Abstract
Angiopoietin 1 (Angpt1) and angiopoietin 2 (Angpt2) are the ligands of tyrosine kinase (Tie) receptors, and they play important roles in vessel formation and the development of inflammatory diseases, such as atherosclerosis. Porphyromonas gingivalis is a Gram-negative periodontal bacterium that is thought to contribute to the progression of cardiovascular disease. The aim of this study was to investigate the role of P. gingivalis infection in the modulation of Angpt1 and Angpt2 in human aortic smooth muscle cells (AoSMCs). We exposed AoSMCs to wild-type (W50 and 381), gingipain mutant (E8 and K1A), and fimbrial mutant (DPG-3 and KRX-178) P. gingivalis strains and to different concentrations of tumor necrosis factor (TNF). The atherosclerosis risk factor TNF was used as a positive control in this study. We found that P. gingivalis (wild type, K1A, DPG3, and KRX178) and TNF upregulated the expression of Angpt2 and its transcription factor ETS1, respectively, in AoSMCs. In contrast, Angpt1 was inhibited by P. gingivalis and TNF. However, the RgpAB mutant E8 had no effect on the expression of Angpt1, Angpt2, or ETS1 in AoSMCs. The results also showed that ETS1 is critical for P. gingivalis induction of Angpt2. Exposure to Angpt2 protein enhanced the migration of AoSMCs but had no effect on proliferation. This study demonstrates that gingipains are crucial to the ability of P. gingivalis to markedly increase the expressed Angpt2/Angpt1 ratio in AoSMCs, which determines the regulatory role of angiopoietins in angiogenesis and their involvement in the development of atherosclerosis. These findings further support the association between periodontitis and cardiovascular disease.
INTRODUCTION
Cardiovascular atherosclerotic disease is a major cause of global morbidity and mortality. The pathological characteristics of atherosclerosis include inflammation, proteolysis, and arterial remodeling processes, such as apoptosis and angiogenesis (1). Angiogenesis is a prominent feature of atherosclerosis and refers to the growth of new blood vessels, following an organized genetic program of vascular sprouting, vessel assembly, and organotypic maturation (2). Tie receptors, together with their corresponding angiopoietins, have been identified as the signaling system that plays a particularly central role in vascular remodeling and angiogenesis (3).
There are 4 forms of angiopoietin (Angpt1, Angpt2, Angpt3, and Angpt4) that are produced by many different types of cells (4, 5). Through the activation of the Tie2 receptor signaling pathway, angiopoietins regulate vascular maturation, stability, and remodeling (6). Angpt1 and Angpt2 have opposing effects in the regulation of inflammatory responses. Angpt1 is an anti-inflammatory regulator, whereas Angpt2 functions as a proinflammatory regulator. Transgenically overexpressed Angpt1 in mice results in reduced vessel leakiness in response to permeability-inducing inflammatory agents (7). Mice without Angpt2 fail to elicit a rapid inflammatory response and to produce molecules that lead to leukocyte adherence during stimulation by an inflammatory mediator, such as tumor necrosis factor (TNF) (8). In addition to binding to Tie2, an endothelium-specific receptor, Angpt2 is also able to bind to and activate integrins in Tie receptor-negative fibroblasts and breast cancer cells (9, 10). These findings triggered a new-found interest in analyzing the function of angiopoietins in non-endothelial cells.
Porphyromonas gingivalis, a Gram-negative oral anaerobe, has been identified as one of the main pathogens in the progression of periodontitis and is detected in up to 85% of disease sites (11). By interacting with other periodontal pathogens, such as Filifactor alocis, a Gram-positive, obligate anaerobic rod bacterium, P. gingivalis plays an important role in infection-induced periodontal disease (12, 13). Many epidemiological studies have shown that severe forms of periodontitis are associated with other inflammatory diseases, such as rheumatoid arthritis and cardiovascular disease (14, 15). The DNA of P. gingivalis has been found in coronary stenotic artery plaques of myocardial infarction patients (16, 17). In animal models, P. gingivalis infection directly induces and accelerates the formation of coronary and aortic atherosclerosis in pigs and mice (18, 19).
P. gingivalis produces a number of different virulence factors, such as lipopolysaccharides (LPS), fimbriae, capsule, hemagglutinins, and proteases (gingipains). Gingipains are cysteine proteinases, and they have been divided into two groups: arginine gingipains (Rgp), which include RgpA and RgpB, and lysine gingipain (Kgp) (20). The gingipains have been shown to support biofilm formation, facilitate P. gingivalis invasion, and regulate the defensive response processes of host cells (21). We and others have demonstrated that gingipains modulate the expression of several cytokines in multiple cell types, including endothelial cells, gingival fibroblasts, T cells, and monocytes (22–25). In addition to gingipains, fimbriae are also important for P. gingivalis adhesion to host cells. Many studies have shown that P. gingivalis requires fimbriae to invade endothelial cells (26) and fibroblasts (27) and to induce inflammatory responses (28, 29).
Interestingly, we have previously shown, using microarray techniques, that P. gingivalis ATCC 33277 downregulates the gene expression of Angpt1 while simultaneously upregulating the gene expression of Angpt2 in human aortic smooth muscle cells (AoSMCs) (30). Smooth muscle cells (SMCs) are the main components of the vascular wall, and dysfunction in these cells is directly or indirectly associated with the development of atherosclerosis (31). To further investigate the roles of different virulence factors of P. gingivalis in the modulation of angiopoietins, we infected AoSMCs with wild-type (ATCC 33277, W50, and 381), gingipain mutant (K1A and E8), or fimbrial mutant (DPG3 and KRX178) strains of P. gingivalis.
The aim of this study was to clarify the effects of P. gingivalis in regulating the expression of angiopoietins in AoSMCs.
MATERIALS AND METHODS
Culture of AoSMCs.
Human primary AoSMCs (Invitrogen, Stockholm, Sweden) were cultured in 231 smooth muscle cell culture medium containing essential growth supplements (Gibco, Carlsbad, CA). The cells were cultured in 75-cm2 explant culture flasks (TPP, Trasadingen, Switzerland) and placed in a cell culture incubator at 37°C with 5% CO2 and 95% air until confluent. In this study, cells from passage 5 to 10 were used.
Bacterial culture and preparation.
F. alocis ATCC 35896 (CCUG Culture Collection, University of Göteborg) and P. gingivalis ATCC 33277 (American Type Culture Collection, Manassas, VA), W50 (wild type) and its isogenic mutant strains E8 (Rgp mutant strain) and K1A (Kgp mutant), and 381 (wild type) were grown in fastidious anaerobe broth (29.7 g/liter, pH 7.2). The P. gingivalis 381 corresponding fimbrial mutant strains DPG3 (major fimbria mutant) and KRX178 (minor fimbria mutant) were grown in fastidious anaerobe broth supplemented with 1 μg/ml erythromycin. The different P. gingivalis strains were grown in an anaerobic chamber (80% N2, 10% CO2, and 10% H2 at 37°C) (Concept 400 anaerobic workstation; Ruskinn Technology Ltd., Leeds, United Kingdom). After 72 h of culturing, bacteria were harvested by centrifugation for 10 min at 10,000 rpm and then washed and resuspended in Krebs-Ringer-glucose (KRG) buffer (120 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 1.7 mM KH2PO4, 8.3 mM Na2HPO4, 10 mM glucose, and 1.1 mM CaCl2, PH 7.3).
The concentration of P. gingivalis was determined by counting the CFU of different dilutions of bacteria on blood agar after 5 to 7 days. The optical density (OD) at 600 nm of the bacterial suspension was measured with a spectrophotometer (BioPhotometer plus; Eppendorf AG, Hamburg, Germany) and was correlated with the concentration (CFU/ml) of the bacteria.
Gingipain quantification.
The activities of arginine and lysine gingipains from different strains of P. gingivalis were quantified using arginine and lysine substrates (Peptanova, Sandhausen, Germany). The arginine gingipain substrate peptide sequence was Boc-Phe-Ser-Arg-AMC (t-butyloxycarbonyl-l-phenylalanyl-l-seryl-l-arginine-4-methylcoumaryl-7-amide), and the lysine gingipain substrate peptide sequence was Z-His-Glu-Lys-AMC (benzyloxycarbonyl-l-histidyl-l-glutamyl-l-lysine-4-methylcoumaryl-7-amide). Different strains of washed P. gingivalis (106 CFU) were incubated with either of the substrates at a final concentration of 100 μM for 1 h at 37°C, and the enzyme activity was registered in a fluorescence microplate reader (Fluostar Optima, Ortenberg, Germany) at excitation/emission wavelength settings of 380/460 nm.
Stimulation of AoSMCs with P. gingivalis, LPS, and TNF.
AoSMCs were seeded at a density of 150,000 cells per well in 6-well plates coated with type I collagen (Gibco, Carlsbad, CA), Thereafter, cells were serum starved for 24 h using Dulbecco modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) containing 0.5% fetal bovine serum (FBS) (Sigma, St. Louis, MO), 2 mM l-glutamine, and antibiotics (Gibco, Carlsbad, CA). After being washed and resuspended in fresh DMEM, AoSMCs were challenged with different strains of P. gingivalis at a multiplicity of infection (MOI) of 10 for 2, 8, 16, 24, or 48 h. Because F. alocis was found to coinfect with P. gingivalis, AoSMCs infected with F. alocis for 24 h served as a control in this study. For AoSMCs stimulated with the fimbrial mutants DPG3 and KRX178, AoSMCs treated with 1 μg/ml of erythromycin served as a control. To determine the role of arginine gingipains, P. gingivalis, ATCC 33277, W50, and K1A were incubated with 1 mM leupeptin (Roche Diagnostics Corporation, USA), which is an arginine gingipain inhibitor, for 1 h prior to stimulation of the AoSMCs. AoSMCs were also stimulated with 10 ng/ml or 50 ng/ml of TNF (Sigma-Aldrich, St. Louis, MO) or 10 ng/ml or 100 ng/ml of P. gingivalis LPS (InvivoGen, Toulouse, France) for 24 or 48 h.
Knockdown of ETS1 in AoSMCs.
Knockdown of ETS1 was performed by using human ETS1 small interfering RNA (siRNA) (Perkin-Elmer Applied Biosystems, Foster City, CA) in 6-well plates at 150,000 cells/well. A volume of 4 μl of Lipofectamine 2000 (Life Technologies, Carlsbad, CA) was added to 250 μl of Opti-MEM (Life Technologies, Carlsbad, CA), left for 5 min, and then mixed with 250 μl of Opti-MEM containing 50 pmol of ETS1 siRNA (VHS40614) (Life Technologies, Carlsbad, CA) or 50 pmol nontargeting siRNA (Life Technologies, Carlsbad, CA) as a control. After 20 min at room temperature, the transfection mixture was added to each cell culture well containing 500 ml of Opti-MEM, and cultures were incubated for 6 h. A volume of 1 ml of antibiotic-free growth medium was then added, and cultures were incubated for another 18 h. The cells were then starved for 24 h and treated with P. gingivalis W50 for 24 h.
Quantitative real-time PCR.
Isolation of RNA from AoSMCs was carried out using a Genejet RNA isolation kit (Fermentas, Sweden). cDNA were synthesized using equal amounts of RNA and high-capacity cDNA reverse transcription kits (Perkin-Elmer Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The primer sequences for Angpt1, Angpt2, ETS1, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Eurofins, Ebersberg, Germany) used in this study are listed in Table 1. Quantitative real-time PCR for SYBR green (Fermentas, Sweden) was performed with an ABI Prism 7900HT sequence analyzer. Relative quantification of gene expression was determined using the ΔΔCT method and normalized by the threshold cycle (CT) value of GAPDH.
TABLE 1.
Primer sequences for real-time PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Angpt1 | CAACAGTGTCCTTCAGAAGCAGC | CCAGCTTGATATACATCTGCACAG |
| Angpt2 | ATTCAGCGACGTGAGGATGGCA | GCACATAGCGTTGCTGATTAGTC |
| ETS1 | GAGTCAACCCAGCCTATCCAGA | GAGCGTCTGATAGGACTCTGTG |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
Western blot assay.
Proteins were extracted from AoSMCs that had been stimulated or not stimulated with F. alocis, different strains of P. gingivalis, or TNF for 24 h or 48 h, using radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, St. Louis, MO) mixed with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). After centrifugation at 10,000 rpm for 10 min, the total protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL, USA). An equal amount of each sample (30 μg) was electrophoresed on precast SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules). After blocking in 3% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, MO) for 1 h, membranes were probed overnight at 4°C with goat polyclonal anti-Angpt2 (R&D Systems, United Kingdom) at a concentration of 1 μg/ml. Rabbit polyclonal anti-GAPDH (Santa Cruz Biotechnology, Dallas, TX) at a 1:15,000 dilution was used as a loading control. For Angpt2, blots were incubated with anti-goat IgG (R&D Systems, United Kingdom) at a concentration of 1:3000 for 2 h. For GAPDH, blots were incubated with anti-rabbit IgG (Santa Cruz Biotechnology, Dallas, TX) at a concentration of 1:15,000 for 2 h. The blots were visualized using Luminata Forte Western horseradish peroxidase (HRP) substrate (Millipore, Darmstadt, Germany) and a ChemiDoc MP imager (Bio-Rad, Hercules, CA). Densitometric analysis was performed using Image Lab software (Bio-Rad, Hercules, CA).
ELISA.
Supernatants from AoSMCs that were challenged with P. gingivalis strain ATCC 33277, W50, E8, or K1A for 24 h or 48 h were collected and centrifuged at 1,500 × g for 5 min at 4°C. Thereafter, the supernatants were stored at −80°C until use. TNF was analyzed by enzyme-linked immunosorbent assay (ELISA) (BioLegend, San Diego, CA) according to the manufacturer's instructions.
Proliferation assay.
To investigate proliferation responses, serum-starved AoSMCs were incubated with different concentrations of recombinant human Angpt2 for 24 h, The medium was then replaced with medium containing 0.5% FBS and left for 24 h. The proliferation responses were monitored using an MTT [3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide] assay. MTT (Sigma-Aldrich, St. Louis, MO) was dissolved in cell culture DMEM at a concentration of 500 μg/ml. After different stimulations, the supernatant was aspirated and cells were washed twice with phosphate-buffered saline (PBS), and then 1 ml of MTT medium was added to each well of the plate. After 2 h of incubation at 37°C, the medium was removed and MTT was extracted from viable cells by adding 1 ml dimethyl sulfoxide (DMSO). Measurements of OD at 540 nm were then performed in a microtiter plate reader (SpectraMax 340 microplate reader; Molecular Devices Corp., Sunnyvale, CA).
Wound healing assay.
Studies of the regulation of AoSMC migration by Angpt2 were preformed using a wound healing assay. AoSMCs were seeded into 6-well culture plates and starved for 24 h in DMEM containing 0.5% FBS. The resulting single cell layer was then carefully wounded using a 100-μl pipette tip. Cells were washed twice with Dulbecco's PBS (DPBS) to remove cellular debris, and then 2 ml of DMEM containing 0.5% FBS was added to each well. AoSMCs were then treated with 10 ng/ml, 100 ng/ml, or 500 ng/ml of Angpt2 or DPBS as control. Wounds were photographed immediately (0 h) and 18 h after wounding with an Olympus inverted CKX41 phase-contrast microscope. Migration was evaluated by measuring the reduction in the area of the wound after migration of the cells into the cell-free zone with the NIH software package Image J (ImageJ 1.32j; NIH, Bethesda, MD).
Statistical analysis.
Data are expressed as the average ± standard error of the mean (SEM), with a P value of <0.05 considered to be significant. Student's t test was used for statistical comparisons of two groups, and one-way analysis of variance (ANOVA) with Bonferroni or Dunnett posttests was used for calculating the statistical significance between differences in data groups obtained from the real-time quantitative PCR, Western blot, proliferation, and wound healing experiments that have more than two groups. Statistical analysis was performed using GraphPad Prism software.
RESULTS
Gingipain activity of P. gingivalis.
In this study, we used gingipain substrates and a fluorescence assay to validate the activity of gingipains in different P. gingivalis strains. As expected, E8 showed only Kgp activity and K1A showed only Rgp activity. There was no clear difference in gingipain activity between the P. gingivalis strains ATCC 33277, W50, 381, DPG3, and KRX178. (see Fig. S1A and B in the supplemental material.)
P. gingivalis and its gingipains regulate Angpt1 and Angpt2 production in AoSMCs.
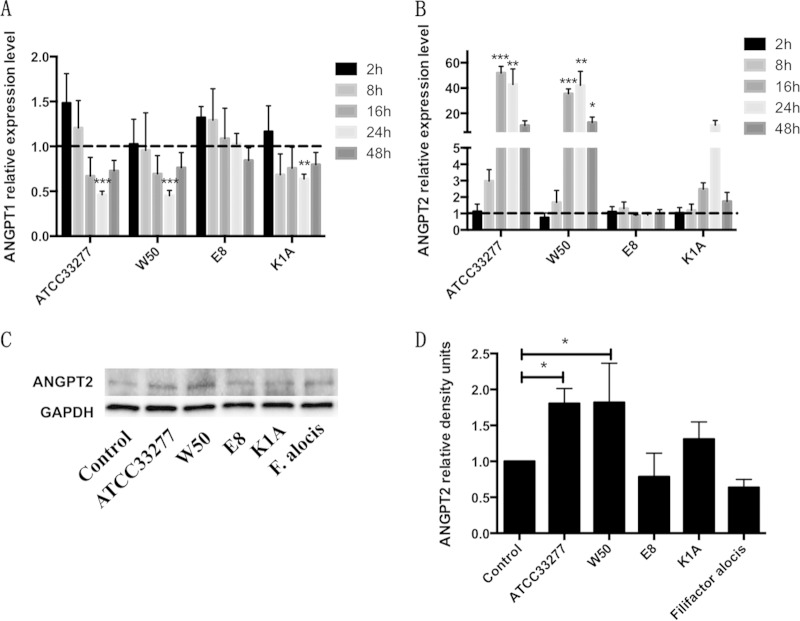
mRNA was collected from samples from each different group and assessed by quantitative real-time PCR for Angpt1 and Angpt2. The results were normalized against GAPDH. For Angpt1, the gene expression level was significantly inhibited by wild-type P. gingivalis ATCC 33277 and W50 after 24 h compared to unstimulated cells, which served as negative control. In contrast, the Rgp-deficient strain E8 had almost no effect on Angpt1 gene expression in AoSMCs. The Kgp-deficient strain K1A also significantly inhibited the expression of Angpt1 after 24 h, but to a lesser extent than the wild-type strains (Fig. 1A). Leupeptin only sparsely reversed the inhibitory effect of wild-type P. gingivalis on Angpt1, but it completely neutralized the inhibition of K1A (see Fig. S2A in the supplemental material).
FIG 1.
P. gingivalis and its gingipains regulate Angpt1 and Angpt2 expression in AoSMCs. (A and B) Quantitative real-time PCR results demonstrate relative transcription levels for Angpt1 (A) and Angpt2 (B) in AoSMCs stimulated with wild-type P. gingivalis (ATCC 33077 and W50), the Rgp mutant (E8), and the Kgp mutant (K1A) at an MOI of 10 for 2 h, 8 h, 16 h, 24 h, and 48 h. All results were normalized to the gene expression level of the GAPDH housekeeping gene. (C) Representative Western blot showing Angpt2 protein expression levels of AoSMCs exposed to different P. gingivalis strains and F. alocis at an MOI of 10 for 48 h. (D) Quantification of Angpt2 protein expression levels by densitometry for cells incubated for 48 h. Angpt2 density signals were normalized to GAPDH signal values. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. (A and B) n = 4 to 7; (C and D) n = 3.
The gene expression of Angpt2 was markedly increased by wild-type P. gingivalis ATCC 33277 after 16 h and 24 h and by W50 after 16 h, 24 h, and 48 h, whereas the Rgp-deficient strain E8 had no effect on the gene expression of Angpt2. K1A also increased the expression of Angpt2; however, the results were not significant (Fig. 1B). Inhibition of Rgp with leupeptin effectively antagonized the stimulatory effect of wild-type and K1A P. gingivalis on Angpt2 (see Fig. S2B in the supplemental material). We also infected AoSMCs with F. alocis, a different bacterial species often found to coinfect with P. gingivalis. However, no significant change was observed in Angpt1and Angpt2 gene expression compared to that of control samples after 24 h (see Fig. S3 in the supplemental material), which indicates that changes to Angpt1/2 are a result of P. gingivalis specifically.
Here, we focused on Angpt2 protein expression because the gene expression of Angpt2 was markedly induced by P. gingivalis. AoSMCs were infected with F. alocis or different strains of P. gingivalis at an MOI of 10 for 24 h and 48 h. The P. gingivalis ATCC 33277 strain significantly increased the Angpt2 protein expression in AoSMCs at 24 h after infection (see Fig. S4A and B in the supplemental material), whereas both ATCC 33277 and W50 significantly increased the Angpt2 protein level at 48 h after infection (Fig. 1C and D). As expected, the Western blot results for 48-h incubations with the bacteria followed the same trend that was observed for Angpt2 gene expression.
Fimbriae and LPS are not involved in P. gingivalis-mediated regulation of Angpt1 and Angpt2 production in AoSMCs.
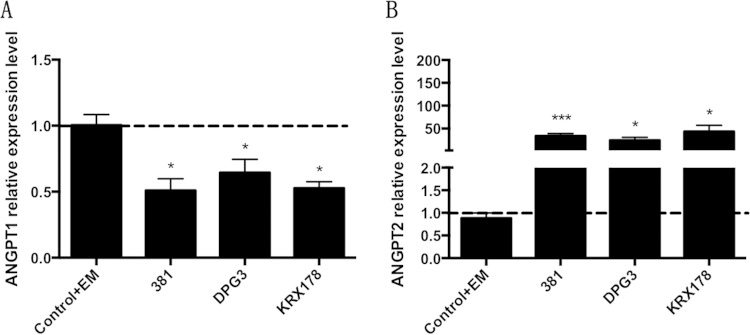
We further investigated the role of fimbriae of P. gingivalis in modulating Angpt1 and Angpt2 expression in AoSMCs. The wild-type strain P. gingivalis 381 and its corresponding major fimbria mutant DPG3 and minor fimbria mutant KRX178 significantly downregulated the gene expression of Angpt1 in AoSMCs after 24 h (Fig. 2A). All three of these strains of P. gingivalis significantly upregulated the gene expression of Angpt2 in AoSMCs after 24 h of incubation (Fig. 2B). We next assessed whether changes to Angpt1/2 were a result of LPS, considering that previous research has demonstrated that the quantity of LPS produced and released differs between various strains (25). Stimulation of AoSMCs with P. gingivalis LPS for 24 h or 48 h resulted in no significant changes in Angpt1 or Angpt2 expression level (see Fig. S5A and B in the supplemental material).
FIG 2.
Fimbriae are not involved in P. gingivalis-mediated regulation of Angpt1 and Angpt2 production in AoSMCs. Quantitative real-time PCR results demonstrate relative transcription levels for Angpt1 (A) and Angpt2 (B) in AoSMCs stimulated with wild-type P. gingivalis 381, the major fimbria mutant (DPG3), the minor fimbria mutant (KRX178) at an MOI of 10, or with medium containing 1 μg/ml of erythromycin (Control+EM) as a control for fimbrial mutants, for 24 h. All results were normalized to the gene expression level of the GAPDH housekeeping gene. Statistically significant differences for the wild-type P. gingivalis strain 381 are shown compared to the negative control. For fimbrial mutants, statistically significant differences are shown compared to group Control+EM. *, P < 0.05; ***, P < 0.005. (A) n = 7; (B) n = 10.
TNF regulates Angpt1 and Angpt2 production in AoSMCs.
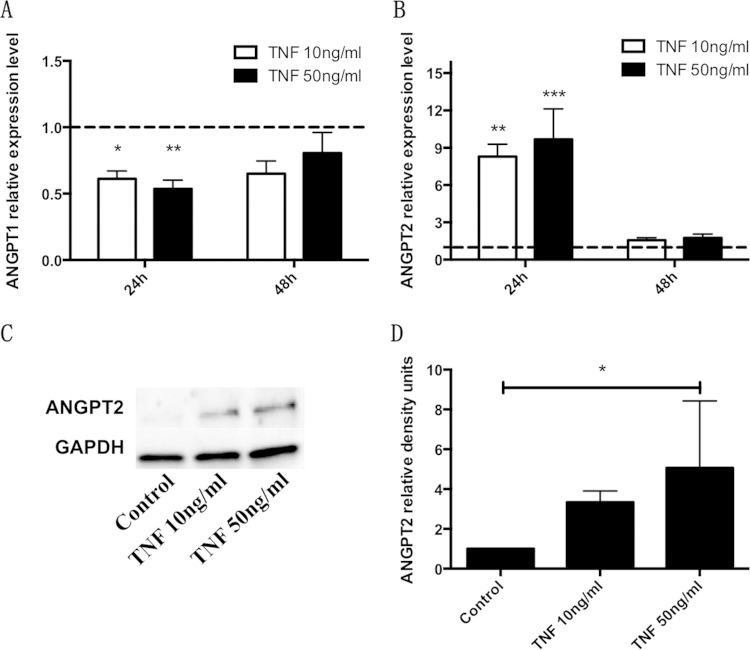
AoSMCs were stimulated with different concentration of TNF for 24 h and 48 h. Gene expression of Angpt1 was significantly reduced (1.64-fold and 1.87-fold) after stimulation for 24 h with 10 ng/ml and 50 ng/ml TNF, respectively (Fig. 3A). The levels of Angpt2 mRNA were significantly increased (8.30-fold and 9.67-fold) after stimulation for 24 h with 10 ng/ml and 50 ng/ml TNF, respectively. Similar to our results for the gene expression of Angpt1, we did not observe a significant change for Angpt2 mRNA expression levels after 48 h (Fig. 3B). With respect to protein expression, TNF stimulation induced Angpt2 after 24 h (see Fig. S4C and D in the supplemental material) and 48 h (Fig. 3C and D), with a significant upregulation induced by stimulation with 50 ng/ml for 48 h.
FIG 3.
TNF regulates Angpt1 and Angpt2 expression in AoSMCs. (A and B) Quantitative real-time PCR results demonstrate relative transcription levels for Angpt1 (A) and Angpt2 (B) of AoSMCs stimulated with 10 ng/ml or 50 ng/ml of TNF for 24 h or 48 h. All results were normalized to the gene expression level of the GAPDH housekeeping gene. (C) Representative Western blot showing Angpt2 protein expression levels in AoSMCs exposed to 10 ng/ml or 50 ng/ml of TNF for 48 h. (D) Quantification of Angpt2 protein expression levels by densitometry. Angpt2 density signals were normalized to GAPDH signal values. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. (A and B) n = 4; (C and D) n = 3.
The TNF ELISA kit detects human TNF levels from 7.8 pg/ml to 500 pg/ml. However, the TNF levels in the supernatants from AoSMCs treated with P. gingivalis strain ATCC 33277, W50, E8, or K1A were undetectable (data not shown).
P. gingivalis and TNF upregulate ETS1 in AoSMCs.
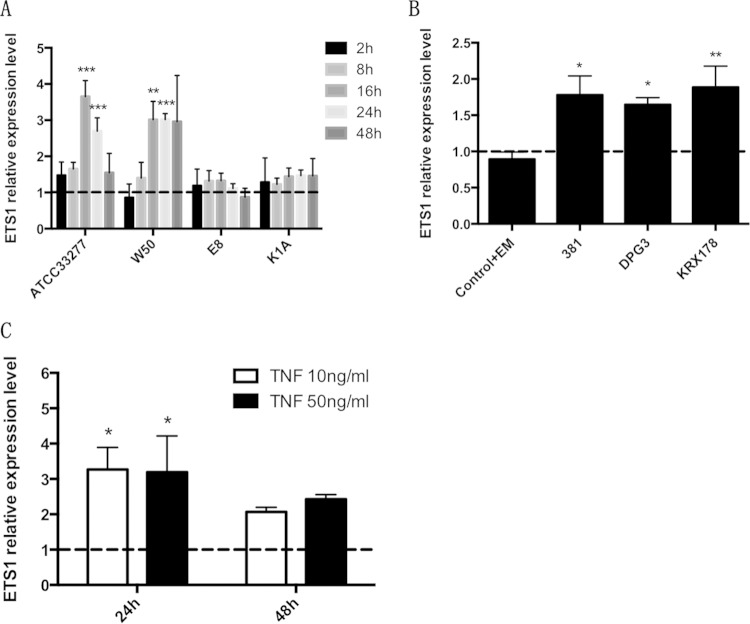
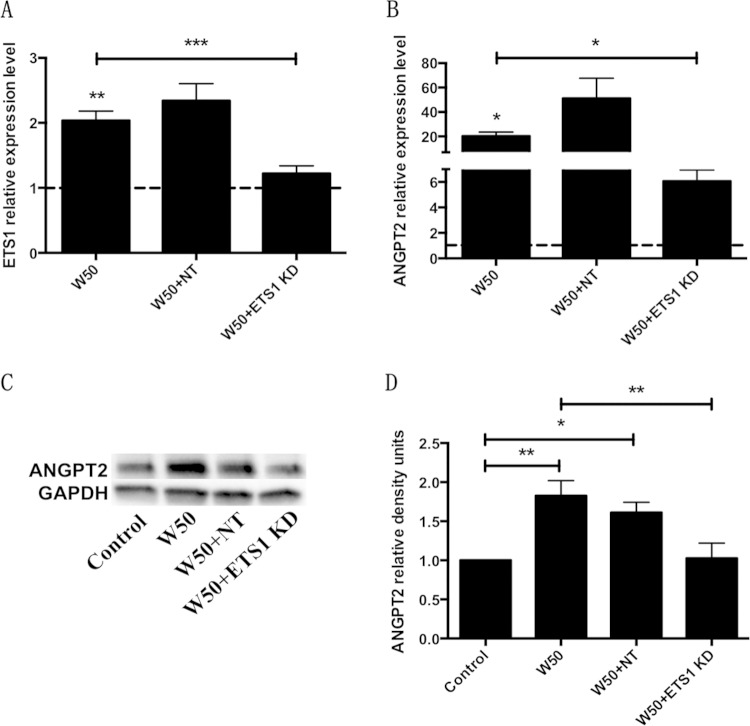
The wild-type P. gingivalis strains ATCC 33277 and W50 significantly increased ETS1 expression in AoSMCs after 16 and 24 h (Fig. 4A). Compared with W50, the Rgp and Kgp mutant strains E8 and K1A, respectively, had no effect on ETS1 gene expression. Inhibition of Rgp with leupeptin significantly antagonized the stimulatory effect of W50 (see Fig. S2C in the supplemental material). The wild-type strain 381 and its fimbrial mutants, DPG3 and KRX178, significantly increased ETS1 expression in AoSMCs after 24 h (Fig. 4B). TNF also significantly increased the expression of ETS1 in AoSMCs after 24 h when applied at a concentration of 10 ng/ml or 50 ng/ml (Fig. 4C). Compared with wild-type P. gingivalis and TNF, F. alocis showed no effect on ETS1 gene expression after 24 h (see Fig. S3 in the supplemental material). To confirm that the upregulation of ETS1 by P. gingivalis infection was correlated with the upregulation of Angpt2 in AoSMCs, we performed an ETS1 siRNA knockdown experiment. We found that the ETS1 siRNA significantly reduced the upregulation of ETS1 in AoSMCs that were treated with P. gingivalis W50 (Fig. 5A). This result confirms our finding by showing that the upregulation of Angpt2 at the mRNA (Fig. 5B) and protein (Fig. 5C and D) levels by W50 is significantly reduced in ETS1 siRNA knockdown AoSMCs.
FIG 4.
P. gingivalis and its gingipains and fimbrial mutants upregulate ETS1 in AoSMCs. Quantitative real-time PCR results demonstrate relative transcription levels of ETS1 in AoSMCs stimulated with wild-type ATCC 33277 and W50 and the corresponding W50 gingipain mutants (E8 and K1A) at an MOI of 10 for 2 h, 8 h, 16 h, 24 h, and 48 h (A), with wild-type 381 and its corresponding fimbrial mutants (DPG3 and KRX178) of P. gingivalis for 24 h (B), or with TNF for 24 h and 48 h (C). All results were normalized to the gene expression level of the GAPDH housekeeping gene. Statistically significant differences for LPS and wild-type P. gingivalis strain 381 are compared to the negative control. For fimbrial mutants, statistically significant differences are compared to the group Control+EM. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. (A) n = 5; (B) n = 4; (C) n = 7.
FIG 5.
P. gingivalis regulates Angpt2 through ETS1 in AoSMCs. AoSMCs were treated with (KD) or without (Control) ETS1 siRNA or nontargeting siRNA (NT). The cells were then infected or not infected with P. gingivalis W50 for 24 h. (A and B) Quantitative real-time PCR results demonstrate the relative transcription levels of ETS1 (A) and Angpt2 (B) in AoSMCs. (C) Protein level of Angpt2 was determined by Western blotting. (D) Quantification of Angpt2 protein expression levels by densitometry. All results were normalized to the GAPDH gene or protein expression level. Asterisks above W50 represent statistical comparisons to the negative control. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. For all panels, n = 5.
Regulation of AoSMC proliferation and migration by Angpt2.
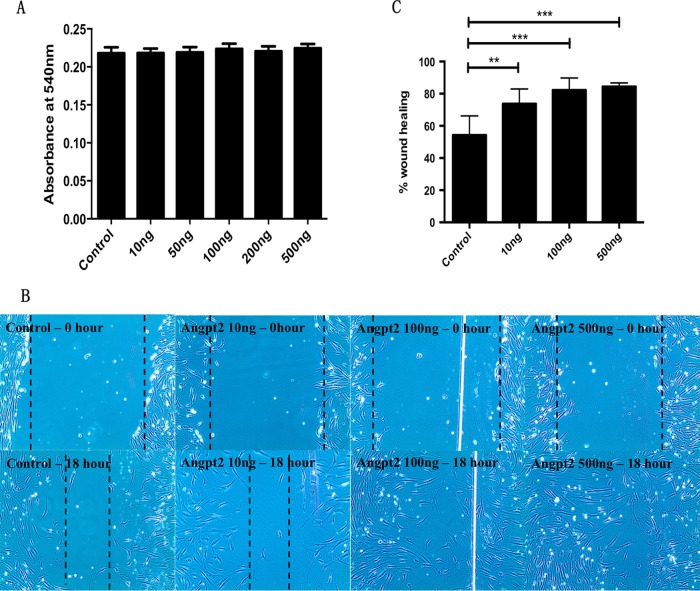
To investigate how Angpt2 regulates the cellular function of AoSMCs, we studied the proliferation and migration of AoSMCs after stimulation with Angpt2 protein. The results of MTT assays showed that Angpt2 has no effect on AoSMCs proliferation (Fig. 6A). However, Angpt2 protein increased the migration of AoSMCs in a dose-dependent manner in cells analyzed with the wound healing assay. After 18 h, cells stimulated with 100 ng/ml or 500 ng/ml of Angpt2 protein covered almost the entire wound area (Fig. 6B and C).
FIG 6.
Angpt2 induces migration, but not proliferation, in AoSMCs. AoSMCs were treated with or without Angpt2 for 24 h. (A) Proliferation of the cells, measured using an MTT assay. (B) The migration of AoSMCs after treatment with Angpt2 or DPBS (Control) was measured in a wound healing assay. The photos were taken after wounding at 0 h (Control) and 18 h. (C) Quantitative results calculated by the reduction in the area of the wound after migration of the cells into the cell-free zone.**, P < 0.005; ***, P < 0.0001. (A) n = 4; (B and C) n = 6.
DISCUSSION
Considerable evidence has indicated that periodontal infection is a mild but significant risk factor for developing cardiovascular disease. The periodontopathogenic bacterium P. gingivalis is considered to be directly or indirectly involved in the development of atherosclerosis and cardiovascular disease. We have previously reported that P. gingivalis causes platelet aggregation, sensitizes platelets to epinephrine, suppresses the inflammatory responses of immunological cells, and modifies low-density lipoprotein (LDL) (32–34).
This study indicates that wild-type P. gingivalis decreases the gene expression level of Angpt1 but increases the gene and protein expression of Angpt2 in AoSMCs. For Angpt1, we did not observe significant differences at the protein level. This might be due to the fluctuations in results observed in Angpt1 gene expression in AoSMCs infected by P. gingivalis at early time points. Endothelial cells, which are stored in the endothelium-specific Weibel-Palade bodies (WPBs), are the main source of Angpt2. Certain stimuli, such as hypoxia, thrombin, or phorbol-12-myristate-13-acetate (PMA), are know to induce the rapid release of Angpt2 from WPBs (35, 36). We found that AoSMCs are also able to produce Angpt2. However, although unstimulated AoSMCs produce Angpt2 at a low level, stimulation with wild-type P. gingivalis dramatically increases the gene expression of Angpt2 in AoSMCs. Elevated levels of Angpt2 have been suggested to be a marker of cardiovascular disease, oral squamous cell carcinoma, and lung cancer (37–39). These results are also relevant because a high angiopoietin-2/angiopoietin-1 ratio is associated with pathogenesis in hemangiomas, atherosclerosis, and hemorrhagic endometrium (40, 41).
The RgpAB mutant E8 was unable to alter the expression of Angpt1 or Angpt2 in AoSMCs. The Kgp mutant K1A downregulated the expression of Angpt1 and upregulated the expression of Angpt2 in AoSMCs, but compared to the wild type, K1A had only small effects on the regulation of these angiopoietins. These results indicate that gingipains, especially Rgp, play important roles in P. gingivalis-induced modulation of angiopoietins in AoSMCs. In support of this idea, inhibition of Rgp with leupeptin antagonized the upregulation of Angpt2 and the downregulation of Angpt1 that was induced by P. gingivalis infection. The infection of AoSMCs with F. alocis, which has a low cysteine protease activity (42), had no effect on the expression of Angpt1, Angpt2, or ETS1. Gingipains are cell surface trypsin-like cysteine proteases that are produced by P. gingivalis. Both Rgp and Kgp are secreted by P. gingivalis and are indispensable for the ability of the bacterium to obtain nutrients from the environment (43). Studies have shown that the RgpAB mutant markedly decreases hemagglutinating activity, whereas the Kgp mutant only slightly affects hemagglutinating activity (44, 45). In contrast to RgpB, which has only a catalytic domain, RgpA and Kgp contain noncovalent complexes that are composed of separate catalytic and adhesion/hemagglutinin domains (46). These results show that RgpA and Kgp are important to the ability of P. gingivalis to acquire hemagglutinins from the host cells through proteolytic processing. Moreover, gingipains have the ability to impair host immune response function through degradation and inactivation of immunoglobulins, such as IgG, IgA, and secretory IgA (47), and proinflammatory cytokines, such as interleukin 6 (IL-6), interleukin 8 (CXCL8), and interleukin-1β (IL-1β) (48). The results of this study suggest that gingipains, especially Rgp, are responsible for P. gingivalis-mediated regulation of Angpt1 and Angpt2 in AoSMCs.
In contrast to gingipains, fimbriae and LPS are not involved in P. gingivalis-mediated regulation of Angpt1and Angpt2 production in AoSMCs. P. gingivalis 381, DPG3, and KRX178 all downregulate Angpt1 and upregulate Angpt2 and ETS1 expression. In our previous study (25), we found that the amount of LPS that was produced and released differed between various bacterial strains. Consequently, we studied how LPS produced by P. gingivalis affects Angpt1 and Angpt2 expression in AoSMCs. The results showed that LPS from P. gingivalis does not modulate the expression of Angpt1 or Angpt2 in AoSMCs. However, different acylated and phosphorylated isoforms of LPS have been reported to display different bioactivity (49), which makes it difficult to compare outcomes when researchers use different isoforms of LPS.
TNF is a key regulator in the pathogenesis and progression of periodontitis and atherosclerosis. This cytokine plays crucial roles in the initiation and maintenance of immune responses to P. gingivalis infection (50–52). TNF can induce the migration, proliferation, and apoptosis of vascular smooth muscle cells (53). Studies have shown that there is an association between plasma levels of Angpt2 and TNF in endotoxemia and sepsis patients (54, 55). To study the role of TNF in the modulation of Angpt1 and Angpt2, we exposed AoSMCs to various doses of TNF. We found that TNF inhibited the expression of Angpt1 but increased the expression of Angpt2 in AoSMCs. These effects are similar to those obtained in AoSMCs that were stimulated with P. gingivalis. However, TNF was not detected in the supernatants of AoSMCs that were stimulated with P. gingivalis, which indicates that P. gingivalis-mediated regulation of Angpt1 and Angpt2 in AoSMCs is independent of TNF.
Because we found that P. gingivalis and TNF dramatically increase the expression of the Angpt2 gene in AoSMCs, we further examined the gene expression of ETS1, the transcription factor of Angpt2 (56, 57). Wild-type and fimbrial mutant strains of P. gingivalis and TNF increased the expression of the ETS1 gene, which was then correlated with the effects on Angpt2. However, the RgpAB mutant E8 was unable to upregulate ETS1 expression, which supports a role for Rgp in P. gingivalis-mediated regulation of Angpt2 in AoSMCs. After knockdown of ETS1 in AoSMCs, the upregulation of Angpt2 by P. gingivalis W50 was inhibited, which reveals that ETS1 is critical for the induction of Angpt2.
To understand the regulatory role of Angpt2 in AoSMCs, we treated AoSMCs with recombinant human Angpt2. Angpt2 did not induce proliferation of AoSMCs, but it significantly induced the migration of the cells in a dose-dependent manner. During the progression of atherosclerosis, smooth muscle cells change from a contractile phenotype to a synthetic phenotype, and the migration of vascular smooth muscle cells in the intima layer marks a key event in the disease pathogenesis (58). Several reports have shown that high levels of Angpt2 are associated with atherosclerosis and coronary heart disease (59, 60).
In summary, we found that P. gingivalis infection induces comparable effects on the expression of Angpt1, Angpt2, and ETS1 in AoSMCs and that gingipains are crucial for this regulation. However, the cardiovascular risk factor TNF is not involved. In combination with observed cellular effects, our findings suggest that Angpt2 plays a role in the association between periodontitis and atherosclerosis.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. Curtis (Barts and The London, Queen Mary's School of Medicine and Dentistry, United Kingdom) for contributing the P. gingivalis wild-type strain W50 and its corresponding gingipain mutants (E8 and K1A) and to R. J. Genco and A. Sharma (School of Dental Medicine, State University of New York at Buffalo) for contributing the P. gingivalis wild-type strain 381 and its corresponding fimbrial mutants (DPG3 and KRX178).
This work was supported by the Swedish Research Council for Medicine and Health (grant 2008-2459), the Swedish Heart and Lung Foundation (grant T77414), the Foundation of Olle Engkvist, the Foundation of Mats Kleberg, and KK-Stiftelsen.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00498-15.
REFERENCES
- 1.Tucka J, Yu H, Gray K, Figg N, Maguire J, Lam B, Bennett M, Littlewood T. 2014. Akt1 regulates vascular smooth muscle cell apoptosis through FoxO3a and Apaf1 and protects against arterial remodeling and atherosclerosis. Arterioscler Thromb Vasc Biol 34:2421–2428. doi: 10.1161/ATVBAHA.114.304284. [DOI] [PubMed] [Google Scholar]
- 2.Trollope AF, Golledge J. 2011. Angiopoietins, abdominal aortic aneurysm and atherosclerosis. Atherosclerosis 214:237–243. doi: 10.1016/j.atherosclerosis.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato TN, Qin Y, Kozak CA, Audus KL. 1993. Tie-1 and tie-2 define another class of putative receptor tyrosine kinase genes expressed in early embryonic vascular system. Proc Natl Acad Sci U S A 90:9355–9358. doi: 10.1073/pnas.90.20.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linares PM, Chaparro M, Gisbert JP. 2014. Angiopoietins in inflammation and their implication in the development of inflammatory bowel disease. J Crohns Colitis 8:183–190. doi: 10.1016/j.crohns.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Richey SL, Hutson TE. 2013. Angiopoietins and non-vascular endothelial growth factor antiangiogenic targets in advanced renal cell carcinoma. Cancer J 19:307–310. doi: 10.1097/PPO.0b013e31829d5d15. [DOI] [PubMed] [Google Scholar]
- 6.Eklund L, Olsen BR. 2006. Tie receptors and their angiopoietin ligands are context-dependent regulators of vascular remodeling. Exp Cell Res 312:630–641. doi: 10.1016/j.yexcr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Thurston G, Suri C, Smith K, McClain J, Sato TN, Yancopoulos GD, McDonald DM. 1999. Leakage-resistant blood vessels in mice transgenically overexpressing angiopoietin-1. Science 286:2511–2514. doi: 10.1126/science.286.5449.2511. [DOI] [PubMed] [Google Scholar]
- 8.Fiedler U, Reiss Y, Scharpfenecker M, Grunow V, Koidl S, Thurston G, Gale NW, Witzenrath M, Rosseau S, Suttorp N, Sobke A, Herrmann M, Preissner KT, Vajkoczy P, Augustin HG. 2006. Angiopoietin-2 sensitizes endothelial cells to TNF-alpha and has a crucial role in the induction of inflammation. Nat Med 12:235–239. doi: 10.1038/nm1351. [DOI] [PubMed] [Google Scholar]
- 9.Carlson TR, Feng Y, Maisonpierre PC, Mrksich M, Morla AO. 2001. Direct cell adhesion to the angiopoietins mediated by integrins. J Biol Chem 276:26516–26525. doi: 10.1074/jbc.M100282200. [DOI] [PubMed] [Google Scholar]
- 10.Imanishi Y, Hu B, Jarzynka MJ, Guo P, Elishaev E, Bar-Joseph I, Cheng SY. 2007. Angiopoietin-2 stimulates breast cancer metastasis through the alpha(5)beta(1) integrin-mediated pathway. Cancer Res 67:4254–4263. doi: 10.1158/0008-5472.CAN-06-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang HW, Huang YF, Chou MY. 2004. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol 75:1077–1083. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- 12.Aruni AW, Zhang K, Dou Y, Fletcher H. 2014. Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect Immun 82:3261–3274. doi: 10.1128/IAI.01727-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, Berning M, Friedmann A, Göbel UB, Moter A. 2010. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol 10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kebschull M, Demmer RT, Papapanou PN. 2010. “Gum bug, leave my heart alone!”—epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J Dent Res 89:879–902. doi: 10.1177/0022034510375281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogrendik M. 2013. Rheumatoid arthritis is an autoimmune disease caused by periodontal pathogens. Int J Gen Med 6:383–386. doi: 10.2147/IJGM.S45929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavrini F, Sambri V, Moter A, Servidio D, Marangoni A, Montebugnoli L, Foschi F, Prati C, Di Bartolomeo R, Cevenini R. 2005. Molecular detection of Treponema denticola and Porphyromonas gingivalis in carotid and aortic atheromatous plaques by FISH: report of two cases. J Med Microbiol 54:93–96. doi: 10.1099/jmm.0.45845-0. [DOI] [PubMed] [Google Scholar]
- 17.Haraszthy VI, Zambon JJ, Trevisan M, Zeid M, Genco RJ. 2000. Identification of periodontal pathogens in atheromatous plaques. J Periodontol 71:1554–1560. doi: 10.1902/jop.2000.71.10.1554. [DOI] [PubMed] [Google Scholar]
- 18.Koizumi Y, Kurita-Ochiai T, Oguchi S, Yamamoto M. 2008. Nasal immunization with Porphyromonas gingivalis outer membrane protein decreases P. gingivalis-induced atherosclerosis and inflammation in spontaneously hyperlipidemic mice. Infect Immun 76:2958–2965. doi: 10.1128/IAI.01572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodala N, Merricks EP, Bellinger DA, Damrongsri D, Offenbacher S, Beck J, Madianos P, Sotres D, Chang YL, Koch G, Nichols TC. 2005. Porphyromonas gingivalis bacteremia induces coronary and aortic atherosclerosis in normocholesterolemic and hypercholesterolemic pigs. Arterioscler Thromb Vasc Biol 25:1446–1451. doi: 10.1161/01.ATV.0000167525.69400.9c. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y, Nguyen KA, Potempa J. 2010. Dichotomy of gingipains action as virulence fators from cleaving substrates with the precision of surgeon's knife to a meat chopper like brutal degradation of proteins. Periodontol 2000 54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bostanci N, Belibasakis GN. 2012. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett 333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 22.Palm E, Khalaf H, Bengtsson T. 2015. Suppression of inflammatory responses of human gingival fibroblasts by gingipains from Porphyromonas gingivalis. Mol Oral Microbiol 30:74–85. doi: 10.1111/omi.12073. [DOI] [PubMed] [Google Scholar]
- 23.Khalaf H, Lönn J, Bengtsson T. 2014. Cytokines and chemokines are differentially expressed in patients with periodontitis: possible role for TGF-β1 as a marker for disease progression. Cytokine 67:29–35. doi: 10.1016/j.cyto.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Nassar H, Chou HH, Khlgatian M, Gibson FC 3rd, Van Dyke TE, Genco CA. 2002. Role for fimbriae and lysine-specific cysteine proteinase gingipain K in expression of interleukin-8 and monocyte chemoattractant protein in Porphyromonas gingivalis-infected endothelial cells. Infect Immun 70:268–276. doi: 10.1128/IAI.70.1.268-276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayaprakash K, Khalaf H, Bengtsson T. 2014. Gingipains from Porphyromonas gingivalis play a significant role in induction and regulation of CXCL8 in THP-1 cells. BMC Microbiol 14:193. doi: 10.1186/1471-2180-14-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshpande RG, Khan MB, Genco CA. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun 66:5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kontani M, Kimura S, Nakagawa I, Hamada S. 1997. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol 24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 28.Amar S, Wu SC, Madan M. 2009. Is Porphyromonas gingivalis cell invasion required for atherogenesis? Pharmacotherapeutic implications. J Immunol 182:1584–1592. [DOI] [PubMed] [Google Scholar]
- 29.Jotwani R, Cutler CW. 2004. Fimbriated Porphyromonas gingivalis is more efficient than fimbria-deficient P. gingivalis in entering human dendritic cells in vitro and induces an inflammatory Th1 effector response. Infect Immun 72:1725–1732. doi: 10.1128/IAI.72.3.1725-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Elmabsout AA, Khalaf H, Basic VT, Jayaprakash K, Kruse R, Sirsjö A. 2013. The periodontal pathogen Porphyromonas gingivalis changes the gene expression in vascular smooth muscle cells involving the TGFbeta/Notch signalling pathway and increased cell proliferation. BMC Genomics 14:770. doi: 10.1186/1471-2164-14-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez D, Owens GK. 2012. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc Res 95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengtsson T, Karlsson H, Gunnarsson P, Skoglund C, Elison C, Leanderson P, Lindahl M. 2008. The periodontal pathogen Porphyromonas gingivalis cleaves apoB and increases the expression of apoM in LDL in whole blood leading to cell proliferation. J Intern Med 263:558–571. doi: 10.1111/j.1365-2796.2007.01917.x. [DOI] [PubMed] [Google Scholar]
- 33.Nylander M, Lindahl TL, Bengtsson T, Grenegård M. 2008. The periodontal pathogen Porphyromonas gingivalis sensitises human blood platelets to epinephrine. Platelets 19:352–358. doi: 10.1080/09537100802056102. [DOI] [PubMed] [Google Scholar]
- 34.Engström KK, Khalaf H, Kälvegren H, Bengtsson T. 2015. The role of Porphyromonas gingivalis gingipains in platelet activation and innate immune modulation. Mol Oral Microbiol 30:62–73. doi: 10.1111/omi.12067. [DOI] [PubMed] [Google Scholar]
- 35.Fiedler U, Scharpfenecker M, Koidl S, Hegen A, Grunow V, Schmidt JM, Kriz W, Thurston G, Augustin HG. 2004. The Tie-2 ligand angiopoietin-2 is stored in and rapidly released upon stimulation from endothelial cell Weibel-Palade bodies. Blood 103:4150–4156. doi: 10.1182/blood-2003-10-3685. [DOI] [PubMed] [Google Scholar]
- 36.Valentijn KM, van Driel LF, Mourik MJ, Hendriks GJ, Arends TJ, Koster AJ, Valentijn JA. 2010. Multigranular exocytosis of Weibel-Palade bodies in vascular endothelial cells. Blood 116:1807–1816. doi: 10.1182/blood-2010-03-274209. [DOI] [PubMed] [Google Scholar]
- 37.David S, Kümpers P, Hellpap J, Horn R, Leitolf H, Haller H, Kielstein JT. 2009. Angiopoietin 2 and cardiovascular disease in dialysis and kidney transplantation. Am J Kidney Dis 53:770–778. doi: 10.1053/j.ajkd.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Sun CJ, Fan JC, Geng N, Li CH, Liao J, Mi K, Zhu GQ, Ma H, Song YF, Tang YL, Chen Y. 2013. Angiopoietin-2 expression is correlated with angiogenesis and overall survival in oral squamous cell carcinoma. Med Oncol 30:571. doi: 10.1007/s12032-013-0571-2. [DOI] [PubMed] [Google Scholar]
- 39.Fawzy A, Gaafar R, Kasem F, Ali SS, Elshafei M, Eldeib M. 2012. Importance of serum levels of angiopoietin-2 and survivin biomarkers in non-small cell lung cancer. J Egypt Natl Canc Inst 24:41–45. doi: 10.1016/j.jnci.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Varughese J, Brown LF, Mulliken JB, Bischoff J. 2001. Increased Tie2 expression, enhanced response to angiopoietin-1, and dysregulated angiopoietin-2 expression in hemangioma-derived endothelial cells. Am J Pathol 159:2271–2280. doi: 10.1016/S0002-9440(10)63077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Post S, Peeters W, Busser E, Lamers D, Sluijter JP, Goumans MJ, de Weger RA, Moll FL, Doevendans PA, Pasterkamp G, Vink A. 2008. Balance between angiopoietin-1 and angiopoietin-2 is in favor of angiopoietin-2 in atherosclerotic plaques with high microvessel density. J Vasc Res 45:244–250. doi: 10.1159/000112939. [DOI] [PubMed] [Google Scholar]
- 42.Aruni AW, Roy F, Fletcher HM. 2011. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis. Infect Immun 79:3872–3886. doi: 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grenier D, Imbeault S, Plamondon P, Grenier G, Nakayama K, Mayrand D. 2001. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect Immun 69:5166–5172. doi: 10.1128/IAI.69.8.5166-5172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem 273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 45.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem 270:23619–23626. [DOI] [PubMed] [Google Scholar]
- 46.Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, Shi Y, Ratnayake DB, Yamamoto K. 2000. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem 128:153–159. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- 47.Kadowaki T, Yoneda M, Okamoto K, Maeda K, Yamamoto K. 1994. Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J Biol Chem 269:21371–21378. [PubMed] [Google Scholar]
- 48.Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. 2009. The host cytokine response to Porphyromonas gingivalis is modified by gingipains. Oral Microbiol Immunol 24:11–17. doi: 10.1111/j.1399-302X.2008.00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol 11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J, Bi L, Yu X, Kawai T, Taubman MA, Shen B, Han X. 2014. Porphyromonas gingivalis exacerbates ligature-induced, RANKL-dependent alveolar bone resorption via differential regulation of Toll-like receptor 2 (TLR2) and TLR4. Infect Immun 82:4127–4134. doi: 10.1128/IAI.02084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komatsu T, Nagano K, Sugiura S, Hagiwara M, Tanigawa N, Abiko Y, Yoshimura F, Furuichi Y, Matsushita K. 2012. E-selectin mediates Porphyromonas gingivalis adherence to human endothelial cells. Infect Immun 80:2570–2576. doi: 10.1128/IAI.06098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holden JA, Attard TJ, Laughton KM, Mansell A, O'Brien-Simpson NM, Reynolds EC. 2014. Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect Immun 82:4190–4203. doi: 10.1128/IAI.02325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinbongard P, Heusch G, Schulz R. 2010. TNFalpha in atherosclerosis, myocardial ischemia/reperfusion and heart failure. Pharmacol Ther 127:295–314. doi: 10.1016/j.pharmthera.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Orfanos SE, Kotanidou A, Glynos C, Athanasiou C, Tsigkos S, Dimopoulou I, Sotiropoulou C, Zakynthinos S, Armaganidis A, Papapetropoulos A, Roussos C. 2007. Angiopoietin-2 is increased in severe sepsis: correlation with inflammatory mediators. Crit Care Med 35:199–206. doi: 10.1097/01.CCM.0000251640.77679.D7. [DOI] [PubMed] [Google Scholar]
- 55.Kümpers P, van Meurs M, David S, Molema G, Bijzet J, Lukasz A, Biertz F, Haller H, Zijlstra JG. 2009. Time course of angiopoietin-2 release during experimental human endotoxemia and sepsis. Crit Care 13:R64. doi: 10.1186/cc7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasegawa Y, Abe M, Yamazaki T, Niizeki O, Shiiba K, Sasaki I, Sasaki I, Sato Y. 2004. Transcriptional regulation of human angiopoietin-2 by transcription factor Ets-1. Biochem Biophys Res Commun 316:52–58. doi: 10.1016/j.bbrc.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 57.Oettgen P. 2010. The role of ets factors in tumor angiogenesis. J Oncol 2010:767384. doi: 10.1155/2010/767384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudijanto A. 2007. The role of vascular smooth muscle cells on the pathogenesis of atherosclerosis. Acta Med Indones 39:86–93. [PubMed] [Google Scholar]
- 59.David S, Kümpers P, Lukasz A, Kielstein JT, Haller H, Fliser D. 2009. Circulating angiopoietin-2 in essential hypertension: relation to atherosclerosis, vascular inflammation, and treatment with olmesartan/pravastatin. J Hypertens 27:1641–1647. doi: 10.1097/HJH.0b013e32832be575. [DOI] [PubMed] [Google Scholar]
- 60.David S, John SG, Jefferies HJ, Sigrist MK, Kümpers P, Kielstein JT, Haller H, McIntyre CW. 2012. Angiopoietin-2 levels predict mortality in CKD patients. Nephrol Dial Transplant 27:1867–1872. doi: 10.1093/ndt/gfr551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.