchers CH, Tonkin ML, Boulanger MJ, Cameron CE. 2014. The multifunctional role of the pallilysin-associated Treponema pallidum protein, Tp0750, in promoting fibrinolysis and extracellular matrix component degradation. Mol Microbiol 91:618–634. doi: 10.1111/mmi.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houston S, Hof R, Honeyman L, Hassler J, Cameron CE. 2012. Activation and proteolytic activity of the Treponema pallidum metalloprotease, pallilysin. PLoS Pathog 8:e1002822. doi: 10.1371/journal.ppat.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95–98. [Google Scholar]
- 27.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Weese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen TN, Brunak S, Von HG, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 29.Juncker AS, Willenbrock H, Von HG, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 32.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- 33.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum-parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilburn TG, Kim KS, Ostrom NE, Byzek KR, Leadbetter JR, Breznak JA. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495–2498. doi: 10.1126/science.1060281. [DOI] [PubMed] [Google Scholar]
- 36.Lukehart SA, Marra CM. 2007. Isolation and laboratory maintenance of Treponema pallidum. Curr Protoc Microbiol Chapter 12:2A.1 doi: 10.1002/9780471729259.mc12a01s7. [DOI] [PubMed] [Google Scholar]
- 37.Ohta K, Makinen KK, Loesche WJ. 1986. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun 53:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott DJ, Moeller MR, Zuerner RL, Goff JP, Waters WR, Alt DP, Walker RL, Wannemuehler MJ. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J Clin Microbiol 41:2522–2529. doi: 10.1128/JCM.41.6.2522-2529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smibert RM. 1991. Anaerobic spirochetes, p 572–578. In Balows A, Hausler WJ Jr., Herrmann KL, Isenberg HD, Shadomy HJ (ed), Manual of clinical microbiology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 40.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 41.Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craig D, Gao M, Schulten K, Vogel V. 2004. Structural insights into how the MIDAS ion stabilizes integrin binding to an RGD peptide under force. Structure 12:2049–2058. doi: 10.1016/j.str.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 43.Gardy JL, Brinkman FS. 2006. Methods for predicting bacterial protein subcellular localization. Nat Rev Microbiol 4:741–751. doi: 10.1038/nrmicro1494. [DOI] [PubMed] [Google Scholar]
- 44.Kay BK, Williamson MP, Sudol M. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14:231–241. [PubMed] [Google Scholar]
- 45.Tompa P. 2002. Intrinsically unstructured proteins. Trends Biochem Sci 27:527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 46.Smajs D, Zobanikova M, Strouhal M, Cejkova D, Dugan-Rocha S, Pospisilova P, Norris SJ, Albert T, Qin X, Hallsworth-Pepin K, Buhay C, Muzny DM, Chen L, Gibbs RA, Weinstock GM. 2011. Complete genome sequence of Treponema paraluiscuniculi, strain Cuniculi A: the loss of infectivity to humans is associated with genome decay. PLoS One 6:e20415. doi: 10.1371/journal.pone.0020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiGiacomo RF, Lukehart SA, Talburt CD, Baker-Zander SA, Giddens WE Jr, Condon J, Brown CW. 1985. Chronicity of infection with Treponema paraluis-cuniculi in New Zealand White rabbits. Genitourin Med 61:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noguchi H. 1922. Venereal spirochetosis in American rabbits. J Exp Med 35:391–407. doi: 10.1084/jem.35.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paster BJ. 2010. Phylum XV: spirochaetes Garrity and Holt 2001, p 471–566. In Parte AC. (ed), Bergey's manual of systematic bacteriology, vol 4 Springer, New York, NY. [Google Scholar]
- 50.Wallace AL, Harris A. 1967. Reiter treponeme: a review of the literature. Bull World Health Organ 36:5–7. [PMC free article] [PubMed] [Google Scholar]
- 51.Walker RL, Read DH, Loretz KJ, Nordhausen RW. 1995. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet Microbiol 47:343–355. [DOI] [PubMed] [Google Scholar]
- 52.Blowey RW, Done SH, Cooley W. 1994. Observations on the pathogenesis of digital dermatitis in cattle. Vet Rec 135:115–117. [DOI] [PubMed] [Google Scholar]
- 53.Moter A, Leist G, Rudolph R, Schrank K, Choi BK, Wagner M, Gobel UB. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459–2467. [DOI] [PubMed] [Google Scholar]
- 54.Wilson-Welder JH, Elliott MK, Zuerner RL, Bayles DO, Alt DP, Stanton TB. 2013. Biochemical and molecular characterization of Treponema phagedenis-like spirochetes isolated from a bovine digital dermatitis lesion. BMC Microbiol 13:280. doi: 10.1186/1471-2180-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dashper SG, Seers CA, Tan KH, Reynolds EC. 2011. Virulence factors of the oral spirochete Treponema denticola. J Dent Res 90:691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh A, Saha DR, Hoque KM, Asakuna M, Yamasaki S, Koley H, Das SS, Chakrabarti MK, Pal A. 2006. Enterotoxigenicity of mature 45-kilodalton and processed 35-kilodalton forms of hemagglutinin protease purified from a cholera toxin gene-negative Vibrio cholerae non-O1, non-O139 strain. Infect Immun 74:2937–2946. doi: 10.1128/IAI.74.5.2937-2946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyoshi S, Shinoda S. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect 2:91–98. doi: 10.1016/S1286-4579(00)00280-X. [DOI] [PubMed] [Google Scholar]
- 58.Moffat JF, Edelstein PH, Regula DP Jr, Cirillo JD, Tompkins LS. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol Microbiol 12:693–705. [DOI] [PubMed] [Google Scholar]
- 59.Kelkar YD, Ochman H. 2013. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193:303–307. doi: 10.1534/genetics.112.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 61.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delvaeye M, Conway EM. 2009. Coagulation and innate immune responses: can we view them separately? Blood 114:2367–2374. doi: 10.1182/blood-2009-05-199208. [DOI] [PubMed] [Google Scholar]
- 63.Levi M, Schultz M, van der Poll T. 2010. Disseminated intravascular coagulation in infectious disease. Semin Thromb Hemost 36:367–377. doi: 10.1055/s-0030-1254046. [DOI] [PubMed] [Google Scholar]
- 64.Levi M, van der Poll T, Buller HR. 2004. Bidirectional relation between inflammation and coagulation. Circulation 109:2698–2704. doi: 10.1161/01.CIR.0000131660.51520.9A. [DOI] [PubMed] [Google Scholar]
- 65.Loof TG, Morgelin M, Johansson L, Oehmcke S, Olin AI, Dickneite G, Norrby-Teglund A, Theopold U, Herwald H. 2011. Coagulation, an ancestral serine protease cascade, exerts a novel function in early immune defense. Blood 118:2589–2598. doi: 10.1182/blood-2011-02-337568. [DOI] [PubMed] [Google Scholar]
- 66.Petaja J. 2011. Inflammation and coagulation. An overview. Thromb Res 127(Suppl 2):S34–S37. doi: 10.1016/S0049-3848(10)70153-5. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, Loseva O, Morgelin M, Ikle J, Cripps RM, Herwald H, Theopold U. 2010. Pathogen entrapment by transglutaminase: a conserved early innate immune mechanism. PLoS Pathog 6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.