Abstract
Genome data has created new opportunities to untangle evolutionary processes shaping microbial variation. Among bacteria, long-term mutualists of insects represent the smallest and (typically) most AT-rich genomes. Evolutionary theory provides a context to predict how an endosymbiotic lifestyle may alter fundamental evolutionary processes - mutation, selection, genetic drift, and recombination - and thus contribute to extreme genomic outcomes. These predictions can then be explored by comparing evolutionary rates, genome size and stability, and base compositional biases across endosymbiotic and free-living bacteria. Recent surprises from such comparisons include genome reduction among uncultured, free-living species. Some studies suggest that selection generally drives this streamlining, while drift drives genome reduction in endosymbionts; however, this remains an hypothesis requiring additional data. Unexpected evidence of selection acting on endosymbiont GC content hints that even weak selection may be effective in some long-term mutualists. Moving forward, intraspecific analysis offers a promising approach to distinguish underlying mechanisms, by testing the null hypothesis of neutrality and by quantifying mutational spectra. Such analyses may clarify whether endosymbionts and free-living bacteria occupy distinct evolutionary trajectories or alternatively, represent varied outcomes of similar underlying forces.
Keywords: genome reduction, endosymbiosis, mutualism, molecular evolution, neutrality, population genetics
Introduction
Reflecting their ~4 billion year history, Bacteria encompass astonishing metabolic diversity, inhabit every imaginable ecological niche, and have played crucial roles in the evolution of other life forms. Because the majority of bacteria have not been cultivated in the lab, molecular approaches have proved invaluable for understanding their distribution and functional significance. Recent years have witnessed an explosion of bacterial genome sequencing and analysis, including genomic studies of uncultured strains and communities. With this data come new opportunities to compare genomes within and across bacterial groups, in order to dissect evolutionary forces that drive molecular changes and contribute to observed variation across ecological contexts.
To date, a clear lesson from bacterial genome comparisons has been the following: lifestyle profoundly affects bacterial genomes. Early comparisons revealed that a host-dependent lifestyle has striking effects on bacterial genome composition, size, and structure. ‘Resident’ genomes that replicate solely within hosts, such as obligate parasites (e.g., Rickettsia and Chlamydia) and mitochondria, were found to exhibit a syndrome called reductive genome evolution, characterized by relatively small genome size (typically <1 Mb), rapid rates of sequence evolution, and typically AT-rich base composition compared to other genomes sequenced at the time, such as Escherichia coli and other bacteria that can replicate outside of hosts.1 The first available genome of Buchnera aphidicola, the long-term endosymbiont of aphids, supported earlier inferences from gene-specific data (e.g., Refs. 2 and 3) that this intracellular mutualist exhibited a similar evolutionary trajectory.4 These early observations suggested distinct paths of genome evolution in host-dependent bacteria versus those that retain a free-living phase to their lifecycle.
More recently, an exponential growth of genomic and metagenome datasets have provided a rich framework to test the generality of initial observations linking bacterial lifestyle to genome features. Among bacteria that replicate within hosts, the extremely constrained lifestyles of long-term, intracellular bacterial mutualists of insects (hereafter called primary endosymbionts, for brevity) make them particularly useful models to study genomic consequences of host-dependence. Primary endosymbionts of insects live exclusively within specialized host cells, undergo maternal transmission to offspring, and are closely integrated into host development and nutritional physiology.5, 6 Since the initial sequencing of Buchnera, genome analyses of primary endosymbionts and related facultative endosymbionts have spanned numerous lineages of bacteria and diverse host groups, thus allowing comparisons across phylogenetic and ecological contexts.
Genomic variation and ecological significance of insect endosymbionts have been discussed in several excellent overviews and syntheses of the literature.7-12 In this review, my goal is to consider patterns of endosymbiont variation in a broader framework of bacterial genome evolution, by highlighting notable differences and surprising parallels with free-living bacteria. I focus on evolutionary mechanisms that may contribute to distinct genome features of host-dependent bacteria, particularly primary endosymbionts of insects. Before delving into specific genome data, I consider several key lifestyle features of long-term insect endosymbionts, and how these features may alter fundamental processes: genetic drift, natural selection, mutation, and recombination. I then highlight recent insights from comparative genomic studies that have informed, and in some cases challenged, our views regarding the genomic consequences of host-dependence. While studies of primary endosymbionts have revealed convergent patterns of genome evolution, including genome reduction and a trend toward AT-richness, they have also uncovered surprises that challenge assumptions about evolution within host cells. On another research front, genomic analysis of free-living bacteria now includes the sequencing of uncultured strains through single-cell genomics technology. Such studies have revealed that many uncultured, free-living bacteria exhibit genome size reduction and a shift toward AT-richness, features once considered typical of host-dependent microbes. These data hint that the distinction between free-living and host-dependent genomes may be blurrier than initially thought, and raises the possibility that similar evolutionary processes could drive genomic changes across lifestyles.
Lifestyle features of primary endosymbionts
Among animals, insects seem especially prone to establishing symbiotic relationships with intracellular bacteria 5. Their intimate bacterial partners include reproductive parasites, transient facultative mutualists that move among host species, and persistent, obligate mutualists that coevolve with a particular insect group. The last category, primary endosymbionts, includes the most highly constrained, stable, and specialized symbioses known in the animal world. These bacteria are essential for host reproduction, are closely integrated with their insect hosts, and occur within specialized host cells called bacteriocytes (Fig. 1) and the female reproductive tract, consistent with their maternal transmission. While the frequency of primary mutualists in insects is difficult to determine since many host groups remain unsampled, such mutualisms are thought to occur in an estimated ~10–15% of insect species.5 Interestingly, intracellular mutualisms are virtually unknown among vertebrates, an animal group in which intracellular microbes are almost always pathogenic.13 The sole exception is a beneficial alga that lives within cells of salamander embryos.14 Among chordates, the only known intracellular bacterial mutualist occurs in tunicates.15
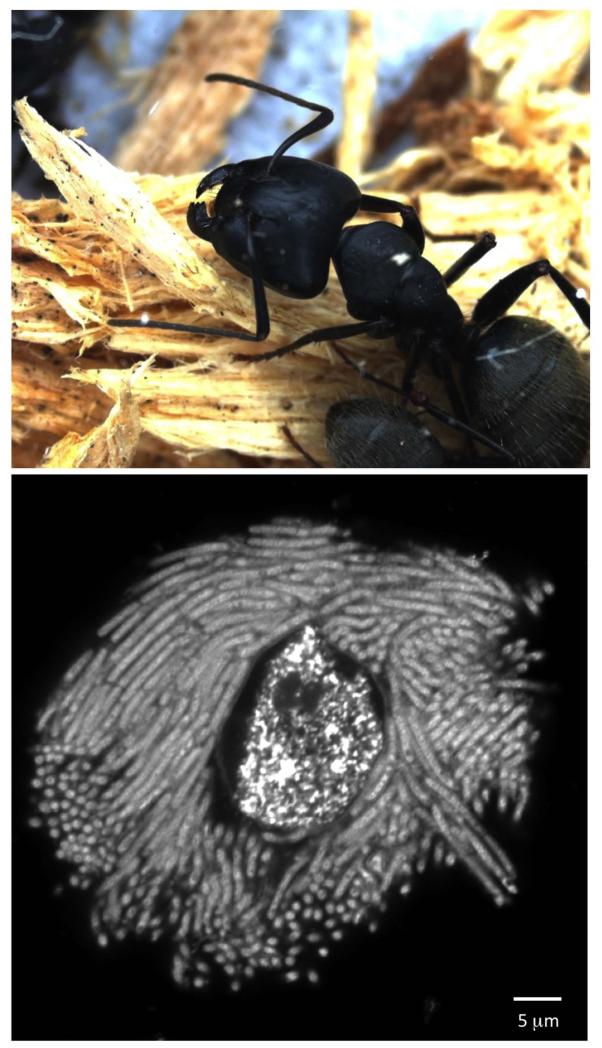
Figure 1.
Camponotine ants rely on a bacterial mutualist that lives exclusively within host cells. Top panel: Wood-nesting Camponotus pennsylvanicus, like other members of the diverse tribe Camponotini, possess an obligate bacterial endosymbiont, Blochmannia. © Adam B. Lazarus. Bottom panel: A single bacteriocyte (specialized host cell) from the ant C. pennsylvanicus, with numerous rod-shaped Blochmannia filling the cytoplasm. Blochmannia has lived exclusively within an intracellular niche for tens of millions of years. Sample was prepared from homogenized ant larvae, fixed, stained with DAPI, and visualized with a fluorescent microscope. Image shows host nucleus at the center. © Erika del Castillo.
Unlike the numerous host-bacteria symbioses that are facultative for one or both partners, primary endosymbioses are mutually obligate. That is, the bacteria apparently lack abilities to replicate outside of host cells, and the insect hosts depend on endosymbiont functions that are often nutritional in nature. Specifically, the bacteria often supplement specific nutrients that are missing in the host’s diet, such as amino acids or vitamins. Not surprisingly, these mutualisms are most common in host groups that feed on unbalanced diets, such as plant sap or vertebrate blood.5 Symbionts may also perform more general functions such as nitrogen recycling, as shown in ants16, 17 and cockroaches.18 By virtue of their bacterial associates, insect hosts can thrive on otherwise inadequate diets.19, 20 Strict host-symbiont cospeciation indicates a high fidelity of maternal transmission, often for tens or hundreds of millions of years since the symbiosis was established.21-23
As a consequence of their intracellular location and stable, long-term vertical transmission, primary endosymbionts have provided valuable models to study the evolutionary consequences of highly specialized host-dependent lifestyles, including shifts in rates and patterns of genome evolution. These mutualists include phylogenetically distinct lineages apparently derived from free-living bacteria, perhaps via a facultative symbiotic stage.24, 25 As such, they offer opportunities to compare independent transitions from a free-living or facultative host-associated lifestyle to an obligately intracellular existence.
Shifts in evolutionary processes: a priori predictions
The transition to a host-dependent lifestyle may be coupled with shifts in the balance among fundamental evolutionary processes, with profound consequences on rates and patterns of genome change. Below, I highlight some predictions we might make a priori, even before considering the genomes of these endosymbionts (Table 1). Such predictions are based on a rich literature exploring the distribution, transmission, ecology, and cell biology of these mutualists, as well as evolutionary theory, and broader insights into mechanisms of bacterial evolution.
Table 1. A priori predictions: Processes that may shift upon transition to an obligately host-dependent lifestyle.
| Process | Rationale for predicted shift | Potential consequence for genome evolution |
|---|---|---|
|
| ||
| Reduced Ne and increased genetic drift |
Population bottlenecks occur upon transmission to host offspring. |
Accelerated fixation of slightly deleterious mutations under drift. |
| Shifts in selection coefficients |
Selection may be relaxed on functions redundant in the intracellular niche. Host- level selection will favor nutritional functions and other host-beneficial traits. |
Shifts in selective constraint across functions encoded. |
| Shifts in mutation rates | Stable intracellular niche may favor reduced mutation rates. However, interaction with host immune system may favor higher rates. |
Shifts in rates of DNA sequence evolution. |
| Small Ne may lead to higher mutation rate, due to reduced efficacy of selection favoring a low mutation rate. |
||
| Constrained gene exchange |
Cellular sequestration may constrain gene exchange with genetically distinct bacteria. |
Asexuality may exacerbate effects of genetic drift. Strong linkage implies that selective sweeps will purge diversity and possibly drive deleterious changes via genetic draft. |
Shifts in Ne
Effective population size (Ne) is strictly defined as the size of a theoretical, idealized Wright-Fisher population 26, 27 that would give the same value of some genetic property (e.g. the magnitude of drift) as the population in question. While this concept can be challenging to apply to bacteria (e.g., due to difficulty in defining population or species boundaries), several studies have explored factors that may influence bacterial Ne. To the extent that members of a population contribute unequally to the next generation, Ne will be reduced compared to the census population size (Nc). For example, bacterial Ne may be reduced by diversity-purging selective sweeps, patch turnover in metapopulations, and population oscillations due to phage predation cycles (reviewed by Fraser et al. 28).
Another process that reduces Ne demographic bottlenecks may be particularly important in obligately host-associated bacteria.1,3 In host-dependent bacteria generally, Ne may be reduced due to physical bottlenecks during transmission to host offspring. The initial infection at the origin of their associations likely purged genetic diversity of these bacteria, if only a small sample of the ancestral species became established in a given host lineage. Vertically transmitted endosymbionts may then experience continual population bottlenecks upon each inoculation of developing host eggs or embryos. Empirical work in Buchnera has demonstrated this bottleneck phenomenon and has shown that the bottleneck size varies across aphid host species.29
While the prediction of reduced Ne in primary endosymbionts of insects is well justified, efforts to quantify Ne have been fairly limited. Currently, estimates across a lifestyle transition are limited to Buchnera versus free-living enteric bacteria such as E. coli. Estimates of Ne for E. coli have relied upon observed patterns of synonymous codon bias,30, 31 sequence variation at binding sites of gene regulatory proteins,32 calculations of the neutral mutation parameter, theta (θ, an estimator of Neμ, where μ is the neutral mutation rate),33 linkage disequilibrium,34 and the distribution of coalescence events in a sequence genealogy.35 Although some studies estimate Ne of E. coli as low as 105 (Refs 31 and 32), many estimates are reassuringly similar and fall in the range of 108–1010. All estimates show that Ne of this species is far less than the 1020 E. coli cells estimated to exist in natural habitats.33 Among primary endosymbionts, species boundaries are defined by the host; that is, strains inhabiting the same host species typically are considered the same endosymbiont species. Estimates of Ne in two Buchnera species based on θ and μ were Ne = 107, suggesting a ~100-fold reduction compared to most estimates for E. coli. 36, 37 This pattern is consistent with a lifestyle-associated reduction in Ne, though additional studies are needed to test whether a similar trend holds across phylogenetically and ecologically diverse endosymbiont lineages. The wide variation of Ne estimates for E. coli (noted above) highlights the challenging nature of this task.
A reduction in Ne has important implications for genome evolution, as it affects the impact of genetic drift. Specifically, under the nearly neutral theory of molecular evolution,38, 39 a balance between selection and genetic drift governs mutations with selective coefficients (s) near the reciprocal of effective population size (Ne). In this range, mutations with mildly harmful fitness consequences may persist under genetic drift, and some fraction of those may become fixed. To the extent that Ne is reduced in endosymbionts, some slightly deleterious mutations that would be eliminated by selection in species with larger Ne (e.g., free-living relatives) could persist and become fixed in endosymbionts.1, 3 Restricted opportunities for gene exchange (see below) may exacerbate the effects of genetic drift.3 Over time, the accumulation of deleterious mutations in endosymbiont lineages could negatively affect the fitness of both the host and the symbiont.
Shifts in natural selection
In bacteria, variation in selection due to habitat shifts may be an important force driving gene content variation. Because bacteria experience a strong mutational bias toward deletions (which are 10-times more frequent than insertions),40, 41 any genes that are not actively maintained by selection will be deleted eventually. Further speeding the removal of superfluous DNA, selection may actually favor the removal of inactivated genes due to the potential toxicity of pseudogenes.42 In light of these processes, selection to retain gene functions likely plays a major role in shaping bacterial gene contents.
Compared to living in an external environment, a strictly intracellular existence likely involves radical changes in selective regime. We might predict relaxed selection on many metabolic functions that are no longer required in a host cellular environment, resulting in the loss of associated genes over time. In addition, because endosymbiont and host fitness are intertwined, endosymbionts experience selection at the level of the insect host. This host-level selection favors the retention of functions that improve host fitness, and may limit the spread of selfish symbiont mutations that compromise host fitness.43
Shifts in mutation rate
The extensive theoretical and empirical studies exploring the evolution of mutation rates across organismal groups, including bacteria, are too vast to summarize here. However, I will highlight two types of models that may have particular relevance to endosymbionts.
Influence of environmental stability on mutation rates
One group of models emphasizes a ‘tension’ between the potential for adaptation through (relatively rare) beneficial mutations, versus the genetic load of deleterious mutations.44, 45 The balance between these factors may be influenced by environmental stability. That is, when populations encounter novel environments, selection for rare, beneficial mutations may outweigh selection against deleterious mutations. Under this scenario, elevated mutation rates may be favored. By contrast, in a stable environment, selection against deleterious mutations may be more important, and lower mutation rates favored. Experimental studies of E. coli support this model by demonstrating the evolution of a hypermutator phenotype (via a mutation in mutT) during adaptation to a novel environment, followed by subsequent evolution of a lower mutation rate (via mutations in mutY) after the potential for adaptation had declined.45
How stable is the intracellular niche? Life within bacteriocyte cytoplasm (or within host-derived vacuoles) may be relatively stable, due to consistent availability of certain nutrients and other compounds. In this sense, we might predict that low mutation rates would be favored in primary endosymbionts. However, the potential for continued adaptation nonetheless exists within this host niche. For example, a high prevalence of mutator strains in pathogens has been attributed to a continued need to evade the host immune system, and pathogen genes mediating host interactions are subject to selection favoring novel variants.46 In long-term mutualisms, the host immune system may likewise regulate endosymbionts.47 In principle, symbiont mutations that overcome host control may be favored by symbiont-level selection. Even stable, vertically-transmitted mutualisms are not immune from such evolutionary conflicts.48-50 In this sense, host interactions could favor elevated mutation rates in mutualists, as is generally accepted for pathogens.
Even if current selective forces do not favor novel mutualist genotypes, the initial transition to an endosymbiotic lifestyle was likely coupled with a significant environmental shift, and this high potential for adaptation could have favored hypermutator phenotypes. If such phenotypes involved the deletion of DNA repair genes, limited opportunities to reacquire the lost repair function (see gene exchange, below) could lead to persistently elevated mutation rates, even if elevated rates are no longer advantageous.
Influence of drift on mutation rates
A second group of models proposes that variation in genetic drift, as influenced by Ne, affects the evolution of mutation rates.51, 52 Across diverse organisms and organelles, an inverse relationship between mutation rate (measured as changes per nucleotide per generation, or μ) and Ne suggests that genetic drift plays a key role.51, 52 Assuming that a lower mutation rate is generally favorable, then mutation rate may be reduced if the selective advantage (s) of that further reduction exceeds the effects of genetic drift (1/Ne for haploids). Under this model, reduced Ne may lower the efficacy of selection to retain DNA repair functions that improve replication fidelity. Thus, to the extent that Ne is reduced in endosymbionts, we might predict an elevation in mutation rates. Even an early, transitory reduction in Ne (e.g., upon initial infection of a given host lineage) could lead to persistent elevation in mutation rates, if disrupted or deleted DNA repair functions cannot be reacquired.
Shifts in gene exchange and recombination
In addition to mutation, gene exchange within and across species can generate significant genomic variation. A recent study of genome flux found widespread horizontal gene transfer among bacterial groups, though rates vary considerably (25-fold) across lineages.53 Such transfer depends on opportunities for close interactions among genetically distinct bacterial strains or species, and/or phage that may shuttle bacterial genes. Such opportunities may be limited for species that are sequestered within host tissues or cells, particularly if the host niche excludes other bacteria. For example, the study above53 noted that certain obligate parasites (e.g., Chlamydia and Rickettsia) showed relatively low levels of horizontal gene transfer, consistent with reduced opportunities for such transfer in a host niche.
Genetic isolation may particularly severe in primary endosymbionts, which can inhabit the same host lineage for tens or hundreds of millions of years. In hosts that harbor just one primary endosymbiont, current evidence suggests the bacteria typically form clonal populations within bacteriocytes and are physically separate from any other bacteria in the host (e.g., separate from Wolbachia, facultative endosymbionts, or gut microbiota). Dual primary endosymbionts, such as those found in leafhoppers and other insect hosts, may also be segregated from each other, with each bacterial species inhabiting a distinct bacteriocyte type.7
Despite this typical pattern of cellular segregation, some primary endosymbionts do occur in physical proximity to other bacterial associates, creating potential opportunities for horizontal gene transfer. In a remarkable example of cellular integration, endosymbionts of mealybugs consist of a gamma-Proteobacterium (Moranella) that lives within the very cells of a beta-Proteobacterium (Tremblaya), which in turn resides within mealybug bacteriocytes.54 In addition, brief but potentially significant interactions between distinct endosymbionts may occur during transmission from host mother to offspring. Microscopy indicates that, in the first stages of endosymbiont transmission from maternal aphid bacteriocytes to developing embryos, cells of Buchnera and a facultative endosymbiont, Serratia, co-occur in the central syncytium,55 though they are separated into distinct host cells later in development. Thus, while primary insect endosymbionts may have reduced chances to exchange genes with other species, this lifestyle does not preclude such opportunities entirely.
The lifestyle of primary endosymbionts may also constrain strain-level recombination within a given mutualist species (i.e., inhabiting a given insect host species). To the extent that population bottlenecks during transmission purge variation, endosymbionts inhabiting a given host individual will be very similar genetically, and diversity within any given bacteriocyte may be further reduced. Thus, any recombination among endosymbionts within a host individual or cell may be unlikely to generate new combinations of alleles. Strict asexuality of this sort can exacerbate effects of genetic drift by reducing the ability to recover wildtype genotypes.3, 56
Along for the ride—selection at linked sites
The above constraints on recombination in endosymbionts may contribute to deleterious substitutions via selection at linked sites. In insect endosymbionts, linkage may be extensive, occurring not only across the entire endosymbiont genome, but also across distinct genomes that undergo stable, maternal inheritance in the host lineage. These genomes include the host mitochondrion and potentially other endosymbionts that experience periods of vertical transmission (e.g., Wolbachia and facultative endosymbionts).
Selection at linked sites may be important in the following ways. First, selection against strongly deleterious mutations reduces variation at linked sites and can speed the fixation of slightly deleterious mutations.57, 58 In addition, hitchhiking with selective sweeps of beneficial mutations may drive the fixation of neutral and deleterious changes at linked sites.59 Theory suggests this contribution of selective sweeps to deleterious evolution, termed genetic draft (named for the practice in cycling of following another’s path in order to reduce wind resistance), may be even more significant than genetic drift, especially in regions with low recombination.60-63 Under this process, positive selection favoring a particular mutation in a primary endosymbiont genome could “drag along” whatever neutral and even deleterious mutations happened to occur elsewhere in the genome. Likewise, selection favoring a particular Wolbachia genotype or mitochondrial variant could drive neutral and deleterious changes in the linked mutualist genome. Though genetic draft has received little attention in studies of endosymbiont evolution, it could potentially play a significant role in driving slightly deleterious substitutions.
In sum, the constrained lifestyle of primary insect endosymbionts may trigger shifts in fundamental processes such as genetic drift, natural selection, mutation, and recombination. Despite the current wealth of genome data, it remains challenging to distinguish the contributions of these processes to endosymbiont genome evolution. Their respective impacts can be difficult to untangle, as they may have shifted simultaneously along the long branches that typically separate primary endosymbionts from their closest non-endosymbiotic relatives.
Below, I consider empirical datasets that shed light on the genomic consequences of adopting an endosymbiotic lifestyle. Recent studies have illustrated important insights that can be gained by combining genomic data with intraspecific (within-species) population genetic analyses, in order to test for weak selective forces and to infer underlying mutational spectra. I focus on genome features for which insect endosymbionts often represent extremes: (1) rates of sequence evolution, (2) genome size and structure, and (3) base compositional biases. Analyses of these genome features have revealed common trajectories among many insect mutualists, surprising ‘exceptions to the rule,’ and unexpected parallels with free-living bacterial species.
Rates of sequence evolution
Early analyses showed that endosymbionts experience a significant acceleration of evolutionary rates at 16S rDNA, when compared to free-living relatives.3, 64 This rate increase involves substitutions that destabilize the structure of the rRNA molecule.65 The rate acceleration observed at 16S rDNA also affects endosymbiont protein-coding genes.3
What processes drive accelerated evolutionary rates in endosymbionts? In part, the observed rate increase could reflect elevated mutation rates in small genomes that have lost many DNA repair capabilities. A significant increase in per-site mutation rates (10-fold higher than any other bacterial group) documented in Buchnera 66 may characterize other endosymbionts. However, a recent study showed that evolutionary rates can vary widely among endosymbiont lineages. Two coresident bacteria, Baumannia and Sulcia, which inhabit the same leafhopper host group, showed dramatically different rates of nonsynonymous and synonymous substitution despite the two endosymbionts sharing a divergence date (i.e., the divergence of the leafhopper host).67 The rate difference is not obviously explained by differences in mutation rate, because the more slowly evolving endosymbiont had lost more DNA repair genes.67 Rather than mutational differences alone, it is possible that as-yet unknown selective forces, including selection on synonymous sites, may influence rates of sequence evolution.
In addition, comparisons of nonsynonymous versus synonymous variation suggest that elevated mutation alone may not entirely explain accelerated rates. In some groups, the rate acceleration affects nonsynonymous sites more severely, leading to an overall elevation in the ratio of nonsynonymous to synonymous substitutions (calculated as Ka/Ks, or dN/dS) compared to free-living relatives.3, 68-70 Increased mutation alone is expected to affect both nonsynonymous and synonymous sites with no expected shift in Ka/Ks. On the premise that nonsynonymous changes are more likely to be harmful than beneficial, elevated Ka suggests relaxed and/or reduced efficacy of purifying selection against these changes.71 It is also possible that increased selective constraint on synonymous changes could suppress Ks and thus elevate Ka/Ks.
In some insect endosymbionts such as Buchnera,68, 69 as well as other obligately host-associated bacteria,72 the elevation in Ka/Ks occurs across most genes sampled and across varied protein functions, suggesting a genome-wide phenomenon. While shifts in selection across most genes is possible, a simpler explanation might be an increased effect of genetic drift, resulting in a genome-wide decrease in the efficacy of purifying selection against nonsynonymous changes.3, 72, 73 This interpretation aligns with the use of genome-wide Ka/Ks as a proxy for the efficacy of selection, and thus a way to infer or confirm differences in bacterial Ne.72,,74
Distinguishing whether accelerated rates of sequence evolution in endosymbionts reflect shifts in selection, drift, or mutation is challenging, since these forces often have similar predicted effects on sequence divergences and may shift simultaneously. Intraspecific, population genetic analyses can more clearly distinguish these processes based on their distinct impacts on patterns of polymorphism within species75 (Figure 2). A neutrality index (NI)76 in excess of 1 indicates an excess of nonsynonymous polymorphisms compared to neutral expectations, suggesting that some portion of nonsynonymous polymorphisms are removed by selection before becoming fixed between species (see Fig. 2 legend). This pattern is consistent with the hypothesis that nonsynonymous changes are slightly deleterious. In addition, quantifying the frequency spectrum of polymorphisms within species can distinguish subtle impacts of selection. For example, an excess of rare alleles compared to that expected under neutrality is consistent with negative selection preventing the spread of slightly deleterious mutations, although this pattern is also consistent with various demographic processes 77.
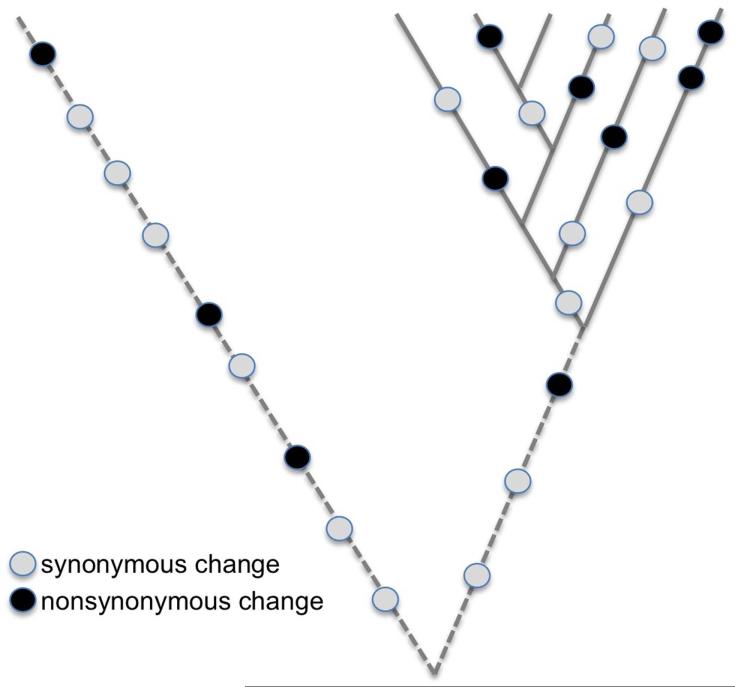
Figure 2.
Schematic of intraspecific sampling to test neutrality. Many population genetic tests rely on sampling intraspecific polymorphisms (marked along solid lines), for comparison to patterns of interspecific substitutions (marked along dashed lines). Such tests often compare two categories of changes with distinct predicted fitness effects, such as nonsynonymous changes (which may be selected for or against) and synonymous changes (typically assumed to be effectively neutral). To test for subtle selection on nonsynonymous changes, nonsynonymous and synonymous polymorphisms and substitutions are compared. Under the null hypothesis that observed nonsynonymous changes are neutral, the ratio of nonsynonymous to synonymous polymorphisms within species (Pn/Ps) will equal the ratio of nonsynonymous to synonymous substitutions between species (Dn/Ds). If, however, nonsynonymous changes are slightly deleterious, then a portion of these changes will be eliminated by selection prior to being fixed between species, leading to a reduction in Dn/Ds when compared to Pn/Ps.137 This schematic figure reflects such a pattern (i.e., a dearth of nonsynonymous substitutions, or in other words, an excess of nonsynonymous polymorphisms). The neutrality index (NI) captures this “ratio of ratios,” by calculating (Pn/Ps)/(Dn/Ds).76 NI>1 indicates an excess of nonsynonymous polymorphisms compared to neutral expectations, consistent with nonsynonymous changes being slightly deleterious. Such results have suggested that nonsynonymous substitutions in endosymbionts are slightly deleterious, consistent with the hypothesis that genetic drift contributes to accelerated rates of protein evolution 37, 36.
Population genetic analyses within Buchnera species (i.e., Buchnera inhabiting the same aphid host species) have revealed NI > 1, as well as an excess of rare nonsynonymous polymorphisms, consistent with nonsynonymous changes being slightly deleterious.37, 36 While the above patterns support a role of genetic drift in this aphid mutualist, current data are too limited to extrapolate across endosymbiont groups. Moreover, population genetic analysis of diverse free-living bacteria suggests that NI > 1 is not particularly unusual.78 Further highlighting the complexity of interpretations, NI < 1 (an excess of nonsynonymous substitutions between species compared to neutral expectations, often interpreted as positive selection) could possibly signal deleterious evolution in bacteria, if this pattern results from the elimination of polymorphisms rather than the fixation of substitutions.78
Moving forward, clarifying the forces shaping endosymbiont sequence evolution will benefit from additional molecular datasets gathered with that goal in mind. The abundance of genome sequences has provided valuable insights, but useful comparisons of Ka/Ks are not always possible, e.g., due to saturation of Ks or uncertainty about the effective neutrality of synonymous changes. Population genetic analyses offer increased power to decipher subtle fitness effects of mutations, thus enabling tests of whether host-dependent genomes are more vulnerable to slightly deleterious changes. An additional, perhaps more challenging task will be developing models that distinguish among processes that may drive deleterious changes, including bottleneck-related genetic drift versus linkage to positively selected sites (genetic draft).
Genome size and structure
Contributing to the exponential increase in available bacterial genomes, numerous host-associated species have been fully sequenced in recent years, including several primary mutualists of insects. These data have revealed a consistent trend of genome reduction, with genome sizes <800 kb, or even much smaller 8 (Fig. 3). Proteins retained by these small genomes may have a widened functional complexity compared to orthologs in larger genomes,79 and small mutualist genomes tend to lose redundant pathways, or duplicate components that can perform the same function to a certain extent.80 Even so, it seems clear that their extensive genome reduction leads to metabolic simplification.
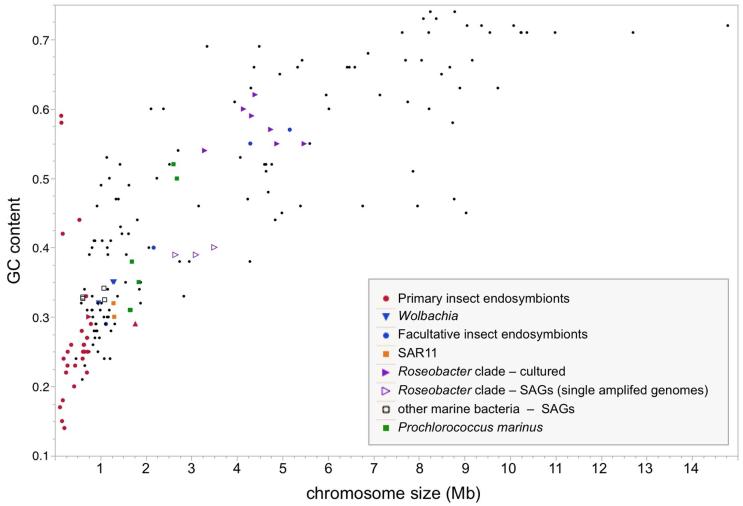
Figure 3.
Chromosome size and genomic GC content across select bacterial genomes. The genomes of primary endosymbionts of insects are severely reduced and, with some notable exceptions, AT-rich. Primary endosymbionts shown here represent long-term, intracellular mutualists (red circles), as well as Serratia symbiotica of the cedar aphid (red upright triangle), which represents a very recent transition to obligate mutualism 138, and Ishikawaella capsulata (red sideways triangle), a nutritional mutualist of the plataspid stinkbug that shows striking genome reduction despite being extracellular.139 Intracellular endosymbionts that transfer among insect groups include Wolbachia and facultative endosymbionts. Because facultative endosymbionts tend to possess a high abundance of insertion sequences and pseudogenes, they have a lower coding capacity than suggested by their moderate chromosome sizes. Genomes less than or near ~1 Mb that are unlabeled (black dots) largely represent intracellular pathogens, such as Mycoplasma, Rickettsia, and Chlamydia. Several groups of free-living marine bacteria with surprisingly small genomes are shown, such as SAR11 and Prochlorococcus marinus. SAGs (single amplified genomes, marked with open symbols) typically represent uncultured strains and include representatives of the Roseobacter clade 120 and other marine bacteria.119 Comparisons within the Roseobacter clade show that SAGs have smaller genomes than related cultured isolates.120 For SAGs, genome size and %GC is typically an estimate that is extrapolated from incomplete genome coverage. Data for fully sequenced genomes were downloaded from JGI’s Integrated Microbial Genomes database (http://img.jgi.doe.gov) on August 15, 2014, and estimates for SAGs were obtained from publications.119, 120
Genome reduction and metabolic integration
Endosymbiont lineages vary in specific genes and pathways that are deleted versus retained, probably reflecting distinct selective pressures across insect hosts and historical contingencies.81 However, broadly speaking, mutualist genomes preferentially retain fundamental cellular processes (e.g., transcription, translation) and biosynthesis of nutrients that are missing from the host’s diet (e.g., amino acids among sap-feeding insects, cofactors among blood-feeders).8, 9 In this sense, genome reduction in primary mutualists reflects functional integration and shared metabolism with the insect host (and sometimes with another co-residing bacterial mutualist, see below). A mossbug mutualist, Evansia, even relies on the insect host for basic housekeeping functions.82 The integration of Buchnera with its aphid hosts involves the targeting of host-encoded proteins to endosymbiont cells.83 Such intimate integration suggests that some insect mutualists have approached, or even crossed into, the zone of organelles.84
Metabolic integration and genome reduction can be particularly extreme co-primary endosymbionts, in which two bacteria form a stable mutualistic association with a given host lineage.7, 8 In these cases, nutrient biosynthesis is often shared among symbiotic players, including the two bacteria and the host.54, 85-91 In light of their functional integration, it is not surprising that these dual endosymbioses include the smallest genome known among cellular life (112 kb genome of Nasuia in leafhoppers.7, 91
Genome stability
Limited gene exchange
Most primary endosymbionts sequenced to date show little, if any, signs of gene acquisition via horizontal gene transfer. Instead, these mutualists tend to be much-streamlined version of their free-living relatives, retaining those functions required to perform basic cellular processes and to fulfill their symbiotic role. Likewise, intraspecific analysis shows identical genealogies across endosymbiont gene regions, arguing against any shuffling of alleles through recombination.92
This striking genome stability may reflect ecological constraints on gene exchange in the intracellular environment and the loss of many DNA repair functions, some of which are also involved in recombination. In addition, the small genomes of primary endosymbionts tend to lack phage, transposable elements, insertion sequences, and other mobile DNA.93, 94 In the absence of frequent recombination with genetically distinct strains, their genomes might not permit parasitic DNA that requires horizontal transfer for its survival.93 In addition, without gene exchange between or within endosymbiont groups, gene losses may be irreversible and constrain the evolutionary potential of the symbiont and host alike.
A surprising exception to this asexuality was discovered in a bacteriocyte-associated, beneficial Wolbachia strain inhabiting bedbugs, which recently acquired an operon encoding the complete biotin biosynthetic pathway.95 This acquisition allows the Wolbachia strain to synthesize and provision vitamin B and likely underlies its recent transition to obligate mutualism. The authors suggest this operon could have been acquired from another endosymbiont, such as Cardinium or Rickettsia, which coinfect the same host.95
Genome rearrangements
In addition to gene exchange, intragenomic changes (genome rearrangements via inversions, duplications, and translocations) constitute an important mechanism of genome flux in many bacteria. Such changes can move or duplicate genes to new chromosomal contexts and place them under the control of different promoters. Promoter capture and gene duplication have been shown to contribute to evolutionary innovation in lab-reared E. coli cultures.96
Current data suggest such intragenomic changes are quite rare in most primary mutualists. Comparisons within endosymbiont groups have revealed that extensive gene loss occurred early in these associations, often before the divergence of genomes sampled.97, 98 Synteny plots of endosymbionts belonging to the same group have revealed few if any changes in gene order and strand orientation across tens or hundreds of millions of years,97, 98 as summarized recently.99 Exceptions to this pattern include extensive rearrangements in a whitefly endosymbiont, Portiera.99 These events are concentrated in a particular lineage of Portiera that also possesses many tandem repeats, suggesting that the unusual persistent of repeats – which are generally missing from endosymbionts – could mediate inversion events.
Young endosymbionts reveal early genome turmoil
The above patterns of genome reduction and stability illustrate profound, long-term genomic consequences of endosymbiosis. To clarify the processes that generate these outcomes, studies of younger endosymbionts have been useful. These bacteria include facultative endosymbionts from which some primary endosymbionts apparently evolved,24, 25 as well as species or strains that adopted an obligately mutualistic lifestyle very recently. These groups offer snapshots of early changes along the road of endosymbiosis.93 Data for numerous facultative endosymbionts and young primary mutualists include the tsetse fly symbiont Sodalis and close relatives,100-102 Cardinium in whiteflies,103 aphid symbionts in the Serratia group,104-106 the aphid secondary symbionts Regiella 107 and Hamiltonella,108 among others. Analyses of these genomes support the notion of genome flux, including abundant pseudogenes suggesting rapid gene inactivation, and an explosion of insertion sequences (IS’s) and other mobile elements that may mediate gene inactivations, large inversions, and deletions.
This turmoil apparently subsides soon after the transition to obligate mutualism. In a young primary mutualist of weevils (SOPE), the ‘epidemic of transposition’ in its Sodalis relatives may be waning, consistent with a prediction of increased stability in primary mutualists.102 Interestingly, in SOPE, IS-mediated duplications include a duplication of co-chaperones groES and groEL, which mediate protein-folding, are constitutively overexpressed in many primary endosymbionts 109, 110 and may buffer the fitness effects of deleterious mutations.111 Duplications of these chaperonins in SOPE could represent an adaptive change mediated by early genome flux.102 Consistent with further genome reduction and a calming of turmoil in obligate mutualists, a newly-obligate, co-primary Serratia endosymbiont in cedar aphids has experienced more extensive gene losses and a depletion of mobile elements upon its recent transition to obligate mutualism.105, 106
Untangling forces driving genome reduction
Despite insights from early snapshots of genome reduction, dissecting the evolutionary forces driving gene loss in endosymbionts is challenging. While IS’s and other mobile DNA apparently act as molecular mechanisms that contribute to early gene inactivation and deletion, the evolutionary mechanisms driving this process are less certain. Relaxed selective constraint on many gene functions may lower selective coefficients against element insertion, thus broadening the genomic space for neutral insertion of mobile elements. Alternatively or simultaneously, reduced Ne of endosymbionts could reduce the efficacy of selection to maintain gene functions. Adaptive changes may also occur in this period of turmoil, such as duplications (as suggested in SOPE, above) and gene losses. Studies of free-living bacteria have contributed to a model of adaptive genome streamlining, which could affect endosymbionts in principle. Perhaps it is most likely that all three mechanisms (neutral processes, deleterious evolution, and even adaptation) contribute to genome changes. Distinguishing among these drivers of gene loss is difficult and often relies on untested assumptions about the fitness consequences of losing particular functions.
Neutral gene loss, due to relaxed selection in the intracellular niche
Many patterns of gene loss in endosymbionts suggest relaxed selection on numerous gene functions in an intracellular niche. The underlying deletion bias in bacteria implies that genes that are not actively maintained by selection will be eventually deleted.40, 111 Relaxed selection may explain striking patterns of differential gene loss in endosymbionts, such as the tendency to lose genes for metabolic diversity yet retain basic housekeeping functions. This process can also explain uncanny metabolic collaboration, including the retention of specific host-beneficial biosynthesis functions and exquisite sharing of metabolic pathways between co-resident primary endosymbionts.
Deleterious gene loss
References to genome reduction in insect endosymbionts often attribute gene loss to genetic drift in small populations,113 and certain patterns of gene loss seem consistent with deleterious changes. For instance, reconstruction of gene deletion events revealed that many Buchnera genes were lost in large, early deletion events involving diverse functions,114 a process that is difficult to reconcile with neutral loss of specific functions that are no longer required within host cells. In addition, certain losses of biosynthetic functions in established mutualist lineages, such as the loss of cys genes in Buchnera of the green bug aphid Schizaphis graminum 98 and the loss of glnA in Blochmannia of the ant Camponotus vafer,115 are surprising. Such losses could represent deleterious deletions that reduce the fitness of the host-symbiont partnership. However, even in these cases, it is difficult to rule out a scenario of relaxed selective constraint and neutral gene deletion, or symbiont-level selection favoring the erosion of host-beneficial functions. While earlier studies support the accumulation of slightly deleterious changes in endosymbiont sequences (e.g., elevated Ka/Ks and patterns of intraspecific variation, see above), this does not necessarily imply that gene losses are deleterious, since purifying selection against the loss of a gene function could be much stronger than selection against nonsynonymous changes in existing genes.
Adaptive genome reduction—models from free-living bacteria
Recent genome and metagenome datasets for free-living bacteria have revealed a surprising prevalence of genome reduction 113 (Fig. 3). While genome reduction in free-livers is not as severe as that found in primary endosymbionts, its discovery has prompted a wider interest in evolutionary mechanisms shaping genome size. Free-living bacteria with small genomes discovered to date tend to live in nutrient-poor marine environments. Even cultured isolates can have small genomes, including strains of Prochlorococcus marinus (the abundant photosynthetic species with genomes as small as 1.64 Mb116) and culturable members of SAR11, a diverse clade characterized by small (average 1.34 Mb), AT-rich genomes.117, 118 Single-cell genomics of bacteria in oligotrophic environments have revealed that uncultured representatives have even smaller (e.g., an estimated 0.61 Mb for strain AAA076-M08 119) and, interestingly, more AT-rich genomes than related cultured isolates. Likewise, genomes of uncultured Roseobacter are smaller (2.64–3.1 Mb, range) and more AT-rich than cultured isolates (typically > 4 Mb) 120 (Fig. 3). A recent synthesis of these data indicates that genome reduction in free-living bacteria is far more common than previously appreciated, may explain why most bacteria cannot be cultured in the lab, and may contribute to the interconnectedness of microbial communities.113 As more uncultured bacteria are sequenced, our notion of a ‘typical’ genome size range for free-living bacteria will likely shift downward.
While diverse mechanisms may underlie genome reduction in free-living bacteria,116 adaptation has been emphasized. Under a model of adaptive genome reduction, often phrased as genome streamlining, smaller genomes may be advantageous, particularly in nutrient-poor environments where fitness is determined by competition for scarce resources. Possible selective advantages of a smaller genome may relate to metabolic efficiency and fewer requirements for energy and nutrients such as phosphorus during replication,121 though evidence for elemental-based streamlining has been critiqued.122 In addition, to the extent that smaller genomes correlate with smaller cell size, increased surface to volume ratios associated with smaller cells may be advantageous.113, 116 Experimental evolution studies have showed fitness benefits of deleting DNA,123, 124 though the trait favored by selection (e.g., loss of gene functions, or reduction in the number of DNA base pairs) is difficult to dissect.
In support of an adaptive explanation to genome reduction, molecular evolutionary analyses of Prochlorococcus showed that the period of extensive gene loss was associated with increased intensity of purifying selection across the genome, as evidenced by reduced dN/dS.116 This pattern is consistent with an expansion of Ne, leading to increased efficacy of selection across genes. The distribution of deleted genes across many functional categories suggests that gene losses were not due to ecological specialization and relaxed selection on particular functions (in which case only a subset of functions would be affected). These results highlight the complex relationship between Ne and genome size. That is, genome reduction could conceivably result from declines in Ne (reduced efficacy of selection to maintain functions, an hypothesis often invoked to explain small endosymbiont genomes) as well as elevations in Ne (increased efficacy of selection favoring metabolic efficiency, which may contribute to small genomes of some marine bacteria).
It remains uncertain whether selection favoring smaller genomes per se offers a general explanation for gene loss in free-living lineages. Studies of Roseobacter suggest some gene losses may reflect relaxed selection on particular functions 125, 126 and thus represent neutral (rather than adaptive) losses. Moreover, an accelerated rate of nonconservative amino acid changes in uncultured Roseobacters with reduced genomes is consistent with a model of increased genetic drift.120 Though, recent comparisons of genome-wide dN/dS suggest that Roseobacter generally have a larger Ne than the more abundant SAR11 clade.74 This result highlights the complex relationship between Ne and Nc (census population size) in bacteria.
Base compositional biases: what does mutation not explain?
With some notable exceptions (recently highlighted 8), genomes of primary endosymbionts tend to be extremely AT-rich (Figure 3). To understand which evolutionary processes drive such low %GC, it is useful to consider the broader context of base compositional variation in bacteria, which spans from 13.5 to >75% GC. In principle, several mechanisms (or combinations thereof) may explain this wide variation, as recently reviewed.127 First, base composition of a given genome may be neutral and shaped by underlying mutational biases. Observations that %GC at 4-fold degenerate sites (GC4) is even more extreme than genome-wide values support a mutation hypothesis, on the basis that selection at degenerate sites is weak or nonexistent so that GC4 reflects underlying mutation more closely than other categories of sites.128 Under this model, the low GC content of most endosymbionts could be explained by more extreme AT mutational bias in these lineages.
As an alternative view, variation in GC content could reflect differences in the strength or direction of selection on base composition. This potential for selection on GC content has important implications, as degenerate sites are often treated as effectively neutral in molecular evolution studies. Selection on base composition may relate to differences between AT and GC pairs in their stability, flexibility, availability, or different metabolic costs of synthesizing GC versus AT pairs. In principle, mutational bias could be comparable across bacterial lineages, but selection on GC content may vary in its direction (favoring AT bases in some genomes, but GC in others) or intensity (stronger selection in some lineage than others). Under this model, the low %GC of most intracellular bacteria could be explained by selection favoring AT base pairs, e.g., since ATP is relatively abundant in an intracellular niche.129 A third model suggests that higher GC content is generally favored by selection, but such selection is less effective in endosymbionts due to their small Ne.
Intraspecific datasets have proved invaluable for testing these various models. The reconstruction of GC→AT and AT→GC changes within endosymbiont species 66, 130, 131 and across diverse bacteria 132, 133 has shed light on underlying mutational pressure. Such studies quantify the mutational spectra as the absolute numbers of GC→AT versus AT→GC changes within species (Fig. 4A). This comparison tests whether a current sequence is at mutational equilibrium, in which case the two values are equal. In addition, calculation of the relative per-site mutational bias quantifies underlying mutational biases, allowing direct comparisons across genomes (Fig. 4B). This per-site value also estimates the %GC predicted under mutational equilibrium (often termed GCeq or GCpred). Comparing this predicted value to the current, observed GC content tests the extent to which mutation can explain current base composition. If GCpred differs from observed GC content, then selection on base composition could play an important role.
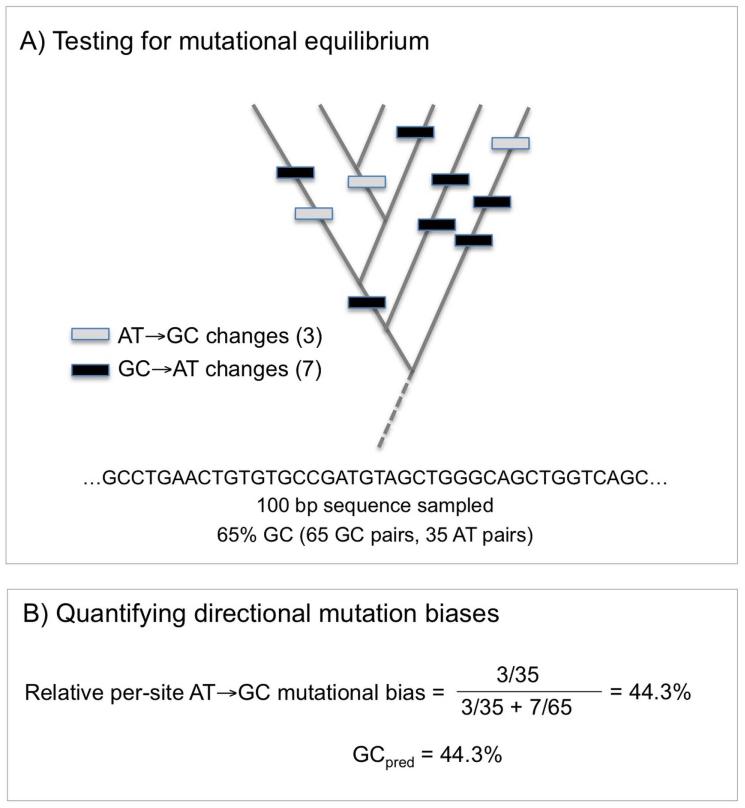
Figure 4.
Simple example of quantifying mutational spectra. Intraspecific analyses of directional base changes allow estimations of underlying mutational processes. Schematic shows a simple analysis of a 100-bp sequence. (A) Testing for mutational equilibrium addresses the question: Is current base composition explained by mutation alone? Comparing the absolute numbers of GC→AT versus AT→GC changes within species sheds light on underlying mutational pressure. If the number of AT→GC changes does not equal the number of GC→AT changes along a given sequence, then under mutation alone, the base composition of the sequence is expected to shift. By contrast, if GC→AT and AT→GC changes are equal, this implies the sequence is at mutational equilibrium. Lack of equilibrium may result from a recent change in mutational pressure or the action of natural selection favoring either GC→AT or AT→GC changes. In this example, GC→AT changes far outnumber AT→GC changes, so the sequence is not at mutational equilibrium. Rather, under mutation alone, it would become more AT-rich. (B) Quantifying directional mutation biases addresses the question: Is mutation AT or GC biased? Per-site rates of AT→GC and GC→AT changes (v and u, respectively) are obtained by dividing the absolute number of inferred changes by the frequency of the original nucleotides. In this example, v = 3/35, and u = 7/65. From these per-site rates, one can then calculate the relative AT→GC per-site mutational bias as v/(v + u). In this example, this relative per-site bias is 44.3%. Since it is less than 50%, this implies a slight GC→AT mutational bias (any given GC pair is more likely to mutate to AT, than vice versa). This value equals the base composition that would be expected under mutation alone (GCpred). In this example, the sequence is expected to be slightly AT-rich (44.3% GC) under mutation alone. The fact that the sequence is actually 65% GC suggests that selection favors AT→GC mutations.
Recent intraspecific analyses indicate that selection shapes base composition in many bacterial genomes, but that directional mutational biases also contribute to observed variation in %GC. Evidence for selection includes observations that many bacterial genomes are not at mutational equilibrium; that is, mutation alone cannot explain base composition.131-133 In particular, many moderate to high %GC bacteria are actually more GC-rich than expected under mutation alone, suggesting that selection favors AT→GC changes over GC→AT changes in these species 131-133 (Fig. 5).
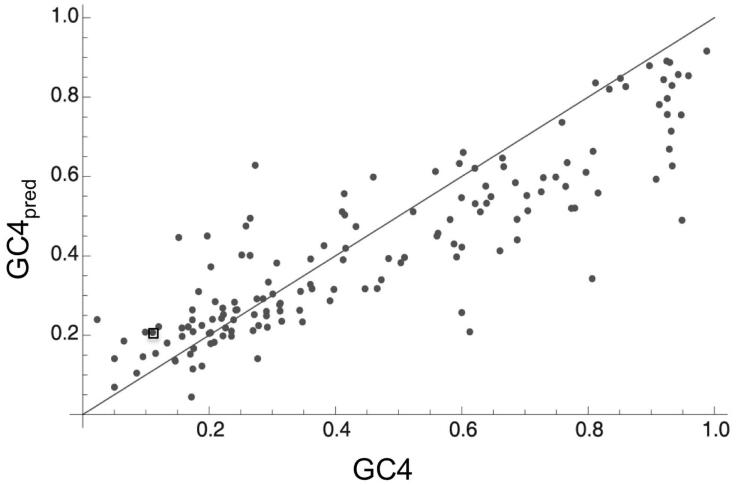
Figure 5.
The equilibrium GC content under the mutation bias model, reproduced from Hildebrand et al.133 Plot compares GC4 (current GC content at 4-fold degenerate sites) to GC4pred (GC content at 4-fold degenerate sites predicted under mutational equilibrium, estimated from intraspecific analysis), across numerous bacterial genomes spanning eight phyla. Estimates of GC4pred are based on the general approach described in Figure 4, but calculated at 4-fold generate sites only. The wide range of GC4pred across bacterial genomes suggests variation in relative per-site mutational bias. Deviation between GC4pred and GC4 indicates that mutation alone cannot explain current base composition and suggests that selection play an important role. original reference 133 for a discussion of alternative explanations to selection, such as biased gene conversion (BGC), and evidence against these alternatives. While this figure suggests that AT-rich bacteria also show a deviation between GC4pred and GC4, and in the opposite direction (less GC-rich than expected under mutation), this deviation was not significant among AT-rich genomes after adjusting for the infinite sites model.133 The point corresponding to the aphid mutualist Buchnera is enclosed in a square, for reference. Other AT-rich bacteria shown include intracellular pathogens.
How might selection act on GC content? While several possible mechanisms have been proposed, few have been tested experimentally. One exception is a recent experimental study in E. coli showing that higher GC4 of an introduced gene corresponded with faster growth rates.134 Growth rates were slightly faster when the introduced gene was transcribed (but not translated), and were significantly faster when the gene was also translated. Observed differences were unrelated to the codon adaptation index. While the specific mechanism is uncertain, selection may favor increased structural stability of the transcript through G:C pairings. Such selection on GC4 may explain why, in most bacterial genomes, GC4 exceeds %GC at intergenic regions.134 Interestingly, in very AT-rich genomes, including most endosymbionts, the opposite is true: GC at intergenic regions exceeds GC4, suggesting selection might act to lower GC4 in some AT-rich genomes, perhaps by further enrichment of A and U, which can engage in A:U pairings in already AU-rich transcripts.134
Intraspecific analysis of the unusually GC-rich genome of the cicada endosymbiont, Hodkinia, revealed that current %GC (%58 genome-wide) exceeds the value expected under mutation alone (~%42), suggesting that AT→GC changes are selectively favored in this mutualist.131 This evidence for selection on GC content is fascinating, as weak selection is often thought to be ineffective in endosymbionts under the hypothesis they have reduced Ne. It is possible this endosymbiont has a relatively large Ne, or that selection on %GC is particularly strong.
The lack of equilibrium in many bacterial genomes suggests that mutation alone does not explain base composition; however, this does not negate the possibility that variation in underlying mutational biases may contribute to base composition variation. Strikingly, GC4 content predicted under mutation (GC4pred) varies considerably and positively associates with observed GC4 133 (Fig. 5), suggesting that directional mutational biases may explain an large portion of the observed base composition variation. GC4pred for many GC-rich genomes exceeds 50%,133 suggesting that some taxa may experience a GC-biased relative per-site mutational bias (i.e., any given AT base pair is more likely to change to a GC pair than vice versa). As a caveat, however, underlying mutational spectra are inherently challenging to estimate, and 4-fold degenerate changes may be subject to weak selection favoring GC pairs, even when comparisons are based on closely related bacterial strains.132
By contrast to the variable and often moderate GC4pred of many bacteria, the exceptional AT-richness of most endosymbiont genomes may largely reflect extreme per-site AT mutational biases in these genomes, evidenced by their exceptionally low GC4pred (Fig. 5). This pattern implicates mutation as a major driving factor and agrees with earlier work showing that Buchnera experiences a strong underlying AT mutational bias and that its AT-rich genome is at mutational equilibrium.66, 130 Such biases may arise due to the frequent loss from endosymbionts (and other small, AT-rich bacterial genomes) of DNA repair genes that are known to repair GC→AT transitions, a common type of mutation in bacteria.
Future directions
The constrained lifestyle of primary insect endosymbionts may profoundly affect fundamental evolutionary mechanisms, such as genetic drift, natural selection, mutation, and recombination. The respective impacts of these forces on genome evolution are often difficult to untangle. When exploring mechanisms driving endosymbiont evolution, it is humbling to recognize that shifts in some or all of these forces may occur simultaneously upon transitions to a host-dependent lifestyle and contribute to the distinct genome features of these mutualists.
A major question in studies of long-term endosymbionts remains: to what extent are observed genomic changes neutral, deleterious, or adaptive? Addressing this question will benefit from comparisons within endosymbiont groups, in order to infer processes that drive shifts in sequence evolution, gene content, and base composition. While the first genome sequenced for an uncharacterized endosymbiont group offers novel insights into diverse outcomes of this lifestyle, comparisons among multiple genomes within a group provide valuable data to untangle mechanisms generating these outcomes, including the roles of mutation, genetic drift, and selection.
Population genetic analyses based on intraspecific samples are particularly well suited to distinguish underlying processes shaping sequence variation. Sequencing multiple genomes of the same endoysmbiont species (i.e., several strains inhabiting one host species) is increasingly feasible, given the ever-increasing throughput and decreasing cost of DNA sequencing. Such analyses provide insights into the dynamics of mutations within population and allow the detection of subtle selection coefficients. As noted above, statistical tests of neutrality can distinguish subtle fitness effects of nonsynonymous changes. In studies of base compositional biases, intraspecific data sheds light on underlying mutational spectra, in order to assess whether observed compositions can be explained by mutation alone, or whether we must invoke additional processes such as selection. While such tests are not without assumptions (e.g., neutrality of synonymous sites), and challenges (e.g., the need for random sampling within species), these approaches can provide unique insights into the evolutionary forces that drive and constrain molecular evolution of endosymbionts.
The same principles may be applied to understand the fitness effects of gene loss, by testing whether polymorphic gene disruptions and losses are effectively neutral or, alternatively, influenced by selection. For example, if gene disruptions were polymorphic within a species, then analyzing the frequency distribution of these ‘alleles’ could potentially distinguish whether disruptions are neutral vs. selected. When interpreting patterns of genome reduction in bacteria (both endosymbionts and free-living species), neutrality serves well as the simplest null model. A large portion of genome reduction may simply reflect the neutral loss of genes that are no longer needed. Under this model, the extent and patterns of genome reduction are expected to vary across lineages; because environments differ, so do selective coefficients to maintain particular gene functions. Relaxed selection may reflect availability of particular compounds in the environment, including metabolites available from other community members. Bacteriocytes represent simple, enclosed, and unusually persistent ‘communities’ consisting of bacteria and the host, in which interdependency and associated gene losses can be extreme. Clearly, neutral, adaptive, and deleterious processes may contribute to genome reduction, but conclusive inference of such processes requires more data than is currently available for most bacterial groups.
Ideally, such comparative approaches to assess mechanisms of genome change will be coupled with genetic manipulation, to test effects of observed variation (e.g., protein sequence variation, gene disruptions) on fitness in experimental systems. While such genetic manipulation has not been performed for most endosymbionts, including long-term primary mutualists of insects, genetic manipulation of uncultured bacteria is an area of active research, and progress has been made in secondary symbionts of insects 135, 136. Expanding comparative genomic and experimental approaches across bacteria, including uncultured free-living groups, will shed light on whether endosymbiotic and free-living species indeed occupy distinct evolutionary trajectories, or if shared processes contribute to genome changes across lifestyles.
Acknowledgements
I thank Richard R. Lawler for helpful discussion, Adam B. Lazarus and Erika del Castillo for their contributions to Figure 1, and Adam Eyre-Walker for permission to reproduce Figure 5. The writing of this review was supported by grants from NSF (MCB-1103113) and NIH (R01GM062626).
Footnotes
Conflicts of interest
The author declares no conflicts of interest.
References
- 1.Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 2.Brynnel EU, et al. Evolutionary rates for tuf genes in endosymbionts of aphids. Mol Biol Evol. 1998;15:574–582. doi: 10.1093/oxfordjournals.molbev.a025958. [DOI] [PubMed] [Google Scholar]
- 3.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigenobu S, et al. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 5.Buchner P. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, Inc; New York: 1965. [Google Scholar]
- 6.Ishikawa H. Insect Symbiosis: An Introduction. In: Bourtzis K, Miller TA, editors. Insect Symbiosis. CRC Press; Boca Raton: 2003. pp. 1–21. [Google Scholar]
- 7.Moran NA, Bennett GM. The Tiniest Tiny Genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 8.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 9.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 10.Gosalbes MJ, et al. Genomics of intracellular symbionts in insects. Int J Med Microbiol. 2010;300:271–278. doi: 10.1016/j.ijmm.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Feldhaar H, Gross R. Insects as hosts for mutualistic bacteria. Int J Med Microbiol. 2009;299:1–8. doi: 10.1016/j.ijmm.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zientz E, Dandekar T, Gross R. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev. 2004;68:745–770. doi: 10.1128/MMBR.68.4.745-770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFall-Ngai M. Adaptive immunity: Care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 14.Kerney R, et al. Intracellular invasion of green algae in a salamander host. Proc Natl Acad Sci U S A. 2011;108:6497–6502. doi: 10.1073/pnas.1018259108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwan JC, Schmidt EW. Bacterial endosymbiosis in a chordate host: long-term co-evolution and conservation of secondary metabolism. PLoS One. 2013;8:e80822. doi: 10.1371/journal.pone.0080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil R, et al. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci U S A. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldhaar H, et al. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 2007;5:48. doi: 10.1186/1741-7007-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabree ZL, Kambhampati S, Moran NA. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci U S A. 2009;106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akman-Gündüz E, Douglas A. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 2009;276(1658):987–91. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldhaar H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecological Entomology. 2011;36:533–543. [Google Scholar]
- 21.Munson MA, et al. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark MA, et al. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution. 2000;54:517–525. doi: 10.1111/j.0014-3820.2000.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 23.Sauer C, et al. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol. 2000;50(Pt 5):1877–1886. doi: 10.1099/00207713-50-5-1877. [DOI] [PubMed] [Google Scholar]
- 24.Wernegreen JJ, et al. One nutritional symbiosis begat another: phylogenetic evidence that the ant tribe Camponotini acquired Blochmannia by tending sap-feeding insects. BMC Evol Biol. 2009;9:292. doi: 10.1186/1471-2148-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husnik F, Chrudimsky T, Hypsa V. Multiple origins of endosymbiosis within the Enterobacteriaceae (gamma-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 2011;9:87. doi: 10.1186/1741-7007-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher R. The genetical theory of natural selection. Oxford University Press; 1930. [Google Scholar]
- 27.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser C, et al. The bacterial species challenge: making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 29.Mira A, Moran NA. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb Ecol. 2002;44:137–143. doi: 10.1007/s00248-002-0012-9. [DOI] [PubMed] [Google Scholar]
- 30.Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berg OG. The evolutionary selection of DNA base pairs in gene-regulatory binding sites. Proc Natl Acad Sci U S A. 1992;89:7501–7505. doi: 10.1073/pnas.89.16.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Wilson A. Evolutionary history of the enteric bacteria. In: Neidhardt F, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Amer. Soc. Microbiol.; Washington, D.C: 1987. pp. 1649–1654. [Google Scholar]
- 34.Selander RK, Caugant DA, Whittam TS. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Amer. Soc. Microbiol.; Washington, D.C.: 1987. pp. 1625–1648. [Google Scholar]
- 35.Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbot P, Moran NA. Extremely low levels of genetic polymorphism in endosymbionts (Buchnera) of aphids (Pemphigus) Mol Ecol. 2002;11:2649–2660. doi: 10.1046/j.1365-294x.2002.01646.x. [DOI] [PubMed] [Google Scholar]
- 37.Funk DJ, Wernegreen JJ, Moran NA. Intraspecific variation in symbiont genomes: Bottlenecks and the aphid-Buchnera association. Genetics. 2001;157:477–489. doi: 10.1093/genetics/157.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta T, Kimura M. On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1971;1(1):18–25. doi: 10.1007/BF01659391. [DOI] [PubMed] [Google Scholar]
- 39.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 40.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 41.Kuo CH, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19(8):1450–4. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo CH, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010;6:e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller’s ratchet with two levels of selection. American Naturalist. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 44.de Visser JA. The fate of microbial mutators. Microbiology. 2002;148:1247–1252. doi: 10.1099/00221287-148-5-1247. [DOI] [PubMed] [Google Scholar]
- 45.Wielgoss S, et al. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc Natl Acad Sci U S A. 2013;110:222–227. doi: 10.1073/pnas.1219574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raynes Y, Sniegowski PD. Experimental evolution and the dynamics of genomic mutation rate modifiers. Heredity. 2014;113:375–380. doi: 10.1038/hdy.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyholm SV, Graf J. Knowing your friends: Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulvestad E. Cooperation and conflict in host-microbe relations. APMIS. 2009;117:311–322. doi: 10.1111/j.1600-0463.2009.02457.x. [DOI] [PubMed] [Google Scholar]
- 49.Sachs JL, et al. Host control over infection and proliferation of a cheater symbiont. J Evol Biol. 2010;23:1919–1927. doi: 10.1111/j.1420-9101.2010.02056.x. [DOI] [PubMed] [Google Scholar]
- 50.Sachs JL, Simms EL. Pathways to mutualism breakdown. Trends Ecol Evol. 2006;21:585–592. doi: 10.1016/j.tree.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 51.Sung W, et al. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci U S A. 2012;109:18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puigbo P, et al. Genomes in turmoil: Quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 2014;12:66. doi: 10.1186/s12915-014-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Dohlen CD, et al. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature. 2001;412:433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- 55.Koga R, et al. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci U S A. 2012;109:E1230–1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller J. The relation of recombination to mutational advance. Mutat. Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 57.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charlesworth B. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet Res. 1994;63:213–227. doi: 10.1017/s0016672300032365. [DOI] [PubMed] [Google Scholar]
- 59.Maynard Smith J, Haigh J. The hitch-hiking effect of a favorable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 60.Gillespie JH. Is the population size of a species relevant to its evolution? Evolution. 2001;55:2161–2169. doi: 10.1111/j.0014-3820.2001.tb00732.x. [DOI] [PubMed] [Google Scholar]
- 61.Gillespie JH. The neutral theory in an infinite population. Gene. 2000;261:11–18. doi: 10.1016/s0378-1119(00)00485-6. [DOI] [PubMed] [Google Scholar]
- 62.Gillespie JH. The role of population size in molecular evolution. Theor Popul Biol. 1999;55:145–156. doi: 10.1006/tpbi.1998.1391. [DOI] [PubMed] [Google Scholar]
- 63.Hudson RR, Turelli M. Stochasticity overrules the “three-times rule”: Genetic drift, genetic draft, and coalescence times for nuclear loci versus mitochondrial DNA. Evolution. 2003;57:182–190. doi: 10.1111/j.0014-3820.2003.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 64.Woolfit M, Bromham L. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol Biol Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]
- 65.Lambert JD, Moran NA. Deleterious mutations destabilize ribosomal RNA in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1998;95:4458–4462. doi: 10.1073/pnas.95.8.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 67.Bennett GM, et al. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. MBio. 2014;5:e01697–01614. doi: 10.1128/mBio.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clark MA, Moran NA, Baumann P. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- 69.Wernegreen JJ, Moran NA. Evidence for genetic drift in endosymbionts (Buchnera): Analyses of protein-coding genes. Mol Biol Evol. 1999;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]
- 70.Fry AJ, Wernegreen JJ. The roles of positive and negative selection in the molecular evolution of insect endosymbionts. Gene. 2005;355:1–10. doi: 10.1016/j.gene.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 71.Ohta T. Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J Mol Evol. 1995;40:56–63. doi: 10.1007/BF00166595. [DOI] [PubMed] [Google Scholar]
- 72.Kuo CH, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19:1450–1454. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woolfit M. Effective population size and the rate and pattern of nucleotide substitutions. Biol Lett. 2009;5:417–420. doi: 10.1098/rsbl.2009.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo H, et al. Comparing effective population sizes of dominant marine alphaproteobacteria lineages. Environ Microbiol Rep. 2014;6:167–172. doi: 10.1111/1758-2229.12129. [DOI] [PubMed] [Google Scholar]
- 75.Akashi H. Within- and between-species DNA sequence variation and the ‘footprint’ of natural selection. Gene. 1999;238:39–51. doi: 10.1016/s0378-1119(99)00294-2. [DOI] [PubMed] [Google Scholar]
- 76.Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- 77.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hughes AL, et al. Synonymous and nonsynonymous polymorphisms versus divergences in bacterial genomes. Mol Biol Evol. 2008;25:2199–2209. doi: 10.1093/molbev/msn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kelkar YD, Ochman H. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics. 2013;193:303–307. doi: 10.1534/genetics.112.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendonca AG, Alves RJ, Pereira-Leal JB. Loss of genetic redundancy in reductive genome evolution. PLoS Comput Biol. 2011;7:e1001082. doi: 10.1371/journal.pcbi.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pal C, et al. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 82.Santos-Garcia D, et al. Small but powerful, the primary endosymbiont of moss bugs, Candidatus Evansia muelleri, holds a reduced genome with large biosynthetic capabilities. Genome Biol Evol. 2014;6:1875–1893. doi: 10.1093/gbe/evu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakabachi A, et al. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr Biol. 2014;24:R640–641. doi: 10.1016/j.cub.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 84.McCutcheon JP, Keeling PJ. Endosymbiosis: protein targeting further erodes the organelle/symbiont distinction. Curr Biol. 2014;24:R654–655. doi: 10.1016/j.cub.2014.05.073. [DOI] [PubMed] [Google Scholar]
- 85.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sloan DB, Moran NA. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 2012;29:3781–3792. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koga R, Moran NA. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. Isme J. 2014;8:1237–1246. doi: 10.1038/ismej.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bennett GM, Moran NA. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 2013;5:1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Funk DJ, et al. Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc R Soc Lond B Biol Sci. 2000;267:2517–2521. doi: 10.1098/rspb.2000.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Newton IL, Bordenstein SR. Correlations between bacterial ecology and mobile DNA. Curr Microbiol. 2011;62:198–208. doi: 10.1007/s00284-010-9693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nikoh N, et al. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 2014;111:10257–10262. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blount ZD, et al. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Degnan PH, Lazarus AB, Wernegreen JJ. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 99.Sloan DB, Moran NA. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol Evol. 2013;5:783–793. doi: 10.1093/gbe/evt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toh H, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clayton AL, et al. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 2012;8:e1002990. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oakeson KF, et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 2014;6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Santos-Garcia D, et al. The genome of Cardinium cBtQ1 provides insights into genome reduction, symbiont motility, and its settlement in Bemisia tabaci. Genome Biol Evol. 2014;6:1013–1030. doi: 10.1093/gbe/evu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lamelas A, et al. Evolution of the secondary symbiont “Candidatus Serratia symbiotica” in aphid species of the subfamily Lachninae. Appl Environ Microbiol. 2008;74:4236–4240. doi: 10.1128/AEM.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Manzano-Marin A, et al. Comparative genomics of Serratia spp.: Two paths towards endosymbiotic life. PLoS One. 2012;7:e47274. doi: 10.1371/journal.pone.0047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Degnan PH, et al. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 2010;12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Degnan PH, et al. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aksoy S. Molecular analysis of the endosymbionts of tsetse flies: 16S rDNA locus and over-expression of a chaperonin. Insect Mol Biol. 1995;4:23–29. doi: 10.1111/j.1365-2583.1995.tb00004.x. [DOI] [PubMed] [Google Scholar]
- 110.Sato S, Ishikawa H. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J Bacteriol. 1997;179:2300–2304. doi: 10.1128/jb.179.7.2300-2304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maisnier-Patin S, et al. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat Genet. 2005;37:1376–1379. doi: 10.1038/ng1676. [DOI] [PubMed] [Google Scholar]
- 112.Kuo CH, Ochman H. Deletional bias across the three domains of life. Genome Biol Evol. 2009;1:145–152. doi: 10.1093/gbe/evp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. Isme J. 2014;8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-research0054. RESEARCH0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Williams LE, Wernegreen JJ. Unprecedented loss of ammonia assimilation capability in a urease-encoding bacterial mutualist. BMC Genomics. 2010;11:687. doi: 10.1186/1471-2164-11-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun Z, Blanchard JL. Strong genome-wide selection early in the evolution of Prochlorococcus resulted in a reduced genome through the loss of a large number of small effect genes. PLoS One. 2014;9:e88837. doi: 10.1371/journal.pone.0088837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grote J, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. MBio. 2012;3:e00252–12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 119.Swan BK, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A. 2013;110:11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luo H, et al. Evolutionary analysis of a streamlined lineage of surface ocean Roseobacters. Isme J. 2014;8:1428–1439. doi: 10.1038/ismej.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hessen DO, et al. Genome streamlining and the elemental costs of growth. Trends Ecol Evol. 2010;25:75–80. doi: 10.1016/j.tree.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 122.Vieira-Silva S, Touchon M, Rocha EP. No evidence for elemental-based streamlining of prokaryotic genomes. Trends Ecol Evol. 2010;25:319–320. doi: 10.1016/j.tree.2010.03.001. author reply 320-311. [DOI] [PubMed] [Google Scholar]
- 123.Lee MC, Marx CJ. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 2012;8:e1002651. doi: 10.1371/journal.pgen.1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koskiniemi S, et al. Selection-driven gene loss in bacteria. PLoS Genet. 2012;8:e1002787. doi: 10.1371/journal.pgen.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo H, et al. Genome reduction by deletion of paralogs in the marine cyanobacterium Prochlorococcus. Mol Biol Evol. 2011;28:2751–2760. doi: 10.1093/molbev/msr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luo H, et al. Evolution of divergent life history strategies in marine Alpha-Proteobacteria. MBio. 2013;4:e00373–13. doi: 10.1128/mBio.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rocha EP, Feil EJ. Mutational patterns cannot explain genome composition: Are there any neutral sites in the genomes of bacteria? PLoS Genet. 2010;6:e1001104. doi: 10.1371/journal.pgen.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Muto A, Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 130.Wernegreen JJ, Funk DJ. Mutation exposed: a neutral explanation for extreme base composition of an endosymbiont genome. J Mol Evol. 2004;59:849–858. doi: 10.1007/s00239-003-0192-z. [DOI] [PubMed] [Google Scholar]
- 131.Van Leuven JT, McCutcheon JP. An AT mutational bias in the tiny GC-rich endosymbiont genome of Hodgkinia. Genome Biol Evol. 2012;4:24–27. doi: 10.1093/gbe/evr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6:e1001107. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci U S A. 2012;109:14504–14507. doi: 10.1073/pnas.1205683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pontes MH, Dale C. Lambda red-mediated genetic modification of the insect endosymbiont Sodalis glossinidius. Appl Environ Microbiol. 2011;77:1918–1920. doi: 10.1128/AEM.02166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weiss BL, et al. An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc Natl Acad Sci U S A. 2008;105:15088–15093. doi: 10.1073/pnas.0805666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 138.Lamelas A, et al. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011;7:e1002357. doi: 10.1371/journal.pgen.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nikoh N, et al. Reductive evolution of bacterial genome in insect gut environment. Genome Biol Evol. 2011;3:702–714. doi: 10.1093/gbe/evr064. [DOI] [PMC free article] [PubMed] [Google Scholar]