Abstract
Genome-wide association studies have identified multiple risk variants and loci that show robust association with schizophrenia. Nevertheless, it remains unclear how these variants confer risk to schizophrenia. In addition, the driving force that maintains the schizophrenia risk variants in human gene pool is poorly understood. To investigate whether expression-associated genetic variants contribute to schizophrenia susceptibility, we systematically integrated brain expression quantitative trait loci and genome-wide association data of schizophrenia using Sherlock, a Bayesian statistical framework. Our analyses identified ZNF323 as a schizophrenia risk gene (P = 2.22×10–6). Subsequent analyses confirmed the association of the ZNF323 and its expression-associated single nucleotide polymorphism rs1150711 in independent samples (gene-expression: P = 1.40×10–6; single-marker meta-analysis in the combined discovery and replication sample comprising 44123 individuals: P = 6.85×10−10). We found that the ZNF323 was significantly downregulated in hippocampus and frontal cortex of schizophrenia patients (P = .0038 and P = .0233, respectively). Evidence for pleiotropic effects was detected (association of rs1150711 with lung function and gene expression of ZNF323 in lung: P = 6.62×10–5 and P = 9.00×10–5, respectively) with the risk allele (T allele) for schizophrenia acting as protective allele for lung function. Subsequent population genetics analyses suggest that the risk allele (T) of rs1150711 might have undergone recent positive selection in human population. Our findings suggest that the ZNF323 is a schizophrenia susceptibility gene whose expression may influence schizophrenia risk. Our study also illustrates a possible mechanism for maintaining schizophrenia risk variants in the human gene pool.
Key words: schizophrenia, ZNF323, association, eQTL, hippocampus, positive selection
Introduction
Schizophrenia is a multifactorial and polygenic mental disorder with strong genetic heterogeneity and high heritability.1,2 To date schizophrenia remains an evolutionary-genetic paradox as fecundity of schizophrenia patients (ie, strongly negative fitness) seemingly contradicts its stable prevalence of approximately 1% across all human cultures.3–5 One potential explanation for this conundrum suggests recent positive selection based on compensatory effects on other human traits as (among others) the driving force in maintaining schizophrenia risk loci in the gene pool.4–6 In line with this hypothesis, recent studies have identified pleiotropic effects of schizophrenia risk variants.7,8
Multiple promising risk variants and loci for schizophrenia have been identified by genome-wide association studies (GWAS) of schizophrenia.1,9–11 Nevertheless, how these variants confer risk to schizophrenia remains elusive. Moreover, the identified susceptibility variants only explain a small fraction of the disease variance. A recent study has suggested that more than 8000 common loci are involved in the etiology of schizophrenia,11 thus indicating that many common risk loci with potentially small effect sizes and/or with infrequent risk alleles are still to be detected.
Although more schizophrenia risk loci remain to be identified, accumulating evidence strongly suggests that changes in gene expression rather than alterations in protein function play a key role in the pathogenesis of schizophrenia.12–14 Accordingly, recent studies have used integrative strategies to combine results from association studies and gene expression analyses.15,16 The successful identification of schizophrenia susceptibility genes by these integrative methods supports the important roles of gene expression alteration in the pathogenesis of schizophrenia. However, currently there is no study that systematically integrated brain expression quantitative trait loci (eQTL) data and GWAS of schizophrenia to identify the potential schizophrenia risk genes.
In this study, we followed an integrated data analysis approach using Sherlock 17 and analyzed eQTL from normal human brain samples18 together with association data from a large GWAS for schizophrenia.11 To further characterize the potential role of our identified loci in schizophrenia etiology, we followed up and refined our results in independent gene expression and large-scale genetic datasets. Moreover, we subsequently tested for pleiotropic effects (ie, a simultaneous effect of our identified risk loci on other human traits) and tried to shed light on the potential presence of positive natural selection (as a consequence of compensatory effects on the traits implicated by the pleiotropic analyses).
Methods
Sherlock integration analysis
Considering that most of the disease-associated variants identified by recent GWAS are located in noncoding regions,19 it is likely that these risk variants affect gene expression rather than protein function. To infer genes whose expression changes may contribute to the etiology of complex diseases, He et al17 developed a statistical framework (named Sherlock) to identify the potential disease-associated genes through matching eQTL and single-nucleotide polymorphism (SNP) associations from GWAS. The underlying assumption is that the expression level of a specific gene(s) may influence the risk of a disease (eg, schizophrenia). Therefore, genetic variation (both in cis, ie, close to the gene, and in trans, ie, distant to the gene or on different chromosomes) that perturbs gene expression may affect the risk of this disease. For a given gene there may be several variants (expression-associated SNPs [eSNPs]) in the genome that act together to regulate the expression level of this gene. Genetic variation at these eSNPs would influence the expression level of this gene, which in turn might affect schizophrenia susceptibility. Hence, cumulative evidence from both eQTL and GWAS findings is indicative of association with schizophrenia. Sherlock first searches for all eSNPs of each gene using the whole genome eQTL data from the human frontal cortex.18 For each eSNP, Sherlock will then evaluate its association with schizophrenia using genome-wide association (GWA) data of schizophrenia.11 There are three different scenarios: (1) If the eSNP of a specific gene is also associated with schizophrenia in GWAS, a positive score would be given; (2) If the eSNP of this gene is not associated with schizophrenia, a negative score would be assigned; and (3) association only in GWAS (ie, non-eSNPs) does not alter the score. The total score of a gene increases along with the increase of the number of SNPs with combined evidence (SNPs that are associated with schizophrenia and expression). Finally, Sherlock identifies gene-disease associations through matching genetic signatures of gene expression traits to that of disease.17 Statistical inference was performed using Bayes statistical framework17 and Bayes factor (BF, the probability of the observed data under a specific model) of each SNP was calculated separately. For each gene, Sherlock computes individual logarithm of BF (LBF) for each SNP pair in the alignment, and the sum of these constitutes the final LBF score for the gene. The value of the LBF score of a gene reflects the strength of evidence (ie, a larger LBF represents higher probability that this gene is associated with the disease). Sherlock utilizes moderately associated cis and trans eSNPs (without introducing many false signals) instead of solely focusing on eSNPs that meet stringent cutoff criteria for both eQTL and GWAS results. P values of the BFs were calculated with simulation using Bayes/non-Bayes compromise.20 False discovery rate (FDR) was calculated using Benjamini–Hochberg procedure21 at P threshold of 10–5. More detailed information about the statistical model and the underlying algorithm is provided in the original paper by He et al.17
Brain Expression (eQTL) Data
Considering that schizophrenia is a mental disorder that mainly originates from abnormal brain function, it is reasonable to assume that the most appropriate tissue for an integrative identification of schizophrenia risk genes is the human brain. Therefore, we used brain expression data (human prefrontal cortex [PFC]) from a previous study on 193 neuropathologically normal human brain samples.18 All individuals enrolled in this study were of self-reported European ancestry. Genotyping was performed on an Affymetrix Human Mapping 500K Array Set and expression profiles were identified using an Illumina HumanRefseq-8 Expression BeadChip. Cis eSNPs were defined as SNPs situated within the transcript of the respective gene or within 1Mb of either the 5′ or the 3′ end of the transcript. More information about statistical analyses and procedures is provided in the original study.18
Schizophrenia GWAS Data
We used summary statistics from a large-scale schizophrenia GWAS.11 In brief, the study represents a meta-analysis of a recent schizophrenia GWAS from the Psychiatric Genomics Consortium (PGC), hereafter referred to as “PGC1,”9 and independent data from a Swedish schizophrenia GWAS (“SWE”).11 The study comprised a total of 13833 schizophrenia cases and 18310 controls (“SWE + PGC1”)11 and identified 22 risk loci for schizophrenia. Most of the subjects were of European ancestry. DNA was extracted from peripheral blood and samples were genotyped using Affymetrix (5.0 and 6.0) and Illumina OmniExpress chips according to the manufacturers’ instructions. The association analysis was first performed in SWE samples (5001 cases and 6243 controls) with PLINK22 using imputed SNP dosages and the principal components as covariates. Then the results from SWE samples were meta-analyzed with the results from PGC1 (8832 cases and 12067 controls) using an inverse-weighted fixed-effects model.23 Genome-wide genetic association information (P values of a total of 9898078 SNPs) from SWE + PGC1 was used as input in this study. Detailed information about sample ascertainment, diagnosis, genotyping quality control, and statistical analyses can be found in the original study and PGC website (https://pgc.unc.edu).11
Replication and Refinement of Gene Expression Results
Four well-characterized expression databases were used to follow-up on the gene expression results that contributed to identification of ZNF323 as our top schizophrenia risk gene. In addition to the replication, the follow-up comprised a deeper characterization of gene expression patterns for ZNF323 in the brain (eg, healthy individuals vs schizophrenia patients) and identification of additional eSNPs of ZNF323.
A brief description of the gene expression resources is provided below; more detailed information can be found in the original studies.24–27 (1) BrainCloud.24 BrainCloud contains genetic information and whole transcriptome expression data from postmortem dorsolateral PFC (DLPFC) of 269 normal human subjects (ie, without neuropathological or neuropsychiatric diagnosis). The data in BrainCloud is aimed at exploring temporal dynamics and genetic control of transcription in the DLPFC across the lifespan, ie, from fetal development through ageing. (2) The Genotype-Tissue Expression project (GTEx).25 Compared with BrainCloud, which focuses on the DLPFC, the GTEx contains information at the level of both genetic variation and gene expression from a diverse set of human tissues including different brain regions. So far, 3797 tissues from 150 postmortem donors have been collected and subsequently analyzed using a RNA sequencing (RNA-seq)–based gene expression approach. (3) The Stanley Neuropathology Consortium Integrative Database (SNCID).26 The SNCID contains gene expression information for different brain regions (including hippocampal tissue and PFC) of 61 individuals, including 20 bipolar disorder patients, 20 schizophrenia cases, and 21 healthy controls. (4) The Human Brain Transcriptome (HBT) database.27 The HBT comprises the transcriptome of 16 regions: the cerebellar cortex, mediodorsal nucleus of the thalamus, striatum, amygdala, hippocampus, and 11 areas of the neocortex.
Replication and Refinement of GWAS Results
In order to replicate the GWAS results that contributed to our integrated results from the discovery step and in order to follow-up on newly identified eSNPs in the eQTL replication step, we utilized four well-characterized GWAS datasets:
(1) Independent Samples From the German MooDS SCZ Working Group.28
This sample contained 1332 schizophrenia cases and 866 controls. It represents the nonoverlapping part (PGC, “SGENE-Bonn”) of the MooDS SCZ Consortium sample that has been described in detail in study of Priebe et al.28 Thus, there is no overlap between individuals from our German replication sample and subjects in the discovery step from the Ripke et al study (PGC1 + SWE).11 The study was approved by the ethics committees of all study centers and written informed consent was obtained from all participants prior to inclusion. All patients were recruited from consecutive admissions to psychiatric inpatient units and of German descent according to self-reported ancestry. Venous blood samples were obtained from all participants and genotyping was performed using Illumina’s BeadChips, including HumanHap550v3, Human610-Quadv1, and Human660W-Quad. More detailed information on sample description, genotyping, quality control of controls utilized in this study can be found in the paper of Muglia et al.29
(2) Independent Family–Based Replication Sample From a Recent Study.30
This sample consisted of 6298 individuals (including 3286 schizophrenia cases) from 1811 nuclear families. There were three independent family–based samples included in this family-based replication sample, the European family–based sample (2740 individuals, 794 families, and 1420 schizophrenia cases), the Asian family–based sample (2296 individuals, 579 families, and 1222 schizophrenia cases), and the African family–based sample (1262 individuals, 438 families, and 644 schizophrenia cases). Because three subsamples included in this family-based samples, genetic association analysis in nuclear families was performed within each ancestral group using UNPHASED program31 to minimize the risk of population stratification effects. The results were then combined to obtain an overall P value. All of the individuals from this family-based sample do not overlap with the subjects from the discovery step of Ripke et al (PGC1 + SWE sample11). More detailed information about sample description (recruitment of schizophrenic patient and healthy controls, diagnosis), genotyping, quality control, population stratification analysis, and statistical analysis can be found in the original paper.30
(3) Two Independent Danish GWAS Datasets.32
This sample composed of two datasets with a total of 1735 schizophrenia cases (876 and 859, respectively) and 1749 controls (871 and 878, respectively). There is no overlap between individuals from our Danish replication sample and subjects in the discovery step from the Ripke et al study (“PGC1 + SWE”11). A detailed description of the Danish study samples is provided elsewhere.32 In brief, the samples used in this study here comprised the discovery step of a Danish schizophrenia GWAS32 and a larger part of its initial replication samples that was in the meantime genotyped using Illumina’s HumanCoreExome BeadChip. All involved samples were initially employed in two matched case-control designs (see above) and identified through the Danish Psychiatric Central Register.33 The DNA was accessed through the Danish Newborn Screening Biobank34 and was extracted from dried blood spots using Extract-N-Amp Blood PCR kit (Sigma Aldrich, Seelze, Germany) and subsequently whole genome-amplified in triplicates using the RepliG kit (Qiagen, Venlo, The Netherlands).35 Subsequent genotyping for the first sample (discovery) was performed using Illumina’s Human 610-quad beadchips (Illumina) and the second sample as described above. Diagnosis of all schizophrenia cases was based on ICD-10-DCR (The ICD-10 Classification of Mental and Behavioural Disorders Diagnostic Criteria for Research; F20). Summary statistics of all four studies (comprising a total of 6353 schizophrenia patients) were meta-analyzed using PLINK.22 There is no overlap between individuals from our replication samples and subjects in the PGC1 + SWE.
Pleiotropic Effects Analyses of the Identified Risk Loci
The National Human Genome Research Institute’s (NHGRI) GWAS Catalog36,37 comprises a curated list of results from all GWAS in humans to date (as of April 16, 2014, 1902 publications and 13156 SNPs are listed) and therefore provides a valuable resource to study pleiotropic effects. We first explored whether ZNF323 and its eSNPs were associated with other traits. We next investigated whether the observed associations of eSNPS of ZNF323 with schizophrenia and lung function could have been the result of confounding. More detailed information about the pleiotropy analyses can be found in supplementary methods.
Bioinformatic Analysis and Functional Prediction of rs1150711
We examined the potential chromatin interactions between the genomic region containing rs1150711 and its neighboring regions using the Hi-C chromatin interactions (from the study of Dixon et al).38 In addition, we performed bioinformatic analyses to predict the potential functional consequences of rs1150711 using Regulome DB (http://www.regulomedb.org).39 Multiple types of data (eg, ChIP-seq, DNase-seq, and eQTLs) from the Encyclopedia of DNA Elements (ENCODE) project40 and other sources were integrated into the Regulome DB to estimate the possible function of candidate SNPs.
Population Genetic Analyses
We utilized different approaches to investigate whether our top eSNPs and/or ZNF323 underwent recent positive selection in human populations. (1) Analysis of extended haplotype homozygosity (EHH): Haplotypes with unusually high EHH and high population frequency suggest recent positive selection that has driven rapid increase in frequency of new variants or haplotypes in the population.41 We used Sweep,41 the rehh package in R42 and data from the HapMap project43 and the 1000 Genomes project44 to estimate the EHH of the region surrounding the eSNP of ZNF323. (2) Calculation of integrated haplotype score (iHS): The iHS derived from EHH45 represents an indicator of recent positive selection. Large negative or positive values indicate unusually long haplotypes (that carry the derived or the ancestral allele), whereas small values of iHS indicates no obvious recent positive selection. An |iHS| > 2.0 indicates a significant (corresponding to P < .05) positive selection signal.45 We used Haplotter45 and data from the HapMap project43 and the 1000 Genomes project44 to calculate iHS. (3) The Composite of Multiple Signals (CMS) test 46: The CMS integrates information from both EHH and iHS (together with Fst and two additional frequency-spectra–based tests) and is thought to have greater power compared with the two aforementioned tests. (4) Level of allelic differentiation in different populations (Fst): Alleles, which experienced positive selection in a specific population or geographic region, have larger allelic differentiation (frequency differences) between populations compared with neutrally evolved alleles. We compared allele frequencies of our eSNPs in global populations by using genotype information from the Human Genome Diversity Project (HGDP) selection browser (http://hgdp.uchicago.edu/cgi-bin/gbrowse/HGDP/),47 which contains the allele frequency data of 53 worldwide populations. To perform neutrality tests (including Tajima’s D,48 Fu and Li’s D49 tests, and Fay and Wu’s H test50), we obtained sequencing data (including 99 Europeans, 103 Han Chinese, and 108 Africans) encompassing the entire ZNF323 gene (chr6:28,292,515-28,304,152, hg19) from the 1000 Human Genomes Project.44 Neutrality tests were carried out using DnaSP software (version 5).51 Fst (a measure of population differentiation) of rs1150711 was calculated using the method described in study of Cheng et al.52 For clarity, a workflow diagram illustrating these analyses is provided in supplementary figure S1.
Results
Sherlock Integrative Analyses (Discovery Step) Identified ZNF323 as a Novel Schizophrenia Risk Gene
Through systematic integration of brain eQTL18 and SNP associations (a total of 9898078 SNPs) from a large GWAS of schizophrenia (PGC1 + SWE, including 13833 schizophrenia cases and 18310 controls)11 using Sherlock 17 (see “Methods” section and supplementary figure S1), ZNF323 (6p22.1) showed the most significant association with schizophrenia (LBF = 7.09, P sher = 2.2×10–6, FDR = 3.69×10–3) (table 1). One eSNP of ZNF323 (rs2859365) showed significant association with ZNF323 expression (P eQTL = 5.90×10–8) and suggestive evidence for association with schizophrenia (P GWAS = 2.07×10–7) (table 2). Homozygous carriers of the risk allele (A) of rs2859365 showed a reduced expression of ZNF323. Results for all genes with LBF > 5.0 and P-value < 10–4 are listed in table 1. Supporting SNPs for top predicted genes for schizophrenia are listed in supplementary table S1.
Table 1.
Integrative Analysis (Sherlock) of Schizophrenia GWAS and Brain eQTL Reveals That ZNF323 Had the Most Significant Association With Schizophrenia
| Gene symbol | LBFa | P-valueb | Supporting SNPc (cis or trans) | PeQTL d | P GWAS e | FDRf |
|---|---|---|---|---|---|---|
| ZNF323 | 7.09 | 2.22×10–6 | rs2859365 (cis) | 5.90×10–8 | 2.07×10–7 | 3.69×10–3 |
| NAPSA | 7.04 | 2.22×10–6 | rs10226475 (trans) | 6.74×10–6 | 2.02×10−10 | 3.69×10–3 |
| LOC378135 | 6.76 | 2.22×10–6 | rs134169 (trans) | 6.17×10–6 | 2.50×10–5 | 3.69×10–3 |
| EPB41L2 | 6.66 | 2.22×10–6 | rs7752195 (trans) | 1.17×10–6 | 1.82×10–5 | 3.69×10–3 |
| GLT8D1 | 5.83 | 4.43×10–6 | rs2240920 (cis) | 5.22×10–16 | 7.30×10–6 | 7.35×10–3 |
| RCN1 | 5.83 | 4.43×10–6 | rs3800917 (trans) | 6.56×10–7 | 6.27×10−10 | 7.35×10–3 |
| ALMS1 | 5.56 | 1.11×10–5 | rs6753344 (cis) | 1.37×10–9 | 5.00×10–5 | 1.84×10–2 |
| EIF4A2 | 5.39 | 1.11×10–5 | rs17002034 (trans) | 4.00×10–7 | 1.40×10–6 | 1.84×10–2 |
| MPV17L2 | 5.30 | 1.11×10–5 | rs7752195 (trans) | 1.54×10–6 | 1.82×10–5 | 1.84×10–2 |
| LSAMP | 5.13 | 1.11×10–5 | rs4976976 (trans) | 4.53×10–7 | 6.23×10–6 | 1.84×10–2 |
| CSNK2B | 5.11 | 1.11×10–5 | rs707939 (cis) | 5.35×10–6 | 8.23×10–5 | 1.84×10–2 |
| PLCD4 | 5.09 | 1.55×10–5 | rs2160567 (trans) | 7.40×10–7 | 8.29×10–6 | 2.57×10–2 |
aLBF (Log Bayes Factors for each gene) evaluates whether the combined evidence from GWAS and expression studies support a gene being associated with schizophrenia. For example, a LBF of 4.6 means that the alternative hypothesis (associated with Schizophrenia) is 100 times (exp[4.6] = 100) more likely than the null hypothesis (no association). High Bayes factors tend to correlate with low P-values. Several eSNPs may be associated with the expression of a gene. For each of these eSNP, if it is also associated with schizophrenia in GWAS, a positive score would be given; otherwise, a negative score. Association only in GWAS (ie, non-eSNPs) does not alter the score. The total score of a gene increases with the number of SNPs with combined evidence (SNPs that are associated with schizophrenia and expression). For each gene, Sherlock computes individual LBFs for each SNP pair in the alignment, and the sum of these constitutes the final LBF score for the gene.
b P-value from Sherlock integrative analysis.
cSNP with the highest LBF score.
d P-value for eQTL SNP from the gene expression study.18
e P-value for eQTL SNP from the GWAS of schizophrenia.11
fFDR was calculated at P = 10–5 threshold. It should be noted that some genes with different LBF scores may have the same P-values. This is because that Sherlock computes P-values by finite permutations. If the number of permutation is not extremely high, it is possible that two different values of LBF correspond to the same P-values (their rankings are the same). Genes with LBF > 5.0 and P-value < 10–4 are listed.
Table 2.
Multiple eSNPs of ZNF323 Are Significantly Associated With Schizophrenia in the PGC1 + SWE Sample (13833 Cases and 18310 Controls)
| Gene symbol | SNP ID | Chromosome: Position | eQTL P valuea | GWAS P valueb |
|---|---|---|---|---|
| ZNF323 | rs1150711 | 6:28208535 | 1.40×10–6 | 2.76×10–9 |
| rs2859365 | 6:28391465 | 1.60×10–7 | 2.07×10–7 | |
| rs853684 | 6:28294550 | 9.52×10–7 | 4.73×10–7 | |
| rs213240 | 6:28315875 | 2.89×10–6 | 5.40×10–7 | |
| rs1997660 | 6:28269663 | 9.99×10–6 | 6.23×10–8 | |
| rs853693 | 6:28282648 | 9.99×10–6 | 6.04×10–8 | |
| rs12214383 | 6:28223731 | 1.48×10–5 | 2.56×10–6 | |
| rs916403 | 6:28408258 | 4.63×10–5 | 3.24×10–6 | |
| rs9380069 | 6:28203300 | 4.76×10–5 | 2.31×10–7 | |
| rs9461446 | 6:28227633 | 5.77×10–6 | 0.78 | |
| rs9461448 | 6:28263721 | 3.87×10–5 | 0.83 |
Boldface letters were used to highlight the genetic variants that we are interested in.
aeQTL P value shows the association significance between this SNP and ZNF323 expression level in human brain (Brain eQTL data is from the study of Colantuoni et al24 ).
bGWAS P value represents the association significance between this SNP and schizophrenia (Based on association data from the PGC1 + SWE samples, included 13833 schizophrenia cases and 18310 controls11 ).
Replication and Refinement of eQTL and GWAS Results for ZNF323
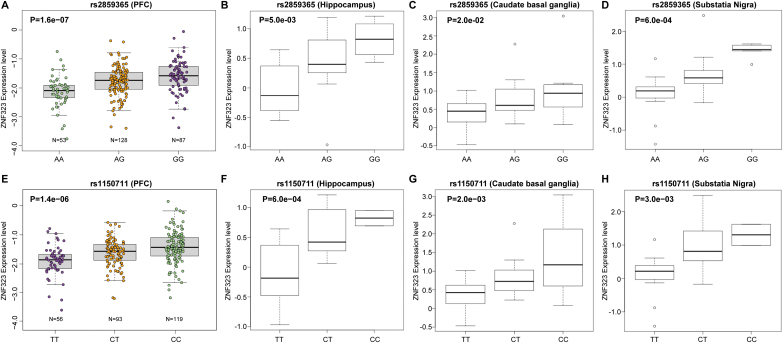
To replicate our results, we tested the association between rs2859365 and expression of ZNF323 in an independent gene expression dataset, the BrainCloud.24 The result confirmed the significant association of rs2859365 with ZNF323 expression in the PFC (P = 1.60×10–7) (figure 1A). Again, homozygous risk allele carriers have a reduced ZNF323 expression compared with the nonrisk allele individuals (figure 1A).
Fig. 1.
rs2859365 and rs1150711 are significantly associated with ZNF323 expression in human prefrontal cortex (PFC). (A) rs2859365 is significantly associated with the expression level of ZNF323 in the PFC of normal human subjects (n = 268) across the lifespan (P = 1.60×10–7). The individuals with AA genotype have lower ZNF323 expression level. (B–D) rs2859365 is also significantly associated with the ZNF323 expression in the hippocampus (B), the caudate basal ganglia (C), and the substatia nigra (D). (E) rs1150711 is significantly associated with the expression level of ZNF323 in the PFC of normal human subjects (n = 268) across the lifespan (P = 1.40×10–6). Individuals carrying the risk rs1150711 TT genotype (risk allele: T allele) have significant lower ZNF323 expression level than CC genotype carriers. (F–H) rs1150711 is also significantly associated with the ZNF323 expression in the hippocampus (F), the caudate basal ganglia (G), and the substatia nigra (H).
We further tested whether rs2859365 is associated with ZNF323 expression in brain regions other than the PFC in a third independent expression dataset (the GTEx).25 We found that rs2859365 showed significant association with ZNF323 expression in the hippocampus (P = 5.0×10–3) (figure 1B), the caudate basal ganglia (P = 2.0×10–2) (figure 1C), and the substantia nigra (P = 6.0×10–4) (figure 1D). Consistent with the findings from the PFC, we found that homozygous risk allele carriers for rs2859365 did show a reduced expression of ZNF323 in the hippocampus, caudate basal ganglia and the substantia nigra (figures 1B–D).
Similar to our validations on the eQTL results, we also attempted to replicate the GWAS signal (PGC1 + SWE study) for rs2859365 (P = 2.07×10–7) in independent schizophrenia datasets. Meta-analysis of six independent datasets comprising 6353 schizophrenia patients and 5618 controls yielded a P-value of .111 (overall combined P-value for PGC1 + SWE and the replication was 1.40×10–7) with the same effect allele (A) and effect direction as in the PGC1 + SWE study (table 3).
Table 3.
Discovery and Replication Results of rs1150711 and rs2859365 in Schizophrenia Samples Comprising a Total of 44123 Individuals
| SNP ID | Sample (cases) | Allelea | ORb | P valuec | |
|---|---|---|---|---|---|
| EA | OA | ||||
| rs2859365 | European sample (1420) | A | G | 1.030 | 0.659 |
| Asian sample (1222) | A | G | 1.077 | 0.266 | |
| African sample (644) | A | G | 1.008 | 0.941 | |
| German sample (1332) | A | G | 1.155 | 0.025 | |
| Danish sample 1 (876) | A | G | 0.964 | 0.609 | |
| Danish sample 2 (859) | A | G | 1.027 | 0.702 | |
| Combined replication (6353) | A | G | 1.047 | 0.111 | |
| PGC1 + SWE (13833) | A | G | 1.095 | 2.07×10–7 | |
| Overall combined (20 186) | A | G | 1.082 | 1.40×10–7 | |
| rs1150711 | European sample (1420) | T | C | 1.109 | 0.140 |
| Asian sample (1222) | T | C | 1.057 | 0.413 | |
| African sample (644) | T | C | 1.134 | 0.329 | |
| German sample (1332) | T | C | 1.209 | 0.007 | |
| Danish sample 1 (876) | T | C | 0.920 | 0.289 | |
| Danish sample 2 (859) | T | C | 1.009 | 0.908 | |
| Combined replication (6353) | T | C | 1.068 | 0.037 | |
| PGC1 + SWE (13833) | T | C | 1.119 | 2.76×10–9 | |
| Overall combined (20 186) | T | C | 1.105 | 6.85×10−10 | |
Boldface letters and italics were used to highlight the genetic variants that we are interested in.
aEA: effect allele, OA: other allele.
bOR (odds ratio) is based on effect allele.
cTwo-tailed P values. Combined analyses are based on a fixed effects meta-analysis of individual study results.
Evidence for Additional eSNPs of ZNF323
Considering that the expression of ZNF323 is significantly associated with schizophrenia, we hypothesized that other eSNPs of ZNF323 might also be associated with schizophrenia. We therefore identified the other eSNPs of ZNF323 by using BrainCloud.24 Only eSNPs with a P-value less than 1.0×10–4 were considered. In total, we identified 11 SNPs that showed an association with ZNF323 expression at the aforementioned P-value threshold (table 2). We further investigated the linkage disequilibrium (LD) pattern among the 11 eSNPs in Europeans based on genotype data from the 1000 Genomes project.44 Our analysis revealed two LD blocks (supplementary figure S2) with the initial lead eSNP rs2859365 (that was identified through the integrated analysis) located in haplotype block 2. Among the eSNPs in haplotype block 1, rs1150711 showed association with ZNF323 expression in the BrainCloud dataset (P = 1.40×10–6) (figure 1E) and reached genome-wide significance in the PGC1 + SWE GWAS data (P = 2.76×10–9) (table 2). Further investigation of rs1150711 in the GTEx expression dataset revealed that rs1150711 is significantly associated with ZNF323 expression in the hippocampus (P = 6.0×10–4), the caudate basal ganglia (P = 2.0×10–3), and the substantia nigra (P = 3.0×10–3) (figures 1F–H). Consistent with results of rs2859365, homozygous risk allele (T) carriers have a reduced ZNF323 expression compared with the nonrisk allele individuals (figures 1E–H). Additional support was gained from meta-analysis of our independent replication samples for rs1150711 (P = 0.037; overall combined P-value for PGC1 + SWE and the replication was 6.85×10−10) (table 3). To explore if rs1150711 is a cis or trans eQTL of ZNF323, we examined the genomic location of rs1150711 and ZNF323 and found that rs1150711 is located in ~84-kb downstream of ZNF323. As rs1150711 and ZNF323 are located closely on same chromosome, according to the criteria described in previous papers,17,53–55 rs1150711 is a cis eQTL of ZNF323.
SNP rs1150711 is located in the MHC region, considering the high LD among genes in this region, it is possible that the association between rs1150711 and schizophrenia is attributed to another causal variant that is highly linked with rs1150711. To test if there are SNPs that are in high linkage disequilibrium with rs1150711, we conducted LD analysis using genotype data from the 1000 Genomes project.44 LD analysis revealed that 7 SNPs are highly linked (r 2 > 0.80) with rs1150711 (supplementary figure S3 and supplementary table S2). We investigated the association between these SNPs and schizophrenia and found that rs1150711 showed the most significant association with schizophrenia (supplementary figure S3 and supplementary table S2). Collectively, these results suggest that SNP rs1150711 may represent an independent schizophrenia-associated variant. However, given the degree of LD in this region (there are many SNPs showed moderate LD (r 2 > 0.6) with rs1150711), we could not completely rule out the potential effect of synthetic association.
Pleiotropic Analyses for rs1150711
Our above data showed a significant association between rs1150711 and ZNF323 expression in human brain tissues. To further explore if rs1150711 is also associated with expression of other genes adjacent to ZNF323, we investigated the pleiotropic effects of rs1150711 using BrainCloud24 and NCBI eQTL browser (which contains well-characterized brain eQTL dataset).56 Both of these two datasets used brain tissues from the frontal cortex, a region associated with schizophrenia.57,58 We found that rs1150711 showed the most significant association with ZNF323 expression in BrainCloud database. SNP rs1150711 is absent in the NCBI eQTL browser, we therefore queried rs2859365. Again, we found the only eQTL gene for rs2859365 is ZNF323 (P = 2.93×10–8). These results suggest that ZNF323 may represent the most possible biological candidate gene behind rs1150711 (or rs2859365).
Bioinformatic Analysis of rs1150711 and Functional Prediction
To explore the potential mechanisms underlying the significant association between rs1150711 and ZNF323 expression, we performed bioinformatic analyses. Recent studies showed that the chromatin interaction plays an important role in regulating gene expression.38,59 We therefore extracted the chromatin interaction data generated by Dixon et al (http://yuelab.org/hi-c/).38 We found there was chromatin interaction (topologically associated domains [TAD]) between the genomic region containing rs1150711 and its neighboring regions (supplementary figure S4). However, due to the fact that TAD spans about 700kb and there are many genes in this region, more work is needed to elucidate if rs1150711 can directly regulate the expression of ZNF323. We also utilized the ENCODE data to predict the potential functional consequences of rs1150711 using Regulome DB (http://www.regulomedb.org).39 We found that SNP rs1150711 have RegulomeDB score of 3a (supplementary figure S5), indicating that at least three lines of evidence support this SNP is functional. First, ChIP-seq data revealed that rs1150711 is located in a transcription factor (TF) binding region. TFs MAFK and RFX3 can bind to the genomic sequence containing rs1150711. Second, rs1150711 is located in a well-defined motif region (Evi-1) and nucleotide substitution at rs1150711 may affect TFs binding. Third, DNase-seq data showed that rs1150711 is located in DNase peaks. These results implied that rs1150711 may have potential functional role in regulating ZNF323 expression. However, more work is needed to test whether SNP rs1150711 is functional.
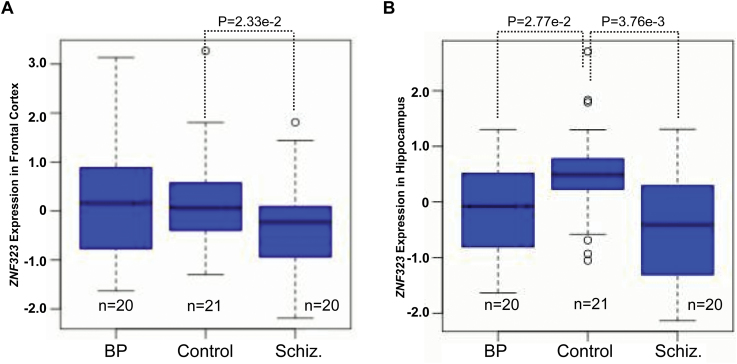
Significant Downregulation of ZNF323 in the Frontal Cortex and Hippocampus of Schizophrenia Patients
Sherlock integrative analysis revealed that the expression changes of ZNF323 may contribute to schizophrenia risk. To further validate this, we tested whether ZNF323 expression in the frontal cortex and/or hippocampus is dysregulated in schizophrenia patients using the SNCID data.26 We found that ZNF323 expression is significantly downregulated in schizophrenia patients compared with normal controls in both frontal cortex (P = .023) and hippocampus (P = .00376) (figure 2). Interestingly, we noticed that the T allele of rs1150711, which is associated with lower ZNF323 expression in PFC and hippocampus, is significantly enriched in schizophrenia cases (table 3). We further examined if other neighboring genes of rs1150711 are differentially expressed in schizophrenia cases and controls using the data from the SNCID.26 Genes located in a 100-kb window surrounding rs1150711 were examined. In addition to ZNF323, six genes (ZNF192, ZNF193, ZNF187, ZNF427, NKAPL, PGBD1) are located in the 100-kb window surrounding rs1150711. We found that these genes did not show differential expression in frontal cortex of schizophrenia cases and controls. Collectively, these findings provide further evidence that support the potential involvement of ZNF323 expression in schizophrenia susceptibility.
Fig. 2.
ZNF323 is significantly downregulated in the frontal cortex and hippocampus of schizophrenia patients. (A) Compared with healthy controls, ZNF323 expression was significantly downregulated in frontal cortex of schizophrenia patients. (B) Expression of ZNF323 was significantly decreased in hippocampus of schizophrenia cases. BP, patients with bipolar disorder, Schiz., patients with schizophrenia.
Pleiotropic Analyses for ZNF323
We explored whether ZNF323 is also associated with other human traits. A query of the NHGRI GWAS catalog (accession date April 01, 2014) for ZNF323 (or its alias names) revealed that the only entry listed was a GWAS for lung function.60 The trait(s) analyzed in this GWAS were the ratio of “forced expiratory volume in 1 second (FEV1)” to “forced vital capacity (FVC)” as well as FEV1. The strongest association for a SNP in ZNF323 in the aforementioned GWAS was rs6903823 (FEV1/FVC: P = .001 for joint meta-analysis of all stages; FEV1: P = 2.18×10−10) (supplementary table S3). Analyses of genetic correlation between rs6903823 and rs2859365/rs1150711 based on 1000 Genomes data (phase 1)44 in CEU-revealed moderate LD (r 2: 0.480 and 0.448, respectively; D′: 0.871 and 1.000, respectively). We observed evidence for association of rs1150711 in the aforementioned lung function GWAS (FEV1: P = 6.62×10–5) (supplementary table S3). The association for rs6903823 in the PGC1+SWE schizophrenia GWAS was significant (P = 2.29×10−10). It is of note that in both SNPs (rs1150711 and rs6903823) the risk allele for schizophrenia (T and A, respectively) shows a protective effect in the lung function GWAS60 (supplementary table S3).
Encouraged by this finding and the observation that ZNF323 is expressed in the lung, we tested whether our eSNPs for ZNF323 in the brain also showed association with ZNF323 expression in the lung. Based on 119 individuals included in the GTEx dataset,25 we found significant association for both rs2859365 and rs1150711 with ZNF323 expression in the lung (P = .008 and P = 9×10–5) (supplementary figure S6). Furthermore, we observed that for both markers the carriers of the schizophrenia risk alleles (rs2859365:A; rs1150711:T), ie, carriers of the allele that confers protective effects to lung function, did show a reduced expression of ZNF323 (in brain and lung) (supplementary figure S6).
Evidence for simultaneous association of genetic variation with different traits (pleiotropy) is sometimes based on confounding factors (hidden factors associated with the genetic variation and at the same time with the seemingly pleiotropic traits). Utilizing results from the TAG GWAS on smoking behavior (see “Methods” section of this manuscript for the analysis rationale), we found no evidence for association of our top eSNPs with smoking behavior as a potential confounding factor (for rs2859365 and rs1150711, all corrected P-value > .05; supplementary table S4).
Population Genetics Analyses for rs1150711
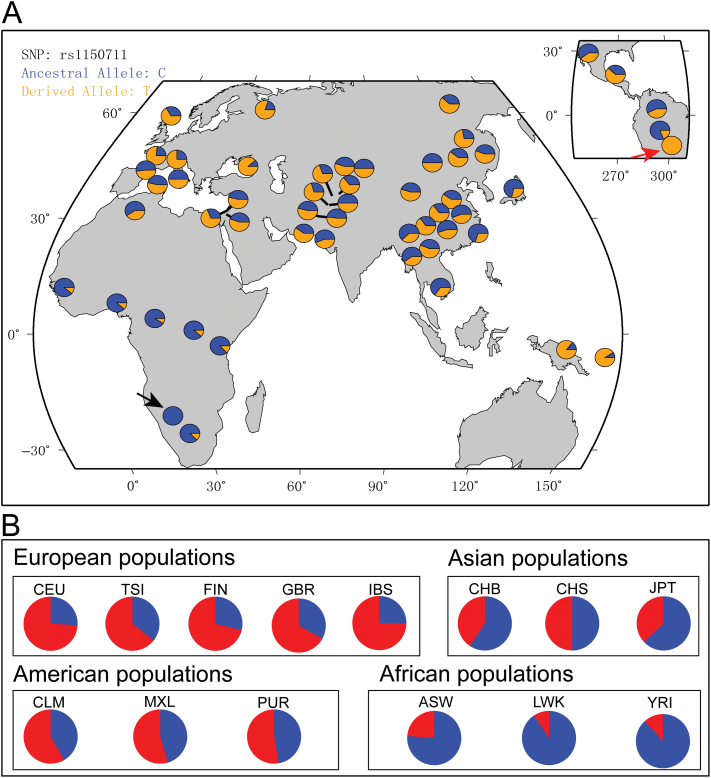
Based on our findings for schizophrenia and lung function, we investigated the evolutionary pattern of rs1150711 and ZNF323 in human populations using population genetic analyses. We first detailed the distribution of rs1150711 allele frequency in worldwide populations using data from the HGDP47 and the 1000 Genomes project.44 We found that the frequency of the T allele of rs1150711 (derived and risk allele) is highly variable in world-wide populations ranging from lower frequencies in most African populations to higher frequencies in other populations (figure 3). At the extremes we found that rs1150711 is fixed (either for the ancestral or the derived allele) in some populations (figure 3). Fst analysis also indicates a high degree of population differentiation in African, European, and Asian populations for rs1150711 (Fst = 0.44).
Fig. 3.
Global distribution of the rs1150711 risk allele (T allele) in world populations. (A) Global distribution of the risk allele (T allele) of rs1150711 in world populations. The frequency of the risk allele is highly variable in world populations. For example, it is absent in San population from Africa (black arrow), nevertheless, the risk allele (T allele) is completely fixed (frequency = 1.0) in Surui population from Brazil (red arrow). (B) Allele frequency distribution of the risk allele (T allele) of rs1150711 in four continental populations from the 1000 genomes projects. The frequency of the risk allele of rs1150711 is low in African populations. However, the risk allele is prevalent in other populations, including European, Asian, and American populations.
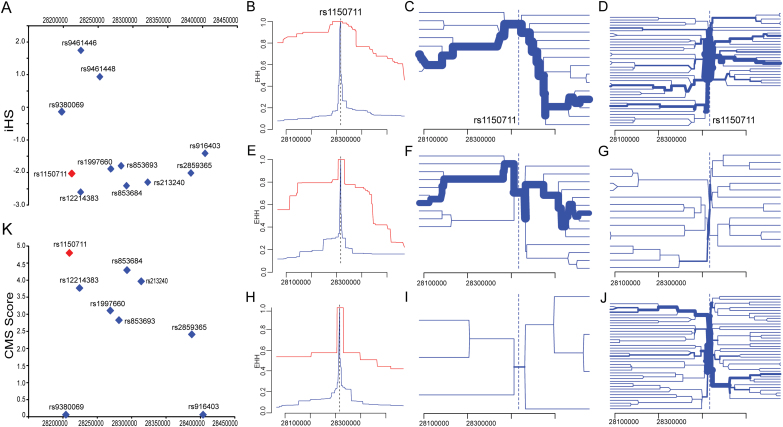
The dramatic differences in allelic frequency strongly suggested that rs1150711 might have experienced recent positive selection in human populations. To test this, we calculated the iHS45 of rs1150711 in world populations. The iHS analysis (figure 4A and supplementary table S5) revealed that the derived allele of rs1150711 (T) has experienced strong recent positive selection in Europeans (CEU; iHS = −2.034, P < .05) and Asians (iHS = −2.206, P < .05), while we did not observe evidence for significant positive selection in Africans (iHS = -0.617, P > .05). This finding was supported by the EHH41 analyses across all three studied populations, the haplotypes carrying the T allele (derived allele, is also the risk allele) of rs1150711 decay slower for the derived alleles than for the ancestral alleles (figures 4B, 4E, and 4H).
Fig. 4.
Recent positive selection of rs1150711 risk allele in human populations. (A) iHS value of the 11 single nucleotide polymorphisms (SNPs) that showed significant association with schizophrenia and ZNF323 expression in Europeans. (B, C, D) extended haplotype homozygosity (EHH) and haplotype bifurcation plots of rs1150711 in Han Chinese. (E, F, G) EHH and haplotype bifurcation plots of rs1150711 in Europeans. (H, I, J) EHH and haplotype bifurcation plots of rs1150711 in Africans. (B, E, H) Decay of EHH for the risk allele (T allele) of rs1150711 in three representative world populations (B, CHB, Han Chinese; E, CEU, Europeans; H, YRI, Africans) over physical distance. In each population, the decay of haplotype homozygosity for the risk allele (red) occurs much more slowly than for the protective allele (blue), indicating strong positive selection acting on haplotypes containing this risk allele. (C, D, F, G, I, J) Haplotype bifurcation graphs show that the bifurcation of haplotypes is also slower for the risk allele (T allele) of rs1150711 (C, F, I) compared with the protective allele (C allele) (D, G, J). In all of studied populations (CHB, CEU, and YRI), bifurcation of haplotypes for the risk allele (T allele) of rs1150711 is significantly lower than for the protective allele (C allele). The haplotype thicknesses represent the haplotype frequency. Haplotypes with unusually high EHH and a high population frequency (thicknesses) suggest the presence of recent positive selection that drives rapid increase in frequency of new variants or haplotypes in the population. Usual long and thick haplotypes are observed for the risk allele of rs1150711, but not for the protective allele, indicating recent positive selection of the risk allele. (K) Composite of multiple signals (CMS) score of the 11 SNPs that showed significant association with schizophrenia and ZNF323 expression in Europeans. rs1150711 (marked by red) has the highest CMS score, suggesting it is likely represents the variant that under positive selection. The CMS score of rs9461446 and rs9461448 was not available.
We further plotted haplotype bifurcation diagrams41 for both alleles at rs1150711 (figures 4C, 4D, 4F, 4G, 4 I, and 4J). Consistent with our EHH and iHS results, we found that the bifurcation of haplotypes containing the derived allele (T) was slower in contrast to haplotypes with the ancestral allele (C). Haplotypes carrying the derived allele (T) of rs1150711 have unusual high EHH and high population frequency, indicating that recent positive selection operated on the derived allele and as a result led to a faster increase in its frequency in the human population when compared with a model under neutral evolution. Further analyses using haplotter45 confirmed our findings (supplementary figure S7). Collectively, these results indicate that the derived allele of rs1150711 (T) has experienced recent positive selection in human populations.
We also explored the recent positive selection on rs2859365. Consistent with the results of rs1150711, iHS analysis revealed that the risk allele of rs2859365 (A allele) might have experienced significant recent positive selection in Europeans (iHS = −2.023, P < .05). Nevertheless, we did not observe significant recent positive selection in Chinese and African populations (iHS = −0.547 and −1.051, respectively, P > .05) (supplementary table S6 and supplementary figure S8). The results of EHH showed similar patterns (supplementary table S6 and supplementary figure S8). Tajima’s D, Fu and Li’s D and F tests, and Fay and Wu’s H test indicated that the evolution of ZNF323 did not deviate from neutrality significantly (supplementary table S7), which further suggesting that the positive selection on rs1150711 and ZNF323 is a relatively recent event.
It is conceivable that the positive selection of the derived allele of rs1150711 (T) in the human populations was driven by other SNPs in LD with rs1150711 (“genetic hitchhiking”). To test this hypothesis we utilized the CMS method46 and found that rs1150711 has the highest score among the 11 brain eSNPs for ZNF323 that were identified in the BrainCloud data (figure 4K), suggesting that rs1150711 is likely the site that drives the positive selection.
As our focus is ZNF323 in this study, therefore, we did not follow other top predicted genes. The reasons that we focused on ZNF323 are as follows: (1) Sherlock integration analysis revealed that ZNF323 showed the most significant association with schizophrenia (with the lowest LBF). (2) We noticed that among the top four predicted genes, only the supporting SNP (rs2859365) of ZNF323 is located near the ZNF323 gene (cis eQTL). Generally speaking, cis eQTL is more likely to be functional. Nevertheless, other genes are also deserved to be investigated in the future.
Discussion
Though genetic studies have revealed multiple promising risk variants that show robust association with schizophrenia, how these risk variants contribute to schizophrenia susceptibility remains largely unknown. Given that most of the identified risk variants are located in noncoding regions, it is likely that these risk variants alter the expression of schizophrenia-associated genes rather than protein function. Our study systematically integrated brain eQTL data18 and genetic association findings from a large GWAS of schizophrenia (PGC1 + SWE, with a total of 32143 subjects).11 Our findings revealed that ZNF323 is significantly associated with schizophrenia. We validated the association between eSNPs of ZNF323 (rs2859365 and rs1150711) at both the gene expression level and at the level of the association with schizophrenia. It should be noted that our replication results for association of rs2859365 and rs1150711 with schizophrenia are in line with the findings of a recently published study (“PGC2 SCZ”) that reported schizophrenia-association of 108 genetic loci.61 Although our own results unfortunately could only provide moderate further evidence for association, “PGC2 SCZ” put the evidence for association of rs2859365 and rs1150711 beyond reasonable doubt (1.68×10–11 and 2.62×10–13). In fact, the P values of rs2859365 and rs1150711 were steadily diminished with the increase of sample size (supplementary table S8), strongly suggesting that rs2859365 and rs1150711 are authentic risk variants for schizophrenia. Please note that all but one (“Danish2”) of the studies used for our analyses were included in the bigger “PGC2 SCZ” study. We also performed Sherlock integrative analysis using the PGC2 SCZ data set and found that ZNF323 still ranked in the top 5, which further suggests that ZNF323 may represent an authentic candidate gene.
It should be noted that the P value of rs2859365 did not reach a significant level and P value of rs1150711 showed marginally significant association (P = .037) with schizophrenia in the combined replication samples. Following two reasons may lead to these results: First, compared with other replication samples, we noticed that the Danish sample 1 has a different risk allele, which increased the P value when all of the replication samples were combined. In fact, when the Danish sample 1 was excluded, rs2859365 and rs1150711 showed significant associations with schizophrenia in the replication samples. Second, considering that the sample size of the combined replication samples is still relatively small (N = 6353 cases) and the association trend (the affect allele) in the combined replication samples is same as the discovery sample, we may obtain a more significant P value if the sample size is increased in the replication stage.
ZNF323 (also known as ZSCAN31) encodes SCAN/(Cys)2(His)2 zinc-finger TF that plays an active role in human embryonic development.62 Expression analysis revealed that ZNF323 is extensively expressed in human tissues, including lung, liver, kidney, brain, heart, and pancreas,62 suggesting that is may be involved in the development of multiple embryonic organs (including brain). Recent studies reveal the important role of zinc-finger TFs in schizophrenia. For example, ZNF804A, the first identified gene that showed genome-wide significant association with schizophrenia,63 also encodes a zinc finger protein. In addition to schizophrenia, significant association between ZNF804A and bipolar disorder also has been reported.64 These lines of evidence suggest that zinc-finger TF may play a pivotal role in psychiatric disorders.
ZNF323 is located on the MHC region. This region on chromosome 6 is known to harbor a large number of disease associated variants for traits such as mental disorders (eg, schizophrenia),10 traits related to immune processes (eg, rheumatoid arthritis),65 and many others.66,67 Due to the extended LD in this region, pinpointing the genes of interest remained a challenge in most of the studies. Our integrated analysis design potentially could help to identify more disease relevant genes in the MHC region. Although eQTL studies in general suffer from similar limitations that GWA studies do (large amount of correlated measurements), integrating several layers of gene expression with genetic data and other data might help to pinpoint disease associated genes. In our study only gene expression of ZNF323 was affected by all the 11 eSNPs in the brain (table 2). Moreover, we found evidence for a significantly reduced ZNF323 expression in brains of schizophrenia patients (in brain regions relevant to schizophrenia). More importantly, we noted that lower ZNF323 expression-associated alleles at rs2859365 and rs1150711 were enriched in schizophrenia cases (OR = 1.082 and 1.105, respectively). In addition to ZNF323, several other genes were also be identified by Sherlock (table 1). We noticed that EIF4A2 was reported to be downregulated in brains (anterior temporal lobe) of schizophrenia patients.68 In addition, genetic variants in LSAMP showed significant association with schizophrenia69 and several other mental disorders, including major depressive disorder and panic disorder,70 and male suicide.71 These lines of evidence strongly suggest the validity of Sherlock in identifying disease susceptibility genes.
Not only did we identify a new potential risk gene for schizophrenia but also we could find evidence for recent positive selection for rs1150711 and thus potentially provide a possible explanation to the evolutionary enigma of schizophrenia.4 Although in general this is of no surprise for genetic variation located in the MHC region,11 we were also able to link the evidence for recent positive selection with a reasonable candidate mechanism that could have provided compensatory advantage. Gene expression of ZNF323 in brain and lung is reduced as a consequence of the presence of the derived allele of rs1150711, which at the same time confers risk to schizophrenia and provides a protective effect on lung function. Considering that the protective allele on lung function (rs1150711-T) has experienced positive selection in Europeans, it is possible that lung function (measured as FEV1 and FCV) may show difference among different populations. Interestingly, one study showed that compared with European children, the lung function (FEV1 and FVC) of African and Asian children were significantly lower.72 These results suggest that lung function might have experienced positive selection in Europeans. It is of note that the protective effect on lung function is probably only one of multiple mechanisms that could have helped the risk allele of rs1150711 to escape extinction from the human gene pool (as a consequence of its involvement in schizophrenia susceptibility). Further work is warranted to investigate the mechanism underlying the expansion of the risk allele of rs1150711.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
X.J.L was supported by the 100 Talents Program (BaiRenJiHua) of the Chinese Academy of Sciences. Y.G.Y was supported by the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (XDB02020000). The research leading to these results has received funding from the European Community’s Seventh Framework Programme: FP7/Health-2011-1.1-2 under grant agreement n°279227 and Health-F4-2009–242257 to M.R. (ADAMS project). This study was also supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Genome Research Network (IG) MooDS (Systematic Investigation of the Molecular Causes of Major Mood Disorders and Schizophrenia; grant 01GS08144 to M.M.N. and S.C., grant 01GS08147 to M.R.), under the auspices of the National Genome Research Network plus (NGFNplus), and through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Programme. M.M.N. is a member of the DFG-funded Excellence-Cluster ImmunoSensation and also received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. We thank the grant support from National Institutes of Health grant (R01LM011177).
Supplementary Material
Acknowledgments
We thank the members of the SpiroMeta consortium and members of the CHARGE consortium for providing results for association of rs1150711 and rs6903823 with lung function. We also thank the members of schizophrenia working group in the Psychiatric Genomics Consortium for making their results publicly available. The authors report no financial relationships with commercial interests.
References
- 1. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. [DOI] [PubMed] [Google Scholar]
- 3. Power RA, Kyaga S, Uher R, et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry. 2013;70:22–30. [DOI] [PubMed] [Google Scholar]
- 4. Crespi B, Summers K, Dorus S. Adaptive evolution of genes underlying schizophrenia. Proc Biol Sci. 2007;274:2801–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pearlson GD, Folley BS. Schizophrenia, psychiatric genetics, and Darwinian psychiatry: an evolutionary framework. Schizophr Bull. 2008;34:722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huxley J, Mayr E, Osmond H, Hoffer A. Schizophrenia as a genetic morphism. Nature. 1964;204:220–221. [DOI] [PubMed] [Google Scholar]
- 7. Andreassen OA, Harbo HF, Wang Y, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2015; 20:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andreassen OA, Djurovic S, Thompson WK, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ripke S, Sanders AR, Kendler KS, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ripke S, O’Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bray NJ. Gene expression in the etiology of schizophrenia. Schizophr Bull. 2008;34:412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68; image 45. [DOI] [PubMed] [Google Scholar]
- 14. Bacanu SA, Chen J, Sun J, et al. Functional SNPs are enriched for schizophrenia association signals. Mol Psychiatry. 2014;19:276–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ayalew M, Le-Niculescu H, Levey DF, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He X, Fuller CK, Song Y, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. [DOI] [PubMed] [Google Scholar]
- 19. Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- 22. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Bakker PI, Ferreira MA, Jia X, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Colantuoni C, Lipska BK, Ye T, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S, Webster MJ. The stanley neuropathology consortium integrative database: a novel, web-based tool for exploring neuropathological markers in psychiatric disorders and the biological processes associated with abnormalities of those markers. Neuropsychopharmacology. 2009;35:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Priebe L, Degenhardt F, Strohmaier J, et al. Copy number variants in German patients with schizophrenia. PLoS One. 2013;8:e64035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muglia P, Tozzi F, Galwey NW, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. [DOI] [PubMed] [Google Scholar]
- 30. Aberg KA, Liu Y, Bukszár J, et al. A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiatry. 2013;70:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Børglum AD, Demontis D, Grove J, et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol Psychiatry. 2014;19:325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mors O, Perto GP, Mortensen PB. The Danish psychiatric central research Register. Scand J Public Health. 2011;39:54–57. [DOI] [PubMed] [Google Scholar]
- 34. Nørgaard-Pedersen B, Hougaard DM. Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Dis. 2007;30:530–536. [DOI] [PubMed] [Google Scholar]
- 35. Hollegaard MV, Grauholm J, Børglum A, et al. Genome-wide scans using archived neonatal dried blood spot samples. BMC Genomics. 2009;10:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hindorff L, MacArthur J, Morales J, et al. A Catalog of Published Genome-Wide Association Studies Available at: www.genome.gov/gwastudies.
- 37. Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. [DOI] [PubMed] [Google Scholar]
- 42. Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. [DOI] [PubMed] [Google Scholar]
- 43. International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 44. The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grossman SR, Shlyakhter I, Karlsson EK, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. [DOI] [PubMed] [Google Scholar]
- 47. Pickrell JK, Coop G, Novembre J, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. [DOI] [PubMed] [Google Scholar]
- 52. Cheng F, Chen W, Richards E, Deng L, Zeng C. SNP@Evolution: a hierarchical database of positive selection on the human genome. BMC Evol Biol. 2009;9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bryois J, Buil A, Evans DM, et al. Cis and trans effects of human genomic variants on gene expression. PLoS Genet. 2014;10:e1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wittkopp PJ. Genomic sources of regulatory variation in cis and in trans. Cell Mol Life Sci. 2005;62:1779–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H, Deng H. Systems genetics, bioinformatics and eQTL mapping. Genetica. 2010;138:915–924. [DOI] [PubMed] [Google Scholar]
- 56. Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Perlstein WM, Carter CS, Noll DC, Cohen JD. Relation of prefrontal cortex dysfunction to working memory and symptoms in schizophrenia. Am J Psychiatry. 2001;158:1105–1113. [DOI] [PubMed] [Google Scholar]
- 58. Knable MB, Weinberger DR. Dopamine, the prefrontal cortex and schizophrenia. J Psychopharmacol. 1997;11:123–131. [DOI] [PubMed] [Google Scholar]
- 59. Pope BD, Ryba T, Dileep V, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Soler Artigas M, Loth DW, Wain LV, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pi H, Li Y, Zhu C, et al. A novel human SCAN/(Cys)2(His)2 zinc-finger transcription factor ZNF323 in early human embryonic development. Biochem Biophys Res Commun. 2002;296:206–213. [DOI] [PubMed] [Google Scholar]
- 63. O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. [DOI] [PubMed] [Google Scholar]
- 64. Riley B, Thiselton D, Maher BS, et al. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Su Z, Gay LJ, Strange A, et al. Common variants at the MHC locus and at chromosome 16q24.1 predispose to Barrett’s esophagus. Nat Genet. 2012;44:1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu XQ, Li M, Zhang H, et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2012;44:178–182. [DOI] [PubMed] [Google Scholar]
- 68. Martins-de-Souza D, Gattaz WF, Schmitt A, et al. Alterations in oligodendrocyte proteins, calcium homeostasis and new potential markers in schizophrenia anterior temporal lobe are revealed by shotgun proteome analysis. J Neural Transm. 2009;116:275–289. [DOI] [PubMed] [Google Scholar]
- 69. Koido K, Janno S, Traks T, et al. Associations between polymorphisms of LSAMP gene and schizophrenia. Psychiatry Res. 2014;215:797–798. [DOI] [PubMed] [Google Scholar]
- 70. Koido K, Traks T, Balõtšev R, et al. Associations between LSAMP gene polymorphisms and major depressive disorder and panic disorder. Transl Psychiatry. 2012;2:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Must A, Tasa G, Lang A, et al. Association of limbic system-associated membrane protein (LSAMP) to male completed suicide. BMC Med Genet. 2008;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johnston ID, Bland JM, Anderson HR. Ethnic variation in respiratory morbidity and lung function in childhood. Thorax. 1987;42:542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.