Abstract
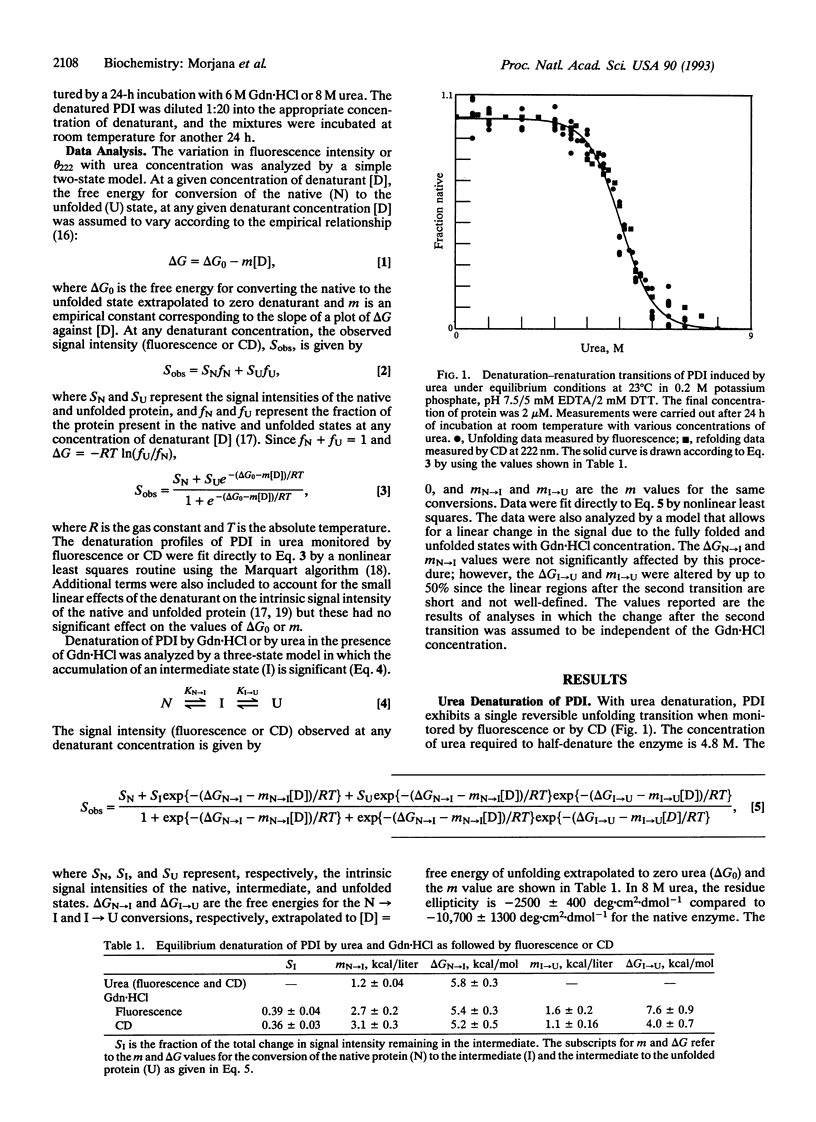
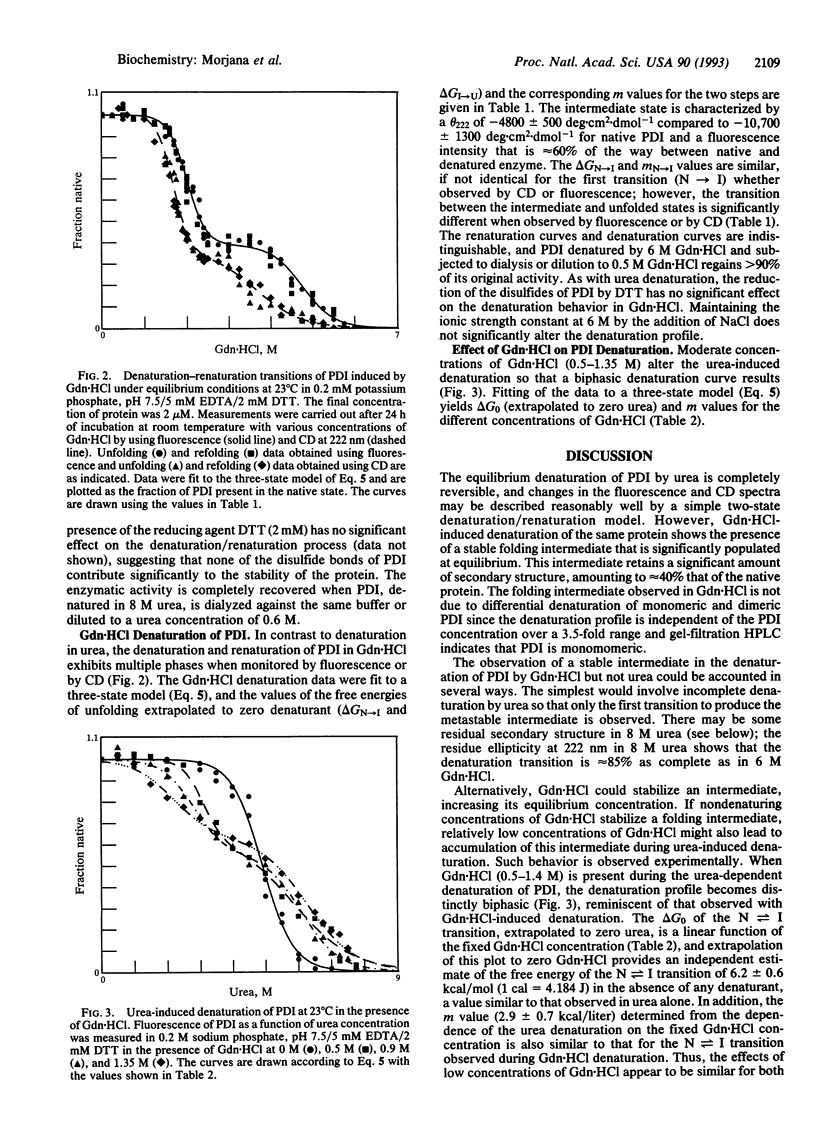
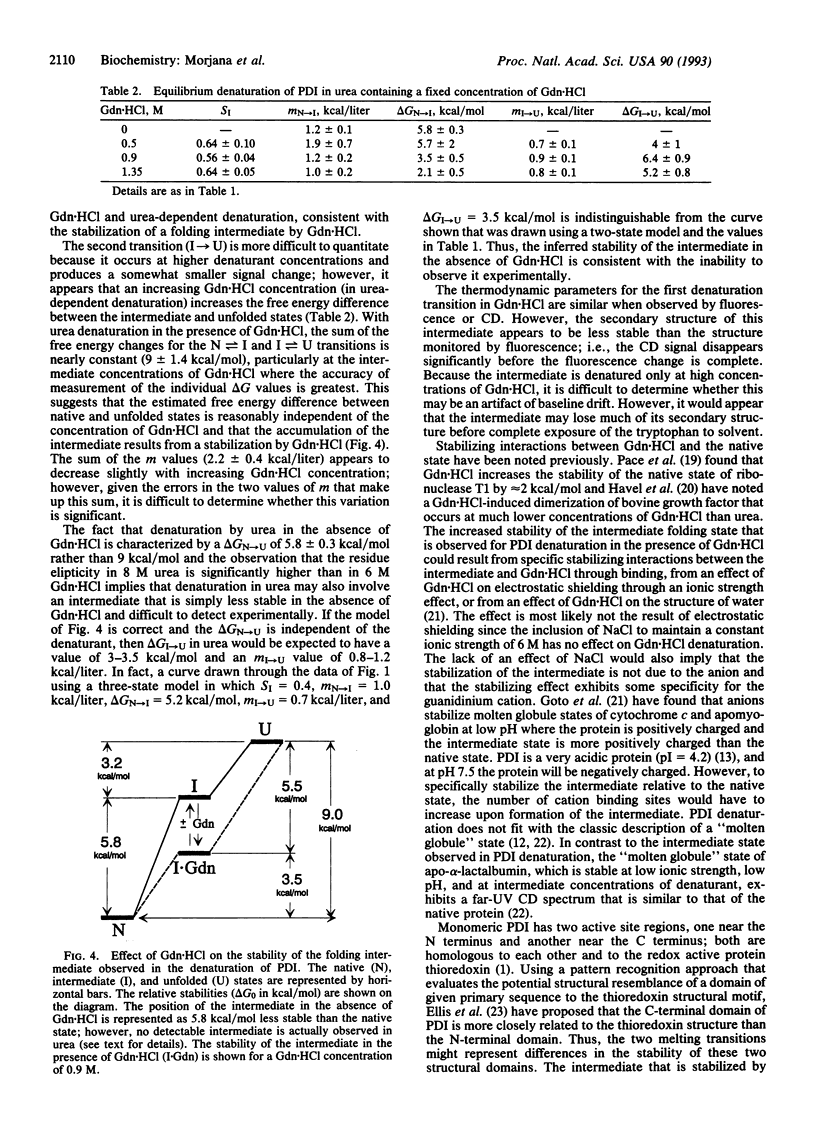
The reversible denaturation of protein disulfide isomerase proceeds through intermediates that are stabilized by interaction with guanidine hydrochloride. At pH 7.5, the equilibrium denaturation by urea is completely reversible and the transition can be reasonably well-described by a two-state model involving only native and denatured forms. In comparison, the equilibrium denaturation by guanidine hydrochloride occurs in two distinct steps. In the presence of a low constant amount of guanidine hydrochloride (0.5-1.4 M), urea denaturation also becomes biphasic, suggesting the accumulation of an intermediate species that is stabilized by specific interaction with guanidine hydrochloride but not by high concentrations of other salts or other denaturants.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Creighton T. E., Hillson D. A., Freedman R. B. Catalysis by protein-disulphide isomerase of the unfolding and refolding of proteins with disulphide bonds. J Mol Biol. 1980 Sep 5;142(1):43–62. doi: 10.1016/0022-2836(80)90205-3. [DOI] [PubMed] [Google Scholar]
- Edman J. C., Ellis L., Blacher R. W., Roth R. A., Rutter W. J. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985 Sep 19;317(6034):267–270. doi: 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- Ellis L. B., Saurugger P., Woodward C. Identification of the three-dimensional thioredoxin motif: related structure in the ORF3 protein of the Staphylococcus aureus mer operon. Biochemistry. 1992 May 26;31(20):4882–4891. doi: 10.1021/bi00135a020. [DOI] [PubMed] [Google Scholar]
- Goto Y., Takahashi N., Fink A. L. Mechanism of acid-induced folding of proteins. Biochemistry. 1990 Apr 10;29(14):3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- Havel H. A., Kauffman E. W., Plaisted S. M., Brems D. N. Reversible self-association of bovine growth hormone during equilibrium unfolding. Biochemistry. 1986 Oct 21;25(21):6533–6538. doi: 10.1021/bi00369a029. [DOI] [PubMed] [Google Scholar]
- Hillson D. A., Lambert N., Freedman R. B. Formation and isomerization of disulfide bonds in proteins: protein disulfide-isomerase. Methods Enzymol. 1984;107:281–294. doi: 10.1016/0076-6879(84)07018-x. [DOI] [PubMed] [Google Scholar]
- Hu C. H., Tsou C. L. C-terminal truncation of bovine protein disulfide isomerase increases its activity. Biochem Biophys Res Commun. 1992 Mar 16;183(2):714–718. doi: 10.1016/0006-291x(92)90541-r. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- Koivu J., Myllylä R. Protein disulfide-isomerase retains procollagen prolyl 4-hydroxylase structure in its native conformation. Biochemistry. 1986 Oct 7;25(20):5982–5986. doi: 10.1021/bi00368a022. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Lambert N., Freedman R. B. Kinetics and specificity of homogeneous protein disulphide-isomerase in protein disulphide isomerization and in thiol-protein-disulphide oxidoreduction. Biochem J. 1983 Jul 1;213(1):235–243. doi: 10.1042/bj2130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles M. M., Gilbert H. F. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991 Jan 22;30(3):613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- Morjana N. A., Gilbert H. F. Effect of protein and peptide inhibitors on the activity of protein disulfide isomerase. Biochemistry. 1991 May 21;30(20):4985–4990. doi: 10.1021/bi00234a021. [DOI] [PubMed] [Google Scholar]
- Pace C. N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V., Thomson J. A. pH dependence of the urea and guanidine hydrochloride denaturation of ribonuclease A and ribonuclease T1. Biochemistry. 1990 Mar 13;29(10):2564–2572. doi: 10.1021/bi00462a019. [DOI] [PubMed] [Google Scholar]
- Santoro M. M., Bolen D. W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988 Oct 18;27(21):8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Vuori K., Myllylä R., Pihlajaniemi T., Kivirikko K. I. Expression and site-directed mutagenesis of human protein disulfide isomerase in Escherichia coli. This multifunctional polypeptide has two independently acting catalytic sites for the isomerase activity. J Biol Chem. 1992 Apr 15;267(11):7211–7214. [PubMed] [Google Scholar]


