Abstract
Ex vivo stability is a valuable protein characteristic but is laborious to improve experimentally. In addition to biopharmaceutical and industrial applications, stable protein is important for biochemical and structural studies. Taking advantage of the large number of available genomic sequences and growth temperature data, we present two bioinformatic methods to identify a limited set of amino acids or positions that likely underlie thermostability. Because these methods allow thousands of homologs to be examined in silico, they have the advantage of providing both speed and statistical power. Using these methods, we introduced, via mutation, amino acids from thermoadapted homologs into an exemplar mesophilic membrane protein, and demonstrated significantly increased thermostability while preserving protein activity.
Introduction
Stable protein is required for biochemical or structural studies and industrial applications. In particular, thermostability is strongly correlated with protein stability in detergent (1–3), in vivo half-life (4), and resistance to denaturation (5). The study of membrane proteins often requires that the proteins are stable upon removal from the native lipid bilayer when solubilized in a detergent micelle. In some cases, the stability of the solubilized membrane protein can be improved by mutations (1), ligand binding (6), or cosolvents such as lipids (7,8). Importantly, increasing the thermal stability of membrane proteins in a reference detergent has led to stability in other, often harsher detergents (1–3). These thermostabilization techniques have played a critical role in achieving the well-diffracting crystals that are necessary for structural studies (6,8–10).
Despite their utility, the identification of thermostable mutants is slow due to the number of possible mutations, which grows exponentially with the sequence length. Experimentally, one can address this problem by either limiting the mutational space (3,11) or increasing the screening rate using high-throughput methods (7,12,13). However, these are still expensive methods in terms of time and effort, and may require a high-affinity ligand, introduce nonnative amino acids, or abolish protein activity.
An alternative is to use a bioinformatics approach. One can take advantage of naturally occurring orthologs that are already adapted to elevated temperatures, although these are often poorly characterized proteins of limited homology to the gene of interest (14,15). In silico stability calculations (4,16) and consensus sequence analysis (17) may also provide insights, but they require significant a priori knowledge or make assumptions about ancestral protein activity.
With the continued expansion of sequenced genomes from diverse organisms, one can identify thousands of homologs from species of varying native environments (including temperature). Here, we utilize the available homologs’ sequences to quickly create a multiple sequence alignment (MSA), sorted by the optimal growth temperature (OGT) of the originating species. We present two methods to analyze this MSA and thereby identify potential thermoadaptive sequence variations. Our findings then limit the screening needed to identify stabilizing mutations of an exemplar protein.
Principle of the methods
Within a protein family, thermostability (18) and optimum enzymatic activity (15) are directly correlated with the OGT of the species of origin. The most common examples are the DNA polymerase enzymes of hyperthermophiles, whose stability and activity at elevated temperatures in vitro allowed for the successful development of PCR (19). Furthermore, the amino acids that drive temperature-dependent stability can be readily transferred between orthologs within a protein family (14,15,20,21). Therefore, in addition to genetic drift and other selective pressures, differences in primary sequence between orthologs with different OGTs often reflect adaptive mutations to the temperature of the native environment of each particular organism. These differences provide a limited set of amino acid changes (mutations) to the protein of interest that likely underlie thermostabilization. However, even close orthologs of a particular gene of interest will likely include many primary sequence differences (22). We use the aggregate sequence differences among many homologs to identify the positions and amino acid differences that drive temperature adaptation. We have developed two methods based on the above assumptions that can be used to compare sequences globally and pairwise to identify family-specific means of thermoadaptation. The most significant amino acids for thermoadaptation can then be introduced, via mutation, into a mesophilic protein of interest for increased thermostability.
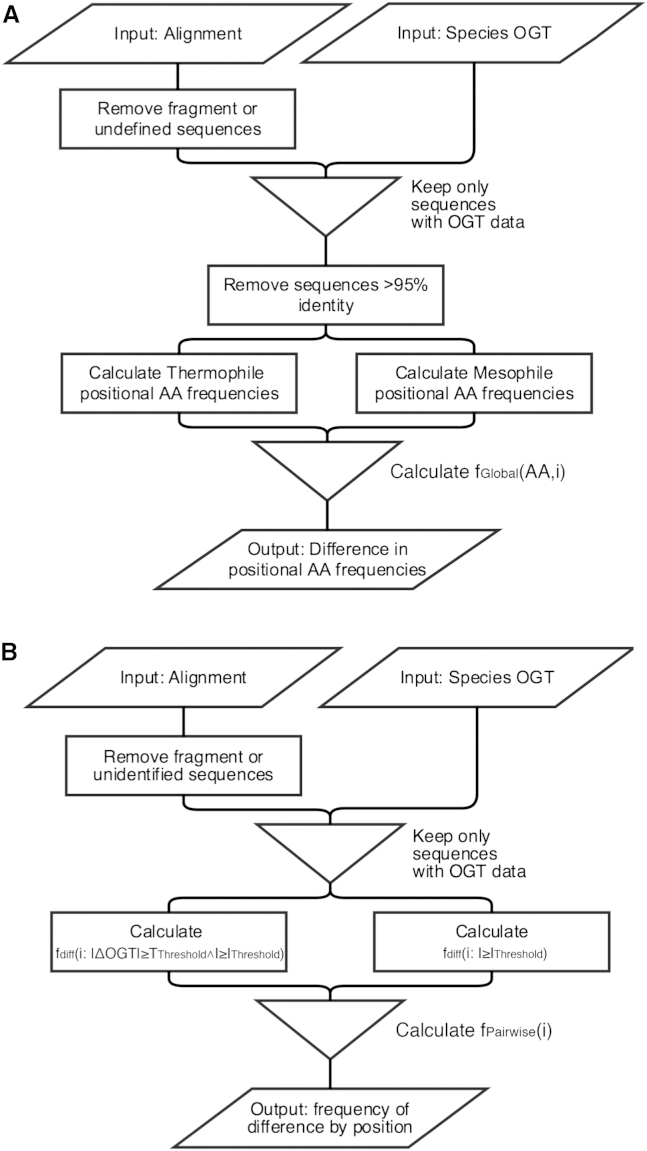
In the global method, the amino acid frequency (AA) at each position (i) between thermophilic (fThermo(AA,i)) and mesophilic (fMeso(AA,i)) orthologs are compared to generate a heatmap of the differential positional amino acid frequency (Fig. 1 A). With these frequency differences (fGlobal(AA,i); Eq. 1), residues that are under- or overrepresented in thermophiles can be quickly identified, suggesting positions and residues that may underlie thermoadaptation and thermostability. Considering absolute OGT adaptation, this method has the advantage of speed and simplicity. However, it can be confounded by shared phylogeny, where apparent differences can also arise from a common ancestry irrespective of temperature adaptation. This is addressed by the exclusion of closely related sequences.
| (1) |
An alternative approach to identify thermoadaptive mutations, termed the pairwise method, is to compare sequences pairwise and specifically take advantage of closely related sequences. Sequence pairs with high identity (I) but significantly different relative OGTs (ΔOGT) can provide valuable insights into thermoadaptation. By utilizing thresholds for relative OGT (TThreshold) and sequence identity (IThreshold), one can note positions that differ between these pairs (Fig. 1 B). By considering all close homologs in the MSA, one can calculate a general frequency of change for close sequence pairs, where each pair meets the criterion (|ΔOGT| ≥ TThreshold ∧ I ≥ IThreshold). A reference frequency without consideration of OGT, where only (I > IThreshold) is required, is then subtracted to account for positional conservation within the family. This yields the frequency of change at each position as a consequence of relative OGT differences (fPairwise(i); Eq. S1 in the Supporting Material). Although it is analogous to previous pairwise analyses (14,15), this method differs in that it leverages thousands of close homologs to identify conserved hotspots of thermoadaptation, at the expense of computational time and amino acid specificity.
Figure 1.
(A and B) Flow chart of the global (A) and pairwise (B) methods.
Materials and Methods
Alignment preparation and OGT assignment
OGTs were approximated from nontorpor body temperatures of endotherms and previously measured OGTs (23–31), or the culture conditions of ATCC (32) and DSMZ (33). Taxa were assigned using the NCBI Taxonomy Database (34). Homologous sequences were collected by BLAST (35) search and aligned in Promals3D (36). OGTs were assigned to sequences based on genus and species annotations in UniProt (37). Sequences without an assigned genus and species, including ambiguous residues, annotated as fragment, or without OGT data for the species, were excluded from further analysis. All bioinformatic analyses were done using Python 2.7 scripts based on Biopython (38), SciPy (39), and Matplotlib (40).
Global method of analysis
Sequences in the MSA with >95% sequence identity were removed. The alignment was then divided based on the species’ OGT into thermophiles (OGT ≥ 45°C) and mesophiles (45°C > OGT ≥ 20°C). The frequency of amino acid by position was calculated for the thermophiles and mesophiles, and the difference in frequency by position was then calculated.
Pairwise method of analysis
Primary sequences were compared pairwise for each sequence of the MSA. Where the difference in OGT was greater than a Tthreshold of 10°C and the overall sequence identity was greater than an Ithreshold of 50%, the positions of difference were recorded. The amino acids of the higher and lower OGT sequences were noted. To account for positional amino acid conservation, the overall differences, irrespective of temperature, were calculated from all pairs with a sequence identity greater than Ithreshold. The difference in frequency, by position, was then calculated. Positions with a >50% gap assignment in the MSA were excluded from further analysis.
Source code
The source code for these analysis methods, with the species-OGT list, is available at http://www.med.nyu.edu/skirball-lab/dwanglab/files/thermostability_v1.1.tar.gz.
Screening of individual mutants for thermostability
Protein was expressed in Escherichia coli strain BL21 (DE3) pLysS as a GFPuv fusion using the pCGFP-BC vector (41). The transporter was expressed by induction (at OD595 ∼1.0) with 1 mM of isopropyl β-D-1-thiogalactopyranoside at 32°C for 2 h. Cells were harvested by centrifugation and cell pellets were frozen at −20°C until use. The cells were resuspended in lysis buffer (50 mM Tris pH 7.5, 300 mM NaCl, 5 mM EDTA) to equivalent density and lysed by sonication. Protein was extracted with 1% n-dodecyl-β-D-maltopyranoside (DDM) for 10 min at 4°C, and lysates were clarified by centrifugation. The lysates were then thermally stressed for 30 min at 4°C and 42°C, centrifuged to remove insoluble material, and injected on a Shodex KW803 analytical size-exclusion chromatography (SEC) HPLC column into a buffer containing 200 mM Na2SO4, 50 mM Tris pH 7.5, 3 mM NaN3, and 0.05% DDM (42,43). Mean and standard deviations were calculated (n = 2–4).
Apparent melting temperatures of thermostabilized mutants
Protein was expressed in Escherichia coli and cells were solubilized as described for the thermostability screen (6,7). Cell lysates were thermally stressed for 30 min at each test temperature, centrifuged to remove insoluble material, and injected on a Shodex KW803 analytical SEC HPLC column into a buffer containing 200 mM Na2SO4, 50 mM Tris pH 7.5, 3 mM NaN3, and 0.05% DDM. The height of the fluorescent BsTetL peak was measured and normalized to the 4°C sample. Mean and standard deviations were calculated (n = 2), and apparent melting temperatures, Tm, were calculated by fitting to the sigmoidal equation using Prism5.
Tetracycline minimum inhibitory concentration
Complementation studies were performed in a manner similar to that previously described (44). Exponential growth phase cultures of thermostabilized constructs transformed into Top10F′ competent cells were used to inoculate LB media with 200 μg/mL of ampicillin and varying concentrations of tetracycline-HCl. Cultures were grown without induction at 37°C and 41°C, and the OD595 was measured at 16.25 h and 23.25 h, respectively. Mean and standard deviations were calculated (n = 3–4).
Results
We applied both the global and pairwise methods to a membrane transporter family and tested the predicted stabilizing mutations.
Tetracycline transporter BsTetL as a case study
TetL from Bacillus subtilis (BsTetL) is an integral membrane protein that induces tetracycline resistance by exporting tetracycline from the bacterial cytoplasm (45). TetL is a member of the major facilitator superfamily, although the TetL subfamily has an atypical number of transmembrane helices (number of helices = 14). Biochemical studies have found that BsTetL couples extrusion of the tetracycline-metal complex to the import of protons. Although it has been studied extensively in vitro and in heterologous expression systems, the structure and transport mechanism of this protein have not yet been described.
Identification of potential thermostabilizing mutations of BsTetL
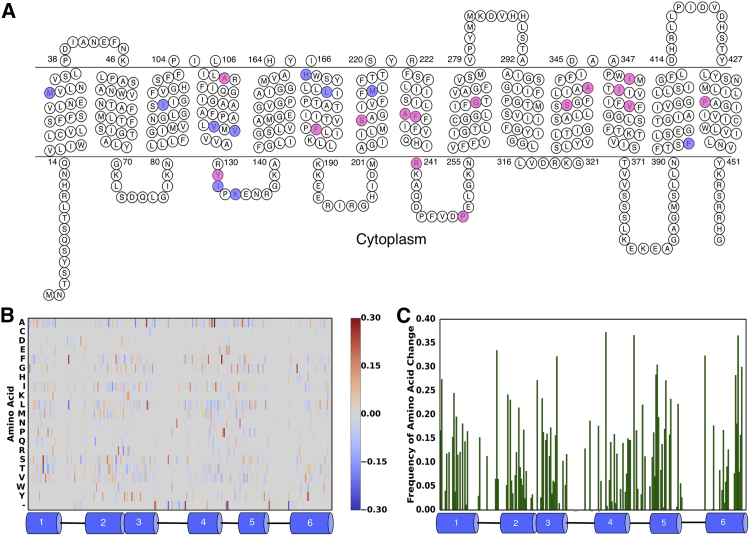
Protein sequences homologous to BsTetL were collected by means of an extensive BLAST search. OGTs were assigned from a list of species-OGT pairs (Fig. S1 C). In total, 2343 sequences were retained after the removal of sequences annotated as fragments, without genus and species, or containing ambiguous residues. For the global analysis, after removing highly similar sequences, we analyzed 1513 sequences, including 140 thermophilic sequences. By comparing the amino acid frequency by position for thermophiles and mesophiles (Figs. 2 B and S2), we selected the 20 most over- and underrepresented amino acids (i.e., those with the greatest absolute frequency difference). From this set, we pursued 10 possible mutations (Table 1; Fig. S3), all of which were highly significant. For each mutation, the native BsTetL residue was mutated to the most common thermophilic amino acid observed. The remaining amino acid differences corresponded to gaps, were coincident positions, or the thermophile-associated residue was already present in BsTetL.
Figure 2.
Identification of positions in TetL correlated with increased native growth temperature. (A) Topology of BsTetL. Positions tested are boxed in blue and magenta for global and pairwise predictions, respectively. (B) Heatmap of positional amino acid conservation by the global method corresponding to helices 1–6; data for the entire protein are shown in Fig. S2. Positive values indicate overrepresented in thermophiles, and negative values indicate underrepresentation (or overrepresented in mesophiles). (C) Frequency of amino acid change as a function of relative OGT differences for helices 1–6; data for the entire protein are shown in Fig. S4. Positions of low conservation (>50% gaps) are hidden. To see this figure in color, go online.
Table 1.
Potential TetL-stabilizing mutations identified by the global method
| Position | Thermophilic AA | fThermo(AA,i) | fMeso(AA,i) | p-Value | TetL Mutation | Successful |
|---|---|---|---|---|---|---|
| 31 | S | 0.393 | 0.149 | 2.44E-10 | M31S | |
| 94 | A | 0.557 | 0.318 | 1.01E-23 | I94A | |
| 124 | A | 0.443 | 0.207 | 1.18E-14 | V124A | |
| 126 | A | 0.614 | 0.308 | 5.28E-23 | V126A | + |
| 132 | F | 0.714 | 0.415 | 5.38E-33 | I132F | |
| 134 | P | 0.550 | 0.307 | 9.80E-23 | K134P | |
| 167 | G | 0.514 | 0.279 | 2.90E-20 | H167G | |
| 173 | Y | 0.414 | 0.173 | 4.36E-12 | L173Y | |
| 215 | L | 0.614 | 0.333 | 4.26E-25 | M215L | + |
| 394 | N | 0.243 | 0.117 | 4.46E-08 | F394N | + |
Overrepresented amino acids by position in the thermophilic sequences are noted with the frequency of that amino acid in both the thermophilic and mesophilic sequences at that same position, with corresponding p-values calculated by the binomial test. A mutant was considered successful if the residual fluorescence, after thermal stress, was significantly greater than that of the wild-type.
The pairwise-method data set contained all 2343 sequences, including 154 thermophilic sequences. By comparing the positions of difference between close homologs, we determined the 18 most frequently different positions (Figs. 2 C and S4). We then examined 15 of these positions (Table 2). Mutations were made in BsTetL to introduce the amino acid that was most frequently seen in the higher-temperature sequences. The remaining three positions were excluded because BsTetL already contained the residue that was most frequently seen in the higher-temperature sequences.
Table 2.
Potential TetL-stabilizing mutations identified by the pairwise method
| Position | fdiff(i: |ΔOGT| ≥ Tthreshold ∧ I ≥ Ithreshold) | fdiff(i: I > Ithreshold) | TetL Mutation | Successful |
|---|---|---|---|---|
| 109 | 0.652 | 0.279 | A109F | |
| 131 | 0.611 | 0.244 | Y131V | |
| 184 | 0.656 | 0.290 | F184L | + |
| 208 | 0.714 | 0.259 | S208T | |
| 232 | 0.795 | 0.448 | A232F | |
| 233 | 0.712 | 0.363 | F233L | |
| 241 | 0.832 | 0.338 | R241K | |
| 250 | 0.569 | 0.183 | P250L | + |
| 270 | 0.614 | 0.248 | G270V | |
| 335 | 0.819 | 0.435 | S335L | + |
| 341 | 0.692 | 0.332 | A341S | + |
| 350 | 0.699 | 0.342 | I350T | |
| 353 | 0.815 | 0.461 | I353L | |
| 358 | 0.641 | 0.219 | V358L | |
| 436 | 0.788 | 0.364 | F436I |
The frequency of a difference at a position is listed for both the presence and absence of a significant OGT difference between pairs. A mutant was considered successful if the residual fluorescence, after thermal stress, was significantly greater than that of the wild-type.
The positions of difference noted are broadly distributed across the protein (Fig. 1 A), suggesting no single mechanism of thermoadaptation. Interestingly, the pairwise and global mutant sets do not overlap, suggesting distinct mechanisms of absolute and relative adaptation to temperature. Notably, the global mutants are largely located in the N-terminal domain of the protein, whereas the pairwise mutants are generally found in the C-terminal domain. The reason for this segregation is unknown.
Screening of BsTetL mutants for thermostability
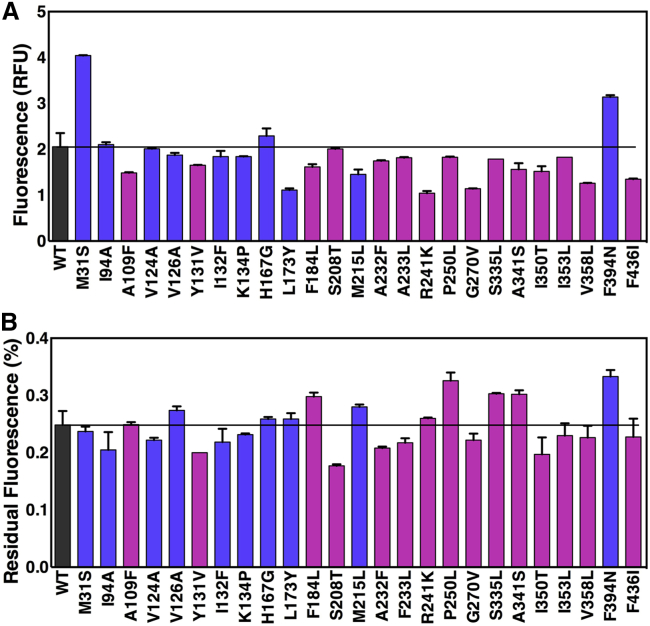
We tested the above predictions experimentally by overexpressing each mutant in E. coli with a C-terminal GFP fusion and solubilized in DDM. The TetL-GFP fusion had an expression level of ∼0.75 mg/L, which is slightly lower than that of the His-tagged construct (45). Lysates were split into two aliquots and incubated at either 4°C or 42°C, and then both samples were examined by analytical SEC with fluorescence detection (41). Total expression levels were approximated by the peak height of the 4°C sample, and thermostability was evaluated based on the peak intensity of the thermally stressed sample normalized to expression. Relative to wild-type BsTetL, seven mutants appeared to thermostabilize significantly and three showed increased expression (Fig. 2). Overall, the mutants exhibited stability that ranged from 70% to 135% that of the wild-type, and expression ranged from 50% to 200% (Fig. S5). Of the seven stabilizing mutations, three came from the global method and four came from the pairwise method, giving a 30% and 26% success rate, respectively.
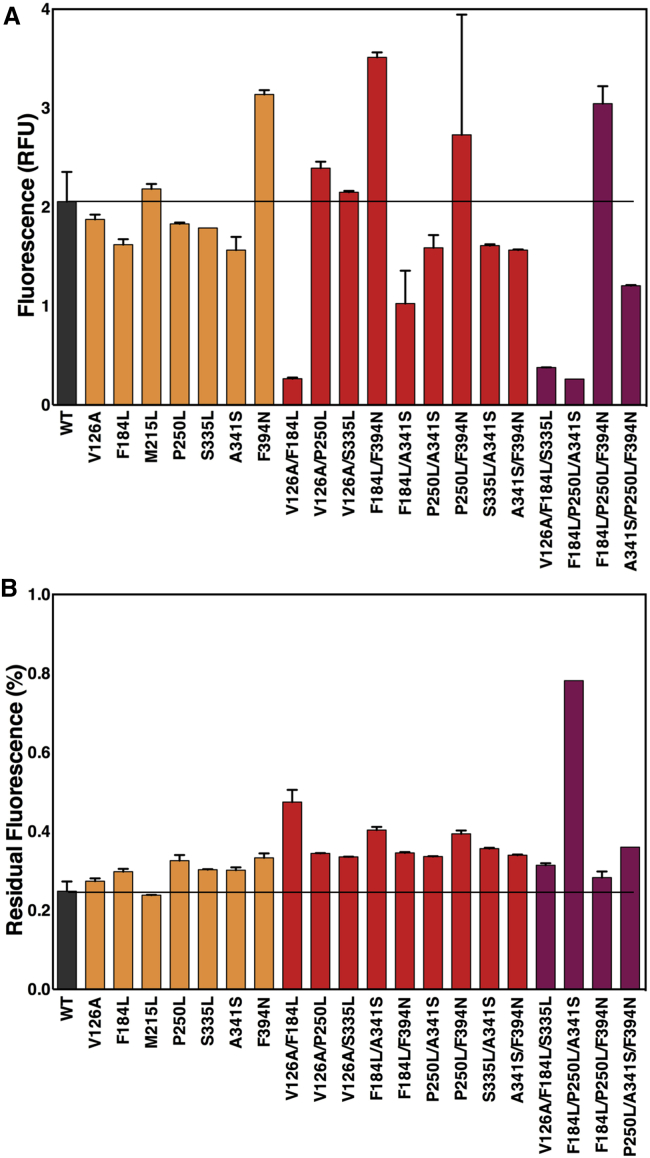
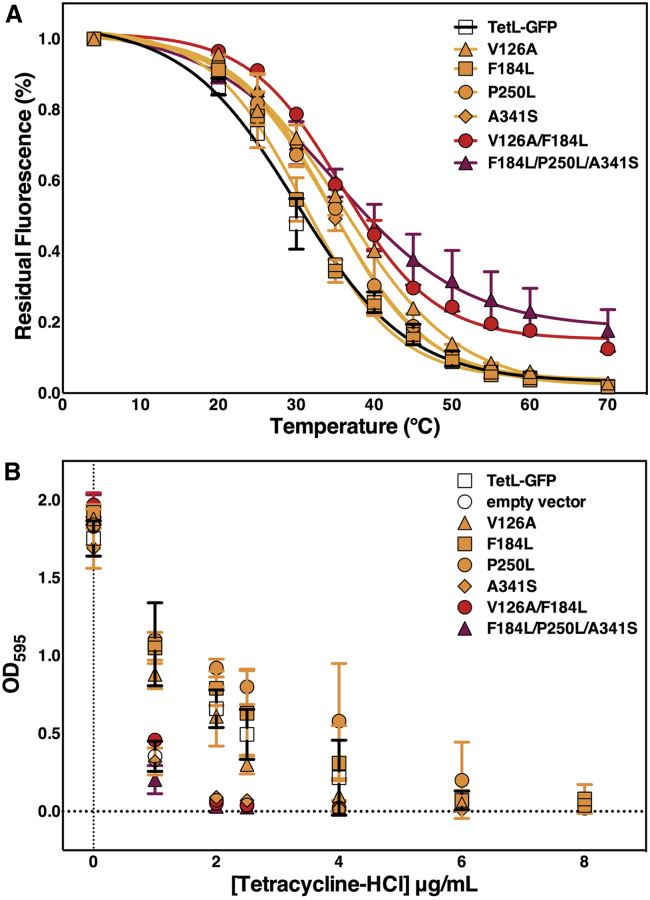
As the effects of thermostabilizing mutations can, in some cases, be combined for further stabilization (3,13,46), we combined single mutants pairwise to look for added stability. We tested the 21 possible pairwise combinations of the seven single mutants, and found that nine had greater stability than either parental mutant. The greatest was V126A/F184L, with an ∼90% increase in stability relative to the wild-type (Fig. 3), albeit with 13% of wild-type expression. The single mutant M215L did not have increased stability in any combination and may represent a false positive from the initial screen; alternatively, however, it may be epistatically forbidden with the other mutants. For those double mutants with increased stability relative to either parental mutant, commutativity was assumed and triple mutants were generated. Among these, and all mutants tested, the triple mutant F184L/P250L/A341S had the greatest thermostability, ∼3-fold higher than the wild-type (Fig. 4).
Figure 3.
Overexpression and stability of single BsTetL mutants. (A) Fluorescence of wild-type BsTetL-GFP and single mutants. Mutants predicted by the global and pairwise methods are colored blue and magenta, respectively. The horizontal bar indicates wild-type BsTetL fluorescence. (B) Residual fluorescence (fractional fluorescence that remains after 42°C thermal stress) of BsTetL-GFP and single mutants. Mutants predicted by the global and pairwise methods are colored blue and magenta, respectively. The stability of wild-type BsTetL is indicated by the horizontal bar. To see this figure in color, go online.
Figure 4.
Overexpression and stability of select double and triple BsTetL mutants. (A) Fluorescence of BsTetL-GFP and mutants. Single, double, and triple mutants are colored orange, red, and maroon, respectively. The horizontal bar indicates wild-type BsTetL-GFP fluorescence. (B) Residual fluorescence of BsTetL-GFP and mutants after 42°C thermal stress. Single, double, and triple mutants are colored orange, red, and maroon, respectively. The stability of wild-type BsTetL is indicated by the horizontal bar. To see this figure in color, go online.
The most stable mutants, V126A/F184L and F184L/P250L/A341S, also had the lowest expression of all mutants screened (Fig. S3). However, it is worth noting that this is not a general trend in the thermostability of BsTetL. Among all the mutants screened, there does not appear to be any overall correlation between expression level and thermostability (Fig. S5). This suggests that although the most promising constructs for stability were among the lowest expressed in this case, expression and stability are not necessarily coupled for BsTetL. This is notable given the varying strong (47), weak (48), or absent (11) correlations between expression and stability seen in stabilized membrane proteins where mutations were identified by other methods. The particular reason for the decreased expression in the V126A/F184L and F184L/P250L/A341S mutants is unknown, but it may reflect the combined effects of decreased expression of the constituent single mutants. Addition of the highly expressing M31S mutation (Fig. 3) was unable to improve expression of these mutants (data not shown).
BsTetL mutants have increased absolute Tm values
To further characterize the contributions of individual mutants to stability, we measured the apparent Tm in solubilized lysates for the most stabilized double (V126A/F184L) and triple (F184L/P250L/A341S) mutants, and the constituent single mutants. Melting curves have the advantage of quantifying stability on an absolute scale and describing the cooperativity of unfolding. Although it is not a measure of thermodynamic stability, this irreversible melting provides a convenient measure of resistance to thermal denaturation. The wild-type protein had a Tm of 30.0°C, whereas the single mutants had apparent Tm values of 35.8°C, 31.0°C, 34.5°C, and 33.6°C for V126A, F184L, P250L, and A341S, respectively (Fig. 5 A). The V126A/F184L and F184L/P250L/A341S mutants had Tm values of 35.8°C and 35.6°C. Notably, although the cooperativity of unfolding did not significantly increase in any mutant, it was clearly reduced in the triple mutant.
Figure 5.
In vitro stability and tetracycline resistance of BsTetL mutants. (A) Thermal denaturation of BsTetL and mutants. (B) Growth curve of BsTetL and mutants in the presence of tetracycline at 37°C. Single, double, and triple mutants are colored orange, red, and maroon, respectively. Wild-type and empty vector control are indicated by open squares and circles, respectively. To see this figure in color, go online.
Stabilized BsTetL mutants retain tetracycline transport activity
Both of the above-described bioinformatics methods identify stabilizing mutations that are conservative within the protein family and therefore likely preserve protein function. To validate the transport activity of the TetL thermostabilizing mutations, we examined tetracycline resistance in E. coli expressing the same stabilized TetL-GFP mutants at 37°C. The two most stable mutants, V126A/F184L and F184L/P250L/A341S, had no apparent activity by comparison with an empty vector and wild-type controls (Fig. 5 B). Among the parental single mutants, V126A had reduced tetracycline resistance relative to the wild-type, whereas A341S had apparent activity. Interestingly, F184L and P250L both showed increased tetracycline resistance. The single mutants clearly show that the mutations suggested by both methods can preserve protein function. The absence of activity in the double and triple mutants may be a consequence of their extremely low expression levels or the dominant effects of the V126A and A341S mutations. The varying activity of the single mutants, where total expression is likely similar to that of the wild-type (Fig. 3), may reflect changes in the substrate-binding site or enzyme kinetics. Growth at 41°C was insufficient to recover transport activity in the A341S mutant (Fig. S6). However, the increased growth temperature did increase the relative activity of V126A, suggesting that this particular mutation alters the energetics of the transport cycle.
Discussion
For biomedical studies or industrial applications of a particular protein, the protein must be stable in vitro. Although orthologous proteins from extremophiles are often stable in harsher conditions than can be tolerated by mesophiles, they are frequently less well characterized and therefore may be of less (or unknown) utility for the desired application. In this study, we used sequence differences between thermophiles and mesophiles to identify the positions and amino acids that are critical for thermoadaptation. Based on these findings, we introduced amino acids from thermoadapted homologs into a mesophilic protein of interest, and demonstrated an increase in thermostability. Here, we used a membrane protein, BsTetL; however, these methods are applicable to any protein family with enough available sequences for statistical significance.
The mechanism underlying the increased thermostability of the BsTetL mutants is unknown, although a majority of the mutations (e.g., the V126A and F184L mutants) introduced smaller side chains. Also, most of the mutations, including the P250L and S335L variants, introduced or maintained a hydrophobic side chain. These observations suggest that the thermostabilization arises from altered protein packing rather than from the introduction of hydrogen bonds or electrostatic interactions. However, novel interactions can be accomplished by indirect means, as was recently modeled in stabilized Neurotensin Receptor 1 (49). Thermostabilization may result from a protein directly favoring a folded state or slowing down the enzyme kinetics and thereby limiting entry into the states preceding unfolding (50,51). Slowed enzyme kinetics, in particular, could correspond to the decreased transport activity of some mutants. In a homology model based on the distantly related glycerol-3-phosphate (52) and peptide transporters (53), equivalent positions were found to be broadly distributed across the protein (Fig. S7). Although these findings do not provide a clear mechanism, they suggest that thermostabilization is not achieved through one or a few structural points. Notably, among the tested mutants, there did not appear to be a correlation between tetracycline transport and the distance to the substrate-binding pocket or domain-domain interface.
Bioinformatic analysis quickly identifies thermostabilizing mutants
Although previous computational analyses of thermostability have addressed whole proteome amino acid distributions, such analyses often have limited utility for a particular protein family (29,54). Experimental methods for identifying thermostabilizing mutants of a particular protein have relied upon efficiency to overcome the large mutational space. Alanine scanning limits the mutational space, with a success rate of 2–14% (2,11). Alternatively, all-versus-all mutations can be coupled with fluorescence-activated cell sorting to identify stabilizing and well-expressed mutants (13,47). Although the success rate is vanishing low, given the large number of mutants screened, the increased throughput allows one to sample a vastly enlarged sequence space, including multiple mutations. Both methods reveal similar or greater increases in Tm than those seen here. However, both are constrained to ligand-bound states.
An alternative is to use computational methods for rational identification of thermostabilizing mutants. By considering protein packing efficiency, conformational state, and fold energetics, these methods have success rates of 15–100% (55–57). However, they require a high-resolution crystal structure of the target protein or a homolog for analysis. Additionally, both screening and in silico methods are insensitive to evolutionary pressures on function and may sample nonconservative mutations, yielding inactive proteins.
The methods presented here provide a speed advantage by applying an in silico analysis, without requiring a crystal structure, with a success rate of 26–30%. Further, these methods identify thermostabilizing mutations that are conservative within a protein family and thus are likely biased toward preserving protein function. This is validated by the tetracycline resistance found in most of the single mutants tested. However, the energetics of transport had clearly shifted in some mutants, which had adapted to an elevated growth temperature.
Although these methods are not limited to any particular protein family, the significance of the noted amino acid differences is dependent upon the OGT and sequence identity ranges of the input MSA. The global method is typically limited by the number of thermophiles described (Fig. S1 A). Increasing the number of sequences by including more distant homologs allows for greater theoretical significance, at the expense of specificity to a particular protein of interest. However, as the number of sequenced genomes and characterized species increases, the sensitivity of these methods will also increase. Additionally, if modeling of growth temperature is used (30,58,59), many more OGTs can be approximated. Increasing the OGT and sequence data will improve the statistical power of both methods and allow investigators to study exclusively eukaryotic genes, for which few thermophilic orthologs are available (Fig. S1 B).
Furthermore, thermosensitive mutants are very useful for in vivo identification of gene function. Using the same analysis methods, but comparing mesophiles and psychrophiles, might allow one to identify temperature-sensitive mutants of a gene of interest without having to perform extensive phenotypic screening or exhaustive mutagenesis.
Further considerations of thermostability by bioinformatics
Although it was not considered in this study, it is possible that epistasis between positions explains why many of the single mutants screened did not show an appreciable increase in stability. A statistical analysis (60) of the family would likely provide insight into the coevolving positions within the primary sequence of the protein family, and indicate which thermostabilizing mutations might be epistatically linked. Alternatively, mutations that did not stabilize the protein may be a consequence of an indirect association with the OGT related to other properties of thermoadaptation, notably including adaptations to the distinct membrane environment and lipids of thermophiles (61).
Although insertions and deletions were noted in the calculation, they were specifically excluded from testing in the BsTetL case study to simplify the analysis. There appears to be a discrepancy in the thermoadaptation of loop length between soluble (62) and membrane proteins (29), and testing such mutations may provide additional thermostabilizing mutations.
Author Contributions
D.B.S. designed the bioinformatics methods. D.B.S., N.K.K., and J.S. performed the research. D.B.S., N.K.K., and D.-N.W. analyzed the data and wrote the manuscript.
Acknowledgments
The authors thank Saba Abdul-Hussein, Stephen Helms, Terry Krulwich, and Avner Schlessinger for helpful discussions regarding the experimental design and analysis. We also thank Jennifer Marden for discussion and a critical review of the manuscript.
This work was financially supported by the National Institutes of Health (R01-GM093825, R01-DK099023, R01-DK073973, and R01-MH083840).
Editor: Jose Faraldo-Gomez.
Footnotes
Seven figures and one equation are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00733-X.
Contributor Information
David B. Sauer, Email: david.sauer@med.nyu.edu.
Da-Neng Wang, Email: da-neng.wang@med.nyu.edu.
Supporting Material
References
- 1.Zhou Y., Bowie J.U. Building a thermostable membrane protein. J. Biol. Chem. 2000;275:6975–6979. doi: 10.1074/jbc.275.10.6975. [DOI] [PubMed] [Google Scholar]
- 2.Magnani F., Shibata Y., Tate C.G. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc. Natl. Acad. Sci. USA. 2008;105:10744–10749. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serrano-Vega M.J., Magnani F., Tate C.G. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. Sci. USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao D., Narasimhan D.L., Zhan C.G. Thermostable variants of cocaine esterase for long-time protection against cocaine toxicity. Mol. Pharmacol. 2009;75:318–323. doi: 10.1124/mol.108.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acharya P., Rajakumara E., Rao N.M. Structural basis of selection and thermostability of laboratory evolved Bacillus subtilis lipase. J. Mol. Biol. 2004;341:1271–1281. doi: 10.1016/j.jmb.2004.06.059. [DOI] [PubMed] [Google Scholar]
- 6.Mancusso R., Karpowich N.K., Wang D.N. Simple screening method for improving membrane protein thermostability. Methods. 2011;55:324–329. doi: 10.1016/j.ymeth.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattori M., Hibbs R.E., Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long S.B., Campbell E.B., Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 9.Li D., Lyons J.A., Caffrey M. Crystal structure of the integral membrane diacylglycerol kinase. Nature. 2013;497:521–524. doi: 10.1038/nature12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penmatsa A., Wang K.H., Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503:85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdul-Hussein S., Andréll J., Tate C.G. Thermostabilisation of the serotonin transporter in a cocaine-bound conformation. J. Mol. Biol. 2013;425:2198–2207. doi: 10.1016/j.jmb.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asial I., Cheng Y.X., Cornvik T. Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell. Nat. Commun. 2013;4:2901. doi: 10.1038/ncomms3901. [DOI] [PubMed] [Google Scholar]
- 13.Sarkar C.A., Dodevski I., Plückthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc. Natl. Acad. Sci. USA. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perl D., Mueller U., Schmid F.X. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 2000;7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- 15.Sayed A., Ghazy M.A., El-Dorry H. A novel mercuric reductase from the unique deep brine environment of Atlantis II in the Red Sea. J. Biol. Chem. 2014;289:1675–1687. doi: 10.1074/jbc.M113.493429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz J.E., Lin C.S., Weiss G.A. Computational design and selections for an engineered, thermostable terpene synthase. Protein Sci. 2011;20:1597–1606. doi: 10.1002/pro.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann M., Pasamontes L., Wyss M. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta. 2000;1543:408–415. doi: 10.1016/s0167-4838(00)00238-7. [DOI] [PubMed] [Google Scholar]
- 18.Hart K.M., Harms M.J., Marqusee S. Thermodynamic system drift in protein evolution. PLoS Biol. 2014;12:e1001994. doi: 10.1371/journal.pbio.1001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiki R.K., Gelfand D.H., Erlich H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 20.Ashenberg O., Gong L.I., Bloom J.D. Mutational effects on stability are largely conserved during protein evolution. Proc. Natl. Acad. Sci. USA. 2013;110:21071–21076. doi: 10.1073/pnas.1314781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano-Vega M.J., Tate C.G. Transferability of thermostabilizing mutations between beta-adrenergic receptors. Mol. Membr. Biol. 2009;26:385–396. doi: 10.3109/09687680903208239. [DOI] [PubMed] [Google Scholar]
- 22.Sheridan P.P., Panasik N., Brenchley J.E. Approaches for deciphering the structural basis of low temperature enzyme activity. Biochim. Biophys. Acta. 2000;1543:417–433. doi: 10.1016/s0167-4838(00)00237-5. [DOI] [PubMed] [Google Scholar]
- 23.Dutta A., Chaudhuri K. Analysis of tRNA composition and folding in psychrophilic, mesophilic and thermophilic genomes: indications for thermal adaptation. FEMS Microbiol. Lett. 2010;305:100–108. doi: 10.1111/j.1574-6968.2010.01922.x. [DOI] [PubMed] [Google Scholar]
- 24.Dyer B.D., Kahn M.J., Leblanc M.D. Classification and regression tree (CART) analyses of genomic signatures reveal sets of tetramers that discriminate temperature optima of archaea and bacteria. Archaea. 2008;2:159–167. doi: 10.1155/2008/829730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo H., Robb F.T. A modulator domain controlling thermal stability in the Group II chaperonins of Archaea. Arch. Biochem. Biophys. 2011;512:111–118. doi: 10.1016/j.abb.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Maheshwari R., Bharadwaj G., Bhat M.K. Thermophilic fungi: their physiology and enzymes. Microbiol. Mol. Biol. Rev. 2000;64:461–488. doi: 10.1128/mmbr.64.3.461-488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald J.H. Temperature adaptation at homologous sites in proteins from nine thermophile-mesophile species pairs. Genome Biol. Evol. 2010;2:267–276. doi: 10.1093/gbe/evq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mcnab B.K. An analysis of body temperatures of birds. Condor. 1966;68:47–55. [Google Scholar]
- 29.Meruelo A.D., Han S.K., Bowie J.U. Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci. 2012;21:1746–1753. doi: 10.1002/pro.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savage V.M., Gilloly J.F., Charnov E.L. Effects of body size and temperature on population growth. Am. Nat. 2004;163:429–441. doi: 10.1086/381872. [DOI] [PubMed] [Google Scholar]
- 31.White C.R., Seymour R.S. Mammalian basal metabolic rate is proportional to body mass2/3. Proc. Natl. Acad. Sci. USA. 2003;100:4046–4049. doi: 10.1073/pnas.0436428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ATCC. 2012. http://www.atcc.org/. Accessed October 11, 2012.
- 33.DSMZ. 2012. https://www.dsmz.de/. Accessed October 24, 2012.
- 34.Sayers E.W., Barrett T., Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S.F., Gish W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Pei J., Kim B.H., Grishin N.V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cock P.J., Antao T., de Hoon M.J. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Walt S., Colbert S.C., Varoquaux G. The numpy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011;13:22–30. [Google Scholar]
- 40.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 41.Kawate T., Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Auer M., Kim M.J., Wang D.N. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry. 2001;40:6628–6635. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 43.Wang D.N., Safferling M., Li X.D. Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim. Biophys. Acta. 2003;1610:23–36. doi: 10.1016/s0005-2736(02)00709-5. [DOI] [PubMed] [Google Scholar]
- 44.Jin J., Guffanti A.A., Krulwich T.A. Twelve-transmembrane-segment (TMS) version (DeltaTMS VII-VIII) of the 14-TMS Tet(L) antibiotic resistance protein retains monovalent cation transport modes but lacks tetracycline efflux capacity. J. Bacteriol. 2001;183:2667–2671. doi: 10.1128/JB.183.8.2667-2671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safferling M., Griffith H., Wang D.N. TetL tetracycline efflux protein from Bacillus subtilis is a dimer in the membrane and in detergent solution. Biochemistry. 2003;42:13969–13976. doi: 10.1021/bi035173q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibata Y., Gvozdenovic-Jeremic J., Tate C.G. Optimising the combination of thermostabilising mutations in the neurotensin receptor for structure determination. Biochim. Biophys. Acta. 2013;1828:1293–1301. doi: 10.1016/j.bbamem.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlinkmann K.M., Hillenbrand M., Plückthun A. Maximizing detergent stability and functional expression of a GPCR by exhaustive recombination and evolution. J. Mol. Biol. 2012;422:414–428. doi: 10.1016/j.jmb.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 48.Tate C.G. A crystal clear solution for determining G-protein-coupled receptor structures. Trends Biochem. Sci. 2012;37:343–352. doi: 10.1016/j.tibs.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Lee S., Bhattacharya S., Vaidehi N. Structural dynamics and thermostabilization of neurotensin receptor 1. J. Phys. Chem. B. 2015;119:4917–4928. doi: 10.1021/jp510735f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Závodszky P., Kardos J., Petsko G.A. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc. Natl. Acad. Sci. USA. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi E.Y., Krishnan S., Carpenter J.F. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 2003;20:1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y., Lemieux M.J., Wang D.N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 53.Guettou F., Quistgaard E.M., Löw C. Structural insights into substrate recognition in proton-dependent oligopeptide transporters. EMBO Rep. 2013;14:804–810. doi: 10.1038/embor.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warren G.L., Petsko G.A. Composition analysis of alpha-helices in thermophilic organisms. Protein Eng. 1995;8:905–913. doi: 10.1093/protein/8.9.905. [DOI] [PubMed] [Google Scholar]
- 55.Chen K.Y., Zhou F., Barth P. Naturally evolved G protein-coupled receptors adopt metastable conformations. Proc. Natl. Acad. Sci. USA. 2012;109:13284–13289. doi: 10.1073/pnas.1205512109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharya S., Lee S., Vaidehi N. Rapid computational prediction of thermostabilizing mutations for G protein-coupled receptors. J. Chem. Theory Comput. 2014;10:5149–5160. doi: 10.1021/ct500616v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korkegian A., Black M.E., Stoddard B.L. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke A., Rothery P., Isaac N.J. Scaling of basal metabolic rate with body mass and temperature in mammals. J. Anim. Ecol. 2010;79:610–619. doi: 10.1111/j.1365-2656.2010.01672.x. [DOI] [PubMed] [Google Scholar]
- 59.Zheng H., Wu H. Gene-centric association analysis for the correlation between the guanine-cytosine content levels and temperature range conditions of prokaryotic species. BMC Bioinformatics. 2010;11(Suppl 11):S7. doi: 10.1186/1471-2105-11-S11-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lockless S.W., Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 61.Koga Y. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea. 2012;2012:789652. doi: 10.1155/2012/789652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson M.J., Eisenberg D. Transproteomic evidence of a loop-deletion mechanism for enhancing protein thermostability. J. Mol. Biol. 1999;290:595–604. doi: 10.1006/jmbi.1999.2889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.