Abstract
The bacterial cytoplasm lies within a multilayered envelope that must be protected from internal and external hazards. This protection is provided by cell envelope stress responses (ESRs), which detect threats and reprogram gene expression to ensure survival. Pathogens frequently need these ESRs to survive inside the host, where their envelopes face dangerous environmental changes and attack from antimicrobial molecules. In addition, some virulence genes have become integrated into ESR regulons. This might be because these genes can protect the cell envelope from damage by host molecules, or it might help ESRs to reduce stress by moderating the assembly of virulence factors within the envelope. Alternatively, it could simply be a mechanism to coordinate the induction of virulence gene expression with entry into the host. Here, we briefly describe some of the bacterial ESRs, followed by examples where they control virulence gene expression in both Gram-negative and Gram-positive pathogens.
Keywords: antimicrobial peptide, bacterial pathogens, cell envelope, gene regulation, peptidoglycan, phospholipid, stress response, teichoic acid, virulence gene
Abbreviations
- BFP
bundle-forming pilus
- CAMP
cationic antimicrobial peptide
- ESR
envelope stress response
- CF
cystic fibrosis
- ECF
extracytoplasmic function
- EPEC
enteropathogenic E. coli
- HMV
hypermucoviscosity
- IM
inner membrane
- LPS
lipopolysaccharide
- LTA
lipoteichoic acids
- OM
outer membrane
- OMP
outer membrane protein
- PG
phosphatidylglycerol
- T(2/3/4)SS
type II/III/IV secretion system
- UPEC
uropathogenic E. coli
- WTA
wall teichoic acids
Introduction
The environment that bacterial pathogens encounter within the host poses many threats to the integrity of their cell envelope. These include changes in temperature, pH and osmolarity, as well as direct attack by molecules such as surfactants and cationic antimicrobial peptides (CAMPs). Therefore, it is not surprising that cell envelope stress response systems (ESRs) and their role in maintaining envelope integrity/functions are often important for virulence. This aspect of ESRs in bacterial virulence has been the subject of other reviews and will not be repeated extensively here.1,2 However, this review focuses specifically on the regulation of virulence genes by these ESRs. This focus presents something of a problem because “virulence genes” and “envelope stress response” are 2 microbiological terms that are difficult and perhaps even contentious to define. It is not our intention to contribute our own entries into that debate here, but we must begin by providing the admittedly subjective and loose definitions that we have chosen solely for this article. We will define virulence genes as those that encode both defensive and offensive functions thought to be required exclusively or at least predominantly in the host. For example, toxins, attachment factors, some protein export systems, and factors involved in resistance to CAMPs. By cell envelope stress response, we refer to transcriptional regulation systems known/suspected to be activated by, and/or determining resistance to, stimuli associated with adverse effects on the cell envelope, such as misfolded, mislocalized or aggregated proteins, perturbation of the cell envelope itself, or the presence of molecules that can cause such perturbation. This is not a comprehensive review of all bacterial ESRs, or of all the impacts that ESRs have on bacterial virulence. Rather, the purpose of this article is to briefly outline some of the ESRs found in Gram-negative and Gram-positive bacteria, and then to focus on describing a few selected examples of virulence gene regulation by ESRs in various pathogens.
Regulation of Virulence Genes by ESRs in Gram-Negative Bacteria
Introduction to Gram-negative ESRs
The cell envelope of Gram-negative bacteria is made up of 2 membranes and the periplasmic space between them, which also contains the peptidoglycan cell wall.3 The outer membrane (OM) is an asymmetric bilayer made up of phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet. The OM provides a major permeability barrier, although it does contain integral β-barrel porin proteins that allow small molecules to pass through by diffusion. The periplasmic space is home to various enzymes, transporters and regulatory proteins. It also contains a thin but rigid layer of peptidoglycan that provides resistance against the inner turgor pressure and maintains cell shape. The inner membrane (IM) is a symmetric phospholipid bilayer with integral α-helical proteins responsible for a broad range of activities that include transport, generation of electrochemical gradients, and the synthesis of ATP. Both membranes also have lipoproteins anchored to them by N-terminal lipid moieties.
The cell envelope is a critical but vulnerable structure and bacteria must perceive and respond to a variety of internal and external threats that might compromise it. This is vital when pathogenic species enter their host, where the cell envelope provides an important barrier against dangerous environmental conditions. The cell envelope is also a target of attack for some components of the innate immune system. To deal with these dangers, bacteria have evolved specialized response systems that detect problems within the cell envelope and communicate with cytoplasmic DNA-binding proteins to alter gene expression and combat the threat. These are known as cell envelope stress responses (ESRs).1,2,4 The best-characterized ESRs in Gram-negative bacteria consist of an alternative sigma factor, 3 different 2-component regulatory systems, and the phage shock protein response. Much, but by no means all, of the basic characterization of these systems has been done in Escherichia coli K-12 and its close relatives.
The extracytoplasmic function (ECF) sigma factor, RpoE or σE, orchestrates the most extensively studied Gram-negative ESR. It is also an ESR with an extremely well studied connection to virulence gene control in Pseudomonas aeruginosa. A proteolytic cascade degrades an inhibitory antisigma factor to control RpoE activity and this cascade is triggered by misfolded/mislocalized OM proteins and by so-called “off-pathway” LPS molecules.5,6 RpoE is essential in some species, with E. coli being a notable example.7 This indicates that it also plays a critical role during normal growth conditions.8 Nevertheless, the stress-induced increase of RpoE activity triggers an extensive alteration in gene expression, with target genes involved in protein folding and degradation, cell envelope biogenesis and various other aspects of metabolism, and in particular the OM.9,10 In fact, overall it seems that the broad role of the RpoE response is to help maintain OM homeostasis.
One of the 2-component ESRs is the conjugative pilus expression (Cpx) response.11,12 Its core components are the IM histidine kinase CpxA and the cytoplasmic response regulator CpxR. Cpx inducing conditions include exposure to alkaline pH, overproduction and accumulation of pilus subunits in the periplasm, mutations that impact periplasmic protein folding, and exposure to environmental conditions that alter the composition of the bacterial cell envelope.13-17 Upon induction, CpxR regulates many genes that are involved in maintaining cell envelope homeostasis, with the most recent data suggesting that IM maintenance might be its most important function.18,19 The Cpx response is also one of the Gram-negative ESRs that regulates virulence genes.
Another Gram-negative ESR is restricted to the Enterobacteriaceae and is a non-canonical 2-component system known as the regulation of capsular synthesis (Rcs) phosphorelay. It is induced by conditions that are deleterious to the cell envelope such as osmotic shock, desiccation, and overproduction of envelope proteins.20,21 The Rcs regulon includes genes involved in diverse cellular processes including maintenance of cellular integrity, motility, biofilm formation and capsular polysaccharide production. Once again, this is a ESR that has been implicated in virulence gene regulation.
The third 2-component ESR is the bacterial adaptive response (Bae). It is induced by exposure to membrane damaging agents including indole and some other antimicrobial compounds and is thought to provide protection against them. Specifically, it promotes the increased expression of a group of linked genes encoding a multidrug transporter that might exclude toxic compounds from the cell.22,23 However, there are no links to virulence gene control and so this ESR will not be discussed further.
The final Gram-negative ESR is the phage shock protein (Psp) system, which can be induced by extreme heat or osmotic shock, and by exposure to some organic solvents and proton ionophores.24-28 One potent and highly specific activator of the Psp response is the mislocalization of pore forming “secretin” proteins into the inner membrane.29,30 The Psp response is thought to react to, and mitigate, specific events that compromise the integrity of the inner membrane. A striking feature of the Psp system is that it is remarkably autonomous. Studies in 3 different species have shown that the genes encoding the Psp proteins are the only ones that change their expression level when a secretin mislocalizes.31,32 Together with other work, this suggests that the Psp proteins detect the inducing trigger and increase only their own production to mitigate the stress. Therefore, although the Psp system itself is essential for virulence in Yersinia enterocolitica and Salmonella enterica serovar Typhimurium (S. Typhimurium), it does not regulate other virulence genes and will not be described further here.28,33,34
All of the ESRs outlined above are known or likely to be induced by one or more conditions found in the host, and their general stress tolerance and envelope homeostasis roles can have broad impacts on virulence.1,2 Their effects on cell envelope functions and integrity can influence many important phenotypes such as adaptation to host environmental conditions (e.g. temperature and osmolarity) as well as resistance to CAMPs, surfactants and reactive oxygen species. However, as stated in the Introduction, this article will focus on their roles in controlling virulence gene expression.
The RpoE ESR
ECF sigma factors are common in both Gram-negative and Gram-positive bacteria and often play a role in ESR.35–37 RpoE is one ECF sigma factor that is widely conserved in Gram-negative bacteria. Numerous studies, mostly in E. coli and P. aeruginosa, have uncovered how RpoE activity is controlled (Fig. 1).38,39 The cytoplasmic domain of the bitopic IM antisigma factor, RseA, sequesters RpoE to inhibit its activity.40,41 However, an inducing condition initiates a proteolytic cascade that degrades RseA. The first protease in this cascade is the IM protein DegS. Misfolded outer membrane proteins (OMPs) interact with DegS, which activates it to cleave the periplasmic domain of RseA. However, the RseA periplasmic domain is also bound by the RseB protein, which protects it from cleavage by activated DegS.40,41 Therefore, 2 signals are required to initiate the RseA proteolytic cascade, activation of DegS by its interaction with misfolded OMPs, and a second signal that disrupts the RseB-RseA complex. Recent work has revealed that this second signal is off-pathway LPS molecules (i.e. intermediates in LPS transport to the OM), which bind to RseB to antagonize its interaction with RseA (Fig. 1).6 Both of these RpoE-inducing signals are likely to be generated simultaneously if defects in LPS biogenesis cause problems in OMP biogenesis and vice versa. Indeed, mutational disruption of LPS biosynthesis also generates the OMP inducing signal for DegS.6 Furthermore, the OMP LptD is part of the machinery that assembles LPS in the OM, and LptD requires the same assembly machinery as other OMPs.42 Thus, defects in OMP biogenesis will disrupt LptD function, and consequently LPS transport and assembly.
Figure 1.
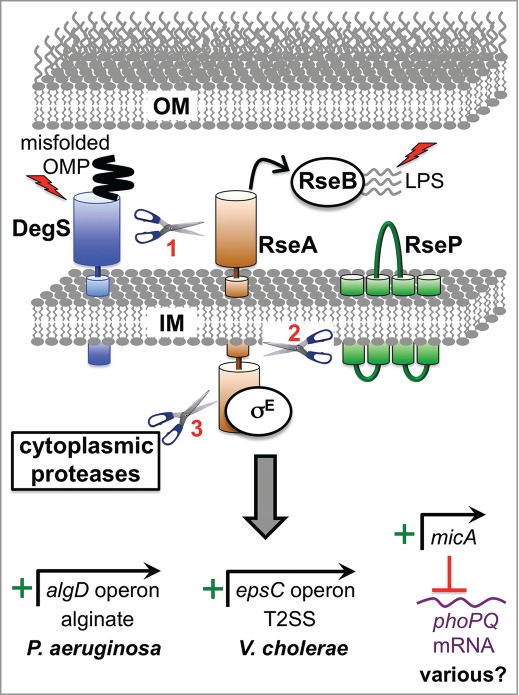
Examples of direct virulence gene regulation by the RpoE ESR. Two inducing signals (shown as lightening bolts) are required for activation. One is binding of a misfolded outer membrane protein (OMP) to DegS (AlgW in P. aeruginosa), which causes a conformational change that induces its proteolytic activity. The other is off-pathway LPS molecules, which bind to RseB (MucB in P. aeruginosa) and dissociate it from the antisigma factor RseA (MucA in P. aeruginosa). Both signals are required in order for DegS to cleave the periplasmic domain of RseA. The truncated RseA is then cleaved by the IM protease RseP (MucP in P. aeruginosa). This releases the cytoplasmic domain of RseA in complex with RpoE (σE; AlgU/T in P. aeruginosa), which is degraded by cytoplasmic proteases to release RpoE. RpoE-containing RNA polymerase binds directly to the algD promoter to induce alginate biosynthesis in P. aeruginosa and to the epsC promoter to induce T2SS production in V. cholerae. In E. coli K-12 the micA promoter is RpoE-dependent and drives the production of a non-coding sRNA that inhibits phoP translation (RpoE also induces 2 other sRNAs, RybB and MicL, which are not shown; see text). The predicted base pairing between MicA sRNA and phoPQ mRNA is conserved in pathogens including Salmonella, Shigella and Enterobacter. Therefore, RpoE might downregulate the virulence-associated PhoPQ regulon in multiple species. Mucoid P. aeruginosa strains from the lungs of individuals with cystic fibrosis most often have mutations that destroy antisigma factor RseA/MucA function, which leads to constitutive activation of RpoE, high algD operon expression and the production of large amounts of the exopolysaccharide, alginate.
After the periplasmic domain of RseA has been cleaved by DegS the remaining part of RseA becomes susceptible to cleavage by RseP, another IM protease, which then cleaves the RseA transmembrane segment (Fig. 1). Interestingly, in S. Typhimurium acid stress has been reported to induce RseP to cleave RseA independent of the misfolded OMP signal and DegS, and this might be relevant during host infection.43 Regardless, after cleavage by RseP the RseA N-terminal domain is released into the cytoplasm, still bound to RpoE, where cytoplasmic proteases degrade it and free RpoE to induce genes involved in mitigating cell envelope stress.9,10,41,44-46 Importantly, this proteolytic cascade and the dual-signal OMP/LPS activation mechanism are evolutionarily conserved, having also been described in the distant E. coli relative, Pseudomonas aeruginosa.6
RpoE control of alginate production in Pseudomonas aeruginosa
The best-studied example of RpoE regulating virulence genes is the control of alginate biosynthesis in the opportunistic pathogen P. aeruginosa. The lungs of people suffering from cystic fibrosis (CF) become colonized by P. aeruginosa, which leads to respiratory decline and a poor prognosis.47 The initially colonizing strains are susceptible to clearance by the immune response or antibiotic treatment. However, over time the bacteria in the lung undergo a phenotypic conversion to a mucoid appearance.48,49 The environmental conditions in the lung, including the inflammatory immune response, are thought to impose a selection for P. aeruginosa mutants that produce the mucoid exopolysaccharide alginate constitutively. Alginate facilitates biofilm formation and promotes resistance to host defenses and antibiotics.50 The alginate biosynthesis operon is positively controlled by RpoE, which is known as AlgU (or AlgT) in P. aeruginosa.51 A proteolytic cascade analogous to that in E. coli is triggered to degrade the antisigma factor RseA (called MucA in P. aeruginosa) when misfolded OMPs interact with its DegS homolog AlgW, and LPS interacts with it RseB homolog, MucB.6,52 However, the mucoid isolates from CF lungs almost always have mutations that inactivate MucA/RseA.52,53 The great majority of these mutations are predicted to cause an aberrant form of MucA (MucA22) with the C-terminal periplasmic domain truncated, which renders it susceptible to proteolysis.54-57
In addition to its role in controlling alginate biosynthesis, there are reasons to suspect that the P. aeruginosa RpoE/AlgU response might function to maintain cell envelope homeostasis as well. First, as mentioned above, the P. aeruginosa RpoE/AlgU system requires the OMP and LPS activation signals. Second, mucoid conversion and AlgU activation can be induced by inactivation of the AlgU-dependent gene mucD. This gene encodes a homolog of the E. coli periplasmic protease DegP, which degrades abnormal cell envelope proteins.58 This suggests that accumulation of aberrant envelope proteins in the absence of DegP/MucD generates a stress that triggers the RpoE/AlgU response.59,60 Third, supplementing growth medium with both ammonium metavanadate, which increases palmitoylation of the lipid A component of LPS, and the cell wall antibiotic triclosan, induces RseA/MucA degradation and causes a reversible mucoid phenotype.61 Fourth, treatment with other cell wall antibiotics also induces RpoE/AlgU activation, although not enough to cause mucoidy.59,62 Finally, a global survey of the RpoE/AlgU-dependent regulon induced by D-cycloserine, which interferes with peptidoglycan cross-linking, identified a large number of genes predicted to be required for recovery from cell envelope stress.60 Therefore, while there are some differences in the activation of the E. coli and P. aeruginosa RpoE responses, and in the genes they control, it seems that both play a roles in stress response.
Some of the conditions that induce RpoE/AlgU in the laboratory mimic aspects of the inflammatory environment in the lung. These include treatment with hydrogen peroxide, osmotic shock and oxygen limitation.50,54,59,60,63-66 This raises the intriguing question of whether or not the RpoE/AlgU regulon, and alginate production, are induced immediately after environmental P. aeruginosa enters the lung. Indeed, there is evidence to support this.67 The resulting relatively low-level production of alginate, and products of other genes within the RpoE/AlgU regulon, might help to buffer P. aeruginosa from the hostile host environment. This could be important to help the bacteria to gain an initial foothold. Ultimately, this could allow time to select for mutants in which the RpoE/AlgU regulon has been induced to a higher level by RseA/MucA inactivation. The copious production of alginate that results, perhaps along with favorable contributions by other RpoE/AlgU regulon members, would then give P. aeruginosa relative long-term immunity to the host immune system and, unfortunately, to therapeutic intervention.
RpoE/AlgU has also been implicated in modulating the expression of other virulence factors in P. aeruginosa, although the mechanism might be indirect in most cases. Notably, microarrays revealed that AlgU downregulated several genes involved in the transcriptional control of flagella and the type IV pilus.60,68 This is consistent with most isolates from the lungs of long-term colonized CF patients being non-motile.69 Although the flagellum is important for attachment and the establishment of infection, its production and use requires a great deal of energy. Furthermore, it is not required in biofilms, and flagellin is a potent trigger of the innate inflammatory response. Therefore, its downregulation by RpoE/AlgU provides another advantage that favors the long-term survival of P. aeruginosa in the lung.68,69
RpoE control of type II secretion in Vibrio cholerae
Gram-negative pathogens use complex systems to secrete toxic proteins.70 One is the type II secretion system (T2SS), a multi-protein apparatus with components in both membranes.71 T2SSs export various substrates including proteases, lipases, cellulases, phospholipases and toxins. Secretion occurs in 2 stages, with Sec- or Tat-dependent translocation into the periplasm, where the protein folds and/or complexes with its partner(s), being followed by translocation across the OM by the T2SS.72 T2SSs in several organisms are involved in pathogenesis.73 One is in Vibrio cholerae, which causes the life-threatening diarrheal disease cholera. V. cholerae employs a T2SS to export at least 19 proteins, including its major virulence factor cholera toxin that is responsible for the profuse watery diarrhea that defines the disease.74,75
The epsC-N operon and the pilD gene are required for production of the V. cholerae T2SS.73,76 Aside from its export role, some studies have also implicated this T2SS in maintaining cell envelope integrity.77,78 Removal of the eps genes compromises the OM, which is characterized by leakage of periplasmic components, the reduction of specific OM proteins, increased sensitivity to membrane-perturbing agents, and activation of the RpoE response.77,78 Activation of RpoE is consistent with removal of the T2SS compromising the cell envelope and might result, at least in part, from T2SS substrates accumulating in the periplasm. However, a particularly interesting and recent finding is that one of the promoters driving expression of the epsC-N operon is RpoE-dependent.79 One possible rationale for this control is to ensure increased T2SS production and virulence factor export when V. cholerae enters its host, where it encounters RpoE-inducing stimuli such as CAMPs and altered environmental conditions. An alternative rationale, suggested by removal of the T2SS compromising envelope integrity, is that increased production of the T2SS is an important component of the RpoE-dependent envelope stress-reduction mechanism. These 2 rationales are not mutually exclusive.
RpoE modulation of the PhoPQ regulon
The tightly packed LPS in the outer leaflet of the Gram-negative OM is a critical permeability barrier. However, LPS has some disadvantages for pathogens. One is that it is a potent inducer of inflammation. Another is that the anionic components of LPS provide an attractive force for CAMPs.80 However, Gram-negative bacteria can modify their LPS to change its properties and increase resistance to the host antimicrobial molecules they encounter. The PhoP (regulator)/PhoQ (sensor) 2-component system, which has been studied extensively in Salmonella, is pivotal in controlling these modifications.81 Its activation by low concentrations of divalent cations or the presence of CAMPs leads to PhoP-dependent regulation of genes involved in Mg2+ transport, resistance to acid, resistance to CAMPs, LPS modifications and virulence.82,83 The PhoPQ regulon is important for virulence and there is evidence from work in E. coli that the RpoE ESR can modulate it (Fig. 1).84 This discovery provides a regulatory connection between an ECF sigma factor and a 2-component system and significantly extends the mechanisms by which the RpoE response can remodel the cell envelope and control virulence gene expression.
Small RNAs (sRNAs) base pair with mRNAs to trigger their degradation and/or inhibit their translation. In E. coli, RpoE induces expression of the RybB, MicA and MicL sRNAs.85,86 MicL mitigates envelope stress by reducing production of the OM Braun lipoprotein, which is one of the most abundant proteins in the cell, and serves as a receptor for CAMPs.85 The Braun lipoprotein-encoding lpp mRNA appears to be the only direct target of MicL.85 In contrast, RybB and MicA have multiple targets and downregulate the production of every major OM porin, preventing their accumulation in conditions of envelope stress.87 Furthermore, RybB and MicA also repress the production of other proteins, some of which have been implicated in virulence-associated processes such as LPS modification (LpxT), and acid survival (Asr).87 MicA was also found to reduce production of the OmpT protease, but only in phoP+ cells. This led to the discovery that MicA base pairs with phoPQ mRNA to inhibit phoP translation, which then reduces ompT gene expression from its PhoP-induced promoter.84,87 Therefore, the RpoE response can downregulate the PhoPQ regulon via MicA-dependent inhibition of PhoP synthesis. Importantly, the predicted base pairing between MicA and phoPQ mRNA is conserved in several enterobacteria, including Salmonella, Shigella and Enterobacter.
This regulatory interaction between RpoE and PhoPQ makes some physiological sense because the PhoPQ regulon includes genes encoding secretion system and flagellar components, membrane transport systems, and proteins that are probable structural components of the OM.81 Reducing their production might help to reduce envelope stress. However, downregulation of the PhoPQ regulon by RpoE is a potential problem for a pathogen. If CAMP-induced damage activates the RpoE ESR, why use this response to decrease expression of the PhoPQ regulon that protects against CAMPs? The answer might be that RpoE does not shut down the PhoPQ regulon, but fine-tunes it to strike an appropriate balance between its beneficial effects and its potential to contribute to envelope stress. In support of a fine-tuning role, production of MicA only modestly downregulates the PhoP regulon.84 ompT expression was reduced 30-fold by a phoP null mutation but only 3-fold when MicA RNA was overproduced from a plasmid, and only 1.2-fold when MicA was produced from the chromosome of an rseA (RpoE anti sigma factor) null mutant.84 These effects are indeed modest, although it remains possible that some conditions might drive higher MicA production and a larger effect on the PhoP regulon.
The Cpx ESR
Gram-negative pathogens assemble pili and protein export systems in their cell-envelope, but problems arising from the production of these complex structures can trigger ESRs. Perhaps more interestingly, ESRs can also regulate the production of these virulence factors directly. The Cpx response provides several examples of this (Fig. 2). This response is a 2-component system consisting of an inner membrane histidine kinase (CpxA) and a cytoplasmic response regulator (CpxR).4,8,12 The small periplasmic protein CpxP, encoded immediately upstream of the cpxRA operon, inhibits the system, perhaps by forming an inhibitory complex with CpxA, although this remains in question.11 This negative regulation might be relieved when CpxP is degraded, perhaps in association with misfolded proteins. However, CpxP is not required for activation of the Cpx response by any known inducing signals, including misfolded proteins.88 Upon activation, CpxA primarily phosphorylates CpxR, which then binds to its target promoters. One promoter controls cpxP, and the CpxP protein has a second function as a chaperone that facilitates protein degradation by another CpxR regulon member, the periplasmic protease DegP.89 In addition, CpxR regulates other genes important for dealing with protein misfolding, transport functions, IM associated functions and many of unknown function.18,19
Figure 2.
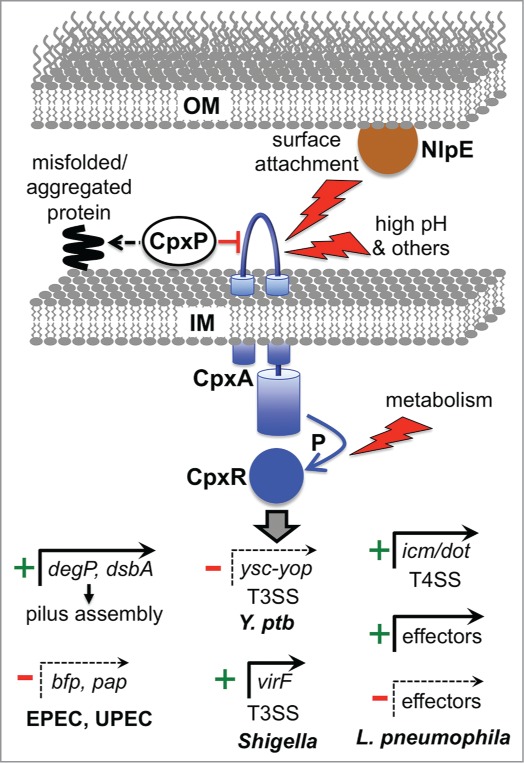
Examples of direct virulence gene regulation by the Cpx ESR. The Cpx response can be induced by various signals, with inputs entering at different points (surface attachment via NlpE to CpxA, external environmental changes via CpxA, and metabolic changes that alter the phosphorylation state of CpxR independently from CpxA). CpxP acts as a chaperone for misfolded proteins, directing them to destruction by the DegP protease (see text) and also negatively regulates the system, perhaps by interacting with CpxA. Phosphorylated CpxR activates the promoters of protein degradation factors (e.g., degP), protein-folding factors (e.g. dsbA) ands in multiple species, as well as many other genes associated with inner membrane functions. DegP and DsbA can reduce stress by promoting efficient assembly of the bundle forming pilus and Pap pilus in EPEC and UPEC, respectively. In addition, CpxR represses the promoters of the bfp and pap operons encoding these pili. In Y. pseudotuberculosis (Y. ptb) CpxR represses expression of several components of the Ysc-Yop T3SS. In Shigella sonnei CpxR directly activates the promoter of the gene encoding the major virulence regulator VirF, which leads to increased T3SS production because one VirF-induced target is the gene encoding the T3SS positive regulator NlpE/VirB (see text). CpxR also controls the T4SS in L. pneumophila by inducing genes encoding structural components (icm/dot) and by activating or repressing genes encoding some of the effectors exported by the T4SS.
Regulation of pili by the Cpx response
In strains of pathogenic E. coli, pili are critical for attachment to host cells and in some cases they are influenced by the Cpx response. One of these is the uropathogenic E. coli (UPEC) Pap or P pilus, which is involved in attachment to kidney cells.90,91 When the P pilus assembly genes were expressed in an E. coli laboratory strain the Cpx response was induced and at least 2 members of the CpxR regulon facilitated pilus assembly.15 These were DegP, which degrades “off-pathway” subunits that aggregate in the periplasm and DsbA, which assists pilus protein folding. Consequently, when the Cpx pathway was inactivated by mutation, laboratory strains of E. coli provided with the pap genes could produce only short, aberrant pili.15
CpxR also controls pap operon expression directly. This operon is regulated by a phase variation mechanism that switches between OFF (not expressed) or ON (expressed) states. A full description of this complicated mechanism is far beyond the scope of this review, but others have covered it extensively.92,93 In brief, the global regulator Lrp (leucine-responsive regulatory protein) binds to distal activating or proximal repressing sites within the pap regulatory region. The Lrp binding location is influenced by the methylation state of GATC sequences, which is influenced by Dam methylase and the local regulatory proteins PapB and PapI.93 It is thought that phase switching requires DNA replication to dissociate Lrp-DNA complexes and to generate a hemimethylated GATC site. Regardless, CpxR competes with Lrp for binding to both promoter proximal and distal binding sites. CpxR binds to these sites in a methylation-insensitive manner and locks the pap operon promoter into the OFF state by preventing Lrp-dependent activation.91 Another study had suggested that CpxR activated the pap operon, but it was later suggested this was due to multicopy pap sequences being used, which disrupted the normal phase variation mechanism.90,91 Taking all of this together, the Cpx response functions in 2 ways, presumably to reduce stress associated with P pilus production. First, it facilitates efficient pilus assembly by assisting protein folding and degrading mislocalized intermediates. Second, it reduces P pilus gene expression.
A similar relationship between a pilus and the Cpx response occurs in the intestinal pathogen Enteropathogenic E. coli (EPEC), where its bundle-forming pilus (BFP) is critical for attachment to host cells.94 The components required for BFP assembly are encoded by the bfp operon.95 Introduction of the bfp operon into E. coli K-12 strain HB101 resulted in pilus production, but this did not happen in E. coli K-12 strain MC4100.96 The authors speculated that the Cpx response could facilitate BFP assembly but its activity might have been insufficient in MC4100. This was supported when mutational activation of the MC4100 Cpx system allowed BFP production.96 Furthermore, BFP production and attachment to host cells were disrupted when the Cpx response was inactivated in EPEC.96 This defect was attributed to a requirement for CpxR regulon members, including DegP and DsbA, for efficient BFP assembly.97
The Cpx response also reduces bfp operon transcription.97 This is probably due to reduced expression of perA, which encodes a bfp transcriptional activator. However, it is not known if CpxR directly represses the perA promoter, which lacks obvious CpxR DNA binding sequences.97 Regardless, like the UPEC P pilus, the Cpx response both facilitates efficient BFP assembly and also reduces BFP pilus gene expression. Again, both mechanisms could be strategies to reduce stress associated with pilus production.
Regulation of protein secretion systems by the Cpx response
The Cpx response also regulates protein export systems associated with virulence (Fig. 2).12 For example, mutational activation of the Cpx response reduced expression of the genes encoding the Ysc T3SS of Yersinia pseudotuberculosis.98 The mechanism appears to be direct because phosphorylated CpxR bound to the control regions of Ysc T3SS-encoding genes in vitro.99 However, although T3SS gene expression was repressed by constitutive activation of the Cpx response, a cpxR null mutation had no effect.98 Therefore, when or if the endogenous Cpx response might regulate the Y. pseudotuberculosis T3SS is not yet known.
Another example of direct regulation of T3SS gene expression by CpxR comes from Shigella sonnei, although in this case the regulation is positive rather than negative. In this species VirF is the master regulator of virulence and one gene it induces is nlpE (virB), which encodes an activator of T3SS gene expression.100 A cpxR null mutation almost abolished virF expression.101 CpxR bound to the virF control region in vitro and this was enhanced by CpxR phosphorylation. Furthermore, phosphorylated CpxR activated virF transcription in vitro.101 The situation in S. sonnei is even more complex because it was later found that a cpxA null mutation reduced the translation, but not the transcription, of nlpE.102 The underlying mechanism of this translational effect, including whether CpxR is involved, is not clear.
Legionella pneumophila is a facultative intracellular respiratory pathogen that causes Legionnaires’ disease. Its ability to replicate inside macrophages requires the Icm/Dot type IV secretion system (T4SS), which translocates approximately 300 effector proteins into host cells.103 A screen for regulators of icmR expression, which encodes a chaperone component of the T4SS, identified CpxR as a positive regulator.104 CpxR bound to the icmR control region in vitro, which contains a conserved CpxR-binding sequence, and a cpxR null mutation reduced expression of an icmR-lacZ translational fusion.104 A later study found that other genes encoding several Icm/Dot components and secreted effectors are also regulated by direct CpxR-DNA binding.105 CpxR induced the genes encoding T4SS structural components. However, genes encoding secreted effectors were either induced or repressed by CpxR, which might indicate that different effectors are produced at different times during infection.105 Despite all of this, a cpxR null mutation did not affect L. pneumophila survival inside macrophages.104 This could be due to redundancy between CpxR-regulated and non-regulated effectors, or because the CpxR-activated effectors are only important in non-human hosts.105 It is more difficult to explain why reduced expression of genes encoding T4SS structural components would not affect intracellular growth. One possibility is that there might be compensatory control by other regulators inside macrophages. Alternatively, any reduced production of the T4SS might be below the threshold required for a phenotype to manifest in this ex vivo experimental system.
The Rcs ESR
Another Gram-negative ESR linked to virulence gene regulation is the Rcs response. It is conserved throughout the Enterobacteriaceae, induced by mutations or conditions that can disrupt the cell envelope, and thought to be important for controlling the production of macromolecular envelope structures and for envelope homeostasis.20,21 Activation of the Rcs response is mediated by a complex phosphorelay that can involve accessory factors and signal sensing by different components (Fig. 3). Regardless, all sensing pathways converge on the IM sensor kinase RcsC. RscC phosphorylates the intermediary IM protein RcsD, which then phosphorylates the cytoplasmic response regulator RcsB.106,107 Phosphorylated RcsB can bind DNA as a homodimer or as a heterodimer with the unstable accessory cytoplasmic protein RcsA or with some other accessory regulators.20,108 Target genes affect many processes including cell division, osmoregulation, cell envelope homeostasis, motility and virulence.20,21,109
Figure 3.
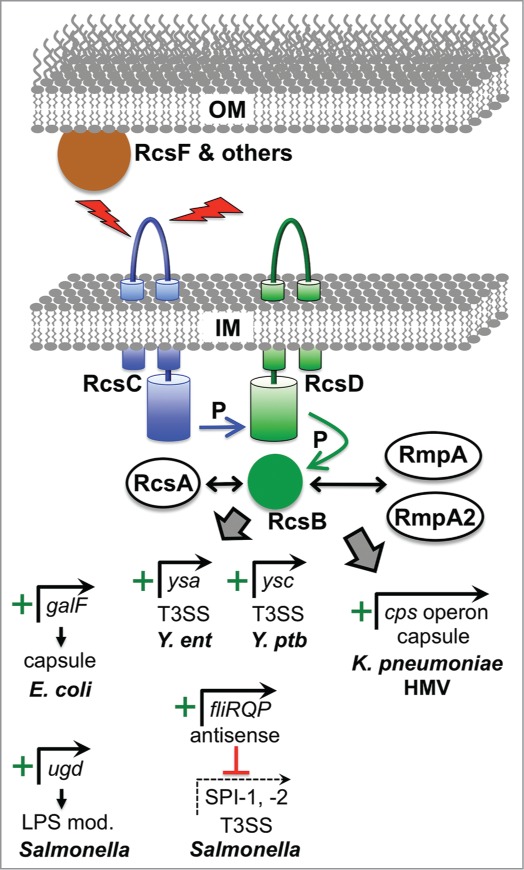
Examples of virulence gene regulation by the Rcs ESR. The Rcs system is induced by various signals deleterious to the outer membrane such as osmotic shock, desiccation, and overproduction of envelope proteins. Detection of some signals involves accessory proteins such as RcsF and others not shown. All signals converge of the IM histidine kinase RcsC, which transfers phosphate (P) to the cytoplasmic response regulator RcsB via the intermediary IM protein RcsD. RcsB can regulate target promoters as a homodimer or as a heterodimer with RcsA, or with the RcsA homologues RmpA and RmpA2 that are found in hypermucoviscous (HMV) strains of K. pneumoniae. Positively controlled target promoters include galF in encapsulated E. coli K strains, which increases precursors available for Group I capsule production. Similarly, ugd is an activated target in Salmonella, which is needed to make a precursor for LPS modification that increases CAMP resistance. In Y. enterocolitica (Y. ent) RcsB is involved in activation of the major operon encoding regulatory and structural components of the Ysa T3SS, but it is not known if this regulation is direct. Very recent data from Y. pseudotuberculosis (Y. ptb) has also shown that RcsB activates genes encoding the Ysc T3SS via direct induction of the lcrF gene, encoding the master Ysc T3SS regulator (see text). In Salmonella, RcsB drives the production of an antisense transcript encompassing the distal end of the fliLMNOPQR operon, which is involved in downregulating multiple virulence genes including those encoding components of the SPI-1 and SPI-2 T3SS systems. In HMV K. pneumoniae, RcsB, most likely as a heterodimer with RmpA or RmpA2, activates the cps operon responsible for capsular polysaccharide production.
Rcs-regulation of polysaccharide capsule production
The Rcs response was discovered as a regulator of E. coli genes required for biosynthesis of the extracellular polysaccharide colanic acid (the cps operon, later renamed the wca operon). However, colanic acid is most highly produced below 30°C, which means that it is probably important only in the environment.110 Nevertheless, polysaccharide capsules are widely conserved and in some cases they are virulence factors associated with resistance to phagocytosis and to host antimicrobial molecules.111–113 Furthermore, the Rcs response has been implicated in regulating production of E. coli group I capsules, which are important virulence determinants. In some E. coli strains the production of group I capsule was induced by overexpression of rcsB.114 However, an rcsB null mutation did not affect capsule production. Furthermore, neither an rcsB null mutation nor rcsB overexpression affected expression of the group I capsule-encoding (cps) operon.114 This suggested that the effect of rcsB overexpression on group I capsule production was indirect. Indeed, it was found that rcsB overexpression induced the galF gene involved in producing sugar nucleotide precursors, and that there was a putative RcsAB-binding sequence (RcsAB box) upstream of galF.115 Independent overexpression of galF also increased capsule production without affecting expression of the cps operon.115 Thus, the Rcs system might be able to induce group I capsule by increasing galF expression and influencing the pool of precursors. This does not explain why an rcsB null mutation did not affect capsule production. Of course, it is possible that the impact of rcsB overexpression was non-physiological. Alternatively, perhaps the appropriate environmental conditions were not used to detect an effect of the rcsB null mutation.115 Finally, an analogous situation also occurs in Salmonella. In this case, the ugd gene, encoding an enzyme that converts UDP-d-glucose to UDP-d-glucuronic acid, is regulated by the Rcs system. Ugd activity is important for both colanic acid production and for an LPS modification that increases CAMP resistance.116,117
Klebsiella pneumoniae is an important nosocomial pathogen that causes various diseases. Clinical isolates generally produce a capsular polysaccharide that confers resistance to serum and phagocytosis and is related to E. coli group I capsule. Interestingly, the RcsAB box upstream of galF is conserved in K. pneumoniae, suggesting that the phenomenon described above is not restricted to E. coli.118 In addition, copious capsule production, or hypermucoviscosity (HMV), has recently become associated with devastating invasive disease.119 The HMV strain K. pneumoniae CG43 has a plasmid with 2 homologous genes, rmpA and rmpA2, which encode transcription factors in the same family as RcsA and RcsB. The presence of these rmpA genes has been correlated with the HMV phenotype in K. pneumoniae isolates, and RmpA and RmpA2 regulate capsule production in strain CG43.120-122 Furthermore, RmpA or RmpA2, in addition to RcsA, can interact with RcsB and are thought to work in concert with RcsB as accessory proteins to regulate the capsular biosynthesis genes (cps; Fig. 3).120 This is analogous to the RscAB heterodimers described in E. coli. Indeed, RmpA2 binds to the cps control region, which contains an RcsAB box, and it is hypothesized that this interaction might be enhanced for an RmpA2-RcsB complex.121
Rcs regulation of T3SSs
A fascinating example of Rcs-mediated regulation has been reported in S. Typhimurium. This organism has 2 T3SSs encoded by Salmonella pathogenicity islands SPI-1 and SPI-2. The SPI-1 T3SS is involved in remodeling the host cell cytoskeleton to induce bacterial uptake, whereas the SPI-2 T3SS promotes intracellular survival by inhibiting phagolysosome maturation. During infection, Salmonella transitions to a non-motile state. RcsB represses the flhDC operon, encoding the master positive regulators of flagella production, via an RcsB box in the flhDC promoters of both E. coli and S. Typhimurium.123,124 A microarray survey of the S. Typhimurium RcsB regulon revealed that the SPI-1 genes were also strongly repressed by RcsB. However, SPI-2 gene expression (and some other virulence genes) was decreased in an rscB null mutant, suggesting positive regulation by RscB, but repressed when rcsB was overexpressed, suggesting negative regulation.123 This confounding regulatory pattern has not been explained, but it has been hypothesized that it might indicate that the Rcs system either represses or induces SPI-2 gene expression depending on the stage of infection.123
An intriguing finding from the microarray survey was that RscB induced the mRNA levels of fliP -Q and -R, whereas all other flagella genes were repressed.123 This was even more surprising because downregulation of flhDC should have prevented fliPQR expression. fliP -Q and -R are the last 3 genes of an operon that is located 281 bp upstream of rcsA and transcribed in the same direction (fliLMNOPQR – rcsA). A clue to what was happening is that the RcsB-induced levels of fliP -Q and -R mRNA occurred in a reverse gradient (fliR > fliQ > fliP).123 The explanation is that RcsB binds to an RcsAB box upstream of rcsA to activate its promoter and in doing so it also induces an antisense transcript that encompasses fliRQP.125 This might be due to RcsB changing the topological state of the fliR-rcsA intergenic region. This fliRQP antisense transcript affects the expression of many Rcs regulon members positively or negatively, and downregulates SPI-1 and -2 genes (Fig. 3).125 Further analysis indicated that this might involve the translation of a small open reading frame in the fliRQP antisense RNA, although this predicted protein was not detected.125
Another example of regulation of a T3SS by the Rcs response occurs in Y. enterocolitica. In this case RcsB positively regulates expression of the Ysa T3SS-encoding genes, which are found in highly pathogenic biogroup B1 strains and might be important in mammals or insects.126-128 RcsB is involved in activating expression of the ysaE operon, which encodes a master regulator of the ysa locus and structural components of the T3SS. However, the mechanism by which this occurs is unclear and it is not yet known if RcsB binds to the ysaE promoter. Regulation of the Ysa T3SS by RcsB was covered extensively in another recent review.109
Very recently, in Y. pseudotuberculosis is has been reported that RcsB positively regulates the Ysc T3SS.129 RcsB binds to an RcsB box upstream of the virG-lcrF operon promoter, with lcrF encoding the master activator of the Ysc T3SS, and this RcsB box is conserved in Y. pestis and Y. enterocolitica. Consistent with this regulatory role, activation of RcsB enhanced Ysc T3SS-dependent delivery of Yop proteins into immune cells and promoted bacterial viability. Thus, RcsB positively controls at least 2 different T3SS in Yersinia species (Ysa and Ysc; Fig. 3). Furthermore, these findings provide an example of a single T3SS being regulated oppositely by different ESRs, as demonstrated by negative or positive regulation of the Y. pseudotuberculosis Ysc T3SS by the Cpx and Rcs systems, respectively Figs. 2 and 3).
Regulation of Virulence Genes by ESRs in Gram-Positive Bacteria
Introduction to Gram-positive ESRs
The discussion changes upon moving from Gram-negative to Gram-positive ESRs. The fundamental differences between their cell envelopes is one obvious reason for this.3 Compared to Gram-negative bacteria, the defining features of the Gram-positive cell envelope are the lack of an outer membrane, a much thicker cell wall, and the presence of teichoic acids.130 The thickness of the cell wall comes from its composition as a multi-layered mesh of highly cross-linked peptidoglycan, in contrast to the single layer found in Gram-negative bacteria. Teichoic acids are phosphate containing anionic polymers anchored to the cytoplasmic membrane or covalently linked to the peptidoglycan.131,132 This complex cell wall must be continuously synthesized, degraded and recycled. These processes must be carefully controlled to maintain cell shape, to increase cell size and for cells to divide. The cell wall also provides Gram-positive cells with resistance to extremely high turgor pressures. Therefore, it is critical for survival and Gram-positive bacteria go to great lengths to maintain it.
Another reason the discussion changes is because Gram-positive ESRs have been defined differently to Gram-negative ESRs. This was discussed in a highly recommended review of Gram-positive ESRs by Jordan et al.133 Those authors noted that many Gram-positive ESRs were discovered due to their role in resisting or responding to cell wall antibiotics. This is in contrast to the defining feature of most Gram-negative ESRs, which is detection or response to mislocalized or misfolded envelope proteins (although some Gram-negative ESRs have also been implicated in resistance to antibiotics, including some that target the cell wall).18,134,135 Therefore, Jordan et al suggested that ESRs of Gram-positive bacteria are defined by their detection or response to the presence of cell wall antibiotics or other cell envelope-damaging molecules. These definitions are not completely satisfactory, comprehensive, or immune to pitfalls, but they are reasonable based on current understanding.
Much of the characterization of Gram-positive ESRs has been done in the non-pathogenic Bacillus subtilis, the model species for low G+C Gram-positive bacteria (Firmicutes). However, many of the systems are conserved in other species including some high G+C Gram-positive bacteria (Actinobacteria). The Gram-positive ESRs have been divided into 3 categories.133 The first responds to one specific compound, or group of closely related compounds. They regulate one target that is usually closely linked to the regulatory genes and involved in conferring resistance to the inducing agent alone. Therefore, this category will not be discussed further.
The second category is induced by cell wall antibiotics or other damaging agents and is involved in counteracting the damage. These are perhaps the most deserving of the ESR title. They include at least 4 ECF σ factors in B. subtilis (σM, σW, σX and σV) that respond to antibiotics and CAMPs to control numerous target genes.136-140 However, relatively little is known about ECF σ factors in other Gram-positive bacteria. Other members of this category include 2 component regulatory systems. For example, LiaRS-like systems respond to a variety of cell envelope-damaging agents and usually, but not always, confer resistance to them.133,141 However, the identity and extent of their target genes varies considerably between species. Another example more pertinent for this review is the ApsXRS/GraXRS system, studied most in staphylococci and streptococci, which responds to CAMPs.142,143 It is critical for resistance to these agents and also one of the few Gram-positive ESRs that has been closely linked to virulence gene regulation (Fig. 4).
Figure 4.
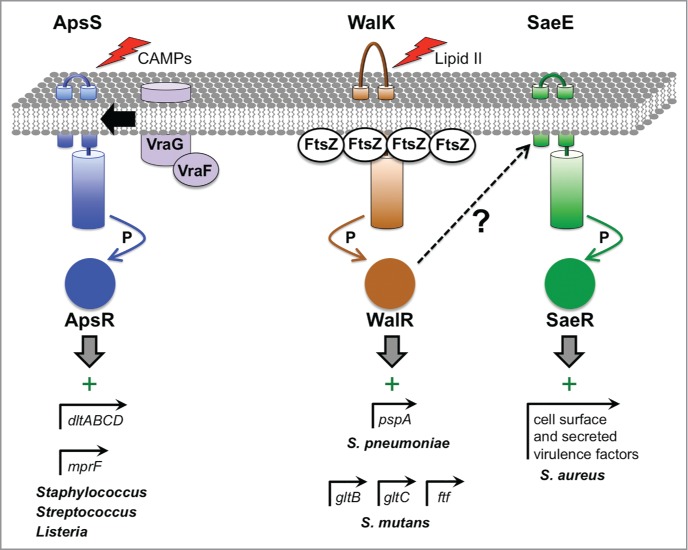
Virulence gene regulation by 2-component ESRs in Gram-positive bacteria. The ApsSR (also known as GraSR) system is activated by a variety of antimicrobial molecules, which are detected by ApsS. This leads to transfer of phosphate (P) from ApsS to ApsR, which can then bind to its target promoters. In some pathogens, ApsR targets include the promoters of dltABCD and mprF, which encode proteins involved in modifying teichoic acids and phospholipids, respectively, to reduce the net negative charge of the cell envelope and increase CAMP resistance. Another ApsR-dependent promoter drives vraFG expression, which encodes an ABC transporter that might be involved in efflux of toxic molecules out of the cell and/or their detection. The activity of the WalKR system is hypothesized to depend on the level of the peptidoglycan biosynthesis intermediate Lipid II, which might provide a report of cell wall synthesis activity and a healthy status of replicating cells. Furthermore, in B. subtilis WalK has been shown to interact with FtsZ at the division septum where cell wall metabolism is highly active, which might promote high WalK activity. WalK phosphorylates the response regulator WalR, which has many targets involved in cell division and cell wall metabolism. In. S. pneumoniae, one WalR-dependent promoter controls the pspA gene, which encodes the well-characterized virulence factor Pneumococcal Surface Protein A that is involved in complement evasion. In S. mutans, the WalKR system affects genes important for biofilm formation and attachment to tooth surfaces, with WalR binding directly to the promoters of at least 3 of them (gtfB, gtfC and ftf). In S. aureus, constitutive activation of WalR generates an inducing signal for the SaeSR system, which increases the production of many cell surface and secreted virulence factors. However, the SaeSR-inducing signal that is generated by WalR activation is not known.
The third category of Gram-positive ESRs is not induced by cell-wall antibiotics but does play a role in controlling cell envelope integrity. The former property perhaps makes their assignment as stress response systems tenuous.133 However, some of them might detect consequences of damaging agents (e.g., decreased membrane potential) or their inducing signals are not yet known. These systems are the LytSR-like, MtrAB-like, and YycGF-like 2 component regulatory systems. In Staphylococcus aureus LytSR works along with another regulator (CidR) to control genes encoding a holin (LrgAB) and antiholin (CidAB) that affect the access of a murein hydrolase to the cell wall.144 This impacts autolysis and has been described as an altruistic programmed cell death system important in biofilms.144,145 LytSR responds to reduced membrane potential and also plays a role in resisting CAMPs.146,147 MtrAB are conserved in the Actinobacteria and implicated in ESR in Corynebacterium glutamicum and Mycobacterium smegmatis.148,149 However, note that Mycobacteria are not Gram-positive (or Gram-negative) because the Gram stain procedure does not work with their cells. Furthermore, both Corynebacteria and Mycobacteria are now known to have an OM, although it is distinct from that of Gram-negative bacteria.150,151 Regardless, MtrAB is essential in Mycobacterium, an unusual property for a 2 component system. Both the essentiality of MtrAB, and its link to ESR, has led to it being described as the Actinobacterial functional equivalent of YycGF/WalKR.152 YycGF/WalKR, first described in B. subtilis, are conserved in most Firmicutes and probably essential in almost all of them.133 It is another of the Gram-positive ESRs implicated in the control of virulence gene expression (Fig. 4).
Gram-positive ESRs regulate cell envelope charge
One important phenotype that is regulated by several different Gram-positive ESRs, and closely connected to virulence, is the net negative charge of the bacterial cell surface. The negative charge arises from 2 features. First, from the anionic membrane phospholipids phosphatidylglycerol (PG) and cardiolipin, which are common to Gram-negative and Gram-positive bacteria. Second, from the teichoic acids, which are unique to Gram-positive bacteria. A negatively charged cell surface represents a significant problem for pathogens because it attracts CAMPs.80 Gram-positive bacteria use separate mechanisms to reduce the negative charge of both their phospholipids and their teichoic acids, and both mechanisms are regulated by ESRs.
Teichoic acids are linear polymers of repeating glycerol-phosphate or ribitol-phosphate residues substituted with N-acetylglucosamine and D-alanine. Wall teichoic acids (WTA) are covalently attached to peptidoglycan, whereas a glycolipid moiety anchors lipoteichoic acids (LTA) to the cytoplasmic membrane.130-132,153 Their negative charge comes from the anionic phosphodiester linkages connecting the monomers. However, when D-alanine is incorporated via an ester bond its amino group remains free and positively charged, which reduces the net negative charge of the polymer.131,132 The products of the dltABCD operon incorporate D-alanine into LTAs, which can then be transferred to WTAs by transacetylation.154,155 Therefore, inactivation of the dlt operon increases the net negative charge of the cell envelope. Consistent with this, dlt null mutants of Gram-positive pathogens have increased sensitivity to CAMPs, increased killing by human neutrophils and reduced virulence in animal models of infection.156-164 The dlt operon is also one of the best examples of virulence genes controlled by ESRs in Gram-positive bacteria. Even though many Gram-positive ESRs are widely conserved, their target genes can vary significantly between species.133 However, the dlt operon is almost always the target of one or more ESRs in any particular species. The ESRs involved include ECF σ factors, LiaRS, LytRS, and ApsXRS/GraXRS.133,142,143,165 As a result, dlt is one of relatively few defining loci of ESR in Gram-positive positive bacteria because it is almost always connected to the envelope stress response.133
A second mechanism for reducing cell surface negative charge involves aminoacylation of anionic membrane phospholipids, a phenomenon first described in S. aureus decades ago.166,167 Most commonly it is positively charged lysine that is attached to PG, although other amino acids linked to PG have been detected including alanine and arginine.168 Furthermore, Listeria species lysylate cardiolipin as well as PG.169,170 The enzyme responsible transfers amino acids from aminoacyl-tRNA and is encoded by mprF (multi peptide resistance factor). MprF was discovered in S. aureus but is conserved in many Gram-positive bacteria (mostly Firmicutes and Actinobacteria) and also Gram-negative proteobacteria.168,171 MprF family members can be specific for lysl-tRNA or alanyl-tRNA, although the Listeria MprF can use both.168,172 A S. aureus mprF null mutant is more sensitive to CAMPs and is also killed more effectively by human neutrophils and attenuated in mouse and rat models of infection.163,171 Similar phenotypes have been reported for a Listeria monocytogenes mprF null mutant.169 mprF expression is controlled by the ApsXRS/GraXRS ESR in S. aureus and S. epidermidis and by the homologous VirRS system in L. monocytogenes.142,143,165 These ESRs also control the dlt operons in these species (see above and Fig. 4). Originally, it was suggested that the lack of phenotypes unrelated to CAMP resistance in a S. aureus mprF null mutant might indicate that the only purpose of MprF is to increase resistance to host defenses.80 If so, why don't the major virulence regulators of Staphylococci or Listeria control mprF? The answer might come from recent findings that lysyl-PG in L. monocytogenes has broad roles in maintaining membrane integrity beyond CAMP resistance.173 Thus, mprF might be considered as both a virulence gene and an envelope stress tolerance gene. This makes it a good fit for control by the ApsXRS/GraXRS system because this ESR also affects the expression of other virulence genes and other stress-tolerance genes, as described next.
ApsXRS/GraXRS
A family of ESR 2-component systems known as BceRS-like systems typically respond to one compound, or group of closely related compounds, to induce linked ABC transporter genes that exclude the inducing agent, or play a role in signal sensing.133,174-176 For example, the BceRS system of B. subtilis responds to bacitracin and promotes resistance to it.177,178 However, a BceRS-like system in S. aureus and S. epidermidis known as GraSR (glycopeptide resistance associated) or ApsSR (antimicrobial peptide-sensing), respectively, has several unusual features.142,143 First, although the regulatory genes are adjacent to ABC transporter-encoding genes (vraFG), which they regulate, this is not their only target. Second, the system detects and confers resistance to a relatively broad range of structurally unrelated antimicrobial agents, although the range is narrower in S. aureus than in S. epidermidis. Third, the apsX/graX gene immediately upstream of apsSR/graSR encodes a putative cytoplasmic protein of unknown function that is required for the regulatory response, making this a 3-component system. In fact, it is even more complex because the VraFG transporter might be involved in signal sensing rather than detoxification, leading to this being described as a 5-component system.176,179
As mentioned above the staphylococcal ApsXSR/GraXSR system is a ESR that regulates virulence genes because it controls the dlt operon and mprF, decreasing cell surface negative charge and promoting resistance to CAMPs (Fig. 4). This regulation is probably direct because a conserved inverted repeat upstream of dlt, mprF and vraFG is a likely GraR-binding site (although it has not been possible to purify active GraR to confirm this in vitro).180 S. aureus transcriptome analysis has also revealed GraXRS-dependent upregulation of lysine biosynthetic genes, which could provide substrate for MprF-dependent lysylation of membrane phospholipids.142 Other S. aureus microarray analyses have expanded the influence of GraXSR to the differential expression of 248 genes in one case and 424 in another.180,181 Among the most highly upregulated were those encoding virulence factors. For example, the agrBDCA operon, encoding a virulence-regulating quorum-sensing system, was expressed up to 21-fold higher in graSR+ vs. ΔgraSR cells grown with colistin.180 The gene encoding delta-hemolysin showed 80-fold higher expression in the graSR+ cells. Other virulence-related genes also demonstrated GraSR-dependent induction, albeit of less impressive magnitude, including those encoding SarA, SarX, SarS (virulence regulators) and Hla (α hemolysin).180 Of course many of these effects are likely to be indirect rather than due to direct binding of GraR to promoters. Nevertheless, whether direct or indirect, the fact that all of these genes are affected by the GraXRS system is consistent with it being required for S. aureus resistance to CAMPs and to killing by human neutrophils and for virulence in mouse models of infection.142,179,182 The GraXRS regulon has also been reported to overlap extensively with another S. aureus ESR implicated in virulence gene control, the WalKR system (see below).142
L. monocytogenes, another Gram-positive pathogen, has a homologous system called VirRS. Its genomic context is different and it lacks an ApsX/GraX homolog.165 Nevertheless, in common with the staphylococcal and streptococcal systems, its regulon includes mprF and dlt as well as genes encoding an ABC transporter. The control regions of these genes, as well as other VirR-induced genes identified by microarray analysis, share a conserved inverted repeat that was bound by VirR in vitro.165 Furthermore, the virR null strain was attenuated in a mouse model of infection and at least one of its regulatory targets (mprF) was required for CAMP resistance.165,169
WalKR
A 2-component system originally identified in B. subtilis as YycGF has since been described by other names in different species.183,184 It was renamed WalKR when it became clear that its major role was to regulate cell wall metabolism.185,186 WalKR are widely conserved in Firmicutes and essential in most species tested.184,187 It is not unprecedented for a ESR to be essential, as exemplified by RpoE in some Gram-negative bacteria. However, it must be acknowledged up front that others have suggested that the assignment of WalKR as a ESR is highly questionable. This is because it is most active during normal growth conditions, is not induced by cell envelope damage, and is actually deactivated by some conditions and antibiotics usually associated with inducing ESR.184 In fact, the signal that induces WalKR has been proposed to emanate from normal cell wall metabolism, leading to it having been described as the antithesis of cell wall stress.185 Nevertheless, in common with others we have included WalKR as a ESR in this review because of its fundamental role in ensuring cell envelope function and integrity.133 We have also included it because it can influence virulence gene expression in both staphylococci and streptococci (Fig. 4).
A survey of cell wall antibiotics that activate or deactivate WalKR led to the proposal that Lipid II, an intermediate in peptidoglycan biosynthesis, might be the inducing signal.184 Most antibiotics that deactivate WalKR deplete Lipid II by inhibiting a step prior to its formation, whereas antibiotics that activate WalKR cause Lipid II to accumulate by inhibiting a downstream step. Lipid II levels could reflect cell wall synthetic activity, which fits with the hypothesis that WalKR monitors a signal derived from normal cell wall metabolism. It is also consistent with the genes controlled by WalKR. Although there is substantial variation between the WalKR regulons of different species, genes involved in cell wall metabolism, membrane integrity and also cell division are a common theme.133,184,187 Putting this together, Dubrac et al. proposed that WalKR monitors the healthy status of replicating cells and adjusts cell wall metabolism to meet growth requirements.184 Furthermore, the B. subtilis WalK sensor interacts with cell division protein FtsZ at the division septum (Fig. 4), and WalKR also induces ftsAZ operon expression.188,189 Association with the septum, where cell wall metabolism is highly active, might promote high WalKR activity and ensure the induction of genes involved in initiating cell division (e.g. ftsAZ) and other functions required for division and growth, such as cell wall hydrolases. In contrast, WalK would not associate with division septa in stationary phase, leading to downregulation of genes required for cell wall plasticity. For more extensive descriptions of this intriguing regulatory system we recommend other reviews that focused upon it specifically.184,187
Early work linking WalKR to virulence came from streptococcal species, where the system is known as VicKR. In Streptococcus, genetic analysis was facilitated because the sensor protein VicK/WalK is not essential. Microarray studies revealed that VicKR/WalKR affected the expression of several S. pneumoniae virulence genes.190-192 These include genes encoding the PiaBCDA ABC transporter involved in iron uptake; Pneumococcal Surface Protein A, which is one of the most studied S. pneumoniae virulence factors and is involved in complement evasion (PspA, not to be confused with the unrelated phage shock protein A); and some other surface proteins linked to virulence. However, when different growth conditions and genetic manipulations were compared, only a subset of genes showed strong dependence on VicKR/WalKR in all cases.192 Nevertheless, this subset included pspA. Importantly, VicR/WalR bound to the control region of pspA in vitro, and also to the control regions of some other genes, and in each case the binding region encompassed a sequence similar to a previously proposed VicR/WalR binding site consensus.192 Therefore, VicR/WalR is a direct regulator of pspA, which encodes the most extensively studied S. pneumoniae virulence factor (Fig. 4). In another Streptococcus that is associated with dental caries, S. mutans, the VicKR/WalKR system affects genes important for biofilm formation and attachment to the tooth surface, and VicR/WalR binds directly to the promoters of at least 3 of them (gtfB, gtfC and ftf; Fig. 4).193 Finally, VicKR/WalKR have also been linked to virulence in S. pyogenes but in that case it has not been determined if virulence gene regulation is involved.194
In S. aureus, a phosphomimetic mutation in WalR (WalR D55E), which rendered it constitutively active, increased the expression of genes encoding major cell surface or secreted virulence factors (e.g., leukotoxins, matrix binding proteins and proteases).195 Also upregulated was the saePQRS operon, the last 2 genes of which encode the SaeRS 2-component system, which is a major virulence gene regulator.196,197 This is significant because there was extensive overlap between the WalR and SaeSR regulons, suggesting that the effect of WalR D55E on virulence genes was mediated through upregulation of saeQPRS. This was confirmed when a ΔsaeRS mutation prevented WalR from inducing 8 out of 8 virulence genes selected for individual analysis.195 WalR is not a direct activator of saePQRS operon expression because the operon does not have any predicted WalR binding sites, and ΔsaeRS also prevented upregulation of saeP expression by WalR D55E. This suggests that constitutive WalR activity generates an inducing signal that triggers the SaeRS 2-component system, leading to activation of the SaeRS regulon, which includes saePQRS (Fig. 4). However, it is not known what SaeRS-activating signal WalR might generate. Possibilities include a change in the level of one or more molecules involved in cell wall metabolism, or overproduction of other WalR regulon members that causes membrane perturbation and changes the conformation and activity of SaeS. The authors of this study also reported that the constitutively active WalR mutant had decreased virulence in a murine sepsis model.195 This might seem surprising because activation of the SaeSR regulon is normally associated with increased virulence and immune evasion. However, the WalR regulon also includes genes involved in cell wall degradation and turnover. Therefore, the increased release of peptidoglycan fragments in the constitutive WalR mutant probably triggers the innate immune inflammatory response, resulting in increased bacterial clearance. In wild type bacteria, careful control of WalKR activity during host infection might maintain an appropriate balance between its effect on the SaeSR regulon and on cell wall metabolism enzymes. Alternatively, the wild type WalKR system might not modulate SaeSR activity and the effect could have been a consequence of using a constitutively active WalR mutant.
Concluding Remarks
The survival of all bacteria depends on their ability to sense and respond to threats against their cell envelope. Many of these threats are prevalent when pathogens enter their host, which means that ESRs are often critical to maintain envelope integrity during infection. This relationship between ESRs and virulence has been extended by the integration of some virulence genes into ESR regulons, in both Gram-negative and Gram-positive bacteria. These virulence genes might have functions related to cell envelope stress relief, or their only apparent role might be in facilitating the host-pathogen interaction. Sometimes, the dividing line between these possibilities is blurred, as demonstrated by RpoE-dependent expression of T2SS genes in V. cholerae. This T2SS is obviously important for virulence factor export, but also seems to be important for cell envelope integrity. We have not discussed every known example of virulence gene regulation by ESRs, but the specific cases we have highlighted will hopefully demonstrate how widespread and varied this phenomenon is. One obvious rationale for controlling virulence genes by a ESR is when the genes are involved in protecting the cell envelope from damage by the host (e.g. CAMP resistance). A second rationale is to reduce envelope stress by moderating the production of virulence structures that might themselves damage the cell envelope (e.g., downregulating T3SS or pili genes). A final rationale is simply to coordinate the induction of virulence gene expression with entry into the host, where ESR-inducing signals are encountered. Understanding the rationale is complicated by examples of the same or similar virulence systems being regulated differently by ESRs (e.g. positive or negative regulation of a T3SS by the Cpx system in Yersinia and Shigella, or by the Rcs system in Yersinia and Salmonella, and positive or negative regulation of the Ysc T3SS by Rcs and Cpx, respectively, in Y. pseudotuberculosis; Figs. 2 and 3). Furthermore, components of the same system can also be regulated differently by a single ESR in a single species, with the T4SS effectors of L. pneumophila providing the example (Fig. 2). It seems almost certain that future research will continue to uncover additional direct relationships between ESRs and virulence gene expression. This will undoubtedly increase our understanding of how this happens, why this happens, and whether or not it might eventually be exploited for therapeutic intervention.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the anonymous reviewers for taking the time to provide detailed, thoughtful and helpful comments.
Funding
Work in our laboratory is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI052148. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. AJD holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
References
- 1. Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol 2005; 56:1119-28; PMID:15882407; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04625.x [DOI] [PubMed] [Google Scholar]
- 2. Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol 2006; 4:383-94; PMID: 16715050; http://dx.doi.org/ 10.1038/nrmicro1394 [DOI] [PubMed] [Google Scholar]
- 3. Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2010; 2:a000414; PMID:20452953; http://dx.doi.org/ 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raivio TL, Silhavy TJ. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol 1999; 2:159-65; PMID:10322173; http://dx.doi.org/ 10.1016/S1369-5274(99)80028-9 [DOI] [PubMed] [Google Scholar]
- 5. Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev 1993; 7:2618-28; PMID:8276244; http://dx.doi.org/ 10.1101/gad.7.12b.2618 [DOI] [PubMed] [Google Scholar]
- 6. Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013; 340:837-41; PMID:23687042; http://dx.doi.org/ 10.1126/science.1235358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Las Penas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol 1997; 179:6862-4; PMID:9352942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacRitchie DM, Buelow DR, Price NL, Raivio TL. Two-component signaling and gram negative envelope stress response systems. Adv Exp Med Biol 2008; 631:80-110; PMID:18792683; http://dx.doi.org/ 10.1007/978-0-387-78885-2_6 [DOI] [PubMed] [Google Scholar]
- 9. Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli sigma E regulon. J Biol Chem 2001; 276:20866-75; PMID:11274153; http://dx.doi.org/ 10.1074/jbc.M100464200 [DOI] [PubMed] [Google Scholar]
- 10. Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol 2006; 4:e2; PMID:16336047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raivio TL. Everything old is new again: An update on current research on the Cpx envelope stress response. Biochim Biophys Acta 2014; 1844:1529-41; PMID:NOT_FOUND; http://dx.doi.org/ 10.1016/j.bbamcr.2013.10.018 [DOI] [PubMed] [Google Scholar]
- 12. Vogt SL, Raivio TL. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 2012; 326:2-11; PMID:22092948; http://dx.doi.org/ 10.1111/j.1574-6968.2011.02406.x [DOI] [PubMed] [Google Scholar]
- 13. Danese PN, Oliver GR, Barr K, Bowman GD, Rick PD, Silhavy TJ. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J Bacteriol 1998; 180:5875-84; PMID:9811644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 1998; 180:831-9; PMID:9473036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones CH, Danese PN, Pinkner JS, Silhavy TJ, Hultgren SJ. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J 1997; 16:6394-406; PMID:9351822; http://dx.doi.org/ 10.1093/emboj/16.21.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mileykovskaya E, Dowhan W. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol 1997; 179:1029-34; PMID:9023180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakayama S, Watanabe H. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol 1995; 177:5062-9; PMID:7665485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raivio TL, Leblanc SK, Price NL. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol 2013; 195:2755-67; PMID:23564175; http://dx.doi.org/ 10.1128/JB.00105-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol 2009; 191:1798-815; PMID:19103922; http://dx.doi.org/ 10.1128/JB.00798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clarke DJ. The Rcs Phosphorelay: Biofilm Formation and Virulence in the Enterobacteriaceae. In: Gross R, Beier D, eds. Two-Component Systems in Bacteria. Norfolk, UK: Caister Academic Press, 2012:333-54. [Google Scholar]
- 21. Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 2005; 59:379-405; PMID:16153174; http://dx.doi.org/ 10.1146/annurev.micro.59.050405.101230 [DOI] [PubMed] [Google Scholar]
- 22. Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol 2002; 184:4168-76; PMID:12107134; http://dx.doi.org/ 10.1128/JB.184.15.4168-4176.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 2005; 55:1113-26; PMID:15686558; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04449.x [DOI] [PubMed] [Google Scholar]
- 24. Darwin AJ. The phage-shock-protein response. Mol Microbiol 2005; 57:621-8; PMID:16045608; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04694.x [DOI] [PubMed] [Google Scholar]
- 25. Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MPH, Buck M. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev 2010; 34:797-827; PMID:20636484 [DOI] [PubMed] [Google Scholar]
- 26. Yamaguchi S, Darwin AJ. Recent findings about the Yersinia enterocolitica phage shock protein response. J Microbiol 2012; 50:1-7; PMID:22367931; http://dx.doi.org/ 10.1007/s12275-012-1578-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Model P, Jovanovic G, Dworkin J. The Escherichia coli phage-shock-protein (psp) operon. Mol Microbiol 1997; 24:255-61; PMID:9159513; http://dx.doi.org/ 10.1046/j.1365-2958.1997.3481712.x [DOI] [PubMed] [Google Scholar]
- 28. Darwin AJ. Stress relief during host infection: The phage shock protein response supports bacterial virulence in various ways. PLoS Pathog 2013; 9:e1003388; PMID:23853578; http://dx.doi.org/ 10.1371/journal.ppat.1003388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J 2006; 25:5241-9; PMID:17082772; http://dx.doi.org/ 10.1038/sj.emboj.7601402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horstman NK, Darwin AJ. Phage shock proteins B and C prevent lethal cytoplasmic membrane permeability in Yersinia enterocolitica. Mol Microbiol 2012; 85:445-60; PMID:22646656; http://dx.doi.org/ 10.1111/j.1365-2958.2012.08120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seo J, Savitzky DC, Ford E, Darwin AJ. Global analysis of tolerance to secretin-induced stress in Yersinia enterocolitica suggests that the phage-shock-protein system may be a remarkably self-contained stress response. Mol Microbiol 2007; 65:714-27; PMID:17608794; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05821.x [DOI] [PubMed] [Google Scholar]
- 32. Lloyd LJ, Jones SE, Jovanovic G, Gyaneshwar P, Rolfe MD, Thompson A, Hinton JC, Buck M. Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG). J Biol Chem 2004; 279:55707-14; PMID:15485810; http://dx.doi.org/ 10.1074/jbc.M408994200 [DOI] [PubMed] [Google Scholar]
- 33. Darwin AJ, Miller VL. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol Microbiol 2001; 39:429-44; PMID:11136463; http://dx.doi.org/ 10.1046/j.1365-2958.2001.02235.x [DOI] [PubMed] [Google Scholar]
- 34. Karlinsey JE, Maguire ME, Becker LA, Crouch ML, Fang FC. The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol Microbiol 2010; 78:669-85; PMID:20807201; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07357.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bashyam MD, Hasnain SE. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect Genet Evol 2004; 4:301-8; PMID:15374527; http://dx.doi.org/ 10.1016/j.meegid.2004.04.003 [DOI] [PubMed] [Google Scholar]
- 36. Mascher T. Signaling diversity and evolution of extracytoplasmic function (ECF) sigma factors. Curr Opin Microbiol 2013; 16:148-55; PMID:23466210; http://dx.doi.org/ 10.1016/j.mib.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 37. Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 2002; 46:47-110; PMID:12073657; http://dx.doi.org/ 10.1016/S0065-2911(02)46002-X [DOI] [PubMed] [Google Scholar]
- 38. Ades SE. Control of the alternative sigma factor sigmaE in Escherichia coli. Curr Opin Microbiol 2004; 7:157-62; PMID:15063853; http://dx.doi.org/ 10.1016/j.mib.2004.02.010 [DOI] [PubMed] [Google Scholar]
- 39. Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol 2004; 52:613-9; PMID:15101969; http://dx.doi.org/ 10.1111/j.1365-2958.2003.03982.x [DOI] [PubMed] [Google Scholar]
- 40. De Las Penas A, Connolly L, Gross CA. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol 1997; 24:373-85; PMID:9159523; http://dx.doi.org/ 10.1046/j.1365-2958.1997.3611718.x [DOI] [PubMed] [Google Scholar]
- 41. Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol 1997; 24:355-71; PMID:9159522; http://dx.doi.org/ 10.1046/j.1365-2958.1997.3601713.x [DOI] [PubMed] [Google Scholar]
- 42. Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 2009; 9:2432-43; PMID:19343722; http://dx.doi.org/ 10.1002/pmic.200800794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muller C, Bang IS, Velayudhan J, Karlinsey J, Papenfort K, Vogel J, Fang FC. Acid stress activation of the sigma(E) stress response in Salmonella enterica serovar Typhimurium. Mol Microbiol 2009; 71:1228-38; PMID:19170886; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06597.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 2002; 16:2156-68; PMID:12183369; http://dx.doi.org/ 10.1101/gad.1008902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev 2002; 16:2147-55; PMID:12183368; http://dx.doi.org/ 10.1101/gad.1002302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA. Design principles of the proteolytic cascade governing the sigmaE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev 2007; 21:124-36; PMID:17210793; http://dx.doi.org/ 10.1101/gad.1496707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosenstein BJ, Zeitlin PL. Prognosis in cystic fibrosis. Curr Opin Pulm Med 1995; 1:444-9; PMID:9363080; http://dx.doi.org/ 10.1097/00063198-199511000-00003 [DOI] [PubMed] [Google Scholar]
- 48. Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 1996; 60:539-74; PMID:8840786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 2002; 15:194-222; PMID:11932230; http://dx.doi.org/ 10.1128/CMR.15.2.194-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol 2005; 175:7512-8; PMID:16301659; http://dx.doi.org/ 10.4049/jimmunol.175.11.7512 [DOI] [PubMed] [Google Scholar]
- 51. Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol 1994; 176:6007-14; PMID:7928961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol 2012; 84:595-607; PMID:22497280; http://dx.doi.org/ 10.1111/j.1365-2958.2012.08049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 1993; 90:8377-81; PMID:8378309; http://dx.doi.org/ 10.1073/pnas.90.18.8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 1999; 145:1349-57; PMID:10411261; http://dx.doi.org/ 10.1099/13500872-145-6-1349 [DOI] [PubMed] [Google Scholar]
- 55. Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, Hoiby N. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology 2008; 154:103-13; PMID:18174130; http://dx.doi.org/ 10.1099/mic.0.2007/010421-0 [DOI] [PubMed] [Google Scholar]
- 56. Rowen DW, Deretic V. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol 2000; 36:314-27; PMID:10792719; http://dx.doi.org/ 10.1046/j.1365-2958.2000.01830.x [DOI] [PubMed] [Google Scholar]
- 57. Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology 2005; 151:2251-61; PMID:16000715; http://dx.doi.org/ 10.1099/mic.0.27772-0 [DOI] [PubMed] [Google Scholar]
- 58. Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol 2011; 193:286-91; PMID:21036998; http://dx.doi.org/ 10.1128/JB.01132-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol 2006; 62:412-26; PMID:17020580; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05390.x [DOI] [PubMed] [Google Scholar]
- 60. Wood LF, Ohman DE. Use of cell wall stress to characterize sigma 22 (AlgTU) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 2009; 72:183-201; PMID:19226327; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06635.x [DOI] [PubMed] [Google Scholar]
- 61. Damron FH, Davis MR, Jr., Withers TR, Ernst RK, Goldberg JB, Yu G, Yu HD. Vanadate and triclosan synergistically induce alginate production by Pseudomonas aeruginosa strain PAO1. Mol Microbiol 2011; 81:554-70; PMID:21631603; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07715.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Hoiby N. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob Agents Chemother 2004; 48:1175-87; PMID:15047518; http://dx.doi.org/ 10.1128/AAC.48.4.1175-1187.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schobert M, Tielen P. Contribution of oxygen-limiting conditions to persistent infection of Pseudomonas aeruginosa. Future Microbiol 2010; 5:603-21; PMID:20353301; http://dx.doi.org/ 10.2217/fmb.10.16 [DOI] [PubMed] [Google Scholar]
- 64. Firoved AM, Ornatowski W, Deretic V. Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect Immun 2004; 72:5012-8; PMID:15321993; http://dx.doi.org/ 10.1128/IAI.72.9.5012-5018.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Firoved AM, Wood SR, Ornatowski W, Deretic V, Timmins GS. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J Bacteriol 2004; 186:4046-50; PMID:15175322; http://dx.doi.org/ 10.1128/JB.186.12.4046-4050.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E). Proc Natl Acad Sci U S A 1995; 92:7941-5; PMID:7644517; http://dx.doi.org/ 10.1073/pnas.92.17.7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bragonzi A, Worlitzsch D, Pier GB, Timpert P, Ulrich M, Hentzer M, Andersen JB, Givskov M, Conese M, Doring G. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis 2005; 192:410-9; PMID:15995954; http://dx.doi.org/ 10.1086/431516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 2005; 187:7955-62; PMID:16291668; http://dx.doi.org/ 10.1128/JB.187.23.7955-7962.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun 1994; 62:596-605; PMID:8300217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thanassi DG, Hultgren SJ. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Opin Cell Biol 2000; 12:420-30; PMID:10873830; http://dx.doi.org/ 10.1016/S0955-0674(00)00111-3 [DOI] [PubMed] [Google Scholar]
- 71. Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 2012; 10:336-51; PMID:22466878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett 2006; 255:175-86; PMID:16448494; http://dx.doi.org/ 10.1111/j.1574-6968.2006.00102.x [DOI] [PubMed] [Google Scholar]
- 73. Sandkvist M. Type II secretion and pathogenesis. Infect Immun 2001; 69:3523-35; PMID:11349009; http://dx.doi.org/ 10.1128/IAI.69.6.3523-3535.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kaper JB, Morris JG, Jr., Levine MM. Cholera. Clin Microbiol Rev 1995; 8:48-86; PMID:7704895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J Biol Chem 2011; 286:16555-66; PMID:21385872; http://dx.doi.org/ 10.1074/jbc.M110.211078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Overbye LJ, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene 1993; 132:101-6; PMID:8406031; http://dx.doi.org/ 10.1016/0378-1119(93)90520-D [DOI] [PubMed] [Google Scholar]
- 77. Sikora AE, Beyhan S, Bagdasarian M, Yildiz FH, Sandkvist M. Cell envelope perturbation induces oxidative stress and changes in iron homeostasis in Vibrio cholerae. J Bacteriol 2009; 191:5398-408; PMID: 19542276; http://dx.doi.org/ 10.1128/JB.00092-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sikora AE, Lybarger SR, Sandkvist M. Compromised outer membrane integrity in Vibrio cholerae Type II secretion mutants. J Bacteriol 2007; 189:8484-95; PMID:17890307; http://dx.doi.org/ 10.1128/JB.00583-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zielke RA, Simmons RS, Park BR, Nonogaki M, Emerson S, Sikora AE. The type II secretion pathway in Vibrio cholerae is characterized by growth-phase dependent expression of exoprotein genes and is positively regulated by sigmaE. Infect Immun 2014; 82:2788-801; PMID:24733097; http://dx.doi.org/ 10.1128/IAI.01292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol 2002; 10:179-86; PMID:11912025; http://dx.doi.org/ 10.1016/S0966-842X(02)02333-8 [DOI] [PubMed] [Google Scholar]
- 81. Dalebroux ZD, Miller SI. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 2014; 17:106-13; PMID: 24531506; http://dx.doi.org/ 10.1016/j.mib.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kato A, Groisman EA, Howard Hughes Medical Institute . The PhoQPhoP regulatory network of Salmonella enterica. Adv Exp Med Biol 2008; 631:7-21; PMID:18792679; http://dx.doi.org/ 10.1007/978-0-387-78885-2_2 [DOI] [PubMed] [Google Scholar]
- 83. Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol 2005; 3:36-46; PMID:15608698; http://dx.doi.org/ 10.1038/nrmicro1068 [DOI] [PubMed] [Google Scholar]
- 84. Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol 2010; 76:467-79; PMID:20345657; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07115.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 2014; 28:1620-34; PMID:25030700; http://dx.doi.org/ 10.1101/gad.243485.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 2006; 9:605-11; PMID:17055775; http://dx.doi.org/ 10.1016/j.mib.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 87. Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A 2011; 108:12875-80; PMID:21768388; http://dx.doi.org/ 10.1073/pnas.1109379108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol 2003; 185:2432-40; PMID:12670966; http://dx.doi.org/ 10.1128/JB.185.8.2432-2440.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Isaac DD, Pinkner JS, Hultgren SJ, Silhavy TJ. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc Natl Acad Sci U S A 2005; 102:17775-9; PMID:16303867; http://dx.doi.org/ 10.1073/pnas.0508936102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hung DL, Raivio TL, Jones CH, Silhavy TJ, Hultgren SJ. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J 2001; 20:1508-18; PMID:11285215; http://dx.doi.org/ 10.1093/emboj/20.7.1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hernday AD, Braaten BA, Broitman-Maduro G, Engelberts P, Low DA. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol Cell 2004; 16:537-47; PMID:15546614; http://dx.doi.org/ 10.1016/j.molcel.2004.10.020 [DOI] [PubMed] [Google Scholar]
- 92. Blomfield IC. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv Microb Physiol 2001; 45:1-49; PMID:11450107; http://dx.doi.org/ 10.1016/S0065-2911(01)45001-6 [DOI] [PubMed] [Google Scholar]
- 93. Hernday A, Krabbe M, Braaten B, Low D. Self-perpetuating epigenetic pili switches in bacteria. Proc Natl Acad Sci U S A 2002; 99 Suppl 4:16470-6; PMID:12202745; http://dx.doi.org/ 10.1073/pnas.182427199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cleary J, Lai LC, Shaw RK, Straatman-Iwanowska A, Donnenberg MS, Frankel G, Knutton S. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 2004; 150:527-38; PMID:14993302; http://dx.doi.org/ 10.1099/mic.0.26740-0 [DOI] [PubMed] [Google Scholar]
- 95. Sohel I, Puente JL, Ramer SW, Bieber D, Wu CY, Schoolnik GK. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol 1996; 178:2613-28; PMID:8626330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nevesinjac AZ, Raivio TL. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J Bacteriol 2005; 187:672-86; PMID:15629938; http://dx.doi.org/ 10.1128/JB.187.2.672-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus. Mol Microbiol 2010; 76:1095-110; PMID:20444097; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Carlsson KE, Liu J, Edqvist PJ, Francis MS. Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect Immun 2007; 75:3913-24; PMID:17517869; http://dx.doi.org/ 10.1128/IAI.01346-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu J, Thanikkal EJ, Obi IR, Francis MS. Elevated CpxR∼P levels repress the Ysc-Yop type III secretion system of Yersinia pseudotuberculosis. Res Microbiol 2012; 163:518-30; PMID:22842077; http://dx.doi.org/ 10.1016/j.resmic.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 100. Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol 1993; 175:6142-9; PMID:7691791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nakayama S, Watanabe H. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J Bacteriol 1998; 180:3522-8; PMID:9657992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mitobe J, Arakawa E, Watanabe H. A sensor of the two-component system CpxA affects expression of the type III secretion system through posttranscriptional processing of InvE. J Bacteriol 2005; 187:107-13; PMID:15601694; http://dx.doi.org/ 10.1128/JB.187.1.107-113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Isaac DT, Isberg R. Master manipulators: an update on Legionella pneumophila IcmDot translocated substrates and their host targets. Future Microbiol 2014; 9:343-59; PMID:24762308; http://dx.doi.org/ 10.2217/fmb.13.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gal-Mor O, Segal G. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J Bacteriol 2003; 185:4908-19; PMID:12897011; http://dx.doi.org/ 10.1128/JB.185.16.4908-4919.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Altman E, Segal G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icmdot components as well as new translocated substrates. J Bacteriol 2008; 190:1985-96; PMID:18192394; http://dx.doi.org/ 10.1128/JB.01493-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC–> YojN–> RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol 2001; 40:440-50; PMID:11309126; http://dx.doi.org/ 10.1046/j.1365-2958.2001.02393.x [DOI] [PubMed] [Google Scholar]
- 107. Chen MH, Takeda S, Yamada H, Ishii Y, Yamashino T, Mizuno T. Characterization of the RcsC–>YojN–>RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci Biotechnol Biochem 2001; 65:2364-7; PMID:11758943; http://dx.doi.org/ 10.1271/bbb.65.2364 [DOI] [PubMed] [Google Scholar]
- 108. Stout V, Torres-Cabassa A, Maurizi MR, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol 1991; 173:1738-47; PMID:1999391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Flores-Kim J, Darwin AJ. Links between type III secretion and extracytoplasmic stress responses in Yersinia. Front Cell Infect Microbiol 2012; 2:125; PMID:23087910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol 1985; 162:1111-9; PMID:3888955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 2004; 72:7107-14; PMID:15557634; http://dx.doi.org/ 10.1128/IAI.72.12.7107-7114.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Farris C, Sanowar S, Bader MW, Pfuetzner R, Miller SI. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J Bacteriol 2010; 192:4894-903; PMID:20675476; http://dx.doi.org/ 10.1128/JB.00505-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 2006; 75:39-68; PMID:16756484; http://dx.doi.org/ 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- 114. Jayaratne P, Keenleyside WJ, MacLachlan PR, Dodgson C, Whitfield C. Characterization of rcsB and rcsC from Escherichia coli O9:K30:H12 and examination of the role of the rcs regulatory system in expression of group I capsular polysaccharides. J Bacteriol 1993; 175:5384-94; PMID:8366025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rahn A, Whitfield C. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol Microbiol 2003; 47:1045-60; PMID:12581358; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03354.x [DOI] [PubMed] [Google Scholar]
- 116. Mouslim C, Groisman EA. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol Microbiol 2003; 47:335-44; PMID:12519186; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03318.x [DOI] [PubMed] [Google Scholar]
- 117. Mouslim C, Latifi T, Groisman EA. Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J Biol Chem 2003; 278:50588-95; PMID:14514676; http://dx.doi.org/ 10.1074/jbc.M309433200 [DOI] [PubMed] [Google Scholar]
- 118. Wehland M, Bernhard F. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 2000; 275:7013-20; PMID:10702265; http://dx.doi.org/ 10.1074/jbc.275.10.7013 [DOI] [PubMed] [Google Scholar]
- 119. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 2013; 4:107-18; PMID:23302790; http://dx.doi.org/ 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 2010; 192:3144-58; PMID:20382770; http://dx.doi.org/ 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 2003; 185:788-800; PMID:12533454; http://dx.doi.org/ 10.1128/JB.185.3.788-800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 2006; 42:1351-8; PMID:16619144; http://dx.doi.org/ 10.1086/503420 [DOI] [PubMed] [Google Scholar]
- 123. Wang Q, Zhao Y, McClelland M, Harshey RM. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J Bacteriol 2007; 189:8447-57; PMID:17905992; http://dx.doi.org/ 10.1128/JB.01198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 2003; 49:823-32; PMID:12864862; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03601.x [DOI] [PubMed] [Google Scholar]
- 125. Wang Q, Harshey RM. Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol Microbiol 2009; 74:71-84; PMID:19703110; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06851.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Venecia K, Young GM. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect Immun 2005; 73:5961-77; PMID:16113317; http://dx.doi.org/ 10.1128/IAI.73.9.5961-5977.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Walker KA, Miller VL. Synchronous gene expression of the Yersinia enterocolitica Ysa type III secretion system and its effectors. J Bacteriol 2009; 191:1816-26; PMID:19124573; http://dx.doi.org/ 10.1128/JB.01402-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Walker KA, Maltez V, Hall JD, Vitko NP, Miller VL. A Phenotype at Last: Essential Role for the Yersinia enterocolitica Ysa Type III Secretion System in a Drosophila S2 cell model. Infect Immun 2013; 81:2478-87; PMID:23630961; http://dx.doi.org/ 10.1128/IAI.01454-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Li Y, Hu Y, Francis MS, Chen S. RcsB positively regulates the Yersinia Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ Microbiol 2014; In press; PMID:25039908; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 130. Foster SJ, Poham DL. Structure and synthesis of cell wall, spore cortex, teichoic acid, S-layers and capsules. In: Sonenshein AL, Hoch JA, Losick R, eds. Bacillus subtilis and its closest relatives: From genes to cells. Washington, DC: ASM Press, 2002:21-41. [Google Scholar]
- 131. Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev 2003; 67:686-723; PMID:14665680; http://dx.doi.org/ 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schneewind O, Missiakas D. Lipoteichoic acids, phosphate-containing polymers in the envelope of gram-positive bacteria. J Bacteriol 2014; 196:1133-42; PMID: 24415723; http://dx.doi.org/ 10.1128/JB.01155-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev 2008; 32:107-46; PMID:18173394; http://dx.doi.org/ 10.1111/j.1574-6976.2007.00091.x [DOI] [PubMed] [Google Scholar]
- 134. Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 2008; 190:2065-74; PMID: 18192383; http://dx.doi.org/ 10.1128/JB.01740-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mahoney TF, Silhavy TJ. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol 2013; 195:1869-74; PMID: 23335416; http://dx.doi.org/ 10.1128/JB.02197-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Cao M, Helmann JD. The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol 2004; 186:1136-46; PMID:14762009; http://dx.doi.org/ 10.1128/JB.186.4.1136-1146.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Eiamphungporn W, Helmann JD. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol 2008; 67:830-48; PMID:18179421; http://dx.doi.org/ 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Guariglia-Oropeza V, Helmann JD. Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J Bacteriol 2011; 193:6223-32; PMID:21926231; http://dx.doi.org/ 10.1128/JB.06023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Helmann JD. Deciphering a complex genetic regulatory network: the Bacillus subtilis sigmaW protein and intrinsic resistance to antimicrobial compounds. Sci Prog 2006; 89:243-66; PMID:17338440; http://dx.doi.org/ 10.3184/003685006783238290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kingston AW, Liao X, Helmann JD. Contributions of the sigma(W), sigma(M) and sigma(X) regulons to the lantibiotic resistome of Bacillus subtilis. Mol Microbiol 2013; 90:502-18; PMID:23980836; http://dx.doi.org/ 10.1111/mmi.12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Jordan S, Junker A, Helmann JD, Mascher T. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol 2006; 188:5153-66; PMID:16816187; http://dx.doi.org/ 10.1128/JB.00310-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 2007; 66:1136-47; PMID:17961141; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05986.x [DOI] [PubMed] [Google Scholar]
- 143. Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A 2007; 104:9469-74; PMID:17517597; http://dx.doi.org/ 10.1073/pnas.0702159104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol 2007; 5:721-6; PMID:17694072; http://dx.doi.org/ 10.1038/nrmicro1743 [DOI] [PubMed] [Google Scholar]
- 145. Rice KC, Bayles KW. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol 2003; 50:729-38; PMID: 14617136; http://dx.doi.org/ 10.1046/j.1365-2958.2003.t01-1-03720.x [DOI] [PubMed] [Google Scholar]
- 146. Patton TG, Yang SJ, Bayles KW. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol Microbiol 2006; 59:1395-404; PMID:16468984; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05034.x [DOI] [PubMed] [Google Scholar]
- 147. Yang SJ, Xiong YQ, Yeaman MR, Bayles KW, Abdelhady W, Bayer AS. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob Agents Chemother 2013; 57:3875-82; PMID:23733465; http://dx.doi.org/ 10.1128/AAC.00412-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Brocker M, Mack C, Bott M. Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J Bacteriol 2011; 193:1237-49; PMID:21183673; http://dx.doi.org/ 10.1128/JB.01032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol Microbiol 2010; 76:348-64; PMID:20233304; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07110.x [DOI] [PubMed] [Google Scholar]
- 150. Bansal-Mutalik R, Nikaido H. Quantitative lipid composition of cell envelopes of Corynebacterium glutamicum elucidated through reverse micelle extraction. Proc Natl Acad Sci U S A 2011; 108:15360-5; PMID:21876124; http://dx.doi.org/ 10.1073/pnas.1112572108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bansal-Mutalik R, Nikaido H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc Natl Acad Sci U S A 2014; 111:4958-63; PMID:24639491; http://dx.doi.org/ 10.1073/pnas.1403078111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Winkler ME, Hoch JA. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J Bacteriol 2008; 190:2645-8; PMID:18245295; http://dx.doi.org/ 10.1128/JB.01682-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fischer W. Physiology of lipoteichoic acids in bacteria. Adv Microb Physiol 1988; 29:233-302; PMID:3289326; http://dx.doi.org/ 10.1016/S0065-2911(08)60349-5 [DOI] [PubMed] [Google Scholar]
- 154. Neuhaus FC, Heaton MP, Debabov DV, Zhang Q. The dlt operon in the biosynthesis of D-alanyl-lipoteichoic acid in Lactobacillus casei. Microb Drug Resist 1996; 2:77-84; PMID:9158726; http://dx.doi.org/ 10.1089/mdr.1996.2.77 [DOI] [PubMed] [Google Scholar]
- 155. Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 1995; 270:15598-606; PMID:7797557; http://dx.doi.org/ 10.1074/jbc.270.26.15598 [DOI] [PubMed] [Google Scholar]
- 156. Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 2002; 43:1-14; PMID:11849532; http://dx.doi.org/ 10.1046/j.1365-2958.2002.02723.x [DOI] [PubMed] [Google Scholar]
- 157. Abi Khattar Z, Rejasse A, Destoumieux-Garzon D, Escoubas JM, Sanchis V, Lereclus D, Givaudan A, Kallassy M, Nielsen-Leroux C, Gaudriault S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J Bacteriol 2009; 191:7063-73; PMID:19767427; http://dx.doi.org/ 10.1128/JB.00892-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Fittipaldi N, Sekizaki T, Takamatsu D, Harel J, Dominguez-Punaro Mde L, Von Aulock S, Draing C, Marois C, Kobisch M, Gottschalk M. D-alanylation of lipoteichoic acid contributes to the virulence of Streptococcus suis. Infect Immun 2008; 76:3587-94; PMID:18474639; http://dx.doi.org/ 10.1128/IAI.01568-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 2006; 188:5797-805; PMID:16885447; http://dx.doi.org/ 10.1128/JB.00336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. McBride SM, Sonenshein AL. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 2011; 157:1457-65; PMID:21330441; http://dx.doi.org/ 10.1099/mic.0.045997-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Poyart C, Pellegrini E, Marceau M, Baptista M, Jaubert F, Lamy MC, Trieu-Cuot P. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol Microbiol 2003; 49:1615-25; PMID:12950925; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03655.x [DOI] [PubMed] [Google Scholar]
- 162. Simanski M, Glaser R, Koten B, Meyer-Hoffert U, Wanner S, Weidenmaier C, Peschel A, Harder J. Staphylococcus aureus subverts cutaneous defense by D-alanylation of teichoic acids. Exp Dermatol 2013; 22:294-6; PMID:23528217; http://dx.doi.org/ 10.1111/exd.12114 [DOI] [PubMed] [Google Scholar]
- 163. Weidenmaier C, Peschel A, Kempf VA, Lucindo N, Yeaman MR, Bayer AS. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect Immun 2005; 73:8033-8; PMID:16299297; http://dx.doi.org/ 10.1128/IAI.73.12.8033-8038.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 1999; 274:8405-10; PMID:10085071; http://dx.doi.org/ 10.1074/jbc.274.13.8405 [DOI] [PubMed] [Google Scholar]
- 165. Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol 2005; 57:1367-80; PMID:16102006; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04776.x [DOI] [PubMed] [Google Scholar]
- 166. Houtsmuller UM, van Deenen LL. On the amino acid esters of phosphatidyl glycerol from bacteria. Biochim Biophys Acta 1965; 106:564-76; PMID: 4956500; http://dx.doi.org/ 10.1016/0005-2760(65)90072-X [DOI] [PubMed] [Google Scholar]
- 167. Houtsmuller UM, Van D. On the Accumulation of Amino Acid Derivatives of Phosphatidylglycerol in Bacteria. Biochim Biophys Acta 1964; 84:96-8; PMID:14124763 [DOI] [PubMed] [Google Scholar]
- 168. Roy H. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life 2009; 61:940-53; PMID:19787708; http://dx.doi.org/ 10.1002/iub.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol 2006; 62:1325-39; PMID:17042784; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05452.x [DOI] [PubMed] [Google Scholar]
- 170. Fischer W, Leopold K. Polar lipids of four Listeria species containing L-lysylcardiolipin, a novel lipid structure, and other unique phospholipids. Int J Syst Bacteriol 1999; 49 Pt 2:653-62; PMID:10408878; http://dx.doi.org/ 10.1099/00207713-49-2-653 [DOI] [PubMed] [Google Scholar]
- 171. Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med 2001; 193:1067-76; PMID:11342591; http://dx.doi.org/ 10.1084/jem.193.9.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci U S A 2008; 105:4667-72; PMID:18305156; http://dx.doi.org/ 10.1073/pnas.0800006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Dare K, Shepherd J, Roy H, Seveau S, Ibba M. LysPGS formation in Listeria monocytogenes has broad roles in maintaining membrane integrity beyond antimicrobial peptide resistance. Virulence 2014; 5:534-46; PMID:24603093; http://dx.doi.org/ 10.4161/viru.28359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Mascher T. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett 2006; 264:133-44; PMID:17064367; http://dx.doi.org/ 10.1111/j.1574-6968.2006.00444.x [DOI] [PubMed] [Google Scholar]
- 175. Hiron A, Falord M, Valle J, Debarbouille M, Msadek T. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraSBraR two-component system (SA2417SA2418) and both the BraDBraE and VraDVraE ABC transporters. Mol Microbiol 2011; 81:602-22; PMID:21696458; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07735.x [DOI] [PubMed] [Google Scholar]
- 176. Falord M, Karimova G, Hiron A, Msadek T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2012; 56:1047-58; PMID:22123691; http://dx.doi.org/ 10.1128/AAC.05054-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 2003; 50:1591-604; PMID: 14651641; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03786.x [DOI] [PubMed] [Google Scholar]
- 178. Ohki R, Giyanto , Tateno K, Masuyama W, Moriya S, Kobayashi K, Ogasawara N. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol Microbiol 2003; 49:1135-44; PMID:12890034; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03653.x [DOI] [PubMed] [Google Scholar]
- 179. Yang SJ, Bayer AS, Mishra NN, Meehl M, Ledala N, Yeaman MR, Xiong YQ, Cheung AL. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun 2012; 80:74-81; PMID:21986630; http://dx.doi.org/ 10.1128/IAI.05669-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Falord M, Mader U, Hiron A, Debarbouille M, Msadek T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 2011; 6:e21323; PMID:21765893; http://dx.doi.org/ 10.1371/journal.pone.0021323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Gotz F. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 2007; 3:e102; PMID:17676995; http://dx.doi.org/ 10.1371/journal.ppat.0030102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kraus D, Herbert S, Kristian SA, Khosravi A, Nizet V, Gotz F, Peschel A. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol 2008; 8:85; PMID:18518949; http://dx.doi.org/ 10.1186/1471-2180-8-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Fabret C, Hoch JA. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol 1998; 180:6375-83; PMID:9829949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 2008; 70:1307-22; PMID: 19019149; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06483.x [DOI] [PubMed] [Google Scholar]
- 185. Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol 2007; 65:180-200; PMID:17581128; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05782.x [DOI] [PubMed] [Google Scholar]
- 186. Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalKWalR (YycGYycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 2007; 189:8257-69; PMID:17827301; http://dx.doi.org/ 10.1128/JB.00645-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Dubrac S, Msadek T. Tearing down the wall: peptidoglycan metabolism and the WalKWalR (YycGYycF) essential two-component system. Adv Exp Med Biol 2008; 631:214-28; PMID: 18792692; http://dx.doi.org/ 10.1007/978-0-387-78885-2_15 [DOI] [PubMed] [Google Scholar]
- 188. Fukushima T, Szurmant H, Kim EJ, Perego M, Hoch JA. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol Microbiol 2008; 69:621-32; PMID:18573169; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06308.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 2000; 146:1573-83; PMID:10878122 [DOI] [PubMed] [Google Scholar]
- 190. Ng WL, Robertson GT, Kazmierczak KM, Zhao J, Gilmour R, Winkler ME. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol 2003; 50:1647-63; PMID:14651645; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03806.x [DOI] [PubMed] [Google Scholar]
- 191. Mohedano ML, Overweg K, de la Fuente A, Reuter M, Altabe S, Mulholland F, de Mendoza D, Lopez P, Wells JM. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J Bacteriol 2005; 187:2357-67; PMID:15774879; http://dx.doi.org/ 10.1128/JB.187.7.2357-2367.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Ng WL, Tsui HC, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol 2005; 187:7444-59; PMID:16237028; http://dx.doi.org/ 10.1128/JB.187.21.7444-7459.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 2005; 187:4064-76; PMID:15937169; http://dx.doi.org/ 10.1128/JB.187.12.4064-4076.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Liu M, Hanks TS, Zhang J, McClure MJ, Siemsen DW, Elser JL, Quinn MT, Lei B. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 2006; 152:967-78; PMID:16549661; http://dx.doi.org/ 10.1099/mic.0.28706-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Delaune A, Dubrac S, Blanchet C, Poupel O, Mader U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon JM, et al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun 2012; 80:3438-53; PMID:22825451; http://dx.doi.org/ 10.1128/IAI.00195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun 2006; 74:4655-65; PMID:16861653; http://dx.doi.org/ 10.1128/IAI.00322-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Steinhuber A, Goerke C, Bayer MG, Doring G, Wolz C. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J Bacteriol 2003; 185:6278-86; PMID:14563862; http://dx.doi.org/ 10.1128/JB.185.21.6278-6286.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]


