Abstract
Transmissible encephalopathies (TSEs), such as CJD and scrapie, are caused by infectious agents that provoke strain-specific patterns of disease. Misfolded host prion protein (PrP-res amyloid) is believed to be the causal infectious agent. However, particles that are stripped of PrP retain both high infectivity and viral proteins not detectable in uninfected mouse controls. We here detail host proteins bound with FU-CJD infectious brain particles by proteomic analysis. More than 98 proteins were differentially regulated, and 56 FU-CJD exclusive proteins were revealed after PrP, GFAP, C1q, ApoE and other late pathologic response proteins were removed. Stripped FU-CJD particles revealed HSC70 (144× the uninfected control), cyclophilin B, an FU-CJD exclusive protein required by many viruses, and early endosome-membrane pathways known to facilitate viral processing, replication, and spread. Synaptosomal elements including synapsin-2 (at 33×) and AP180 (a major FU-CJD exclusive protein) paralleled the known ultrastructural location of 25nm virus-like TSE particles and infectivity in synapses. Proteins without apparent viral or neurodegenerative links (copine-3), and others involved in viral-induced protein misfolding and aggregation, were also identified. Human sCJD brain particles contained 146 exclusive proteins, and heat shock, synaptic and viral pathways were again prominent, in addition to Alzheimer (AD), Parkinson, and Huntington aggregation proteins. Host proteins that bind TSE infectious particles can prevent host immune recognition and contribute to prolonged cross-species transmissions (the species barrier). Our infectious particle strategy, which reduces background sequences by >99%, emphasizes host targets for new therapeutic initiatives. Such therapies can simultaneously subvert common pathways of neurodegeneration.
Keywords: SWATH, synapse, cell-to-cell spread, viral entry and maturation, early endosomes, chaperones, protein folding, Alzheimer's, Parkinson, prion protein, amyloid, bioinformatics
Introduction
Transmissible encephalopathies (TSEs) affect many mammalian species and are caused by a variety of distinct infectious strains which induce the host's prion protein (PrP) to misfold into an amyloid conformation (PrP-res). Unresolved PrP misfolding patterns are presumed to encode strain-specific information. However, misfolded PrP-res, assayed as partially digested truncated bands on Western blots, is indistinguishable in normal mice infected with markedly different TSE strains; moreover, changing the PrP-res band pattern does not alter the agent's strain phenotype and its ability to breed true (1-3). Normally such data would indicate that TSE agents contain a strain-determining nucleic acid genome. As with many microorganisms and viruses, but not host proteins, TSE agents additionally i) have a long latent phase in lymphoid tissues before causing disease, ii) are spread environmentally, iii) reach epidemic proportions and then disappear when the environmental source of infection is removed, (e.g., bovine TSE of the UK and human kuru in New Guinea), and iv) separate as virus-like dense particles of ∼25nm (4). Comparable virus-like particles are found in fixed scrapie and CJD infected brain and cell cultures, but not in uninfected controls (5-8). Unlike PrP-res amyloid fibers, these 25nm particles do not bind PrP antibodies (8). While it is widely believed that host PrP spontaneously misfolds into infectious PrP-res, the evidence for any infectious form of PrP is questionable. Misfolded recombinant PrP has not been reproducibly infectious even though massive amounts of PrP-res are readily generated. In addition, infectivity is preserved even when no PrP is detectable. This is seen both in natural digestion of PrP within the GI tract (9, 10), and after straightforward proteinase K (PK) treatment of brain homogenates (11). In contrast to PrP, many viral particles are built to survive digestive juices and effectively transit beyond the gut epithelium to white blood cells, a route also used by TSE agents (12).
Proteins other than PrP, as well as PCR-verified nucleic acids of >1kb, are present in all infectious brain preparations (13). We previously isolated purified infectious particles from brain to identify i) integral and/or agent protective proteins, and ii) host complexes that are critical for TSE agent spread, replication and maturation. To avoid redistributing host components, 18,000g particle pellets (p18) were made without detergents. FU-CJD p18 particles with markedly reduced host components, as well as those purified by sucrose gradients, retained high infectivity even after all visible host PrP on Western blots was abolished by PK digestion; these PK stripped FU-CJD particles revealed 47 viral motifs not present in parallel uninfected controls and attest to the high degree of infectious particle purification (14). We here report on the host associated proteins present in aliquots of these high titer FU-CJD particles (pre and post-PK) to identify those most tightly bound to agent. A clear PrP signal became undetectable after PK digestion, and other pathologic host response proteins were also simultaneously removed as shown here. The very limited remaining host proteins allowed deeper insight into how TSE particles are processed and spread.
For independent comparison, we also evaluated host particle components from a sCJD infected human brain. Several major human p18 proteins overlapped with FU-CJD proteins despite differences in both agent-strain and species. This underscored their importance. AD and Lewy body linked proteins in the human samples further suggested new therapeutic targets for progressive neurodegeneration. Amyloid and fibril pathology has been recognized for over 50 years as a common thread in many neurodegenerative diseases, and a recent diagnostic marker for AD, Apolipoprotein J (clusterin), was originally shown to be a major component of large PrP amyloid plaques in experimental sCJD in rats (15, 16). As other pathology-associated host proteins, it was not tightly associated with infectious particles.
Materials and Methods
i) Particle preparation, Protein extraction, Tryptic Digestion and Sample Clean-up
Briefly, a pool of 6 individually verified FU-CJD mouse brain homogenates and normal brain pools made without detergents were separated from highly infectious supernatant by a 5,500g × 10min centrifugation in 25% sucrose. This removed the large visible lipid myelin-membrane layer and nuclei from the collected clarified supernatant that was then diluted to 8% sucrose and spun at 10,000g × 5 min to remove lysosomes and debris (14). The supernatant (s10) was digested with RNAse A to allow quantitative sedimentation of infectious particles at 18,000g × 30 min (p18) that retained only ∼12% of the starting protein and PrP sequences (14). For further particle purification, the FU-CJD and normal p18 preparations were digested with PK under conditions that maintained high infectivity (14); these particles contained <1% of the starting protein with no visible PrP peptides despite high cell equivalent (CE) loads (see Fig. 1). Cortical regions (temporal, occipital and parietal) from human sCJD brain, verified histologically and by Western blot for PrP-res at Yale, and confirmed by the CDC prion center, were pooled to make comparable sCJD and control p18 samples. Mouse and human p18 particles (with 30ug protein), and much higher (e7) cell equivalents (CE) of PK digested samples were solubilized in fresh 8M Urea / 0.4M NH4HCO3 with 4mM DTT for 20min at 22°C for 30 min and quenched with 8.3mM IAA. Samples were diluted to <2M urea, digested with 5mg Lys C for 4h at 37°C, followed by sequential trypsin digestions for ∼3h (5mg, with a trypsin to protein ratio of 1:15), and then overnight (+2mg) for 4 h. For p18 post-PK+ digested aliquots trypsin was reduced by ½ because of the markedly reduced protein. Reactions were quenched with 12 μl 20% TFA, peptides desalted by C18 UltraMicroSpin columns, dried, dissolved in 20μl 3:8 v/v 70 % FA/0.1% TFA and protein quantitation determined by hydrolysis and amino acid analysis (AAA). The peptide concentration was adjusted to 0.6 μg/μl for LC-MS.
Fig. 1.
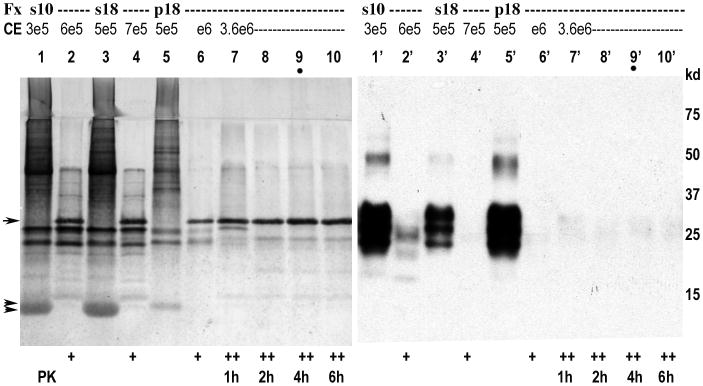
Sequential steps in the rapid isolation of p18 FU-CJD brain particles showing a blot with gold-stained proteins on left panel and corresponding PrP/PrP-res in the right panel. Cytosol from non-detergent treated homogenates was first separated from nuclei and membrane-myelin components and cleared of lysosomes at 10,000g to yield s10 supernatant with >90% of the starting brain infectivity. RNAse added to s10 is seen at double arrows, and subsequent generation of the p18 shows reduced contaminating RNAse. The p18, treated with high PK (++ lanes, residual PK at arrow) which reduced proteins to barely visible amounts despite the high CE load, and PrP was not visible after 2hr PK (<0.05% of starting PrP); this 0.05% signal at ∼29kd is typical of non-specific binding of PrP antibody to PK. The 4 hr sample analyzed had verified high infectivity as previously detailed (14). Standard lower PrP-res bands are seen in lane 2′ and molecular weight markers are indicated.
ii) LC-MS/MS Protein Identification
LC-MS/MS was performed in duplicate on a 5600 TripleTOF (AB Sciex, Framingham, MA) with a nanospray source connected to a nanoACQUITY UPLC (Waters, Milford, MA). The mobile phases for LC separation consisted of water (A) and acetonitrile (B) containing 0.1 % of FA, respectively. Three micrograms of peptides were injected onto a Waters Symmetry C18, 5 μm particle size, 180 μm ID × 20 mm nanoACQUITY UPLC trap column which was connected to a Waters BEH 30 C18, 1.7 μm particle size, 75 μm ID × 150 mm capillary column operated at 45°C. Peptides were trapped for 3 min at 1% B with a flow rate of 5 μl/min. Gradient elution was carried out using a flow rate of 0.5 μl/min with a two-step gradient from 5-40% B in 160 min and 40-85% B in 3.3 min, respectively. The mass spectrometer was operated in IDA mode with a single 250msec high resolution TOF MS survey scan (m/z = 400-1250) followed by up to 20 MS/MS scans (m/z = 100-1500) at > 15,000 resolution and 50 msec accumulation time. Precursor ions exceeding 125 counts were fragmented with a collision energy of 37 eV ± 15 eV using N2 as the collision gas. Fragmented peptides were set on an exclusion list for 10 sec.
iii) Database searching
LC-MS/MS was performed in duplicate on all samples and all MS/MS spectra were searched in-house using the Mascot algorithm (version 2.4.0) (17) after using the AB Sciex MS Data Converter Software (1.0) program to generate Mascot compatible files. Each of the Mascot Compatible Files (mgf) was searched using the SwissProt reviewed database with either a Mouse (16,695 sequences) or Human (20,199 sequences) taxonomy filter for the Mouse and Human samples respectively, and resulting Mascot files inserted into Yale Protein Expression Database (18-20). Each protein had two or more peptides, and each peptide was distinct within a combined either Mouse or Human Taxonomy SwissProt database blast search (21). Search criteria were: Enzyme: Trypsin, allowing 1 missed cleavage. The precursor ion mass tolerance was set to 20 ppm for the precursor and 0.2 Da for fragment ions, respectively. Fixed modifications were Carbamidomethyl at cysteine and the variable modification was oxidation of Met. The overall false discovery rate estimated by MASCOT was < 1% and we selected an arbitrary cut-off of 0.05 FDR to filter protein IDs and utilized a 0.01 FDR rate (22). Mascot results files (*.dat) were transformed into an XML file employing the Mascot's script export_dat_2.pl. The resulting XML files were processed using JAXB (http://jaxb.java.net/) and the Java StAX API (http://stax.codehaus.org/).
iv) YPED Mouse and Human Proteome Spectral Library
To identify proteins from the 5600 TripleTOF Discovery Protein Identification Data, the data were filtered to only include peptides with MASCOT scores greater than or equal to the identify score. The MS/MS spectra with the highest MASCOT score were chosen and all y- and b-ions were matched to an in-silico fragmentation and then sorted by peak height intensity. From there, the list was filtered so that each protein had two or more peptides and each peptide was distinct within a combined either a Mouse or Human Taxonomy SwissProt database blast search (21). The remaining peptides were sorted based on the number of occurrences in the YPED database with the b- or y-ions for all peptides exported for downstream SWATH analysis (26). These SWATH peak extraction transitions along with their retention times were exported as a tab-delimited file (tsv).
v) Data-Independent SWATH Proteome Analysis
Supplemental Figure 1 outlines an LC-MRM Proteome Workflow that begins with an AB SCIEX 5600 TripleTOF that first “sequences” as many peptides as possible from a tryptic digest of the relevant sample. Each sample was processed through the Yale Protein Expression Database (YPED) pipeline and the “learned” peptide sequences were transformed into spectral libraries that were used for data-independent SWATH analysis. Quantitation was accomplished using two (or more) of the most intense fragment ions from each of 2-3 tryptic peptides (e.g., 15 data points with 3 peptides × 5 transitions/peptide). The resulting data were integrated using Peakview (version 2.1), filtered for FDR <0.05 based on mProphet model (23) and exported for data analysis where a comprehensive suite of custom bioinformatics tools as previously described (18). Briefly, Matlab and MSstats R scripts (24) were used to calculate fold-change differences between sample groups after MLR normalization and produce Volcano plots (Supplemental Fig. 2). The raw data and fold-change calculations were then uploaded to YPED for web-accessible data dissemination and archiving in the YPED data repository (19, 20).
vi) Statistics Analysis and Bioinformatics
Differences between groups using non-parametric measures were assessed by a 2-tailed Mann-Whitney test otherwise differences between experimental groups were analyzed using a 2-tailed Student's t-test. Transplant survival between groups was calculated using the log-rank method. Fisher's exact test was employed to compare differences between samples from human heart transplant recipients. No outlying data points were removed. P < 0.05 was considered significant. All error bars represent and +/- in tables represent standard error of the mean. The use of data independent acquisition enabled simultaneous quantitation and confirmation of proteins in complex tissue samples.
Results
TSE infectious particles are maintained inside the cell. Even in cultured cells, only a minute fraction (≤ 1 in 10,000 particles) of cell infectivity can be retrieved from debris-cleared culture supernatants or animal blood, and cell-to-cell contacts are required for physiological infection (12, 25). Hence TSE particles will always be contaminated with a variety of intracellular host components. Our strategy was to first separate highly infectious FU-CJD brain particles from low-infectivity nuclei, myelin-membranes, lysosomes and soluble proteins. We then strip away additional host proteins that are not tightly bound or integral to the infectious particle with proteinase K (PK). As previously quantitated, TSE infectious brain particles are quantitatively separated from the vast majority of starting cellular lipid-membrane components DNA, RNA and proteins. These “p18” particles still contain diverse cell proteins, including 12% of starting host PrP (14) that may or may not be ligands for agent particles. We discovered that FU-CJD brain particles could be further purified, without loss of infectious titer, by digesting PrP and many other host proteins with PK. Proteins with validated viral sequence motifs were then revealed, showing the high sensitivity and depth of analysis (14), and one or more of these may be part of the infectious particle, and/or protective of an agent genome. As in the previous study of viral motifs, when we compared the biological replicate runs, greater then 95% of the proteins showed minimal fold change (less than 1.5).
The initial separation of infectious cytosol from a large lipid-membrane layer and nuclei was accomplished without detergents at 5,500g. Fig. 1 shows a blot of the subsequent sequential FU-CJD particle purification steps from aliquots with high infectivity (14) used for proteomic analysis below. For reference, the p18 before PK contained 12% of the starting protein p18 (lane 5) with an >80% reduction in protein from supernatants s10 and s18 (left panel lanes 1 & 3, gold stained). Several different protein bands become visible in the p18, along with reduced RNAse (double arrows). A series of PK digestions on 2e8 CE (1 to 6 hr) of this p18 were used to reduce and PrP and other cellular proteins, but not infectious titer. In the highly infectious 4hr, lane 9 sample analyzed here, there are minimal visible cellular proteins present despite the >10 fold cell equivalent (CE) load in this lane. Only the added PK (arrow) is prominent. No PrP is visible (right panel) despite the high CE load, as compared to the undigested p18. This lack of PrP was further confirmed below. For reference, the p18 from FU-CJD brain contains 1 LD50/cell by animal assay, with ∼107 CE analyzed by LC-MS, i.e., a minimum of 5 million infectious particles in each duplicate sample, clearly enough to detect exclusive viral-type sequences, as actually identified (14). Moreover, key host sequences that tightly bind infectious particles would typically be present at >1 per infectious particle, and most viruses require >10 particles per LD50 in animals.
Indeed, this PK strategy stripped away many pathologic response proteins from p18 particles as shown below, allowing for the detection of specific host proteins and cell complexes directly involved in agent processing and replication. The majority of host proteins so discovered engage known viral pathways of infection. A second protein set brings together neurodegenerative processes and protein misfolding with conformational changes in viral proteins. In efforts to find commonality among the differentially regulated proteins we gene ontology enrichment analysis for biological process, molecular function, and pathway analysis. The results of this analysis were fairly routine, thus we pursued an alternate approach to possibly uncover some new functionality for the differentially particle associated host proteins. Similarly, Ingenuity Pathway (Supplement Figs. 3,4) illuminated some proteins of interest (as HSPs) but did not clarify important viral links or a separation from less specific neurodegenerative processing, probably because there are few proteomic studies of infectious brain subcellular fractions. We posited that infectious particle isolates should reveal critical host proteins for agent invasion and replication, and are aware of no proteomic reports on subcellular components titered for infectivity. To find even more relevant functional links of Uniprot identified sequences, we systematically queried PubMed using the term neurodegeneration or virus with all the identified protein names. The biologic significance of major proteins is referenced in the Results, and contaminating keratin and most mitochondrial oxidative enzymes are not discussed but included in Tables in order of intensity as found.
Relative representation of proteins in p18 preparations
Prior to PK stripping, relatively few proteins are elevated in less purified p18 particles as compared to uninfected controls, a consequence of relatively impure particles. These mainly relate to neurodegeneration and amyloid formation. Table I shows FU-CJD infectious particles before PK. Only 11 proteins are increased by ≥2.5 fold over the parallel control. Nevertheless, the highest three on the list connect pathological threads common to CJD, Alzheimer's Disease (AD) and other neurodegenerative conditions. Astrocyte increase and hypertrophy is a classical response to many types of neuronal insults and death, here exemplified by astrocyte-specific GFAP. GFAP is major reactive change in CJD and other TSEs (26) and its mRNA rises before PrP converts to PrP-res in both hamster sCJD and rat sCJD infections; at clinical stages it is >15 fold greater than normal (15, 27). The 6.7 fold increase in GFAP protein in p18 particles here is consistent with this GFAP mRNA rise, although only a portion of astrocytic GFAP rich processes co-purifies with p18 infectious particles.
Table 1. Significant Protein Fold Change: Mouse Brain FU-CJD PK- versus Normal PK-.
| N | Protein | Uniprot Accession | Protein name | Gene Name | log2fc | Fold-change | SE | adj_pvalue |
|---|---|---|---|---|---|---|---|---|
| 1 | P03995 | GFAP_MOUSE | Glial fibrillary acidic protein | Gfap | 2.738 | 6.671 | 0.088 | 0.000 |
| 2 | P98086 | C1QA_MOUSE | Complement C1q subcomponent subunit A | C1qa | 2.719 | 6.585 | 0.784 | 0.041 |
| 3 | P08226 | APOE_MOUSE | Apolipoprotein E | Apoe | 2.093 | 4.266 | 0.105 | 0.000 |
| 4 | P20152 | VIME_MOUSE | Vimentin | Vim | 2.017 | 4.048 | 0.135 | 0.000 |
| 5 | Q99L04 | DHRS1_MOUSE | Dehydrogenase/reductase SDR family member 1 | Dhrs1 | 1.864 | 3.641 | 0.250 | 0.002 |
| 6 | P04925 | PRIO_MOUSE | Major prion protein | Prnp | 1.720 | 3.294 | 0.131 | 0.000 |
| 7 | P48036 | ANXA5_MOUSE | Annexin A5 | Anxa5 | 1.560 | 2.949 | 0.200 | 0.000 |
| 8 | P04919 | B3AT_MOUSE | Band 3 anion transport protein | Slc4a1 | 1.542 | 2.911 | 0.184 | 0.000 |
| 9 | O54734 | OST48_MOUSE | Dolichyl-diphosphooligosaccharide--protein glyco | Ddost | 1.506 | 2.840 | 0.171 | 0.000 |
| 10 | P07356 | ANXA2_MOUSE | Annexin A2 | Anxa2 | 1.469 | 2.769 | 0.174 | 0.000 |
| 11 | Q922Q8 | LRC59_MOUSE | Leucine-rich repeat-containing protein 59 | Lrrc59 | 1.314 | 2.487 | 0.092 | 0.000 |
| 12 | Q9DBG6 | RPN2_MOUSE | Dolichyl-diphosphooligosaccharide--protein glyco | Rpn2 | 1.274 | 2.418 | 0.082 | 0.000 |
| 13 | P02535 | K1C10_MOUSE | Keratin, type I cytoskeletal 10 | Krt10 | 1.243 | 2.367 | 0.322 | 0.009 |
| 14 | P62301 | RS13_MOUSE | 40S ribosomal protein S13 | Rps13 | 1.228 | 2.343 | 0.248 | 0.000 |
| 15 | P24270 | CATA_MOUSE | Catalase | Cat | 1.144 | 2.210 | 0.264 | 0.029 |
| 16 | Q91YQ5 | RPN1_MOUSE | Dolichyl-diphosphooligosaccharide--protein glyco | Rpn1 | 1.121 | 2.175 | 0.096 | 0.000 |
| 17 | Q9CR57 | RL14_MOUSE | 60S ribosomal protein L14 | Rpl14 | 1.086 | 2.123 | 0.084 | 0.000 |
| 18 | Q99JY0 | ECHB_MOUSE | Trifunctional enzyme subunit beta, mitochondrial | Hadhb | 1.039 | 2.055 | 0.054 | 0.000 |
| 19 | P09103 | PDIA1_MOUSE | Protein disulfide-isomerase | P4hb | 1.032 | 2.045 | 0.150 | 0.003 |
| 20 | P24369 | PPIB_MOUSE | Peptidyl-prolyl cis-trans isomerase B | Ppib | 1.026 | 2.036 | 0.098 | 0.000 |
|
| ||||||||
| 21 | P31324 | KAP3_MOUSE | cAMP-dependent protein kinase type II-beta regul | Prkar2b | -1.006 | -2.008 | 0.277 | 0.006 |
| 22 | Q61644 | PACN1_MOUSE | Protein kinase C and casein kinase substrate in neu | Pacsin1 | -1.009 | -2.013 | 0.340 | 0.020 |
| 23 | Q91XM9 | DLG2_MOUSE | Disks large homolog 2 | Dlg2 | -1.254 | -2.385 | 0.173 | 0.003 |
| 24 | P62204 | CALM_MOUSE | Calmodulin | Calm1 | -1.303 | -2.467 | 0.313 | 0.000 |
| 25 | P35802 | GPM6A_MOUSE | Neuronal membrane glycoprotein M6-a | Gpm6a | -1.683 | -3.210 | 0.182 | 0.000 |
Complement C1q and APOE identified are also known components of the AD amyloid cascade. APOE mRNA and C1q are only slightly elevated at clinical stages in FU-CJD infected mouse brain, and thus their higher 4.3 and 6.6 fold protein increase here indicates their accumulation in concert with the pathologic PrP-res formation. C1q protein levels in TSE infected whole brain can be ∼10 fold normal (28) in contrast to weak mRNA elevations. A substantial elevation of C1q is also found in AD amyloid neurodegeneration and as an innate immune response to invading microorganisms.
PrP is prominent in less purified p18 preparations and increased 3.3 fold in FU-CJD infectious particles. This corresponds to ∼12% of brain PrP (including PrP-res) that copurifies with FU-CJD infectious particles (14). In contrast to C1q, other innate immune response proteins such as the microglial MIF1α and 1β which are provoked very early in FU-CJD infection and again later in disease (29), were not detectable in p18 particles. This demonstrates that infectious brain particles were well separated from these myeloid components. Clusterin J and other large plaque associated proteins were also not elevated, in accord with the overall reduction of PrP in the untreated p18 particles. However, an ∼3 fold increase in the anion transporter B3AT, an AD and senescence marker (30) was observed. The 2.5 fold increase in the cytosolic leucine rich repeat LRC59 that binds polyA RNA is also involved in pathogen detection (31). In sum, most of the proteins in less purified FU-CJD p18 particles are host pathologic responses and disease markers. The only marker that was decreased (by >3 fold) is GPM6a, a neuronal differentiation membrane glycoprotein, indicative of neurodegeneration and/or loss of function.
In the undigested p18 particles there were also a few FU-CJD exclusive proteins not detectable in control brains. One of these, PPIB, had an intensity of 8,939 and PCYOX, a lysosome and exosome protein also had a substantial intensity of 5,937. While several FU-CJD elevated proteins and lysosome-exosome elements are known to be involved in PrP to PrP-res processing, after PK stripping a very different set of host proteins was revealed. Notably, PCYOX and all other exclusive proteins were no longer detectable, whereas PPIB was represented at >10 times greater intensity (95,445). Moreover, in sharp contrast to Table 1, Table 2 shows the major pathologic proteins GFAP, Complement C1q, Apo E, vimentin and PrP were no longer detectable by independent proteomic analyses. PrP, known to reside in lysosomes, disappeared with lysosomal PCYOX. Supplemental Table I shows all 74 differentially regulated PK+ proteins in FU-CJD. In sum, the infectious particle strategy developed here effectively excluded many components not tightly linked to a TSE particle's ability to productively infect a cell. Not all elevated proteins in Table 1 were obliterated by PK, e.g., Annexin 5 was retained but not substantially increased, unlike PPIB.
Table 2. Significant Protein Fold Change: Mouse Brain FU-CJD PK+ compared to Normal PK+.
| N | Protein | Uniprot ID | Protein Name | Gene Name | log2fc | Fold-change | SE | adj_pvalue |
|---|---|---|---|---|---|---|---|---|
| 1 | P63017 | HSP7C_MOUSE | Heat shock cognate 71 kDa protein | Hspa8 | 7.174 | 144.411 | 0.327 | 0.000 |
| 2 | Q64332 | SYN2_MOUSE | Synapsin-2 | Syn2 | 5.058 | 33.318 | 0.178 | 0.000 |
| 3 | P97429 | ANXA4_MOUSE | Annexin A4 | Anxa4 | 2.738 | 6.670 | 0.220 | 0.000 |
| 4 | Q9JIS5 | SV2A_MOUSE | Synaptic vesicle glycoprotein 2A | Sv2a | 2.552 | 5.866 | 0.448 | 0.000 |
| 5 | P08551 | NFL_MOUSE | Neurofilament light polypeptide | Nefl | 2.419 | 5.350 | 0.421 | 0.000 |
| 6 | Q99104 | MYO5A_MOUSE | Unconventional myosin-Va | Myo5a | 2.221 | 4.661 | 0.248 | 0.000 |
| 7 | O08532 | CA2D 1_MOUSE | Voltage-dependent calcium channel subunit alpha- | Cacna2d1 | 1.902 | 3.737 | 0.451 | 0.000 |
| 8 | P11835 | ITB2_MOUSE | Integrin beta-2 | Itgb2 | 1.873 | 3.663 | 0.170 | 0.000 |
| 9 | Q62261 | SPTB2_MOUSE | Spectrin beta chain, non-erythrocytic 1 | Sptbn1 | 1.829 | 3.554 | 0.240 | 0.000 |
| 10 | P20060 | HEXB_MOUSE | Beta-hexosaminidase subunit beta | Hexb | 1.544 | 2.915 | 0.018 | 0.000 |
| 11 | P62631 | EF1A2_MOUSE | Elongation factor 1-alpha 2 | Eef1a2 | 1.438 | 2.710 | 0.108 | 0.000 |
| 12 | P39053 | DYN1_MOUSE | Dynamin-1 | Dnm1 | 1.325 | 2.505 | 0.533 | 0.024 |
| 13 | P47740 | AL3 A2_MOUSE | Fatty aldehyde dehydrogenase | Aldh3a2 | 1.268 | 2.408 | 0.065 | 0.000 |
| 14 | P09055 | ITB1_MOUSE | Integrin beta-1 | Itgb1 | 1.128 | 2.185 | 0.131 | 0.000 |
| 15 | P55088 | AQP4_MOUSE | Aquaporin-4 | Aqp4 | 1.092 | 2.132 | 0.079 | 0.000 |
| 16 | P48036 | ANXA5_MOUSE | Annexin A5 | Anxa5 | 1.049 | 2.070 | 0.165 | 0.000 |
|
| ||||||||
| 56 | O55125 | NIPS1_MOUSE | Protein NipSnap homolog 1 | Nipsnap1 | -1.838 | -3.575 | 0.425 | 0.000 |
| 57 | Q8BYI9 | TENR_MOUSE | Tenascin-R | Tnr | -1.857 | -3.622 | 0.243 | 0.000 |
| 58 | P50396 | GDIA_MOUSE | Rab GDP dissociation inhibitor alpha | Gdi1 | -1.974 | -3.929 | 0.186 | 0.000 |
| 59 | Q8BG39 | SV2 B_MOUSE | Synaptic vesicle glycoprotein 2B | Sv2b | -1.980 | -3.945 | 0.584 | 0.007 |
| 60 | Q8VDQ8 | SIR2_MOUSE | NAD-dependent protein deacetylase sirtuin-2 | Sirt2 | -1.980 | -3.945 | 0.402 | 0.000 |
| 61 | Q9D051 | ODPB_MOUSE | Pyruvate dehydrogenase E1 component subunit be | Pdhb | -2.282 | -4.864 | 0.228 | 0.000 |
| 62 | Q9CPU4 | MGST3_MOUSE | Microsomal glutathione S-transferase 3 | Mgst3 | -2.323 | -5.004 | 0.305 | 0.000 |
| 63 | Q9D2G2 | ODO2_MOUSE | Dihydrolipoyllysine-residue succinyltransferase co | Dlst | -2.355 | -5.116 | 0.144 | 0.000 |
| 64 | P08113 | ENPL_MOUSE | Endoplasmin | Hsp90b1 | -2.376 | -5.190 | 0.434 | 0.000 |
| 65 | Q9CQC7 | NDUB4_MOUSE | NADH dehydrogenase [ubiquinone] 1 beta subcom | Ndufb4 | -2.559 | -5.893 | 0.166 | 0.000 |
| 66 | P34884 | MIF_MOUSE | Macrophage migration inhibitory factor | Mif | -2.623 | -6.162 | 0.060 | 0.000 |
| 67 | Q9DB20 | ATPO_MOUSE | AT P synthase subunit O, mitochondrial | Atp5o | -2.681 | -6.411 | 0.558 | 0.000 |
| 68 | P12787 | COX5 A_MOUSE | Cytochrome c oxidase subunit 5A, mitochondrial | Cox5a | -2.741 | -6.684 | 0.305 | 0.000 |
| 69 | O55143 | AT2 A2_MOUSE | Sarcoplasmic/endoplasmic reticulum calcium AT P | Atp2a2 | -2.821 | -7.066 | 0.240 | 0.000 |
| 70 | P70704 | AT8A 1_MOUSE | Phospholipid-transporting ATPase IA | Atp8a1 | -3.291 | -9.790 | 0.296 | 0.000 |
| 71 | Q9CQQ7 | AT5F1_MOUSE | AT P synthase F(0) complex subunit B1, mitochon | Atp5f1 | -3.578 | -11.942 | 0.232 | 0.000 |
| 72 | Q8K2B3 | SDH A_MOUSE | Succinate dehydrogenase [ubiquinone] flavoprotei | Sdha | -4.627 | -24.706 | 0.252 | 0.000 |
| 73 | P40124 | CAP1_MOUSE | Adenylyl cyclase-associated protein 1 | Cap1 | -4.813 | -28.115 | 0.356 | 0.000 |
| 74 | Q99N28 | CAD M3_MOUSE | Cell adhesion molecule 3 | Cadm3 | -5.037 | -32.842 | 0.502 | 0.000 |
FU-CJD Particle proteins revealed after stripping by PK
PK+ FU-CJD particles revealed a huge 144-fold elevation of the Heat shock protein 71 (HSP7C, a cognate of HSC70) versus PK+ controls (Table 2). This HSP was detectable only after PK stripping. No one has reported such a major elevation in this or any HSP proteins in TSE infected brain, and its specific appearance indicates it interacts with infectious particles. HSC70 functions as a repressor of transcriptional activation, in the disassembly of clathrin-coated vesicles during transport of membrane components through the cell, and as a chaperone. More specifically, HSC70 directly interacts with many viruses, consistent with its link to TSE infectious particles here. HSC70 is induced by Hantavirus infections to directly suppress virus infection via the viral nucleocapsid (32). HSC70 similarly protects against a neurovirulent Measles virus that causes non-cytolytic neuronal infections (33), and this is especially relevant for TSEs where progressive exponential infection is not cytolytic. Indeed, CJD agents only induce vacuolization at terminal stages of disease in many species (26, 34). In Tobacco Mosaic Virus infections HSC70 associates with aggregated viral coat protein and virus induced membrane structures (35) and this observation also relates well to the late spongiform membrane changes in TSEs. On the other hand, HSC70 can enhance the replication of circoviruses via a specific interaction with the viral cap (36), and circular “Sphinx” DNAs with viral features have been identified in FU-CJD preparations (13). Regardless, HSC70 is part of a common viral response, and HSC70 also connects seemingly disparate amyloid and misfolded protein structures because it is involved in virally induced prion-like aggregates (37), much as TSE particles induce PrP amyloid. Interestingly, proteomic analysis of vesicular stomatitis virus revealed the presence of host HSC70 within highly purified VSV virions (38). Hence one cannot rule out a role for HSC70 as part of the infecting structure of FU-CJD particles, especially in connection with clathrin and early endosome uptake proteins identified below.
An additional 10 proteins were also differentially increased from 2.5 to 33-fold in PK+ FU-CJD particles. Uncovering synapse, vesicle membrane, and neuronal proteins such as SYN2, SV2A, MYO5A and NFL is not surprising because virus-like dense particles of 25nm have been identified by multiple investigators in synaptic regions of TSE, but not uninfected brains (5-7). Moreover, virtually all infectivity in sCJD infected Guinea Pig and hamster brains fractionate with ultrastructurally verified synaptosomes (39).
Annexin 4, elevated 6.7-fold, is involved in intracellular calcium-regulated pathways and related Annexin 2 acts as a receptor for entry and infection by human papilloma virus 16 (40). Integrin β-2 (at 3.7-fold) and other integrins can also interact with viruses and function as transmembrane receptors and bridges for cell-cell transit, including at synapses. Thus this integrin could help spread TSE particles from cell-to-cell (25), possibly via membranes. The APP membrane AD β-amyloid precursor colocalizes with beta-1 integrins at substrate contact sites (41), and both APP and normal membrane PrP are involved in axonal pathfinding and the developmental of dense tight junctions (42, 43). In sum, the presence of integrin further signifies common threads between physiologic cell contacts, viral transit, and neurodegeneration. Finally, DYN1 (#12, Table 2) is strongly involved in endocytic entry of a vast number of different viruses (44), and would be expected to tightly associate with a viral TSE agent.
PK+ particle purification uncovered a longer list of proteins that decreased in FU-CJD. There were 58 proteins underrepresented, and those with the greatest decrement are also shown in Table 2 (Supplement Table I lists all). The most underrepresented FU-CJD proteins are at the bottom, and CADM3 (#74) shows a 33.3-fold decrement, whereas NIPS1 (#56) shows only a 3.6-fold decrease. A 33-fold underrepresentation of CADM3 (synaptic adhesion molecule 3) might be a new molecular target for replacement therapy. Many of the other underrepresented proteins are mitochondrial enzymes, probably indicating an apoptotic response in late infection.
It is also remarkable that HSP90 (#64) is decreased 5.2-fold, in contrast to the 144-fold elevation of HSC70. This demonstrates that the massive HSC70 recruitment to particles is very specific. Several viruses are enhanced by HSP90, and a proteomic analysis showed that Rift Valley Fever virus tightly bound HSP90 to increase viral titers (45). Low HSP90 in TSE particles should repress infection, since chaperone HSP90 is required for viral maturation. HSP90 is also necessary for HIV-1 reactivation from latency (46), further indicating low HSP90 probably inhibits or delays TSE agent activation and maturation. Because HSP90 inhibitors can also ameliorate the burden of the AD abnormal proteins tau and β-amyloid (47), HSP inhibitors could potentially decrease the burden of PrP amyloid and also simultaneously delay exponential agent replication (34).
Proteins found exclusively in PK+ infectious particle but not uninfected controls
Remarkably, the PK treated FU-CJD particles unmasked an even longer list of proteins that were not detectable in the parallel PK+ control brains. This exclusive set contained 56 proteins from 131,858 to 81 in relative intensity. Table 4 lists the highest 29 with intensities >1,500, and Supplement Table 2 shows all 56.
Table 4. Significant Protein Fold Change: Human Brain sCJD compared to Normal (adjusted P-value < 0.05).
| N | Protein | Uniprot Accession | Protein Name | Gene Name | log2fc | Fold-change | SE | adj_pvalue |
|---|---|---|---|---|---|---|---|---|
| 1 | NEDD8 | NEDD 8_HUMAN | NEDD8 | NEDD8 | 4.444 | 21.772 | 0.323 | 0.033 |
| 2 | GON4L | GON4L_HUMAN | GON-4-like protein | GON4L | 3.147 | 8.859 | 0.538 | 0.018 |
| 3 | FUBP3 | FUBP3 _HUMAN | Far upstream element-binding protein 3 | FUBP3 | 2.840 | 7.159 | 0.527 | 0.036 |
| 4 | HPCL4 | HPCL4_HUMAN | Hippocalcin-like protein 4 | HPCAL4 | 2.761 | 6.778 | 0.473 | 0.030 |
| 5 | SYFA | SYFA_HUMAN | Phenylalanine--tRNA ligase alpha subunit | FARSA | 2.252 | 4.762 | 0.473 | 0.018 |
| 6 | SYT2 | SYT2_HUMAN | Synaptotagmin-2 | SYT2 | 2.005 | 4.013 | 0.372 | 0.001 |
| 7 | NDUB9 | NDUB9_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcom | NDUFB9 | 1.965 | 3.903 | 0.386 | 0.001 |
| 8 | UT1 | UT1_HUMAN | Urea transporter 1 | SLC14A1 | 1.913 | 3.765 | 0.545 | 0.030 |
| 9 | QCR7 | QCR7_HUMAN | Cytochrome b-c1 complex subunit 7 | UQCRB | 1.893 | 3.715 | 0.457 | 0.005 |
| 10 | GBRA1 | GBRA1_HUMAN | Gamma-aminobutyric acid receptor subunit alpha- | GABRA1 | 1.795 | 3.471 | 0.555 | 0.049 |
| 11 | NFH | NFH_HUMAN | Neurofilament heavy polypeptide | NEFH | 1.734 | 3.325 | 0.446 | 0.006 |
| 12 | SCN2A | SCN2 A_HUMAN | Sodium channel protein type 2 subunit alpha | SCN2A | 1.466 | 2.763 | 0.317 | 0.001 |
| 13 | MPC2 | MPC2_HUMAN | Mitochondrial pyruvate carrier 2 | MPC2 | 1.435 | 2.703 | 0.424 | 0.032 |
| 14 | SEPT4 | SEPT4_HUMAN | Septin-4 | SEPT4 | 1.399 | 2.637 | 0.409 | 0.044 |
| 15 | UK114 | UK 114_HUMAN | Ribonuclease UK114 | HRSP12 | 1.398 | 2.636 | 0.390 | 0.042 |
| 16 | AP1G1 | AP1G 1_HUMAN | AP-1 complex subunit gamma-1 | AP1G1 | 1.340 | 2.531 | 0.419 | 0.031 |
|
| ||||||||
| 62 | PVRL1 | PVRL1_HUMAN | Nectin-1 | PVRL1 | -1.940 | -3.837 | 0.523 | 0.019 |
| 63 | RPGF2 | RPGF2_HUMAN | Rap guanine nucleotide exchange factor 2 | RAPGEF2 | -2.128 | -4.370 | 0.279 | 0.031 |
| 64 | GSH1 | GSH1 _HUMAN | Glutamate--cysteine ligase catalytic subunit | GCLC | -2.280 | -4.858 | 0.538 | 0.034 |
| 65 | STX7 | STX7_HUMAN | Syntaxin-7 | STX7 | -2.417 | -5.342 | 0.657 | 0.033 |
| 66 | K2C1 | K2 C1 _HUMAN | Keratin, type II cytoskeletal 1 | KRT1 | -3.020 | -8.113 | 0.274 | 0.000 |
| 67 | K1C10 | K1C10_HUMAN | Keratin, type I cytoskeletal 10 | KRT10 | -3.024 | -8.136 | 0.261 | 0.000 |
| 68 | CACB4 | CACB4_HUMAN | Voltage-dependent L-type calcium channel subuni | CACNB4 | -3.149 | -8.873 | 0.549 | 0.012 |
| 69 | OPRK | OPRK_HUMAN | Kappa-type opioid receptor | OPRK1 | -3.568 | -11.859 | 0.490 | 0.034 |
| 70 | KAP0 | KAP0_HUMAN | cAMP-dependent protein kinase type I-alpha regul | PRKAR1A | -4.706 | -26.095 | 0.351 | 0.009 |
| 71 | TM205 | TM205_HUMAN | Transmembrane protein 205 | TMEM205 | -6.243 | -75.728 | 0.293 | 0.018 |
| 72 | ASCC3 | ASCC3_HUMAN | Activating signal cointegrator 1 complex subunit 3 | ASCC3 | -9.206 | -590.547 | 0.657 | 0.002 |
| 73 | CCD87 | CCD8 7_HUMAN | Coiled-coil domain-containing protein 87 | CCDC87 | -10.425 | -1375.252 | 0.351 | 0.000 |
The most intense exclusive protein was copine-3, a protein thought to be involved in membrane trafficking. It can bind polyA RNA and mediate calcium-dependent targeting of proteins to various intracellular locations including the plasma membrane (48) but little is known of its function or ligand specificity in brain or in viral infections.
The second most intense FU-CJD exclusive protein was PPIB (also known as cyclophilin B), a protein that increased in intensity >10 fold with unmasking (unlike all other PK- exclusive proteins). PPIB is a chaperone that accelerates the folding of proteins in the endoplasmic reticulum (ER) and in human papilloma virus 16 infections, the virion undergoes important conformational changes via cyclophilin B. PPIB is also linked to integrin and annexin binding (40), with the dissociation of virus capsid proteins following acidification of endocytic vesicles. PPIB knockdowns impair the propagation of both Japanese B encephalitis and Hepatitis C viruses (49, 50) and Cyclosporin A, a PPIB inhibitor, also decreases Japanese B encephalitis virus. Moreover, Cyclophilin B is critical for the influenza virus life cycle, as well as that for a variety of other viruses (49, 50). Given the tight association of Cyclophilin B with infectious FU-CJD particles, Cyclophilin B inhibitors should be tested for TSE agent inhibition in neuronal cell cultures. Interestingly, Cyclosporin A induces “prion-like” PrP species in uninfected cells (51), and PrP-res accumulation in neuronal cultures dramatically decreases FU-CJD titers by 4 logs. Because PrP-res amyloid can help clear infectious particles (52), Cyclosporin A should increase TSE agent clearance.
Other intense exclusive proteins that can form macromolecular complexes in PK+ FU-CJD particles are known to participate in endocytic, synaptic vesicle and vacuolar sorting processes in brain. Whereas many viruses adhere to early endosomes where acidic fusion leads to viral uncoating in many cell types (44), the synaptic association of CJD particles is agent-specific. The Ca++ channel proteins that were exclusive (CACB4, Table 3) or elevated >2.5-fold in FU-CJD (CA2D1, Table 2) implicate a targeted neuron-specific route of accumulation with trans-synaptic infectious spread. Exclusive VA0D1 (Table 3,#3), part of the integral membrane vacuolar ATPase complex, acidifies a variety of intracellular compartments in eukaryotic cells and is yet another PK+ unmasked protein that regulates cellular receptors and their trafficking via endocytotic and exocytotic pathways, including at the synapse. VA0D1 is also involved in the development of neurodegenerative disorders (53). Thus VA0D1 further links infectious particles to specific neuronal features.
Table 3. Mouse FU-CJD brain proteins exclusive to infectious PK+ particles.
| N | ProteinName | Gene | Name | Intensity |
|---|---|---|---|---|
| 1 | Q8BT60 | CPNE3_MOUSE | copine III | 131,857.6 |
| 2 | P24369 | PPIB_MOUSE | peptidylprolyl isomerase B | 95,445.3 |
| 3 | P51863 | VA0D1_MOUSE | ATPase, H+ transporting, lysosomal V0 subunit D1 | 81,670.2 |
| 4 | Q61548 | AP180_MOUSE | synaptosomal-associated protein 91 | 73,774.6 |
| 5 | P05208 | CEL2A_MOUSE | elastase 2A | 52,675.1 |
| 6 | Q9EP69 | SAC1_MOUSE | SAC1 (suppressor of actin mutations 1, homolog)-like (S. cerevisiae) | 51,093.4 |
| 7 | Q61879 | MYH10_MOUSE | myosin, heavy polypeptide 10, non-muscle | 45,252.7 |
| 8 | Q922Q1 | MOSC2_MOUSE | MOCO sulphurase C-terminal domain containing 2 | 40,681.6 |
| 9 | Q8QZT1 | THIL_MOUSE | acetyl-Coenzyme A acetyltransferase 1 | 34,822.6 |
| 10 | Q8R0S4 | CACB4_MOUSE | calcium channel, voltage-dependent, beta 4 subunit | 25,702.5 |
| 11 | Q9QYR6 | MAP1A_MOUSE | microtubule-associated protein 1 A | 24,937.9 |
| 12 | P12970 | RL7A_MOUSE | 60S ribosomal protein L7a (Surfeit locus protein 3) | 21,714.9 |
| 13 | P28738 | KIF5C_MOUSE | kinesin family member 5C | 21,473.4 |
| 14 | Q5SQX6 | CYFP2_MOUSE | cytoplasmic FMR1 interacting protein 2 | 16,845.7 |
| 15 | Q9CZW5 | TOM70_MOUSE | translocase of outer mitochondrial membrane 70 homolog A (yeast) | 16,111.4 |
| 16 | P63038 | CH60_MOUSE | predicted gene 12141; heat shock protein 1 (chaperonin) | 16,107.1 |
| 17 | P02535 | K1C10_MOUSE | keratin 10 | 12,291.6 |
| 18 | P47963 | RL13_MOUSE | 60S ribosomal protein L13 | 9,587.9 |
| 19 | Q9CXZ1 | NDUS4_MOUSE | NADH dehydrogenase (ubiquinone) Fe-S protein 4 | 8,651.2 |
| 20 | Q8CGK3 | LONM_MOUSE | lon peptidase 1, mitochondrial | 6,686.4 |
| 21 | Q9CZM2 | RL15_MOUSE | 60S ribosomal protein L15 | 6,669.9 |
| 22 | Q6ZQ38 | CAND1_MOUSE | cullin associated and neddylation disassociated 1 | 5,450.2 |
| 23 | Q8BMK4 | CKAP4_MOUSE | cytoskeleton-associated protein 4 | 4,700.5 |
| 24 | Q9JME5 | AP3B2_MOUSE | adaptor-related protein complex 3, beta 2 subunit | 3,688.1 |
| 25 | P62806 | H4_MOUSE | histone cluster 1, H4k; | 3,152.0 |
| 26 | P48722 | HS74L_MOUSE | heat shock protein 4 like | 2,872.9 |
| 27 | Q9JM63 | IRK10_MOUSE | potassium inwardly-rectifying channel, subfamily J, member 10 | 2,231.2 |
| 28 | P80314 | TCPB_MOUSE | chaperonin containing Tcp1, subunit 2 (beta) | 2,069.2 |
| 29 | Q9Z1S5 | SEPT3_MOUSE | septin 3 | 1,848.2 |
AP180, a clathrin assembly protein that is high on the exclusive FU-CJD list (Table 3), is also strongly linked to brain pre-synaptic junctions and viruses. Clathrin facilitates the morphogenesis of retrovirus particles, is exceptionally abundant in highly purified HIV-1 particles, and is recruited with high specificity by the virus (54). Many different viruses engage clathrin complexes. Human rhinovirus is internalized by clathrin-mediated endocytosis (55), early endocytic acidic fusion uncoats many enveloped viruses. Even non-enveloped viruses such as picornaviruses also utilize early endosomes for infection. In sum, infectious TSE particles strongly associate with common endocytic viral processing functions and macromolecular complexes, and additionally display unique links to synaptic proteins and Ca++ channels. These findings are in accord with the ultrastructural synaptic location of ∼25nm dense virus-like TSE particles and accumulation of high infectivity in synaptosomes.
Three other exclusive proteins of high intensity appear with FU-CJD particles. First, MYH10 (#7, Table 3) is involved in neurogenesis and apoptosis in rat brain (56) and may contribute to the neurodegenerative cascade. Second, CH60 (Hsp60, chaperonin, #16), could be a non-specific mitochondrial contaminant. However, mitochondria-associated ER membranes have important cellular functions, including Ca++ signaling, and outer mitochondrial membrane-ER contacts are acted on by viruses. Cytomegalovirus restructures this contact region and requires HSP60, a mitochondrial import protein that prevents misfolding and promotes proper refolding under stress conditions (57). PrP-res also emerges as a stress protein late in disease to arrest replication during the agent plateau phase (34) or eliminate infectious TSE particles (52). The third protein, CAND1 (#21) restricts host defenses through viral rewiring of cullin 3 ubiquitin ligases, thereby increasing viral replication (58), and many viruses hijack members of the Cullin-RING E3 Ligase family which is linked to NEDD; CAND1 locks cullin 3 ligases in a non-substrate binding conformation, i.e., in a viral permissive state. Remarkably, NEDD also appears as part of the human sCJD particle complexes.
Differential representation of p18 proteins in human sCJD brain particles
Because p18 and equivalently gradient purified particles from mouse brain and cell cultures all gave high yields of infectivity with both CJD and scrapie agent strains (59), we assumed sCJD infected human brain would also yield infectious particles with comparable associations. Only non-PK particles were analyzed because quantitative infectious assay for sCJD human samples by same species transmission, even in culture, is not yet possible. Table 5 shows p18 sCJD particle associated proteins differences, and Supplement III shows all 73. As in the FU-CJD samples, one would expect more reactive and neurodegenerative molecules in non-PK treated particles. Nevertheless, some sCJD associated host components such as WASL, were clearly linked both to stress and viral pathways. Several proteins identified here have not heretofore been linked to TSEs.
Table 5. Human brain particle proteins exclusive to sCJD.
| N | ProteinName | Gene | Name | Intensity |
|---|---|---|---|---|
| 1 | WASL | WASL_HUMAN | Wiskott-Aldrich syndrome-like | 169,140.4 |
| 2 | DCTN3 | DCTN3_HUMAN | dynactin 3 (p22) | 157,980.3 |
| 3 | USO1 | USO1_HUMAN | USO1 homolog, vesicle docking protein (yeast) | 126,390.3 |
| 4 | NDUS7 | NDUS7_HUMAN | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH- Q | 88,332.1 |
| 5 | FKBP4 | FKBP4_HUMAN | FK506 binding protein 4, 59kDa | 78,704.9 |
| 6 | COX7R | COX7R_HUMAN | cytochrome c oxidase subunit VIIa polypeptide 2 like | 57,524.1 |
| 7 | HD | HD_HUMAN | huntingtin | 49,592.2 |
| 8 | PPCE | PPCE_HUMAN | prolyl endopeptidase | 40,681.6 |
| 9 | NDUB7 | NDUB7_HUMAN | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa | 39,724.8 |
| 10 | CHM2A | CHM2A_HUMAN | chromatin modifying protein 2A | 39,370.7 |
| 11 | APOO | APOO_HUMAN | apolipoprotein O | 38,975.3 |
| 12 | TTC7B | TTC7B_HUMAN | tetratricopeptide repeat domain 7B | 37,903.9 |
| 13 | VP13D | VP13D_HUMAN | vacuolar protein sorting 13 homolog D (S. cerevisiae) | 37,399.7 |
| 14 | KI21A | KI21A_HUMAN | kinesin family member 21A | 35,181.9 |
| 15 | KGUA | KGUA_HUMAN | guanylate kinase 1 | 31,610.5 |
| 16 | CNN3 | CNN3_HUMAN | calponin 3, acidic | 28,600.5 |
| 17 | NDUS4 | NDUS4_HUMAN | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coe | 27,990.7 |
| 18 | CLAP2 | CLAP2_HUMAN | cytoplasmic linker associated protein 2 | 26,087.2 |
| 19 | ATD3A | ATD3A_HUMAN | ATPase family, AAA domain containing 3A | 24,858.7 |
| 20 | PI42C | PI42C_HUMAN | phosphatidylinositol-5-phosphate 4-kinase, type II, gamma | 24,676.1 |
| 21 | PI42A | PI42A_HUMAN | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | 24,055.0 |
| 22 | PCBP2 | PCBP2_HUMAN | poly(rC) binding protein 2 | 23,668.3 |
| 23 | RS30 | RS30_HUMAN | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously e | 23,633.2 |
| 24 | RL7 | RL7_HUMAN | ribosomal protein L7 pseudogene 26; ribosomal protein L7 pseudogene | 21,737.1 |
| 25 | EF1B | EF1B_HUMAN | eukaryotic translation elongation factor 1 beta 2; eukaryotic translation e | 21,735.4 |
| 26 | GBLP | GBLP_HUMAN | guanine nucleotide binding protein (G protein), beta polypeptide 2-like | 21,492.4 |
| 27 | ROCK2 | ROCK2_HUMAN | Rho-associated, coiled-coil containing protein kinase 2 | 20,987.8 |
| 28 | RAB18 | RAB18_HUMAN | RAB18, member RAS oncogene family | 20,773.8 |
| 29 | S6A11 | S6A11_HUMAN | solute carrier family 6 (neurotransmitter transporter, GABA), member 1 | 20,112.7 |
| 30 | GON4L | GON4L_HUMAN | YY1 associated protein 1; gon-4-like (C. elegans) | 19,555.0 |
| 31 | ITAV | ITAV_HUMAN | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51 | 19,400.2 |
| 32 | 4-Sep | SEPT4_HUMAN | septin 4 | 18,835.6 |
| 33 | TPRGL | TPRGL_HUMAN | tumor protein p63 regulated 1-like | 18,308.7 |
| 34 | LPPRC | LPPRC_HUMAN | leucine-rich PPR-motif containing | 17,899.0 |
| 35 | CPLX1 | CPLX1_HUMAN | complexin 1 | 17,831.0 |
| 36 | TMM65 | TMM65_HUMAN | transmembrane protein 65 | 17,611.7 |
| 37 | SYT12 | SYT12_HUMAN | synaptotagmin XII | 17,491.9 |
| 38 | LIRB2 | LIRB2_HUMAN | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITI | 17,041.4 |
| 39 | APOD | APOD_HUMAN | apolipoprotein D | 16,204.0 |
| 40 | DNJB1 | DNJB1_HUMAN | DnaJ (Hsp40) homolog, subfamily B, member 1 | 16,102.7 |
| 41 | SH3G1 | SH3G1_HUMAN | SH3-domain GRB2-like 1 | 15,567.6 |
| 42 | ECHM | ECHM_HUMAN | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 15,557.8 |
| 43 | DCTN5 | DCTN5_HUMAN | dynactin 5 (p25) | 14,162.7 |
| 44 | RPN1 | RPN1_HUMAN | ribophorin I | 13,949.7 |
| 45 | PSMD1 | PSMD1_HUMAN | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 13,793.4 |
| 46 | RB27B | RB27B_HUMAN | RAB27B, member RAS oncogene family | 13,611.1 |
| 47 | COF2 | COF2_HUMAN | cofilin 2 (muscle) | 13,449.9 |
| 48 | RBGPR | RBGPR_HUMAN | RAB3 GTPase activating protein subunit 2 (non-catalytic) | 13,246.6 |
| 49 | LRC47 | LRC47_HUMAN | leucine rich repeat containing 47 | 13,222.8 |
NEDD 8 showed a substantial 21.8-fold overrepresentation compared to uninfected controls. This is in complete accord with the cullin-ring ligase links above for FU-CJD in mouse brain particles. HIV and SIV defeat antiviral proteins by usurping Cullin-RING E3 ubiquitin ligases, and neddylation plays an important role during HIV infection by overcoming cellular anti-viral defenses (60). NEDD8 in sCJD should have a similar permissive effect since E3 ubiquitin ligases are regulated by the covalent attachment of the NEDD8 in to the cullin subunit (61). NEDD8 additionally forms covalent links to neurodegenerative protein aggregates. Parkinson's disease (PD)-related parkin and PINK1 are both NEDD8 conjugated, and NEDD8 immunoreactivity is found in Lewy bodies of midbrain dopaminergic neurons from Parkinson patients and also is incorporated into their Lewy bodies biochemically (62). NEDD components uncovered in both FU-CJD and sCJD particles solidify infection-induced protein aggregates.
Calponin 3 (CNN3) is expressed in brain but it has not been linked to either viral infection or neurodegeneration, and the biologic significance of exclusive proteins #4- 6 is also indeterminate. The strong 6.8-fold elevation of HPCL4, a neuronal Ca++ sensor superfamily that initiates exocytosis of neurotransmitters, reemphasizes the synaptic and vesicular location of different TSE infectious particle strains. Human USO1, a protein required for transcytotic fusion and/or subsequent binding of vesicles to a target membrane can be critical for sCJD agent spread within and across cells and synapses. Synaptogamin-2 (#11), involved in endoplasmic reticulum-plasma membrane contacts (63), again underscores a cell-to-cell spread of TSE synaptic particles. In sum, human sCJD particle proteins give a highly consistent and convergent picture with the mouse FU-CJD particles. Nevertheless, a few human proteins are distinct outliers. The 4-fold higher representation of Urea transporter 1 (#13) probably reflects end-stage uremia in this terminal sCJD patient. The lack of elevated PrP in the non-PK particle fraction of this verified human sCJD brain also deserves comment. Although this brain homogenate contained diagnostic PrP-res, the sCJD agent does not provoke multiple PrP amyloid aggregates but instead induces a weak and diffuse PrP-res distribution and the p18 particles may not contain higher amounts than the control brain. Moreover, human brain sCJD samples without apparent PrP-res on Western blots have proven to be infectious by positive transmissions to rodents (64), a finding consistent with the lack of PrP elevation in the sCJD brain here, and one that is no longer surprising in view of many other verified infectious samples without detectable PrP (vide supra).
Proteins most underrepresented in human sCJD infected brain are also listed n in Table 4 (#69 to #80), where CCD87 at the bottom shows the most marked change. CCD87 was reduced -1,375-fold as compared to the uninfected control, an extraordinary difference. There is little in the Pub Med or protein database on the function of this coil-coil transcriptional promoter. ASCC3 is also markedly underrepresented at -591-fold in sCJD. This helicase promotes DNA unwinding and silencing of ASCC3 upregulates multiple antiviral interferon stimulated genes (65) to restrict infection by many divergent RNA and DNA viruses. This reduced ASCC3 again emphasizes a blocked interferon response that parallels the incomplete IFN response in FU-CJD infection (66). The paucity of lysosomal components in sCJD particles is also especially remarkable because PrP-res specifically accumulates in late endosomes and lysosomes (67, 68). Low representation of lysosomal components in sCJD particles parallels the loss of lysosomal PCYOX as well as PrP from PK+ FU-CJD particles.
Proteins found exclusively in sCJD but not parallel control brain
Table 5 displays the top 29 sCJD particle associated sequences not detected in the parallel controls, and Supplement IV lists all 146 exclusive proteins in order of intensity. The Neural Wiskott-Aldrich protein (WASL) had the highest intensity of 169,140. This heat shock protein parallels the HSC elevation in murine FU-CJD particles. WASL forms a complex on heat shock promoter elements (HSE) that negatively regulates HSP90 expression. It is also part of an innate immune response, and positively regulates clathrin-mediated endocytosis, underscoring yet another common viral pathway with FU-CJD. Interestingly, HSP90 was detected neither in human sCJD, in accord with the high WASL value, nor was it detected in any FU-CJD mouse-passaged particles.
Dynactin 3 (DCTN3), a motor protein involved in antigen processing and presentation of exogenous peptides also shows a very high relative intensity of 157,980. Dynactin's prominence suggests a host response to a foreign viral antigen, consistent with those previously identified in TSEs (29). DCTN3 can be critical for viral infection and latency. DCTN3 overexpression decreases foot & mouth virus replication, and a viral mutant that could not bind DCTN3 produced a delayed disease in cattle (69). Thus strong DCTN3 here probably indicates host inhibition of sCJD agent replication. Kinesin (#15) and Dynactin 5 in Table 5 again strongly link sCJD particles to microtubules, an intracellular transport route used by many viruses. Its presence parallels MAP1A and kinesin 5C in FU-CJD (#11 & #13, Table 3) and solidifies a common route for TSE agent transit to the synapse. In contrast, prion protein disrupts and reduces microtubule proteins to very low levels in scrapie (70), and forms large intracellular amyloid aggregates that show no microtubule binding ultrastructurally (8). Moreover, misfolded PrP is transported rapidly into the late endosomal pathway, unlike the very slow neuronal transit and cross-synaptic infection displayed by scrapie agents (71).
FKBP4, another high intensity exclusive sCJD protein, is part of an immunophilin and chaperone family associated with AD tau pathology, and binds FK506 which interacts with HSP90. Surprisingly, Huntington protein (#7, HD) also yielded an intense exclusive signal in human sCJD. A proteomic study of a mutant and normal HD in subcellular brain fractions linked this ubiquitous protein to functional changes in translation (72) and in sCJD, it signifies a translational holdup with broad functional consequences. HD is also involved in microtubule-mediated transport, and very recent studies show it is a scaffolding protein required for secretory vesicle fusion at the plasma membrane (73). Other vesicle, endosomal and synaptic proteins are also exclusive to human sCJD versus control brain, such as CHM2A (#10), a core multivesicular endosomal sorting component involved in HIV particle release, VP13D (#13) a vacuolar sorting protein, and P142C (#20) which is involved in endocytic trafficking and Hepatitis C replication (74). Synaptogamin is also quite high at 17,4922, and other synaptic and Ca++ proteins of >9,000 that parallel shared common pathways with FU-CJD are in Supplement Table IV.
A single-stranded DNA binding protein that preferentially binds oligo dC (PCBP2, #22) was also high on the exclusive sCJD list, but had no parallel in FU-CJD, where only RNA binding sequences were detected. PCB2 can also bind poly (rC) for cap independent translation and poliovirus RNA replication (75), and the rabies virus glycoprotein specifically interacts with host PCB2 (76). PCB2 enhances viral and mRNA stability, and negatively regulates cellular antiviral responses and the antiviral mitochondrial protein (MAV) that binds ubiquitin ligase (77). Its presence indicates the escape of sCJD agent from these controls. RS30 (#23) is also fused to a ubiquitin-like protein, further demonstrating that TSE particles adhere to a complex array of host proteins that can modify agent replication and processing to define strain-specific virulence features.
With respect to neurodegenerative processes, ROCK2 (#27), a major regulator of axonal degeneration and regeneration in the CNS (78), can provide a new target to help inhibit neurodegeneration. Another is Septin4 (#33) that physiologically associates with alpha-synuclein and parkin, and colocalizes with neurodegenerative Lewy bodies (79). Another septin (Sept5) was also strong in the Ingenuity mouse neurodegeneration search (Supplemental Fig. 4).
Discussion
The above proteomic results demonstrate that a select group of overlapping host proteins associate with two different infectious CJD agent-strains: the murine passaged Asiatic FU-CJD agent and the human sporadic sCJD agent. After PK stripping, virus size FU-CJD brain particles with verified high infectivity titers (14) here revealed many associated host proteins by deep proteomic analysis. Because PrP was not detectable, these results do not support the infectious PrP (prion protein) hypothesis. Instead, they support the concept that TSE viral particles i) use classical viral pathways for infection and reproduction, ii) require membrane PrP for effective replication in the host cell, and iii) induce PrP-res amyloid as a separable response to stress at late stages of infection [reviewed in (34)]. The fact that PrP is not a necessary component of the infectious particle does not minimize its essential role in cell infection because many viruses require specific host receptors and their associated organelle scaffolds to replicate and mature. PrP is clearly critical for TSE agent susceptibility, and can also function in agent clearance (52).
PK unmasking brought out other host components not previously associated with TSE infectious particles using sensitive proteomic approaches. Even less purified (non-PK treated) particles yielded a very different profile than complex whole brain homogenates. TSE brain homogenates typically show small quantitative PrP homogenate differences (< 3-fold), mainly in pathologic response and plaque proteins, such as C1q complement, GFAP, mitochondrial apoptosis-linked and lysosomal proteins. These proteins, some of which were present in less purified FU-CJD p18 particles were removed by PK, and this resolved massive relative increases in FU-CJD (up to 131,857 fold over control particles), in addition to uncovering many exclusive proteins. Infectious PK+ particles and sCJD particles also lacked strong or exclusive signals for late endosomal and lysosomal components where abnormal PrP is concentrated (67, 68). In sum, infectious particles followed a divergent intracellular pathway from PrP.
Because PK abolished all forms of detectable PrP, but not particle infectivity, at least a few PK unmasked proteins could function in particular cell organelles to either enhance or compromise particle infectivity. Our PK stripping strategy unmasked 56 exclusive host proteins in FU-CJD particles that were not evident in the parallel controls, and many of these proteins are linked to established viral processing pathways. After major pathologic inflammatory response proteins such as astrocytic GFAP, complement C1q, and PrP were removed by PK to undetectable levels by LC-MS/MS, synaptic regional proteins became prominent. Synapses are where both TSE virus-like particles and infectivity collect (5-8, 39). Additionally, early endosomal proteins, clathrin and vesicle-membrane transport components used by many viruses were selectively unmasked. Remarkably, some of these host components can be incorporated by viruses. Several proteins found within vesicular somatitis virus (VSV) were unmasked by a comparable PK stripping strategy to completely purify infectious virion elements for mass spectrometry (38). Aside from independently validating the usefulness of our PK approach to link infectious TSE particles to essential specific host complexes, that study identified HSC70 as well as integrin β1 within purified VSV virions. These two proteins tightly associated with TSE particles here, and they might contribute to a TSE viral structure. However, the abundant HSC70 in TSEs is more likely to have a broader physiological role because HSC70 targets many different viruses to suppress infection. Thus its marked elevation in PK+ FU-CJD particles indicates a strong and specific host response to arrest ongoing agent production.
The HSC70 results underscore three concepts with practical ramifications. First, HSC70 antibodies can be used to further isolate or pull down infectious brain particles from less specific host components. This approach can resolve the major and still unknown structural TSE agent elements. Second, the vast excess of HSP70 with infectious particles can disguise the agent's foreign nature, and hide integral agent molecules from host immune recognition. Other agent-associated proteins identified here such as copine-3, and vesicle-membrane proteins such as SYN2, SV2a and AP180 can further this effect. Third, particle-linked host proteins provide an explanation for the species barrier in TSEs. Although nucleic acid mutation is at the root of viral adaptation to a new species, TSE agents show a high degree of strain-specific conservation across many species and cell types (2, 34), suggesting few major mutations are permissible or retained. However, if a TSE agent is covered by a protein that contains a species-specific amino acid, the first cross-species transmission should be slow because the new host will recognize the former “coating” species sequence as foreign, and more effectively eliminate the infectious complexes. Once the agent attaches to the new host's protein(s), the agent becomes part of the self, and can better replicate by escaping immune surveillance. A pattern of markedly reduced incubation time with the first serial cross-species passage is seen in many TSE agent transmissions, including vCJD and kuru to mice, and sCJD to guinea pigs (1, 3, 34). The huge amount of HSC70, uncovered only after PK treatment in FU-CJD, was not seen in less purified human sCJD particles, so it is difficult to know if the lack of human HSC70 is PK treatment dependent, agent-strain specific, or species determined, but studies of wt mice infected with vCJD, kuru or sheep derived scrapie agents, e.g., (1, 3) can clarify this.
It is also remarkable that another heat shock protein, HSP90, was undetectable in both FU-CJD and sCJD particles, a finding completely in accord with the subverted interferon pathways documented in FU-CJD (66). HSP90 is a chaperone required for the production and maturation of several viruses as detailed above, and is also critical for activation of HIV-1 from latency (46). Thus the absence of HSP90 in TSE particles may help retard TSE infection. Moreover, because HSP90 inhibitors reduce the burden of abnormal protein aggregates, including β-amyloid and tau (47), HSP90 and HSP90 inhibitor pathways should provide important pharmacologic targets to reduce both the infectious titer and the formation of amyloid aggregates in CJD. A second attractive therapeutic target uncovered here is Cyclophilin B (PPIB), a chaperone involved in protein folding in the ER that is required for the lifecycle of a variety of viruses (see Results). Its strong association with PK treated FU-CJD particles, along with the ability of its inhibitor, Cyclosporin A, to induce abnormal PrP in uninfected cells (51), suggests that Cyclosporin A may inhibit TSE infection by at least two avenues: first, by arresting TSE agent uncoating or disassembly needed for replication, and second, by increasing PrP aggregation that can trap and clear infectious particles (52). One can test the effects of Cyclosporin A on infectious titers in stable FU-CJD neuronal cultures, and with more precise Cyclophilin B knockout approaches determine if narrowly targeting this protein is sufficient to reduce or prevent infection.
Destructive targeting of other tightly bound proteins identified here would probably be toxic, such as those that aid TSE agent infection and spread. These include clathrin and early endosomal/vesicle proteins which are common viral infection pathways (44), and synaptic junctions where the scrapie agent crosses (71). On the other hand, the markedly elevated human sCJD associated protein Dynactin 3, if further activated might enhance its anti-infective effect without major sequelae. Notably, this Dynactin approach has been effective in diminishing other viruses (69). Replacement of proteins found to be markedly reduced may also be a useful complementary approach, but greater purification of human brain TSE particles needs to be accomplished before pursuing specific protein modulators because not all proteins identified here were particle associated, e.g, NADH dehydrogenase and urea in human sCJD brain that signified terminal metabolic compromise.
In summary, the above analyses of more purified infectious TSE particles has brought to the forefront a rich set of host proteins most critical for infectious agent propagation and recognition. A consistent cell organelle association, particularly with early endosomal uptake, synapses and membrane junctions has yielded proteomic details of TSE particle processing that were previously concealed using less sensitive methods and/or crude brain homogenates or amyloid fractions. Agent-associated proteins identified here diverge from PrP-res with its known locations, i.e., as amyloid aggregates in the cytosol (8) and in lysosomes (67). Instead, infectious particles not subjected to artifactual detergent lysis emphasize their coincidence with ultrastructural arrays of 25nm TSE virus-like particles in infected cultured cells (8) and in “tubular” or vesicular early endosome-like structures within brain synapses (7).
The select group of proteins identified here also offer new targets for therapy. Several of these targets are strongly and simultaneously linked to both viral and neurodegenerative changes, and are involved in protein folding and aggregation processes. Hence, therapies directed at several of such “bifunctional” proteins may affect both the infectious TSE agent and the formation of amyloid aggregates in TSEs, with beneficial effects as well for neurodegenerative AD and Parkinson's Disease. Vesicular-endosomal targeting by pharmacologic chaperones has shown promise in decreasing AD processing of APP into amyloid (80), and the identification of particular chaperones here further validates our suggestion that such therapies might be beneficial in TSEs (14). Finally, improved purification of infectious particles is now possible using antibodies to specific host proteins to pull down tightly associated agent particles. Such an approach can further resolve the still mysterious infectious agent molecules. It is likely that these molecules will display a set of related sequences that are largely conserved among different TSE agent strains.
Supplementary Material
Acknowledgments
Funding Sources: Portions of this work were supported by NIH NS RO1 012674-34, the William Prusoff Foundation, a Yale OSHE grant, 1S10OD018034-01 (6500 QTrap Mass Spectrometer for Yale University), 1S10RR026707-01 (5500 QTrap Mass Spectrometer for Yale University)
Footnotes
The authors declare no financial/commercial conflicts of interest.
SYN2, HSC70, CPNE3, clathrin, Annexin, DYN1, PPIB (cyclophilin B), VAOD1, AP180, LRC59, MYO5A, beta integrins, Neddylation NEDD8, Cullin Ligases, CAND1, Calcium channels (C2D1, CACB4), Wiskott-Aldrich (WASL), ApoO, ApoE, USO1, DCTN3, Huntingtin (HD), VP13D, CHM2A, SEPT4, FXBP4, FK506, VP13D, NADH dehydrogenase, RL7A.
References
- 1.Manuelidis L, Chakrabarty T, Miyazawa K, Nduom NA, Emmerling K. The kuru infectious agent is a unique geographic isolate distinct from Creutzfeldt-Jakob disease and scrapie agents. Proc Natl Acad Sci U S A. 2009;106:13529–13534. doi: 10.1073/pnas.0905825106. published online Epub Aug 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arjona A, Simarro L, Islinger F, Nishida N, Manuelidis L. Two Creutzfeldt-Jakob disease agents reproduce prion protein-independent identities in cell cultures. Proc Natl Acad Sci USA. 2004;101:8768–8773. doi: 10.1073/pnas.0400158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manuelidis L, Liu Y, Mullins B. Strain-specific viral properties of variant Creutzfeldt-Jakob disease (vCJD) are encoded by the agent and not by host prion protein. J Cell Biochem. 2009;106:220–231. doi: 10.1002/jcb.21988. published online Epub Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manuelidis L. A 25 nm virion is the likely cause of Transmissible Spongiform Encephalopathies. J Cell Biochem. 2007;100:897–915. doi: 10.1002/jcb.21090. [DOI] [PubMed] [Google Scholar]
- 5.Bignami A, Parry H. Aggregations of 35-nanometer particles associated with neuronal cytopathic changes in natural scrapie. Science. 1971;171:389–390. doi: 10.1126/science.171.3969.389. [DOI] [PubMed] [Google Scholar]
- 6.David-Ferreira J, David-Ferreira K, Gibbs C, Morris J. Scrapie in mice: ultrastructural observations in the cerebral cortex. Proc Soc Exp Biol Med. 1968;127:313–320. doi: 10.3181/00379727-127-32680. [DOI] [PubMed] [Google Scholar]
- 7.Liberski P, Jeffrey M. Tubulovesicular structures—the ultrastructural hallmark for transmissible spongiform encephalopathies (prion diseases) Folia Neuropathologica. 2004;42 Supplement B:96–108. [PubMed] [Google Scholar]
- 8.Manuelidis L, Yu ZX, Barquero N, Mullins B. Cells infected with scrapie and Creutzfeldt-Jakob disease agents produce intracellular 25-nm virus-like particles. Proc Natl Acad Sci USA. 2007;104:1965–1970. doi: 10.1073/pnas.0610999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffrey M, Gonzalez L, Espenes A, Press C, Martin S, Chaplin M, Davis L, Landsverk T, MacAldowie C, Eaton S, McGovern G. Transportation of prion protein across the intestinal mucosa of scrapie-susceptible and scrapie-resistant sheep. J Pathol. 2006;209:4–14. doi: 10.1002/path.1962. [DOI] [PubMed] [Google Scholar]
- 10.Scherbel C, Pichner R, Groschup MH, Mueller-Hellwig S, Scherer S, Dietrich R, Maertlbauer E, Gareis M. Infectivity of Scrapie Prion Protein (PrPSc) Following In vitro Digestion with Bovine Gastrointestinal Microbiota. Zoonoses and Public Health. 2007;54:185–190. doi: 10.1111/j.1863-2378.2007.01040.x. [DOI] [PubMed] [Google Scholar]
- 11.Miyazawa K, Emmerling K, Manuelidis L. High CJD infectivity remains after prion protein is destroyed. J Cell Biochem. 2011;112:3630–3637. doi: 10.1002/jcb.23286. published online Epub Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuelidis EE, Gorgacz EJ, Manuelidis L. Viremia in experimental Creutzfeldt-Jakob disease. Science. 1978;200:1069–1071. doi: 10.1126/science.349691. [DOI] [PubMed] [Google Scholar]
- 13.Manuelidis L. Nuclease resistant circular DNAs copurify with infectivity in scrapie and CJD. J Neurovirol. 2011;17:131–145. doi: 10.1007/s13365-010-0007-0. published online Epub Apr. [DOI] [PubMed] [Google Scholar]
- 14.Kipkorir T, Tittman S, Botsios S, Manuelidis L. Highly Infectious CJD Particles Lack Prion Protein but Contain Many Viral-Linked Peptides by LC-MS/MS. J Cell Biochem. 2014;115:2012–2021. doi: 10.1002/jcb.24873. published online Epub Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manuelidis L, Fritch W, Xi YG. Evolution of a strain of CJD that induces BSE-like plaques. Science. 1997;277:94–98. doi: 10.1126/science.277.5322.94. [DOI] [PubMed] [Google Scholar]
- 16.Sattlecker M, Kiddle SJ, Newhouse S, Proitsi P, Nelson S, Williams S, Johnston C, Killick R, Simmons A, Westman E, Hodges A, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Lovestone S, Dobson RJ C. AddNeuroMed. Alzheimer's disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2014;10:724–734. doi: 10.1016/j.jalz.2013.09.016. published online Epub Nov. [DOI] [PubMed] [Google Scholar]
- 17.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/SICI1522-26831999120120:18<3551∷AID-ELPS3551>3.0.CO;2-2. published online Epub Dec. [DOI] [PubMed] [Google Scholar]
- 18.Colangelo CM, Ivosev G, Chung L, Abbott T, Shifman M, Sakaue F, Cox D, Kitchen RR, Burton L, Tate SA, Gulcicek E, Bonner R, Rinehart J, Nairn AC, Williams KR. Development of a highly automated and multiplexed targeted proteome pipeline and assay for 112 rat brain synaptic proteins. Proteomics. 2014 doi: 10.1002/pmic.201400353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shifman MA, Li Y, Colangelo CM, Stone KL, Wu TL, Cheung KH, Miller PL, Williams KR. YPED: a web-accessible database system for protein expression analysis. Journal of proteome research. 2007;6:4019–4024. doi: 10.1021/pr070325f. published online Epub Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colangelo CM, Shifman M, Cheung KH, Stone KL, Carriero NJ, Gulcicek EE, Lam TT, Wu T, Bjornson RD, Bruce C, Nairn AC, Rinehart J, Miller PL, Williams KR. YPED: An Integrated Bioinformatics Suite and Database for Mass Spectrometry Based Proteomics Research. Genomics Proteomics Bioinformatics. 2014 doi: 10.1016/j.gpb.2014.11.002. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-28360580360-2. published online Epub Oct 5. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. published online Epub Oct 15. [DOI] [PubMed] [Google Scholar]
- 23.Reiter L, Rinner O, Picotti P, Huttenhain R, Beck M, Brusniak MY, Hengartner MO, Aebersold R. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nature methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. published online Epub May. [DOI] [PubMed] [Google Scholar]
- 24.Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B, Vitek O. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. published online Epub Sep 1. [DOI] [PubMed] [Google Scholar]
- 25.Nishida N, Katamine S, Manuelidis L. Reciprocal interference between specific CJD and scrapie agents in neural cell cultures. Science. 2005;310:493–496. doi: 10.1126/science.1118155. [DOI] [PubMed] [Google Scholar]
- 26.Manuelidis E, Manuelidis L. In: Progress in Neuropathology. Zimmerman HM, editor. Vol. 4. Raven Press; N.Y: 1979. pp. 1–26. [Google Scholar]
- 27.Manuelidis L, Fritch W. Infectivity and host responses in Creutzfeldt-Jakob Disease. Virology. 1996;216:46–59. doi: 10.1006/viro.1996.0033. [DOI] [PubMed] [Google Scholar]
- 28.Lv Y, Chen C, Zhang BY, Xiao K, Wang J, Chen LN, Sun J, Gao C, Shi Q, Dong XP. Remarkable Activation of the Complement System and Aberrant Neuronal Localization of the Membrane Attack Complex in the Brain Tissues of Scrapie-Infected Rodents. Molecular neurobiology. 2014 doi: 10.1007/s12035-014-8915-2. published online Epub Oct 14. [DOI] [PubMed] [Google Scholar]
- 29.Lu ZH, Baker C, Manuelidis L. New molecular markers of early and progressive CJD brain infection. J Cellular Biochem. 2004;93:644–652. doi: 10.1002/jcb.20220. [DOI] [PubMed] [Google Scholar]
- 30.Kay M. Immunoregulation of cellular life span. Annals of the New York Academy of Sciences. 2005;1057:85–111. doi: 10.1196/annals.1356.005. published online Epub Dec. [DOI] [PubMed] [Google Scholar]
- 31.Clay GM, Sutterwala FS, Wilson ME. NLR proteins and parasitic disease. Immunologic research. 2014;59:142–152. doi: 10.1007/s12026-014-8544-x. published online Epub Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L, Ye L, Zhao R, Liu YF, Yang SJ. HSP70 induced by Hantavirus infection interacts with viral nucleocapsid protein and its overexpression suppresses virus infection in Vero E6 cells. American journal of translational research. 2009;1:367–380. [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MY, Ma Y, Zhang Y, Li J, Shu Y, Oglesbee M. hsp70-dependent antiviral immunity against cytopathic neuronal infection by vesicular stomatitis virus. J Virol. 2013;87:10668–10678. doi: 10.1128/JVI.00872-13. published online Epub Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manuelidis L. Infectious particles, stress, and induced prion amyloids: a unifying perspective. Virulence. 2013;4:373–383. doi: 10.4161/viru.24838. published online Epub Jul 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jockusch H, Wiegand C, Mersch B, Rajes D. Mutants of tobacco mosaic virus with temperature-sensitive coat proteins induce heat shock response in tobacco leaves. Molecular plant-microbe interactions: MPMI. 2001;14:914–917. doi: 10.1094/MPMI.2001.14.7.914. published online Epub Jul. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Bai J, Zhang L, Jiang Z, Wang X, Li Y, Jiang P. Hsp70 positively regulates porcine circovirus type 2 replication in vitro. Virology. 2013;447:52–62. doi: 10.1016/j.virol.2013.08.025. published online Epub Dec. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z, Wu SW, Lei CQ, Zhou Q, Li S, Shu HB, Wang YY. Heat shock cognate 71 (HSC71) regulates cellular antiviral response by impairing formation of VISA aggregates. Protein & cell. 2013;4:373–382. doi: 10.1007/s13238-013-3902-3. published online Epub May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moerdyk-Schauwecker M, Hwang SI, Grdzelishvili VZ. Analysis of virion associated host proteins in vesicular stomatitis virus using a proteomics approach. Virol J. 2009;6:166. doi: 10.1186/1743-422X-6-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manuelidis L, Manuelidis E. Fractionation and infectivity studies in Creutzfeldt Jakob disease. Banbury Rep (Cold Spring Harbor) 1983;15:399–412. [Google Scholar]
- 40.Raff AB, Woodham AW, Raff LM, Skeate JG, Yan L, Da Silva DM, Schelhaas M, Kast WM. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol. 2013;87:6062–6072. doi: 10.1128/JVI.00330-13. published online Epub Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamazaki T, Koo EH, Selkoe DJ. Cell surface amyloid beta-protein precursor colocalizes with beta 1 integrins at substrate contact sites in neural cells. J Neurosci. 1997;17:1004–1010. doi: 10.1523/JNEUROSCI.17-03-01004.1997. published online Epub Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sosa LJ, Bergman J, Estrada-Bernal A, Glorioso TJ, Kittelson JM, Pfenninger KH. Amyloid precursor protein is an autonomous growth cone adhesion molecule engaged in contact guidance. PLoS One. 2013;8:e64521. doi: 10.1371/journal.pone.0064521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyazawa K, Emmerling K, Manuelidis L. Proliferative arrest of neural cells induces prion protein synthesis, nanotube formation, and cell-to-cell contacts. J Cell Biochem. 2010;111:239–247. doi: 10.1002/jcb.22723. published online Epub Sep 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. published online Epub May. [DOI] [PubMed] [Google Scholar]
- 45.Nuss JE, Kehn-Hall K, Benedict A, Costantino J, Ward M, Peyser BD, Retterer CJ, Tressler LE, Wanner LM, McGovern HF, Zaidi A, Anthony SM, Kota KP, Bavari S, Hakami RM. Multi-faceted proteomic characterization of host protein complement of Rift Valley fever virus virions and identification of specific heat shock proteins, including HSP90, as important viral host factors. PLoS One. 2014;9:e93483. doi: 10.1371/journal.pone.0093483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson I, Low JS, Weston S, Weinberger M, Zhyvoloup A, Labokha AA, Corazza G, Kitson RA, Moody CJ, Marcello A, Fassati A. Heat shock protein 90 controls HIV-1 reactivation from latency. Proc Natl Acad Sci U S A. 2014;111:E1528–1537. doi: 10.1073/pnas.1320178111. published online Epub Apr 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao H, Michaelis ML, Blagg BS. Hsp90 modulation for the treatment of Alzheimer's disease. Advances in pharmacology. 2012;64:1–25. doi: 10.1016/B978-0-12-394816-8.00001-5. [DOI] [PubMed] [Google Scholar]
- 48.Perestenko PV, Pooler AM, Noorbakhshnia M, Gray A, Bauccio C, Jeffrey McIlhinney RA. Copines-1, -2, -3, -6 and -7 show different calcium-dependent intracellular membrane translocation and targeting. The FEBS journal. 2010;277:5174–5189. doi: 10.1111/j.1742-4658.2010.07935.x. published online Epub Dec. [DOI] [PubMed] [Google Scholar]
- 49.Kambara H, Tani H, Mori Y, Abe T, Katoh H, Fukuhara T, Taguwa S, Moriishi K, Matsuura Y. Involvement of cyclophilin B in the replication of Japanese encephalitis virus. Virology. 2011;412:211–219. doi: 10.1016/j.virol.2011.01.011. published online Epub Mar 30. [DOI] [PubMed] [Google Scholar]
- 50.Watashi K, Shimotohno K. Chemical genetics approach to hepatitis C virus replication: cyclophilin as a target for anti-hepatitis C virus strategy. Reviews in medical virology. 2007;17:245–252. doi: 10.1002/rmv.534. published online Epub Jul-Aug. [DOI] [PubMed] [Google Scholar]
- 51.Cohen E, Taraboulos A. Scrapie-like prion protein accumulates in aggresomes of cyclosporin A-treated cells. EMBO J. 2003;22:404–417. doi: 10.1093/emboj/cdg045. published online Epub Feb 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyazawa K, Kipkorir T, Tittman S, Manuelidis L. Continuous production of prions after infectious particles are eliminated: implications for Alzheimer's disease. PLoS ONE. 2012;7:e35471. doi: 10.1371/journal.pone.0035471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshansky V, Rubinstein JL, Gruber G. Eukaryotic V-ATPase: novel structural findings and functional insights. Biochimica et biophysica acta. 2014;1837:857–879. doi: 10.1016/j.bbabio.2014.01.018. published online Epub Jun. [DOI] [PubMed] [Google Scholar]
- 54.Zhang F, Zang T, Wilson SJ, Johnson MC, Bieniasz PD. Clathrin facilitates the morphogenesis of retrovirus particles. PLoS Pathog. 2011;7:e1002119. doi: 10.1371/journal.ppat.1002119. published online Epub Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snyers L, Zwickl H, Blaas D. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J Virol. 2003;77:5360–5369. doi: 10.1128/JVI.77.9.5360-5369.2003. published online Epub May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolobov VV, Storozheva ZI, Gruden MA, Sherstnev VV. Regional features of the expression of genes involved in neurogenesis and apoptosis in the brain of adult rats. Bulletin of experimental biology and medicine. 2012;153:746–749. doi: 10.1007/s10517-012-1816-3. published online Epub Sep. [DOI] [PubMed] [Google Scholar]
- 57.Zhang A, Williamson CD, Wong DS, Bullough MD, Brown KJ, Hathout Y, Colberg-Poley AM. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Molecular & cellular proteomics: MCP. 2011;10:M111 009936. doi: 10.1074/mcp.M111.009936. published online Epub Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahon C, Krogan NJ, Craik CS, Pick E. Cullin E3 ligases and their rewiring by viral factors. Biomolecules. 2014;4:897–930. doi: 10.3390/biom4040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun R, Liu Y, Zhang H, Manuelidis L. Quantitative recovery of scrapie agent with minimal protein from highly infectious cultures. Viral Immunol. 2008;21:293–302. doi: 10.1089/vim.2008.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nekorchuk MD, Sharifi HJ, Furuya AK, Jellinger R, de Noronha CM. HIV relies on neddylation for ubiquitin ligase-mediated functions. Retrovirology. 2013;10:138. doi: 10.1186/1742-4690-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hofmann H, Norton TD, Schultz ML, Polsky SB, Sunseri N, Landau NR. Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J Virol. 2013;87:11741–11750. doi: 10.1128/JVI.02002-13. published online Epub Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choo YS, Vogler G, Wang D, Kalvakuri S, Iliuk A, Tao WA, Bodmer R, Zhang Z. Regulation of parkin and PINK1 by neddylation. Human molecular genetics. 2012;21:2514–2523. doi: 10.1093/hmg/dds070. published online Epub Jun. doi:1 10.1093/hmg/dds070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. published online Epub Jun 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manuelidis L, Manuelidis EE. Recent developments in scrapie and Creutzfeldt-Jakob disease. Prog Med Virol. 1985;33:78–98. [PubMed] [Google Scholar]
- 65.Li J, Ding SC, Cho H, Chung BC, Gale M, Jr, Chanda SK, Diamond MS. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio. 2013;4:e00385–00313. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker C, Lu Z, Manuelidis L. Early induction of interferon-responsive mRNAs in Creutzfeldt-Jakob disease. J Neurovirol. 2004;10:1–12. doi: 10.1080/13550280490261761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnold JE, Tipler C, Laszlo L, Hope J, Landon M, Mayer RJ. The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. The Journal of pathology. 1995;176:403–411. doi: 10.1002/path.1711760412. published online Epub Aug. [DOI] [PubMed] [Google Scholar]
- 68.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci U S A. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gladue DP, O'Donnell V, Baker-Bransetter R, Pacheco JM, Holinka LG, Arzt J, Pauszek S, Fernandez-Sainz I, Fletcher P, Brocchi E, Lu Z, Rodriguez LL, Borca MV. Interaction of foot-and-mouth disease virus nonstructural protein 3A with host protein DCTN3 is important for viral virulence in cattle. J Virol. 2014;88:2737–2747. doi: 10.1128/JVI.03059-13. published online Epub Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Y, Gong HS, Zhang J, Xie WL, Tian C, Chen C, Shi Q, Wang SB, Xu Y, Zhang BY, Dong XP. Remarkable reduction of MAP2 in the brains of scrapie-infected rodents and human prion disease possibly correlated with the increase of calpain. PLoS One. 2012;7:e30163. doi: 10.1371/journal.pone.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott J, Fraser H. Transport and targeting of scrapie infectivity and pathology in the optic nerve projections following intraocular infection. Prog Clin Biol Res. 1989;317:645–652. [PubMed] [Google Scholar]
- 72.Culver BP, Savas JN, Park SK, Choi JH, Zheng S, Zeitlin SO, Yates JR, 3rd, Tanese N. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. The Journal of biological chemistry. 2012;287:21599–21614. doi: 10.1074/jbc.M112.359307. published online Epub Jun 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandstaetter H, Kruppa AJ, Buss F. Huntingtin is required for ER-to-Golgi transport and for secretory vesicle fusion at the plasma membrane. Disease models & mechanisms. 2014;7:1335–1340. doi: 10.1242/dmm.017368. published online Epub Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. published online Epub May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chase AJ, Daijogo S, Semler BL. Inhibition of poliovirus-induced cleavage of cellular protein PCBP2 reduces the levels of viral RNA replication. J Virol. 2014;88:3192–3201. doi: 10.1128/JVI.02503-13. published online Epub Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palusa S, Ndaluka C, Bowen RA, Wilusz CJ, Wilusz J. The 3′ untranslated region of the rabies virus glycoprotein mRNA specifically interacts with cellular PCBP2 protein and promotes transcript stability. PLoS One. 2012;7:e33561. doi: 10.1371/journal.pone.0033561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jacobs JL, Coyne CB. Mechanisms of MAVS regulation at the mitochondrial membrane. Journal of molecular biology. 2013;425:5009–5019. doi: 10.1016/j.jmb.2013.10.007. published online Epub Dec 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koch JC, Tonges L, Barski E, Michel U, Bahr M, Lingor P. ROCK2 is a major regulator of axonal degeneration, neuronal death and axonal regeneration in the CNS. Cell death & disease. 2014;5:e1225. doi: 10.1038/cddis.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shehadeh L, Mitsi G, Adi N, Bishopric N, Papapetropoulos S. Expression of Lewy body protein septin 4 in postmortem brain of Parkinson's disease and control subjects. Movement disorders: official journal of the Movement Disorder Society. 2009;24:204–210. doi: 10.1002/mds.22306. published online Epub Jan 30. [DOI] [PubMed] [Google Scholar]
- 80.Mecozzi VJ, Berman DE, Simoes S, Vetanovetz C, Awal MR, Patel VM, Schneider RT, Petsko GA, Ringe D, Small SA. Pharmacological chaperones stabilize retromer to limit APP processing. Nature chemical biology. 2014 doi: 10.1038/nchembio.1508. published online Epub Apr 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


