Abstract
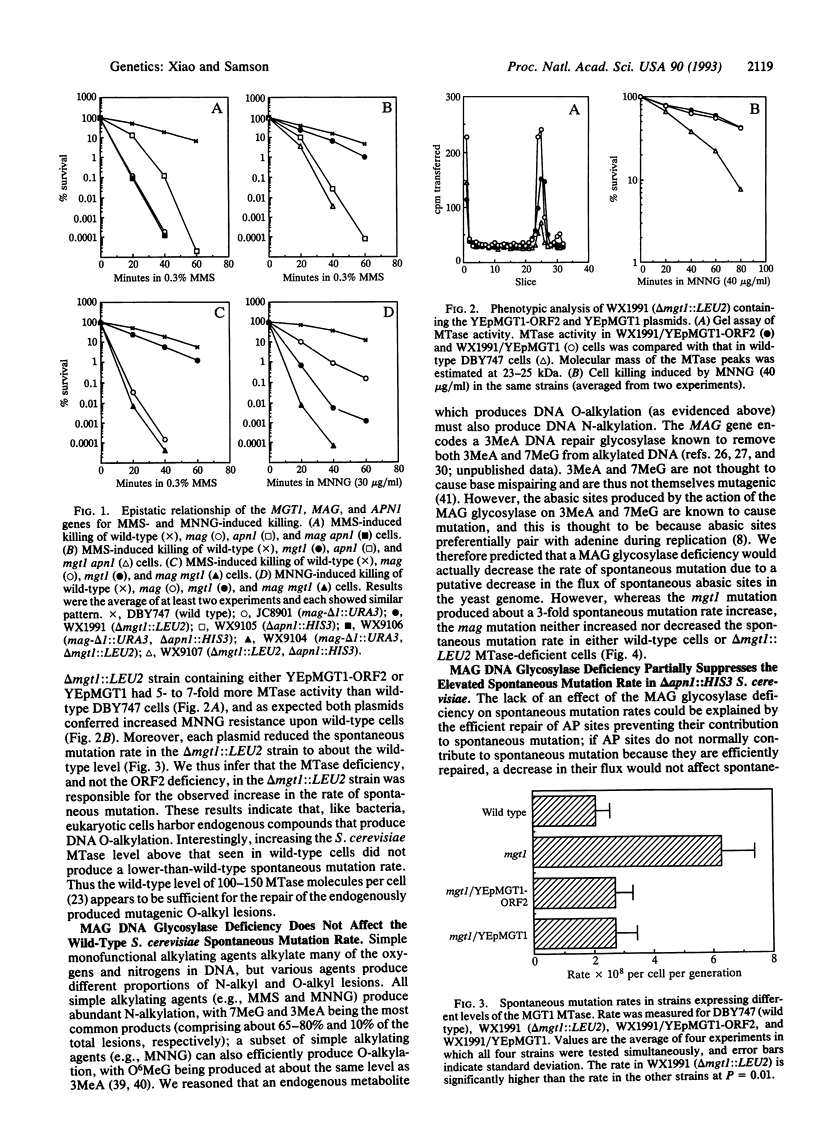
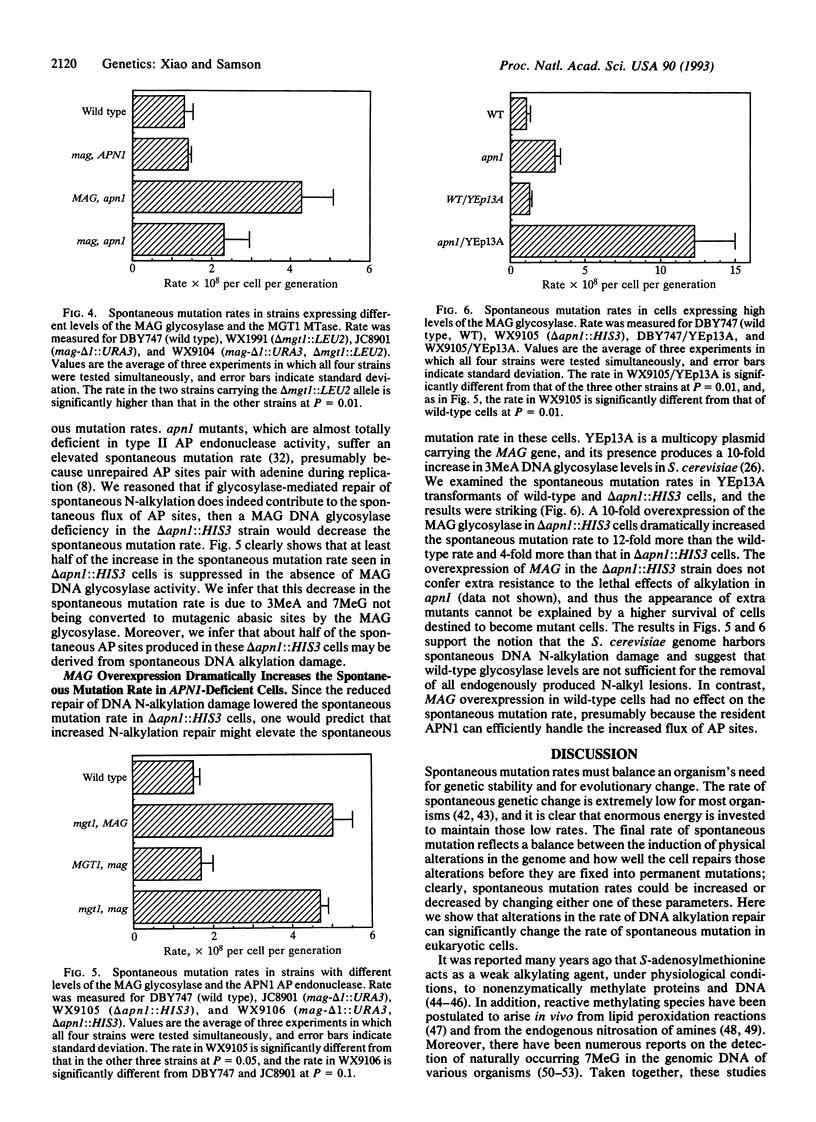
Three genes that participate in the repair of DNA alkylation damage were recently cloned from Saccharomyces cerevisiae: the MGT1 O6-methylguanine DNA methyltransferase gene, the MAG 3-methyladenine DNA glycosylase gene, and the APN1 apurinic/apyrimidinic (AP) endonuclease gene. Altering the expression levels of these three genes produced significant changes in the S. cerevisiae spontaneous mutation rate. Spontaneous mutation increased in the absence of the MGT1 DNA methyltransferase, presumably because unrepaired, spontaneously produced, O6-alkylguanine lesions mispair during replication. Moreover, changing the ratios of the MAG 3-methyladenine DNA glycosylase and the APN1 AP endonuclease had profound effects on spontaneous mutation rates. In the absence of APN1, the overexpression of MAG increased spontaneous mutation, and the underexpression of MAG decreased spontaneous mutation. We infer that the MAG glycosylase acts upon spontaneously produced 3-alkyladenine and 7-alkylguanine DNA lesions to produce mutagenic abasic sites, and that if the repair of these abasic sites is not initiated by the APN1 AP endonuclease they cause mutations during replication. Our results indicate that eukaryotic cells harbor endogenous metabolites that alkylate nuclear DNA at both oxygens and nitrogens.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrows L. R., Magee P. N. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3(3):349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- Beranek D. T. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990 Jul;231(1):11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Berdal K. G., Bjørås M., Bjelland S., Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990 Dec;9(13):4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M., Nghiem Y., Miller J. H. mutM, a second mutator locus in Escherichia coli that generates G.C----T.A transversions. J Bacteriol. 1988 Nov;170(11):5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., Miller S. The origin of mutants. Nature. 1988 Sep 8;335(6186):142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Calmels S., Ohshima H., Crespi M., Leclerc H., Cattoen C., Bartsch H. N-nitrosamine formation by microorganisms isolated from human gastric juice and urine: biochemical studies on bacteria-catalysed nitrosation. IARC Sci Publ. 1987;(84):391–395. [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Samson L. Induction of S.cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic Acids Res. 1991 Dec 11;19(23):6427–6432. doi: 10.1093/nar/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R. S., 3rd, Babich M. A., Yarosh D. B., Scudiero D. A. The role of O6-methylguanine in human cell killing, sister chromatid exchange induction and mutagenesis: a review. J Cell Sci Suppl. 1987;6:333–353. doi: 10.1242/jcs.1984.supplement_6.22. [DOI] [PubMed] [Google Scholar]
- Diller L., Kassel J., Nelson C. E., Gryka M. A., Litwak G., Gebhardt M., Bressac B., Ozturk M., Baker S. J., Vogelstein B. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol. 1990 Nov;10(11):5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Allen E. F., Forsberg S. A., Preparata R. M., Greening E. O. Genetic control of mutation rates in bacteriophageT4. Nature. 1969 Mar 22;221(5186):1128–1132. [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Farr S. B., D'Ari R., Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E. R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990 Jun 1;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Foster P. L. Directed mutation: between unicorns and goats. J Bacteriol. 1992 Mar;174(6):1711–1716. doi: 10.1128/jb.174.6.1711-1716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla E. B., Valentine J. S. Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. J Bacteriol. 1991 Sep;173(18):5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J. T., Demple B. Overproduction of peroxide-scavenging enzymes in Escherichia coli suppresses spontaneous mutagenesis and sensitivity to redox-cycling agents in oxyR-mutants. EMBO J. 1988 Aug;7(8):2611–2617. doi: 10.1002/j.1460-2075.1988.tb03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Loeb L. A. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991 Jun 15;51(12):3075–3079. [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992 Jan 16;355(6357):273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Cruz C., Grollman A. P., Miller J. H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Cruz C., Miller J. H. MutM, a protein that prevents G.C----T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991 Jul 11;19(13):3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Low K. B. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell. 1984 Jun;37(2):675–682. doi: 10.1016/0092-8674(84)90400-8. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Paik W. K., Lee H. W., Kim S. Non-enzymatic methylation of proteins with S-adenosyl-L-methionine. FEBS Lett. 1975 Oct 15;58(1):39–42. doi: 10.1016/0014-5793(75)80220-1. [DOI] [PubMed] [Google Scholar]
- Park J. W., Ames B. N. 7-Methylguanine adducts in DNA are normally present at high levels and increase on aging: analysis by HPLC with electrochemical detection. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7467–7470. doi: 10.1073/pnas.85.20.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff S. C., Spira A. I., Johnson A. W., Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar D., Popoff S. C., Gralla E. B., Demple B. Cellular role of yeast Apn1 apurinic endonuclease/3'-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol Cell Biol. 1991 Sep;11(9):4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck G. W., Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991 Mar;173(6):2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1(2):211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargentini N. J., Smith K. C. Much of spontaneous mutagenesis in Escherichia coli is due to error-prone DNA repair: implications for spontaneous carcinogenesis. Carcinogenesis. 1981;2(9):863–872. doi: 10.1093/carcin/2.9.863. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Dosanjh M. K., Essigmann J. M., Samson L. Relative efficiencies of the bacterial, yeast, and human DNA methyltransferases for the repair of O6-methylguanine and O4-methylthymine. Suggestive evidence for O4-methylthymine repair by eukaryotic methyltransferases. J Biol Chem. 1991 Feb 15;266(5):2767–2771. [PubMed] [Google Scholar]
- Sassanfar M., Samson L. Identification and preliminary characterization of an O6-methylguanine DNA repair methyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990 Jan 5;265(1):20–25. [PubMed] [Google Scholar]
- Shuker D. E., Farmer P. B. Relevance of urinary DNA adducts as markers of carcinogen exposure. Chem Res Toxicol. 1992 Jul-Aug;5(4):450–460. doi: 10.1021/tx00028a001. [DOI] [PubMed] [Google Scholar]
- Smith K. C. Spontaneous mutagenesis: experimental, genetic and other factors. Mutat Res. 1992 Aug;277(2):139–162. doi: 10.1016/0165-1110(92)90002-q. [DOI] [PubMed] [Google Scholar]
- Storz G., Christman M. F., Sies H., Ames B. N. Spontaneous mutagenesis and oxidative damage to DNA in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8917–8921. doi: 10.1073/pnas.84.24.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B. S. The origin of point mutations in human tumor cells. Cancer Res. 1992 Jan 15;52(2):249–253. [PubMed] [Google Scholar]
- Tan B. H., Bencsath F. A., Gaubatz J. W. Steady-state levels of 7-methylguanine increase in nuclear DNA of postmitotic mouse tissues during aging. Mutat Res. 1990 Sep-Nov;237(5-6):229–238. doi: 10.1016/0921-8734(90)90004-b. [DOI] [PubMed] [Google Scholar]
- Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimis J., Yarosh D. B. Adaptive response induction by bacterial catalysis of nitrosation. Environ Mol Mutagen. 1990;15(2):69–70. doi: 10.1002/em.2850150202. [DOI] [PubMed] [Google Scholar]
- Vaca C. E., Wilhelm J., Harms-Ringdahl M. Interaction of lipid peroxidation products with DNA. A review. Mutat Res. 1988 Mar;195(2):137–149. doi: 10.1016/0165-1110(88)90022-x. [DOI] [PubMed] [Google Scholar]
- Von Borstel R. C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- Xiao W., Derfler B., Chen J., Samson L. Primary sequence and biological functions of a Saccharomyces cerevisiae O6-methylguanine/O4-methylthymine DNA repair methyltransferase gene. EMBO J. 1991 Aug;10(8):2179–2186. doi: 10.1002/j.1460-2075.1991.tb07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Samson L. The Saccharomyces cerevisiae MGT1 DNA repair methyltransferase gene: its promoter and entire coding sequence, regulation and in vivo biological functions. Nucleic Acids Res. 1992 Jul 25;20(14):3599–3606. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]


