Significance
Fish bones from the 11,500-y-old Upward Sun River site in interior Alaska represent the oldest evidence for salmon fishing in North America. We used ancient DNA analysis to identify the fish specimens as chum salmon (Oncorhynchus keta), and stable isotope analysis to confirm that the salmon were anadromous (sea-run). The exploitation of salmon at this early date is noteworthy because Paleoindians are traditionally portrayed as big-game hunting specialists. Furthermore, the presence of salmon at Upward Sun River over 1,400 km upriver from the coast shows that spawning runs had been established by the end of the last Ice Age. The early availability and use of anadromous salmon has important implications for understanding Paleoindian economies and expansion into North America.
Keywords: salmon, Paleoindians, Beringia, ancient DNA, stable isotopes
Abstract
Salmon represented a critical resource for prehistoric foragers along the North Pacific Rim, and continue to be economically and culturally important; however, the origins of salmon exploitation remain unresolved. Here we report 11,500-y-old salmon associated with a cooking hearth and human burials from the Upward Sun River Site, near the modern extreme edge of salmon habitat in central Alaska. This represents the earliest known human use of salmon in North America. Ancient DNA analyses establish the species as Oncorhynchus keta (chum salmon), and stable isotope analyses indicate anadromy, suggesting that salmon runs were established by at least the terminal Pleistocene. The early use of this resource has important implications for Paleoindian land use, economy, and expansions into northwest North America.
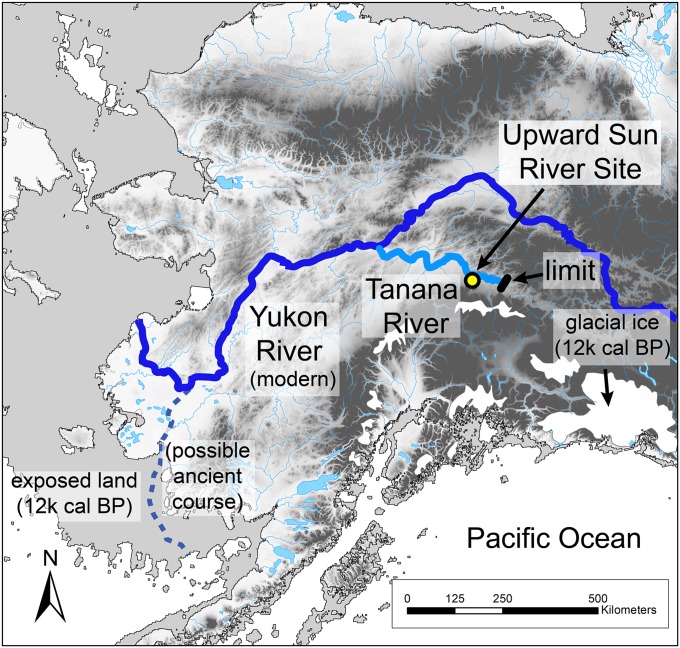
Each year along the Pacific coast of North America, millions of salmon migrate from the ocean to spawn and die in their natal rivers and lakes; however, during the last Ice Age, many of the rivers that today support salmon were blocked by glacial ice, severely restricting salmon ranges (1). A potential glacial refugium for salmon was Beringia, the mostly ice-free landmass that bridged northeast Asia and Alaska (1–3). Evidence for such a refugium comes from studies of present-day diversity and distributions of Pacific salmon (1, 2), but there is little direct evidence of the antiquity of salmon spawning runs in North America. Here we confirm the presence of an anadromous salmon species, Oncorhynchus keta (chum salmon) through ancient DNA (aDNA) and stable isotope analyses of fish remains at the Upward Sun River site located deep in the interior of Alaska, about 50 km downstream from the modern limit of major spawning areas (Fig. 1). These specimens, dating to the terminal Pleistocene, represent the oldest genetically confirmed Pacific salmon species in an archaeological context in North America. These data are important for testing competing models of subsistence strategies and diet breadths of Paleoindian populations in the New World (4, 5), as well as for understanding Beringian ecosystem biodiversity.
Fig. 1.
Location of Upward Sun River Site, course of the Tanana-Yukon River and possible course across the Bering Shelf during lower sea level, and modern chum salmon fall spawning limit along the Tanana River. Details are provided in SI Text.
Oncorhynchus is a salmonid genus that includes several Pacific salmon and Pacific trout species; some species of this genus occur as both freshwater resident and anadromous forms, migrating from the sea to freshwater to spawn (6). The spawning behavior of anadromous Pacific salmon results in massive and predictable runs in freshwater streams over a short period, making these fish a potentially valuable human food resource (7). Salmon are also ecologically important because they transport rich marine-derived nutrients into relatively unproductive interior riparian areas (8). Five species of Pacific salmon and one species of Pacific trout presently occur in central Alaskan waters, including Chinook (Oncorhynchus tshawytscha), coho (Oncorhynchus kisutch), chum (O. keta), sockeye (Oncorhynchus nerka), pink salmon (Oncorhynchus gorbuscha), and rainbow/steelhead trout (Oncorhynchus mykiss) (1).
Pleistocene-aged remains of Pacific salmon from North America are extremely rare in both paleontological and archaeological contexts. This scarcity is related in part to the spawning habitat of most salmon species, leading to death in river gravels where remains are unlikely to be preserved (9) and to the fragility of fish skeletal elements and their small size (inhibiting recovery), resulting in their underrepresentation in the archaeological record (10). Paleontological specimens of Pacific salmon have been recovered from middle Pleistocene sediments in the Skokomish Valley, Washington (United States) (9), and from late Pleistocene sediments at Kamloops Lake, British Columbia (Canada) (11). Although the remains from both of these locales were morphologically identified as O. nerka, carbon stable isotope analysis of specimens from Kamloops Lake suggests that the fish were likely the landlocked form of O. nerka, known as kokanee (11). Other late Pleistocene paleontological fish remains assigned to Pacific salmon derive from two additional sites in British Columbia, including Courtenay (Vancouver Island) and Gaadu Din 1 cave (Haida Gwaii) (12, 13). Specimens from the latter site were genetically identified as “salmon,” but the details of the aDNA analysis were not reported (13).
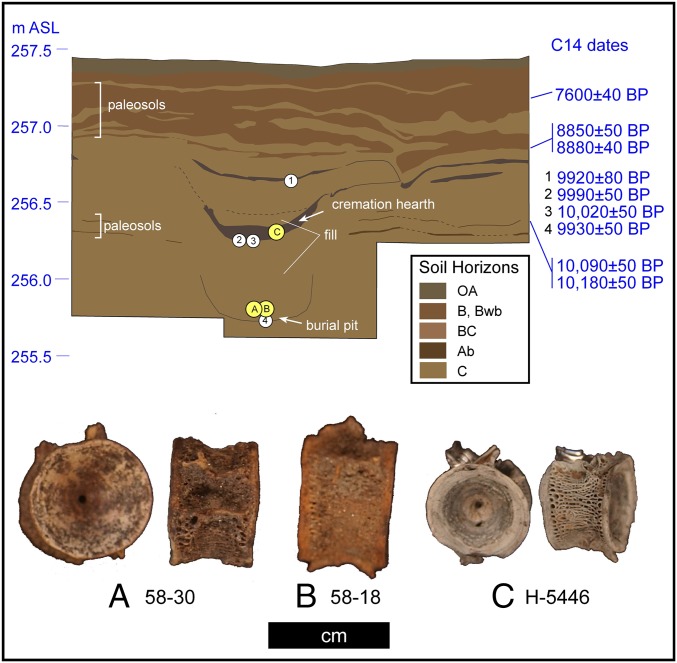
The only report of Oncorhynchus remains from a Pleistocene-age archaeological site in North America comes from Upward Sun River, located adjacent to the Tanana River (a major tributary of the Yukon River) in central Alaska (14) (see also SI Text) (Fig. 1). Here, 308 Oncorhynchus specimens were recovered from the central hearth of a residential feature, also associated with a cremated 3-y-old child (15). A double infant burial with associated grave goods was located directly below (40 cm) this hearth. Radiocarbon and contextual data suggest near contemporaneity between the hearth and the burial pit, with a mean pooled age of 9,970 ± 30 B.P. (11,600–11,270 cal B.P.); thus, these represent the oldest known human remains in the North American Arctic/Subarctic (Fig. 2 and SI Text). A total of 29 additional Oncorhynchus specimens were found within the pit fill. The fish remains were mostly fragmentary and over 90% were burned and calcined. The component and burials are culturally affiliated with the Denali Complex, which was widespread in Eastern Beringia from ∼12,700 cal B.P. to the early Holocene (3, 16).
Fig. 2.
Upward Sun River stratigraphy, chronology, and aDNA and stable isotope bone samples. Details provided in SI Text and Table S1.
The fish vertebrae were morphologically identified as Oncorhynchus using a modern comparative collection based on their large size, fenestration pattern, and characteristic shape (17) (Fig. 2). Based on overall size and occupation season, based on other fauna, the vertebrae resembled O. keta (Fig. S1 and Table S1); however, species-level identifications based on morphology remain ambiguous without genetic confirmation (18, 19). Species distinctions are critical to separate salmon from trout and other salmonids, because although some other members of this family are anadromous, they do not typically form the extensive and massive spawning runs that make salmon such an exceptional resource (6, 20). Additionally, salmon species differ in habitat requirements, run timing and abundance, and body size and fat content, all of which have implications for understanding past human land use and subsistence strategies (18, 19, 21, 22). aDNA analysis provides more accurate identifications of fish remains, and has recently been successfully applied to fish assemblages from Holocene archaeological sites in the Pacific Northwest of North America (18, 19, 23, 24).
Fig. S1.
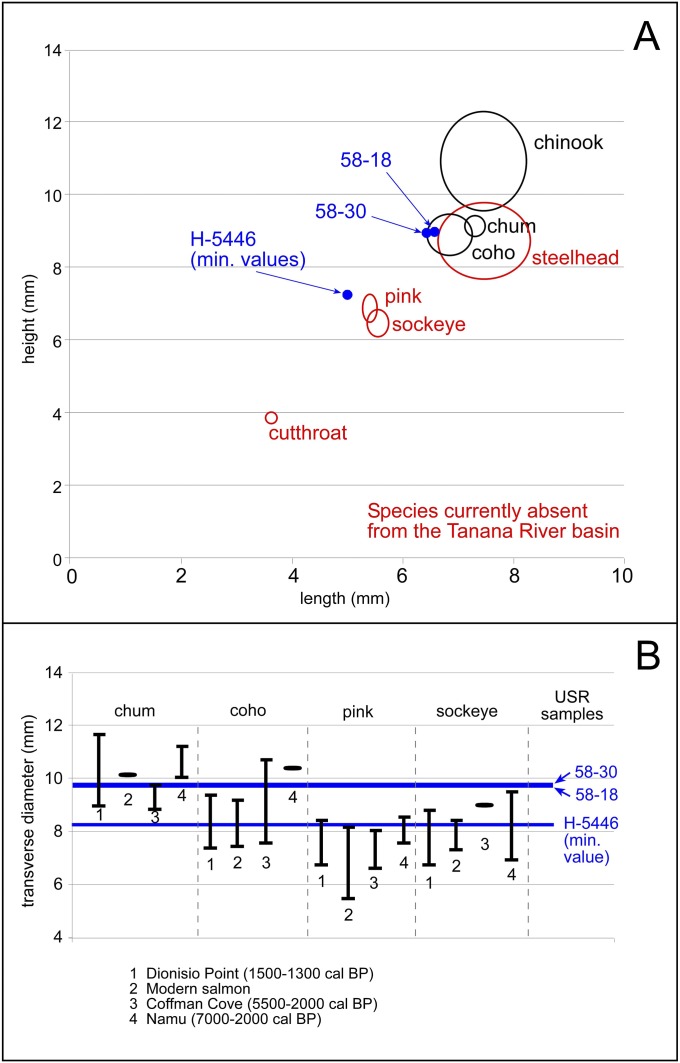
USR osteometric values compared with various genetically identified Oncorhynchus datasets. (A) Fused vertebra (type III) measurements [data from Huber et al. (21)]. (B) Vertebral transverse diameter measurements of genetically identified salmon species [summary data from Moss et al. (18); original data: “1” Grier et al. (19), “2” Orchard and Szpak (20), “3” Moss et al. (18), and “4” Yang et al. (22)].
Table S1.
USR fish vertebrae samples subjected to aDNA analysis
| ID | Location | Provenience | Element | Height (mm) | Length (mm) | Transverse diameter (mm) | Weight (g) |
| 58-18 | Burial pit fill | N501.578, E514.192, −1.226 SD | Caudal vertebra | 8.96 | 6.58 | 9.72 | 0.269 |
| 58-30 | Burial pit fill | N501.495, E514.262, −1.222 SD | Caudal vertebra | 8.95 | 6.44 | 9.71 | 0.242 |
| H-5446 | Cremation hearth | N501.710, E514.360, −0.771 SD | Caudal vertebra | 7.25* | 5.01* | 8.19* | 0.094* |
Minimum values due to burning.
Genetic identification cannot resolve questions about anadromy because salmon life history pattern is variable. Most Pacific salmon stocks are anadromous; however, natural or introduced freshwater stocks also occur for all species except chum, and captive chum have been reared to maturity in fresh water (25). Stable isotope analysis is an appropriate technique for distinguishing between anadromous and nonanadromous salmon because the isotope ratios of carbon (δ13C) and nitrogen (δ15N) are typically elevated in marine compared with freshwater food consumers (26). This technique has been successfully applied to identify life history patterns in modern salmonid stocks (27–30).
To address the questions of taxonomic identification and life history of the Upward Sun River Oncorhynchus specimens, we subjected two unburned vertebral specimens (58-18 and 58-30) to aDNA analysis, one of which (58-18) was also subjected to carbon and nitrogen stable isotope analysis (SI Text and Table S1). An unsuccessful attempt was also made to analyze DNA from a third specimen (H-5446), a nearly complete but calcined vertebra. The three specimens shared the distinctive morphological and overall size characteristics of the larger number of burned and fragmented Oncorhynchus specimens.
Results and Discussion
DNA was extracted from the specimens following established methods (SI Text). The extracts for the unburned specimens were successfully PCR-amplified and sequenced for a region of the mitochondrial 12S gene that is useful for discriminating among Pacific salmonids and other fishes (31) (SI Text). Repeated sequence analysis demonstrated the samples to be Oncorhynchus keta (chum salmon) (Table S2). Additionally, one sample (58-18) was analyzed at an independent aDNA laboratory using discriminating fragments of the d-loop and cytochrome b regions of the salmonid mitochondrial genome (22), confirming the identification made with 12S (SI Text, Figs. S2–S4, and Table S3). Thus, repeatable and independent analyses of mitochondrial DNA (mtDNA) demonstrate that both samples are chum salmon.
Table S2.
Sequencing results obtained at WSU
| Sample | Extract treatment | Sequence span (nps)* | Mutations* |
| 58-18 | None (1.5 μL) | 593–740 | 660T, 713T |
| Resilica extracted (1.5 μL) | 593–711 | 660T | |
| 1:10 dilution (1.5 μL) | 593–740 | 660T, 713T | |
| 1:50 dilution (1.5 μL) | 593–740 | 660T, 713T | |
| 58-30 | None, first amplification (1.5 μL) | 593–740 | 660T, 713T |
| None, second amplification (1.5 μL) | 593–740 | 660T, 713T | |
| H-5446 | None (1.5 μL) | No DNA | NA |
| None (3.0 μL) | No DNA | NA | |
| 1:10 dilution (1.5 μL) | No DNA | NA | |
| 1:50 dilution (1.5 μL) | No DNA | NA |
Relative to NCBI accession number DQ288271.
Fig. S2.
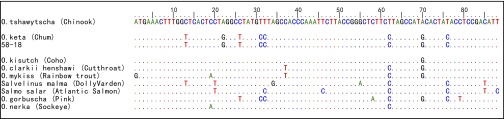
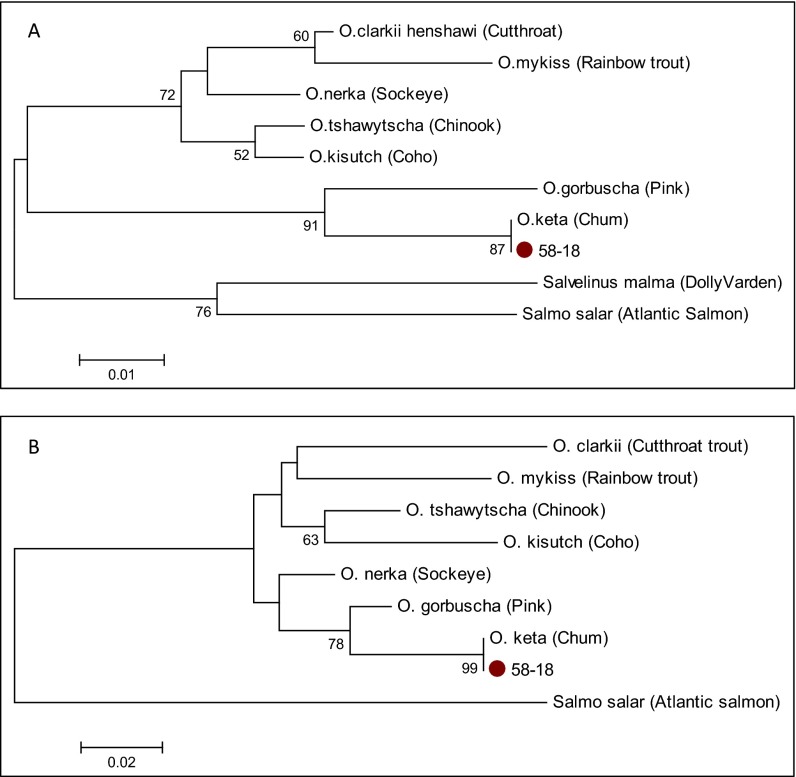
Multiple-alignment of ancient cytochrome b sequences from specimen 58-18 and GenBank reference sequences. The dots indicate identical base pairs compared with the Chinook salmon sequence at the top. Ancient sequences have been truncated to remove primer sequences. GenBank accession numbers: Oncorhynchus clarkii NC006897; Oncorhynchus mykiss NC001717; Oncorhynchus tshawytscha NC002980; Oncorhynchus kisutch NC009263; Oncorhynchus nerka NC008615; O. keta AJ314561; Oncorhynchus gorbuscha NC010959; Salvelinus malma DQ298801; Salmo salar NC001960.
Fig. S4.
Phylogenetic tree displaying the relationships between (A) the obtained cytochrome b haplotype from specimen 58-18 and GenBank reference sequences. GenBank subspecies accession numbers: O. clarkii NC006897; O. mykiss NC001717; O. tshawytscha NC002980; O. kisutch NC009263; O. nerka NC008615; O. keta AJ314561; O. gorbuscha NC010959; S. malma DQ298801; S. salar NC001960. (B) The obtained d-loop haplotype from specimen 58-18 and GenBank reference sequences. GenBank subspecies accession numbers: O. clarkii NC006897; O. mykiss NC001717; O. tshawytscha NC002980; O. kisutch NC009263; O. nerka NC008615; O. keta AJ31456; S. salar NC001960; O. gorbuscha EF455489.1. Trees (NJ with Kimura 2-parameter, 2,000 replications) were composed using Mega 6.0 software, Tamura et al. (60). Bootstrap values <50% not shown.
Table S3.
Primers used in the SFU Laboratory
| Primer | Target | Locus | Primer sequence (5′–3′) | Fragment size (bp) | Source |
| F271 | Bony fish (Osteichthyes) | COI | CYAYCYTACCTGTGGCMAT | 220 | (79) |
| R271 | ACTATAAAGAARATYATWACRAARGCRTG | ||||
| F800 | Salmon (Salmonidae) | d-loop | AACCCCTAAACCAGGAAGTCTCAA | 248 | (57) |
| R1048 | CGTCTTAACAGCTTCAGTGTTATGCT | ||||
| Cyt5 | Salmon (Salmonidae) | Cyt b | AAAATCGCTAATGACGCACTAGTCGA | 168 | (57) |
| Cyt6 | GCAGACAGAGGAAAAAGCTGTTGA |
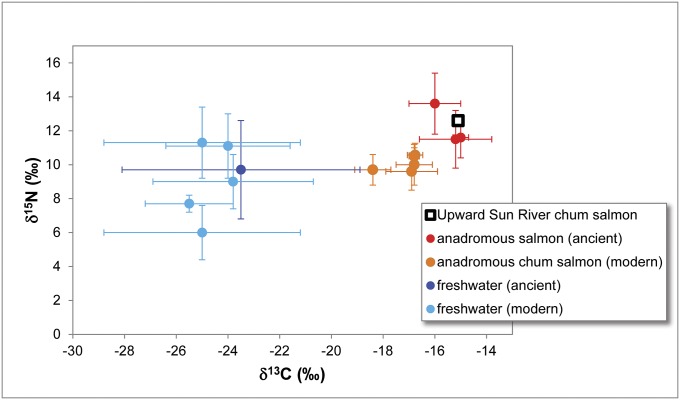
Stable isotope analysis of bone collagen extracted from one of the confirmed O. keta vertebra produced δ13C and δ15N values of −15.1‰ and 12.6‰, respectively (SI Text). These values are consistent with previously reported isotope values for ancient and modern salmon from Alaska, and are elevated over those for ancient and modern freshwater fishes (Fig. 3 and Tables S4 and S5). These data indicate that the Upward Sun River chum salmon specimen was anadromous.
Fig. 3.
Stable isotope values for the Upward Sun River sample compared with published values for ancient and modern anadromous and freshwater fishes in Alaska. Modern values have been adjusted for tissue offsets (muscle to bone collagen) and for the Suess effect (reduction of δ13C in carbon reservoirs from industrial carbon emissions). Details provided in SI Text and Tables S4 and S5.
Table S4.
Published δ13C and δ15N values on bone collagen from prehistoric archaeological Pacific salmon and freshwater fish specimens in Alaska
| Taxon | Alaskan region | Site | Age (cal B.P.) | n | δ13C (‰) | δ15N (‰) | Source |
| Pacific Salmon | |||||||
| Oncorhynchus keta | Tanana basin | Upward Sun River | ∼11,500 | 1 | −15.1 | 12.6 | Present study |
| Oncorhynchus spp. | Sanak, Aleutians | Multiple | ∼4500–150 | 91 | −15.2 ± 1.4 | 11.5 ± 1.7 | (69) |
| Oncorhynchus spp. | Unalaska, Aleutians | Amaknak | ∼3590–2570 | 5 | −15.0 ± 0.3 | 11.6 ± 1.2 | (68) |
| Salmonid | Kuskokwim Bay | Nunalleq | ∼610 | 2 | −16.0 ± 1.0 | 13.6 ± 1.8 | (67) |
| Freshwater | |||||||
| Lota lota (burbot) | Kuskokwim basin | Tochak | ∼430 | 2 | −23.5 ± 4.6 | 9.7 ± 2.9 | (70) |
Table S5.
Published δ13C and δ15N values on tissues from modern chum salmon and freshwater fish from central Alaska
| Taxon | Alaskan Region | n | Original tissue | Original δ13C (‰) | Original δ15N (‰) | Adjusted δ13C (‰)* | Adjusted δ15N (‰)* | Source |
| Oncorhynchus keta (chum) | Central | 4 | Muscle | −20.3 ± 0.2 | 11.5 ± 0.5 | −16.8 | 10.5 | (80) |
| South Central | 39 | Muscle† | −20.4 ± 1.0 | 10.6 ± 1.0 | −16.9 | 9.6 | (74) | |
| Southwest | 23 | Muscle‡ | −21.9 ± 0.7 | 10.7 ± 0.9 | −18.4 | 9.7 | (71) | |
| Southwest | 2 | Bone collagen | na | 11.6 ± 1.1 | – | 11.6 | (75) | |
| Southeast | 3 | Muscle | −20.3 ± 0.3 | 11.6 ± 0.7 | −16.8 | 10.6 | (72) | |
| Southeast | 25 | Muscle | −20.3 ± 0.7 | 11.0 ± 1.2 | −16.8 | 10.0 | (73) | |
| Freshwater fish | ||||||||
| Nonsalmonids | ||||||||
| Cottus cognatus (sculpin) | Central | 47 | Muscle | −28.5 ± 3.8 | 7.0 ± 1.6 | −25.0 | 6.0 | (80) |
| Lota lota (burbot) | Central | 21 | Muscle | −27.5 ± 2.4 | 12.1 ± 1.9 | −24.0 | 11.1 | (80) |
| Salmonids | ||||||||
| Oncorhynchus mykiss (rainbow trout) | Central | 11 | Muscle | −23.0 ± 1.4 | 13.2 ± 1.6 | −19.5 | 12.2 | (80) |
| Salvelinus namaycush (lake trout) | Central | 35 | Muscle | −28.5 ± 3.8 | 12.3 ± 2.1 | −25.0 | 11.3 | (80) |
| Thymallus arcticus (Arctic grayling) | Central | 34 | Muscle | −29.0 ± 1.7 | 8.7 ± 0.5 | −25.5 | 7.7 | (81) |
| Thymallus arcticus (Arctic grayling) | Central | 9 | Muscle | −27.3 ± 3.1 | 10.0 ± 1.6 | −23.8 | 9.0 | (80) |
Isotope values on modern fish tissue were adjusted for differences between tissue types and between ancient and modern tissues due to the Suess effect as follows (after ref. 69; see text for further details.): δ13C prehistoric fish tissue = δ13C modern fish tissue + 1‰; δ13C fish collagen = δ13C fish muscle + 2.5‰; δ15N fish collagen = δ15N fish muscle − 1‰.
Lipid extracted muscle.
Lipid corrected muscle.
The confirmed presence of an anadromous Pacific salmon species, O. keta, at the terminal Pleistocene Upward Sun River site provides the earliest evidence for human use of salmon in North America, and adds to our understanding of Paleoindian adaptations. Previous reports of genetically identified Pacific salmon remains from archaeological sites date to as old as ∼6610–5940 cal B.P. from Namu on the central coast of British Columbia (23), and to as old as 9300–8200 cal B.P. for morphologically identified salmon from the Dalles Roadcut site on the Columbia River (32). Pacific salmon remains have not been previously reported for a late Pleistocene-age site, and, indeed, fish remains of any kind are rare in sites for this period in the Americas. In a broad review of taxa exploited by early Paleoindians (33), only two sites (Aubrey and Shawnee-Minisink) had evidence of fish (with no further taxonomic identification) in strong association with human occupation, though abundance values were low. Recently, Erlandson et al. (34) reported 78 specimens (1.4% of total) of various fish taxa (none are salmon) at three Paleoindian sites on the Channel Islands, California.
In Beringia, there is additional evidence for fish exploitation, particularly during the Younger Dryas (16). In central Alaska, Broken Mammoth Cultural Zone 3 (∼12,080 cal B.P.) contained 28 salmonid specimens (possibly grayling) (35), and Mead Cultural Zone 3 (∼11,990 cal B.P.) contained unidentified fish remains (16). In Kamchatka (Russia), Ushki 1 Level 6 (∼12,160 cal BP) contained an unspecified number of burned fish bones (possibly salmon) within houses (36). The salmon data from Upward Sun River reported here add an important component to Paleoindian diet breadth, with implications for seasonal exploitation and possibly storage that is markedly different from multiseasonal exploitation of freshwater fish.
Although Paleoindians are traditionally portrayed as specialist big-game hunters, evidence from Upward Sun River and other early sites in Beringia increasingly suggests a diversity of subsistence strategies, which included hunting, gathering, and fishing. Although the degree of reliance on salmon by early Beringians is currently unresolved, historically in subarctic Alaska, salmon were taken in great numbers in summer to early fall for drying and storing through the winter (37). The extent of salmon storage among early Beringians is also unknown, but chum are well suited for preservation because of their low oil content relative to other salmon species (23).
Our findings have important implications for the paleoecology and human colonization of Beringia and the Americas. Of the 40 earliest known components in northwest North America (dating between 14,000 and 11,500 cal B.P.) (16), 36 (90%) are located in the interior and the majority (60%) are associated with large interior river bottomlands, suggesting the importance of riverine resources to early foragers. Our data show that at least by 11,500 cal B.P., salmon had established spawning runs in the deep interior of eastern Beringia near their current limit of migration, an upriver distance of over 1,400 km from the current mouth of the Yukon, plus an additional ∼300 km of river on the exposed gradually sloping Bering Sea shelf (38) (Fig. 1). The addition of salmon to the early subsistence record further indicates that interior eastern Beringia was a productive landscape in the terminal Pleistocene, supporting a variety of large and small terrestrial mammals, waterfowl, and other fish. This resource diversity contrasts with the assumptions of very low resource availability in inland ecosystems in debates about New World migration corridors (39, 40).
Modern ocean foraging areas for chum salmon include the Bering Sea and the subarctic North Pacific Ocean. Although suitable salmon habitat was likely available in this region throughout the Late Pleistocene, during intervals within glacial periods habitat was probably suboptimal, and especially so near coastal areas surrounding the Bering Sea and the northern Gulf of Alaska (41, 42). Recent paleoceanographic data show that conditions improved dramatically between ∼17,000 and 13,000 cal B.P., depending on indicator and location, in terms of warming sea-surface temperatures, reduced cover of seasonal sea ice, and increased primary productivity (41, 42). River migration access and spawning habitat in the lower Yukon River basin during the Late Pleistocene was probably not an impediment to salmon, as it was not glaciated, even at maximum glacial extent (38, 43). These observations suggest that ocean habitat and migration barriers to Yukon-Tanana salmon were not substantial during the period of early human colonization. Given this, salmon may have been an important resource to earlier Paleoindian subsistence economies and may have played a role in the biogeographic expansions of humans into northwest North America.
SI Text
Sample Archaeological Context.
All three samples were recovered from a single complex feature associated with Component 3 at the Upward Sun River site (USR), located in the central Tanana River basin (Fig. 2). USR site context, geoarchaeology, site formation, and evaluation of postdepositional disturbance and taphonomy have been previously reported (14, 15, 44, 45). Multiple datasets (including clearly defined and horizontal Ab horizons, thin unimodal distributions of lithics and faunal remains, and lack of bioturbation and cryoturbation) indicate minimal postdepositional disturbance, allowing for exploration of cultural behaviors responsible for site formation. A series of 27 radiocarbon dates securely date all four components, with sterile zones (derived from backscatter plots and level totals) separating the occupations. Component 3 contains 10 hearths and associated artifact and faunal scatters dating to ∼11,500 cal B.P. (14, 15).
The largest quantity of faunal remains is situated directly within two superimposed features: (i) an upper hearth feature associated with a cremated 3-y-old human (14) and (ii) burial pit fill associated with a double infant burial (15). These features are within a residential feature, interpreted to represent a tent structure (14), and the dating indicates they are potentially contemporaneous. Analyses presented in Potter et al. (15) suggest they were created and used within a single season or subsequent seasons by the same population.
Faunal data are provided in Potter et al. (15). Number of identified specimens (NISP) for both features totals 930, with 827 (89%) in the cremation hearth and 103 (11%) in the burial pit fill. Based on NISP, the most abundant taxon is Oncorhynchus spp. (salmon) for both features (337 NISP), comprising 37% of hearth NISP and 36% of pit-fill NISP. Other taxa include Urocitellus parryii (ground squirrel) (242 NISP, 26% of both features), Avricolinae (vole and microtine) (245 NISP, 26%), and small quantities of Lepus americanus (snowshoe hare), Marmota sp. (marmot), Tetraoninae (ptarmigan/grouse), and Paridae (passerine bird). Oncorhynchus spp. is the predominant fish taxa, although 25 specimens of Salmonidae (small salmonid, not Oncorhynchus) were also identified. Taxa associated with the hearth were nearly all burned/calcined (96–100% of NISP per taxon), and burned proportions were lower for burial pit fill specimens (0–29% of NISP per taxon). Given the contextual information presented in Potter et al. (15), two related assemblages were identified: (i) the pit-fill fauna, representing a preburial occupation of the residential feature, and (ii) the cremation hearth, representing a second occupation of the residential feature. Faunal diversity is similar between features, suggesting similar (summer) seasons of occupation and similar economy.
Sample Descriptions and Morphometric Analysis.
Of the very few unburned complete Oncorhynchus spp. vertebrae (n = 6), we selected two vertebrae for destructive analysis on the basis of likely preservation of collagen and sufficient mass for genetic and stable isotope analyses (Table S1). A third vertebra (H-5446) was selected from the cremation material and was one of the very few complete calcined vertebra in the upper assemblage. All three were fused (type III) caudal vertebrae and were typical of the specimens assigned to Oncorhynchus spp. Sample provenience and metric data are provided in Table S1 (see Fig. 2 for stratigraphic location).
Because of current debates over identification of salmon species through osteometric analyses, we took a variety of measurements and compared them with published modern and ancient archaeological salmon assemblages (18, 20–22). Fig. S1A shows the USR samples relative to type III vertebral length and height dimensions for modern Oncorhynchus species (at 1 σ) [data from Huber et al. (21)]. The two unburned specimens (58-18, 58-30) are larger than smaller Oncorhynchus species (sockeye, pink, and cutthroat), smaller than Chinook, but similar to chum, coho, and steelhead.
We also compared USR samples with data from Moss et al. (18), who provide summary statistics of vertebral transverse diameter for modern and genetically identified chum, coho, pink, and sockeye from archaeological assemblages (Fig. S1B). USR sample measurements were consistent with both chum and coho, but were larger than both pink and sockeye. These osteometric analyses demonstrate that genetic analysis is necessary to distinguish Oncorhynchus specimens to the species level.
Relationship of Site Location to Modern Salmon Limit.
The location of the Upward Sun River site and limits of modern major fall spawning areas along the Tanana River are illustrated in Fig. 1. The modern course of the Tanana River and Yukon River (south of the junction with the former) is shown along with a possible ancient course to the southern Bering Sea (38). Glacial ice distributions and exposed continental shelf are derived from Dyke et al. (46) and Manley 2002 (47), respectively. Modern major fall spawning areas of chum salmon are documented to the Delta River (50 km from USR), with only limited spawning to Billy Creek (the limit shown in Fig. 1) (48). Ethnographic use of salmon is limited to lower Tanana clearwater tributaries (49). The geographic position of Upward Sun River salmon relative to the modern extent of major salmon spawning in the Tanana River shows that spawning habitat was established by at least 11,500 cal B.P. The presence of anadromous chum salmon indicates their availability as a resource along the length of the Tanana River and the lower Yukon River, totaling at least 1,400 km.
aDNA Extraction and Analysis (Washington State University).
DNA was first extracted from specimen 58-18 at Washington State University (WSU). Located in a separate building from where PCR and post-PCR activities are conducted, the Ancient DNA Laboratory at WSU is dedicated for the study of highly degraded DNA samples. Precautions to minimize the introduction of contamination are routinely practiced in the laboratory (50).
A subsample of the vertebrae, weighing 34 mg, was removed from the whole with a single-use razor blade. The subsample was submerged in 6% (wt/vol) sodium hypochlorite (bleach) for 4 min to remove possible surface contaminants (51). The bleach was poured off and the sample rinsed twice by submersion in DNA-free water. The sample was then transferred to a 1.5-mL tube, to which an aliquot of 500 μL of EDTA (pH 8.0) was added. An extraction negative accompanied the sample through the extraction procedure to monitor for the possible origination of contamination. DNA was extracted from the sample following the WSU method described by Cui et al. (52). After 48 h of decalcification in EDTA, 60 U of proteinase K (Biobasic) were added to the tubes and incubated at 65 °C for 3 h. The volumes were transferred to 5-mL Falcon tubes, to which 750 μL of 2% (wt/vol) celite in 6 M guanidine HCl, and 250 μL of 6 M guanidine HCl were added. The tubes were vortexed numerous times over a 2-min period. These solutions were pulled across Promega Wizard Minicolumns using Luer-Lok syringes and a vacuum manifold. The silica pellets were rinsed by pulling 3 mL of 80% (wt/vol) isopropanol across the columns. Residual isopropanol was removed from the columns by centrifugation in 1.5-mL tubes at 10,000 g for 2 min. The columns were moved to new 1.5-mL tubes and 50 μL of 65 °C DNA-free ddH2O was added to the column and left for 3 min before centrifugation at 10,000 × g for 30 s, and this step was repeated again resulting in 100 mL of extracted DNA.
The DNA extract was first tested for the presence of PCR inhibitors following Kemp et al. (53). Following determination that the extract was inhibited, 50 μL of the volume was subjected to repeat silica extraction (53) and tested again for inhibition. Following one round of repeat silica extraction, the treated extract was determined not to be inhibited. In addition, 5 μL each of the full concentration extract was diluted with water to 1:10 and 1:50 its original concentration. These dilutions were also determined not to be inhibited. DNA from the full concentration extract, the repeat silica-treated portion of the extract, the 1:10 dilution, and the 1:50 dilution were PCR-amplified and sequenced for a region of the mitochondrial 12S gene (189 bp, minus primers the sequence length is 148 bp) that is useful for discriminating among Pacific salmonids and other fishes (31) using Omni Klentaq LA, as described by Monroe et al. (54). Fifteen-microliter PCRs contained 0.32 mM dNTPs, 1× Omni Klentaq Reaction Buffer, 0.24 μM of each primer, 0.3 U of Omni Klentaq LA, and 1.5 μL of template DNA. Primers were those described by Jordan et al. (31); however, it is important to note that in the original study the authors reported the wrong orientation for the reverse primer. Described correctly, the primer sequences are: (i) “OST12S-F” 5′-GCTTAAAACCCAAAGGACTTG-3′ and (ii) “OST12S-R” 5′-CTACACCTCGACCTGACGTT-3′. PCR cycling conditions consisted of: (i) a 3-min hold at 94 °C, (ii) 60 cycles of 15-s holds at 94 °C, 55 °C, and 68 °C, and (iii) a 3-min hold at 68 °C. Successful amplifications were prepared for sequencing following Monroe et al. (54). Sequences were generated in both directions with an ABI 3730xl sequencer at Elim Biopharm. Aligned against a rainbow trout mtDNA sequence [National Center for Biotechnology Information (NCBI) accession no. DQ288271] in Sequencher (v4.8), sequences generated in this study were used for species determination according to Jordan et al. (31).
Although the full concentration extract was determined to be inhibited as described above, it was successfully amplified. This outcome has been observed in a minority of cases by Kemp et al. (53) and a subsequent test of the full concentration extract in the present study indicated that it was not inhibited. These observations can be explained by the stochastic nature of PCR amplification of low copy-number DNA extracts. The sequence, spanning nucleotides positions (nps) 593–740 relative to the rainbow trout mtDNA reference sequence (NCBI accession no. DQ288271), exhibited the following mutations: 660T and 713T (Table S2). According to Jordan et al. (see table 2 in ref. 31), this sequence corresponds to Oncorhynchus keta (chum salmon). In comparison with the records maintained by the NCBI, the Basic Local Aligment Search Tool (BLAST) reveals five complete matches to this sequence, all of which are chum salmon (accessions nos.: KM273849.1, HQ592292.1, AP010773.1, AP125512.1, and AF113119.1). Both the 1:10 dilution and 1:50 dilution amplified for the 12S fragment and the sequences exhibited the same mutations, matching 100% to chum salmon. The forward sequencing reaction from the repeat silica-treated extract failed, but the reverse sequence was consistent with chum salmon (i.e., the sequence spans nps 593–711 and exhibits the following mutation: 660T). Neither the extraction-negative control nor any of the PCR-negative controls amplified. In total, four PCR amplifications and the resulting sequences from the first extraction of 58-18 demonstrate the sample to be chum salmon. Repeated observations of an identical sequence effectively rules out these results as having been influence by postmortem nucleotide damage (55). Following analysis of 58-18 at WSU, the remainder of the vertebra was sent to Simon Fraser University (SFU) for independent verification (described below).
After the analysis of 58-18, the complete H-5446 (calcined) vertebrae and a powered portion of the 58-30 (unburned) vertebrae (i.e., the portion remaining following isotopic analysis) were sent to the WSU laboratory for genetic species identification. DNA was extracted from a 39-mg portion of H-5446 that was removed from the whole with a one-time razor blade and a 53-mg aliquot of the 58-30 powered vertebrae. The extraction, PCR, and sequencing methods were as described above except that the powered bone was not submerged in bleach nor rinsed with DNA-free water. DNA extracted from sample 58-30 was not inhibited. Determined twice by PCR, the sequence, spanning nps 593–740 relative to the rainbow trout mtDNA reference sequence (NCBI accession no. DQ288271) exhibited the following mutations: 660T and 713T (Table S2). This finding is identical to that determined for specimen 58-18 and, as described above, reveals specimen 58-30 to be of chum salmon.
DNA extracted from sample H-5446 was also not inhibited yet it repeatedly failed to amplify (with 1.5 μL of template DNA from the full concentration extract, 3.0 μL of template DNA from the full concentration extract, 1.5 μL of a 1:10 dilution, and 1.5 μL of a 1:50 dilution) (Table S2). Neither the extraction-negative control nor any of the PCR-negative controls amplified. In total, the results demonstrate 58-18 and 58-30 to be chum salmon and H-5446 to not contain analyzable DNA with the methods used here.
aDNA Extraction and Analysis (Simon Fraser University).
A 25-mg subsample of specimen 58-18 was sent to the Ancient DNA Laboratory at SFU for independent replication. The Ancient DNA Laboratory at SFU is specifically designed for ancient DNA analysis and includes UV-filtered ventilation, positive airflow, bench UV lights, and dedicated equipment. No modern DNA samples have ever been processed in the laboratory and strict contamination control protocols are followed at all times (56).
Sample 58-18 was chemically decontaminated through submersion in 6% (wt/vol) sodium hypochlorite for 7 min before being rinsed twice in ultrapure water and UV-irradiated in a cross-linker for 30 min on two sides. The sample was then manually crushed into powder and incubated overnight in 4 mL of lysis buffer [0.5 M EDTA (pH 8.0), 0.5% SDS, and 0.5 mg/mL proteinase K] in a rotating hybridization oven at 50 °C. The tubes were then centrifuged and 2 mL of supernatant from each sample was concentrated to ≤100 μL using Amicon Ultra-4 Centrifugal Filter Devices (10 KD, 4 mL; Millipore). Concentrated extracts were purified using QIAquick spin columns (Qiagen) following the method developed by Yang et al. (22, 56). Next, 100 μL of DNA from each sample was eluted from each QIAquick column for PCR amplification.
PCR amplifications (60 cycles) were performed on an Eppendorf Mastercycler Gradient using a 30-μL reaction volume containing 1.5× Applied Biosystems Buffer, 2 mM MgCl2, 0.2 mM dNTP, 1.0 mg/mL BSA, 3.0–4.0 μL of template DNA, and 1.5 U AmpliTaq Gold (Applied Biosystems). “Universal” bony fish primers, as well as salmonid-specific primers, targeting mtDNA were amplified separately using previously published protocols (57) (Table S3). PCR cycling conditions consisted of: (i) a 12-min hold at 95 °C; (ii) 60 cycles of a 30-s hold at 95 °C, 30-s hold at 52 °C, and 40-s hold at 70 °C; and (iii) a 7-min hold at 72 °C. Five microliters of PCR product from each sample was separated on a 2% agarose gel and visualized using SYBR Green on dark reader (Clare Chemical Research).
Sample 58-18 was successfully amplified using the universal and salmonid-specific primers for DNA (22, 57). The universal primers targeted a COI DNA fragment (∼220 bp) designed to discriminate between bony fish species. The resulting DNA sequence was identified as O. keta (chum salmon) through the BLAST analysis of GenBank sequences. To strengthen this identification, salmonid-specific primers yielded discriminating fragments of the cytochrome b (168 bps) and d-loop (248 bp) regions of the salmonid mitochondrial genome. The resulting amplicons were sent to Eurofins MWG Operon for sequencing and good-quality sequences were obtained. Sequences were visually edited and base-pair ambiguities were examined using ChromasPro software (technelysium.com.au). Multiple alignments of the ancient sequences and published salmonid reference sequences were achieved using ClustalW (58), through BioEdit (59), and phylogenetic analysis was conducted using Mega 6.0 software (60).
Determination of Species Identity (SFU).
The species identity of sample 58-18 was determined through multiple analytical approaches of the d-loop and cytochrome b salmonid sequences. First, a BLAST search against GenBank was used. All amplifications yielded salmonid sequences, which matched either identically or within a few base pairs with GenBank BLAST reference sequences for O. keta (chum salmon). Second, multiple alignments and phylogenetic comparison of this sample to closely-related species also clearly and explicitly point to the species identity of O. keta (chum salmon). Figs. S2 and S3 display multiple alignments of the ancient salmonid cytochrome b and d-loop sequences, respectively, and reference sequences obtained from GenBank, and Fig. S4 A and B display phylogenetic relationships between the ancient salmonid cytochrome b and d-loop haplotypes, respectively, and reference sequences obtained from GenBank. The accuracy of the phylogenetic trees is enhanced by relatively low intraspecific DNA variation for each Pacific salmon species because of their unique evolutionary history (2).
Fig. S3.
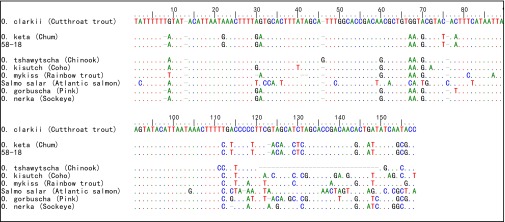
Multiple-alignment of ancient d-loop sequences from specimen 58-18 and reference sequences. The dots indicate identical base pairs, and the dashes represent insertions/deletions compared with the Cutthroat trout sequence at the top. Ancient sequences have been truncated to remove primer sequences. GenBank accession numbers: O. clarkii NC006897; O. mykiss NC001717; O. tshawytscha NC002980; O. kisutch NC009263; O. nerka NC008615; O. keta AJ314561; S. salar NC001960. O. gorbuscha reference sequence from Shedlock et al. (78).
Exclusion of Contamination (SFU).
Contamination can be excluded from the analysis of specimen 58-18. The dedicated aDNA laboratory at SFU follows strict contamination control protocols, such as the separation of the pre-PCR and post-PCR work spaces; the use of aDNA-dedicated equipment, including clothing, equipment, and reagents; and the analysis of blank extraction and negative-PCR controls alongside the aDNA samples. The contamination controls undertaken in this study were successful at eliminating any systematic contamination because no PCR amplification was observed in blank extracts and PCR-negative controls. Furthermore, both cytochrome b and d-loop sequences from the same sample yielded the same species identity.
Bone Collagen Extraction and Stable Isotope Methods.
Bone collagen was extracted from a single unburned salmonid vertebra (58-30) for carbon and nitrogen stable isotope analysis. The specimen was cleaned using a dental pick to remove visible adhering sediment, followed by ultrasonication in ultrapure water (resistivity > 18 MΩ-cm; total oxidizable carbon < 5 ppb) for 15 min (repeated once with a change of water). After air-drying overnight, the sample was pulverized in a ball mill to less than 0.5 mm. Bone collagen was extracted using the Longin method (61) with some modifications. Extraction steps were carried out at room temperature, unless otherwise noted. To demineralize the sample, ∼0.04 g of powdered bone was placed into a 1.5-mL microcentrifuge tube to which ∼1.0 mL of 0.5 M HCl was added. The contents were mixed using moderate shaking (600 rpm) on a laboratory mixer for 30 min, followed by centrifuging and discard of the supernatant. The HCl treatment was repeated twice (for a total of three acid washes), after which the sample was rinsed three times in ultrapure water. To remove base-soluble contaminants, such as humic acids, ∼1.0 mL of 0.1 M NaOH was added to the sample, and the contents were mixed using moderate shaking (600 rpm) on a laboratory mixer for 10 min, followed by centrifuging and discard of the supernatant. The NaOH treatment was repeated once (for a total of two alkaline washes), after which the sample was rinsed three times in ultrapure water. To gelatinize (solubilize) the collagen, ∼1.0 mL of dilute HCl (0.001 M, pH 3) was added to the sample, and the tube was placed on a laboratory mixer at 70 °C with moderate shaking (800 rpm) for 20 h, followed by centrifuging and collection of the supernatant (containing the solubilized collagen) and discard of the pellet. Samples were freeze-dried for ∼48 h.
Bone collagen samples were submitted to the WSU Stable Isotope Core Laboratory (R. Dale Evans, Laboratory Director) for carbon and nitrogen stable-isotope ratio measurement on the Thermo-Finnigan Delta PlusXP continuous flow isotope ratio mass spectrometer, coupled to a Costech Elemental Analyzer (ECS 4010). The ratios of nitrogen stable isotopes (15N/14N) and carbon stable isotopes (13C/12C) are conventionally expressed in δ notation in parts per thousand (per mil; ‰) relative to a standard, as follows:
where Rsample and Rstandard are the ratios of the heavier to the lighter isotope in the sample and the standard, respectively (62, 63). The international standard for nitrogen is atmospheric nitrogen (AIR), and that for carbon is Vienna Pee Dee Belemnite. Within-run reference checks using an animal protein (casein) produced SDs of <0.1‰ for both δ13C and δ15N. The Upward Sun River fish bone collagen sample met the accepted quality standards [%N > 5%, %C > 13%, atomic C:N ratio 2.9–3.6; and collagen yield > 1% (64–66)], as follows: %C = 43.1, %N = 12.6; atomic C:N = 3.2; %Yield = 7.0 [all concentrations are (wt/wt)].
Comparative δ13C and δ15N Values for Ancient and Modern Alaskan Fish Specimens.
There are few published δ13C and δ15N values on bone collagen for prehistoric salmon from Alaska (67–69) and none for prehistoric freshwater fish, except for a small sample (n = 2) of late-prehistoric burbot from the upper Kuskowkim River (70) (Table S4). None of the published ancient salmon were identified to species. Although there are several isotopic studies of modern salmon and freshwater fishes in Alaska (71–75), these analyzed muscle, which is not directly comparable to bone collagen. The δ13C and δ15N offsets between muscle and bone collagen have been published by Misarti et al. (69) for salmon, and are used here for all fish, including freshwater fish, as follows: δ13C for fish bone collagen is 2.5‰ higher than muscle, whereas δ15N for fish bone collagen is 1‰ lower than muscle. All published isotope values for modern fish muscle tissue were adjusted using these offsets (Table S5). Some studies of modern fish tissues extracted lipids before isotopic analysis (74) or corrected for lipid content (71), but we did not adjust isotope values further for this methodological difference.
A further correction is necessary to compare δ13C between ancient and modern tissues to account for the “Suess effect” (the reduction of δ13C in the Earth’s carbon reservoirs as a result of industrial carbon emissions) (76). Here we use the maximum adjustment described by Misarti et al. (69) of 1‰ to be added to modern tissue δ13C values for comparability with preindustrial values (Table S5). Note that the Misarti et al. (69) correction factor is for the oceanic Suess effect in the North Pacific. Because the freshwater Suess effect has not been characterized for Alaska, we use the oceanic correction. The magnitude of the freshwater Suess effect may be slightly larger [e.g., the atmospheric Suess effect is roughly 1.5‰ (77)], but this would not greatly affect the pattern of differences seen in δ13C between chum salmon and freshwater fish in Alaska.
Acknowledgments
We thank the Healy Lake Tribal Council and Tanana Chiefs Conference representatives for their support; Robert Sattler (Tanana Chiefs Conference), MTNT Ltd., and the McGrath Native Village council for their support in the recovery and analysis of prehistoric freshwater fish specimens; Cara Monroe and Lindsay Hilldorfer for assistance in the Washington State University laboratory; and Bobbi Johnson for the initial analysis of the 58-18 morphometric measures. This project was funded in part by National Science Foundation Grants 1138811 and 1223119.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KT693031, KT693032, KT725810, and KT725811).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509747112/-/DCSupplemental.
References
- 1.Lindsey CC, McPhail JD. Zoogeography of fishes of the Yukon and Mackenzie basins. In: Hocutt CH, Wiley EO, editors. The Zoogeography of North American Freshwater Fishes. Wiley; New York: 1986. pp. 639–674. [Google Scholar]
- 2.Waples RS, Pess GR, Beechie T. Evolutionary history of Pacific salmon in dynamic environments. Evol Appl. 2008;1(2):189–206. doi: 10.1111/j.1752-4571.2008.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffecker JF, Elias SA. Human Ecology of Beringia. Columbia Univ Press; New York: 2007. [Google Scholar]
- 4.Surovell TA, Waguespack NM. Human prey choice in the late Pleistocene and its relation to megafaunal extinctions. In: Haynes G, editor. American Megafaunal Extinctions at the End of the Pleistocene. Springer; Dordrecht, The Netherlands: 2009. pp. 77–105. [Google Scholar]
- 5.Cannon MD, Meltzer DJ. Explaining variability in early Paleoindian foraging. Quat Int. 2008;191:5–17. [Google Scholar]
- 6.Quinn TP. The Behavior and Ecology of Pacific Salmon and Trout. Univ of Washington Press; Seattle: 2005. [Google Scholar]
- 7.Schalk RF. The structure of an anadromous fish resource. In: Binford LR, editor. For Theory Building in Archaeology. Academic; New York: 1977. pp. 207–249. [Google Scholar]
- 8.Schindler DE, et al. Pacific Salmon and the ecology of coastal ecosystems. Front Ecol Environ. 2003;1(1):31–37. [Google Scholar]
- 9.Smith GR, Montgomery DR, Peterson NP, Crowley B. Spawning sockeye salmon fossils in Pleistocene lake beds of Skokomish Valley, Washington. Quat Res. 2007;68:227–238. [Google Scholar]
- 10.Lyman RL. Vertebrate Taphonomy. Cambridge Univ Press; Cambridge, UK: 1994. [Google Scholar]
- 11.Carlson CC, Klein K. Late Pleistocene salmon of Kamloops Lake. In: Ludvigsen R, editor. Life in Stone: A Natural History of British Columbia’s Fossils. Univ of British Columbia Press; Vancouver: 1996. pp. 274–280. [Google Scholar]
- 12.Harington CR, Ross RLM, Mathewes RW, Stewart KM, Beattie O. A late Pleistocene Steller sea lion (Eumetopias jubatus) from Courtenay, British Columbia: Its death, associated biota, and paleoenvironment. Can J Earth Sci. 2004;41(11):1285–1297. [Google Scholar]
- 13.Fedje D, Mackie Q, Lacourse T, McLaren D. Younger Dryas environments and archaeology on the Northwest Coast of North America. Quat Int. 2011;242(2):452–462. [Google Scholar]
- 14.Potter BA, Irish JD, Reuther JD, Gelvin-Reymiller C, Holliday VT. A terminal Pleistocene child cremation and residential structure from eastern Beringia. Science. 2011;331(6020):1058–1062. doi: 10.1126/science.1201581. [DOI] [PubMed] [Google Scholar]
- 15.Potter BA, Irish JD, Reuther JD, McKinney HJ. New insights into Eastern Beringian mortuary behavior: A terminal Pleistocene double infant burial at Upward Sun River. Proc Natl Acad Sci USA. 2014;111(48):17060–17065. doi: 10.1073/pnas.1413131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potter BA, Holmes CE, Yesner DR. Technology and economy among the earliest prehistoric foragers in interior eastern Beringia. Paleoamerican Odyssey. Proceedings of the 2013 Paleoamerican Odyssey Conference in Santa Fe, New Mexico. In: Graf KE, Ketron CV, Waters MR, editors. Texas A&M Press; College Station, TX: 2013. pp. 81–103. [Google Scholar]
- 17.Butler VL. Fish remains. In: Hicks BA, editor. Marmes Rockshelter: A Final Report on 11,000 Years of Cultural Use. Washington State Univ Press; Pullman, WA: 2004. pp. 320–337. [Google Scholar]
- 18.Moss ML, Judd KG, Kemp BM. Can salmonids (Oncorhynchus spp.) be identified to species using vertebral morphometrics? A test using ancient DNA from Coffman Cove, Alaska. J Archaeol Sci. 2014;41:879–889. [Google Scholar]
- 19.Grier C, et al. Using ancient DNA identification and osteometric measures of archaeological Pacific Salmon vertebrae for reconstructing salmon fisheries and site seasonality at Dionisio Point, British Columbia. J Archaeol Sci. 2013;40:544–555. [Google Scholar]
- 20.Orchard TJ, Szpak P. Identification of salmon species from archaeological remains on the Northwest Coast. In: Moss ML, Cannon A, editors. The Archaeology of North Pacific Fisheries. Univ of Alaska Press; Fairbanks, AK: 2011. pp. 17–29. [Google Scholar]
- 21.Huber HR, Jorgensen JC, Butler VL, Baker G, Stevens R. Can salmonids (Oncorhynchus spp.) be identified to species using vertebral morphometrics? J Archaeol Sci. 2011;38(1):136–146. [Google Scholar]
- 22.Yang DY, Cannon A, Saunders SR. DNA species identification of archaeological salmon bone from the Pacific Northwest coast of North America. J Archaeol Sci. 2004;31:619–631. [Google Scholar]
- 23.Cannon A, Yang DY. Early storage and sedentism on the Pacific Northwest Coast: Ancient DNA analysis of salmon remains from Namu, British Columbia. Am Antiq. 2006;71(1):123–140. [Google Scholar]
- 24.Speller CF, Yang DY, Hayden B. Ancient DNA investigation of prehistoric salmon resource utilization at Keatley Creek, British Columbia, Canada. J Archaeol Sci. 2005;32:1378–1389. [Google Scholar]
- 25.Clarke WC, Hirano T. Osmoregulation. In: Groot C, Margolis L, Clarke WC, editors. Physiological Ecology of Pacific Salmon. Univ of British Columbia Press; Vancouver: 1995. pp. 317–378. [Google Scholar]
- 26.Schoeninger MJ, DeNiro MJ. Nitrogen and carbon isotopic composition of bone collagen from marine and terrestrial animals. Geochim Cosmochim Acta. 1984;48:625–639. [Google Scholar]
- 27.Ciancio JE, et al. Stable isotope profiles of partially migratory salmonid populations in Atlantic rivers of Patagonia. J Fish Biol. 2008;72:1708–1719. [Google Scholar]
- 28.Swanson HK, et al. Anadromy in Arctic populations of lake trout (Salvelinus namaycush): Otolith microchemistry, stable isotopes, and comparisons with Arctic char (Salvelinus alpinus) Can J Fish Aquat Sci. 2010;67:842–853. [Google Scholar]
- 29.Doucett RR, Power M, Power G, Caron F, Reist JD. Evidence for anadromy in a southern relict population of Arctic charr from North America. J Fish Biol. 1999;55:84–93. [Google Scholar]
- 30.Kline TC, Jr, Wilson WJ, Goering JJ. Natural isotope indicators of fish migration at Prudhoe Bay, Alaska. Can J Fish Aquat Sci. 1998;55:1494–1502. [Google Scholar]
- 31.Jordan LG, Steele CA, Thorgaard GH. Universal mtDNA primers for species identification of degraded bony fish samples. Mol Ecol Resour. 2010;10(1):225–228. doi: 10.1111/j.1755-0998.2009.02739.x. [DOI] [PubMed] [Google Scholar]
- 32.Butler VL, O’Connor JE. 9000 years of salmon fishing on the Columbia River, North America. Quat Res. 2004;62:1–8. [Google Scholar]
- 33.Cannon MD, Meltzer DJ. Early Paleoindian foraging: Examining the faunal evidence for large mammal specialization and regional variability in prey choice. Quat Sci Rev. 2004;23:1955–1987. [Google Scholar]
- 34.Erlandson JM, et al. Paleoindian seafaring, maritime technologies, and coastal foraging on California’s Channel Islands. Science. 2011;331(6021):1181–1185. doi: 10.1126/science.1201477. [DOI] [PubMed] [Google Scholar]
- 35.Yesner DR. Faunal extinction, hunter-gatherer foraging strategies, and subsistence diversity among eastern Beringian Paleoindians. In: Walker RB, Driskell BN, editors. Foragers of the Terminal Pleistocene in North America. Univ of Nebraska Press; Lincoln, NB: 2007. pp. 15–31. [Google Scholar]
- 36.Dikov NN. 1979. Drevnie kul’tury Severo-Vostochnoi Azii (Aziia na styke s Amerikoi v drevnosti) trans Bland RL [Early Cultures of Northeastern Asia] (Nauka, Moscow)
- 37.VanStone JW. Athapaskan Adaptations. Field Museum of Natural History; Chicago: 1974. [Google Scholar]
- 38.Knebel HJ, Creager JS. Yukon River: Evidence for extensive migration during the Holocene transgression. Science. 1973;179(4079):1230–1232. doi: 10.1126/science.179.4079.1230. [DOI] [PubMed] [Google Scholar]
- 39.Fladmark KR. A Paleoecological Model for Northwest Coast Prehistory. National Museums of Canada; Ottawa: 1975. [Google Scholar]
- 40.Surovell TA. Simulating coastal migration in New World colonization. Curr Anthropol. 2003;44(4):580–591. [Google Scholar]
- 41.Caissie BE, Brigham-Grette J, Lawrence KT, Herbert TD, Cook MS. Last Glacial Maximum to Holocene sea surface conditions at Umnak Plateau, Bering Sea, as inferred from diatom, alkenone, and stable isotope records. Paleoceanography. 2010;25:PA1206. [Google Scholar]
- 42.Addison JA, et al. Productivity and sedimentary δ15N variability for the last 17,000 years along the northern Gulf of Alaska continental slope. Paleoceanography. 2012;27:PA1206. [Google Scholar]
- 43.Dyke AS. An outline of North American deglaciation with emphasis on central and northern Canada. In: Ehlers J, Gibbard PL, editors. Quaternary Glaciations-Extent and Chronology. Part II: North America. Elsevier; Amsterdam: 2004. pp. 373–424. [Google Scholar]
- 44.Reuther JD. 2013. Late Glacial and Early Holocene geoarchaeology and terrestrial paleoecology in the lowlands of the Middle Tanana Valley, subarctic Alaska. PhD Dissertation. (University of Arizona, Tucson, AZ)
- 45.Potter BA, Reuther JD, Bowers PM, Gelvin-Reymiller C. Little Delta Dune site: A late-Pleistocene multicomponent site in central Alaska. Current Research in the Pleistocene. 2008;25:132–135. [Google Scholar]
- 46.Dyke AS, Moore A, Robertson L. 2003. Deglaciation of North America. Geological Survey of Canada Open File 1574. Thirty-two digital maps at 1:7,000,000 scale with accompanying digital chronological database and one poster (two sheets) with full map series (Geological Survey of Canada, Ottawa)
- 47.Manley WF. 2002 Postglacial flooding of the Bering Land Bridge: A geospatial animation: INSTAAR, University of Colorado, v1. Available at instaar.colorado.edu/QGISL/bering_land_bridge/. Accessed May 17, 2014.
- 48.Barton LH. A Catalog of Yukon River Salmon Spawning Escapement Surveys. Technical Data Report 121. Alaska Department of Fish and Game, Division of Commercial Fisheries; Juneau, AK: 1984. [Google Scholar]
- 49.McKennan RA. Tanana. In: Sturtevant WC, Helm J, editors. Handbook of North American Indians: Vol. 6: Subarctic. Government Printing Office; Washington DC: 1981. pp. 562–576. [Google Scholar]
- 50.Kemp BM, Glenn Smith D. Ancient DNA methodology: Thoughts from Brian M. Kemp and David Glenn Smith on “Mitochondrial DNA of protohistoric remains of an Arikara population from South Dakota”. Hum Biol. 2010;82(2):227–238. doi: 10.3378/027.082.0207. [DOI] [PubMed] [Google Scholar]
- 51.Barta JL, Monroe C, Kemp BM. Further evaluation of the efficacy of contamination removal from bone surfaces. Forensic Sci Int. 2013;231(1-3):340–348. doi: 10.1016/j.forsciint.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Cui Y, et al. Ancient DNA analysis of mid-Holocene individuals from the Northwest Coast of North America reveals different evolutionary paths for mitogenomes. PLoS One. 2013;8(7):e66948. doi: 10.1371/journal.pone.0066948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kemp BM, Monroe C, Judd KG, Reams E, Grier C. Evaluation of methods that subdue the effects of polymerase chain reaction inhibitors in the study of ancient and degraded DNA. J Archaeol Sci. 2014;42:373–380. [Google Scholar]
- 54.Monroe C, Grier C, Kemp BM. Evaluating the efficacy of various thermo-stable polymerases against co-extracted PCR inhibitors in ancient DNA samples. Forensic Sci Int. 2013;228(1-3):142–153. doi: 10.1016/j.forsciint.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 55.Winters M, Barta JL, Monroe C, Kemp BM. To clone or not to clone: method analysis for retrieving consensus sequences in ancient DNA samples. PLoS One. 2011;6(6):e21247. doi: 10.1371/journal.pone.0021247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105(4):539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 57.Yang DY, Speller CF. Co-amplification of cytochrome b and d-loop mtDNA fragments for the identification of degraded DNA samples. Mol Ecol Notes. 2006;6(3):605–608. [Google Scholar]
- 58.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 60.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Longin R. New method of collagen extraction for radiocarbon dating. Nature. 1971;230(5291):241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 62.Coplen TB. 1995. Reporting of stable carbon, hydrogen, and oxygen isotopic abundances. Reference and Intercomparison Materials for Stable Isotopes of Light Elements. Proceedings of a Consultants Meeting Held in Vienna, 1–3 December 1993, ed International Atomic Energy Agency (IAEA, Vienna), pp 31–34.
- 63.Böhlke JK, Coplen TB. 1995. Interlaboratory comparison of reference materials for nitrogen-isotope-ratio measurements. Reference and Intercomparison Materials for Stable Isotopes of Light Elements. Proceedings of a Consultants Meeting Held in Vienna, 1–3 December 1993, ed International Atomic Energy Agency (IAEA, Vienna), pp 51–66.
- 64.Ambrose SH. Preparation and characterization of bone and tooth collagen for isotopic analysis. J Archaeol Sci. 1990;17(4):431–451. [Google Scholar]
- 65.DeNiro MJ. Postmortem preservation and alteration of in vivo bone collagen isotope ratios in relation to palaeodietary reconstruction. Nature. 1985;317:806–809. [Google Scholar]
- 66.van Klinken GJ. Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J Archaeol Sci. 1999;26:687–695. [Google Scholar]
- 67.Britton K, et al. Maritime adaptations and dietary variation in prehistoric Western Alaska: Stable isotope analysis of permafrost-preserved human hair. Am J Phys Anthropol. 2013;151(3):448–461. doi: 10.1002/ajpa.22284. [DOI] [PubMed] [Google Scholar]
- 68.Byers DA, Yesner DR, Broughton JM, Coltrain JB. Stable isotope chemistry, population histories and Late Prehistoric subsistence change in the Aleutian Islands. J Archaeol Sci. 2011;38(1):183–196. [Google Scholar]
- 69.Misarti N, Finney B, Maschner H, Wooller MJ. Changes in northeast Pacific marine ecosystems over the last 4500 years: Evidence from stable isotope analysis of bone collagen from archeological middens. Holocene. 2009;19:1139–1151. [Google Scholar]
- 70.Halffman CM, Sattler R, Clark JL. Bone collagen stable isotope analysis of three late Holocene humans from Interior Alaska. Am J Phys Anthropol. 2015;156(S60):157. [Google Scholar]
- 71.Johnson SP, Schindler DE. Trophic ecology of Pacific salmon (Oncorhynchus spp.) in the ocean: A synthesis of stable isotope research. Ecol Res. 2009;24(4):855–863. [Google Scholar]
- 72.Ben-David M. 1996. Seasonal diets of mink and martens: Effects of spatial and temporal changes in resource abundance. PhD Dissertation. (University of Alaska Fairbanks, Fairbanks, AK)
- 73.Satterfield FR, Finney BP. Stable isotope analysis of Pacific salmon: Insight into trophic status and oceanographic conditions over the last 30 years. Prog Oceanogr. 2002;53(2):231–246. [Google Scholar]
- 74.Kaeriyama M, et al. Change in feeding ecology and trophic dynamics of Pacific salmon (Oncorhynchus spp.) in the central Gulf of Alaska in relation to climate events. Fish Oceanogr. 2004;13(3):197–207. [Google Scholar]
- 75.Knudson KJ, Frink L. Nitrogen isotope analysis in the Arctic: Identifying fish processing and marine resource use through ethnoarchaeological soil analysis on Nelson Island, Alaska. Alaska Journal of Anthropology. 2011;9(2):17–54. [Google Scholar]
- 76.Keeling CD. The Suess effect: 13Carbon-14Carbon interrelations. Environ Int. 1979;2(4-6):229–300. [Google Scholar]
- 77.McCarroll D, Loader NJ. Stable isotopes in tree rings. Quat Sci Rev. 2004;23:771–801. [Google Scholar]
- 78.Shedlock AM, Parker JD, Crispin DA, Pietsch TW, Burmer GC. Evolution of the salmonid mitochondrial control region. Mol Phylogenet Evol. 1992;1(3):179–192. doi: 10.1016/1055-7903(92)90014-8. [DOI] [PubMed] [Google Scholar]
- 79.Royle TC. 2014. Ancient DNA analysis of Middle and Late Period archaeological fish remains from Kamloops, British Columbia. MA Thesis (Simon Fraser University, Burnaby, BC)
- 80.Piorkowski RJ. 1995. Ecological effects of spawning salmon on several southcentral Alaskan streams. PhD Dissertation. (University of Alaska Fairbanks, Fairbanks, AK)
- 81.Crock JG, Seal RR, II, Gough LP, Weber-Scannell P. 2003. Results of elemental and stable isotopic measurements, and dietary composition of Arctic grayling (Thymallus arcticus) collected in 2000 and 2001 from the Fortymile River Watershed, Alaska. Open-File Report 2003-03-057 (US Department of the Interior, US Geological Survey, Denver)