Abstract
A meropenem-resistant Pseudomonas aeruginosa isolate was obtained from a patient in a medical setting in Hanoi, Vietnam. The isolate was found to have a novel IMP-type metallo-β-lactamase, IMP-51, which differed from IMP-7 by an amino acid substitution (Ser262Gly). Escherichia coli expressing blaIMP-51 showed greater resistance to cefoxitin, meropenem, and moxalactam than E. coli expressing blaIMP-7. The amino acid residue at position 262 was located near the active site, proximal to the H263 Zn(II) ligand.
TEXT
Metallo-β-lactamases (MBLs) confer resistance to all β-lactams, except for monobactams, and are characterized by their efficient hydrolysis of carbapenems (1). Acquired MBLs are produced by Gram-negative bacteria, including Pseudomonas aeruginosa, Acinetobacer spp., and enterobacteria (1). The acquired MBLs are categorized by their amino acid sequences into various types (2–4), including AIM (5), DIM (6), FIM (7), GIM (8), IMPs (9), KHM (10), NDMs (11), SMB (12), SIM (13), SPM (14), TMBs (15) and VIMs (16). The most prevalent types of MBLs are the IMP-, VIM-, and NDM-type enzymes (1, 2, 17). We describe here a novel IMP-type MBL, IMP-51, produced by a clinical isolate of P. aeruginosa in a medical setting in Vietnam.
The P. aeruginosa clinical isolate NCGM3025 was obtained from a sputum sample of a patient in 2013 in an intensive care unit in a medical setting in Hanoi, Vietnam. MICs of various antibiotics were determined using the microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (18). IMP-type MBLs and an aminoglycoside modification enzyme, AAC(6′)-Ib, were detected using immunochromatographic assay kits (19, 20). DNA was extracted from the isolate using DNeasy blood and tissue kits (Qiagen, Tokyo, Japan), and the entire genome was sequenced by MiSeq (Illumina, San Diego, CA). Sequence data were analyzed using CLC Genomics Workbench version 8.0 (CLC bio, Tokyo, Japan). Multilocus sequence typing (MLST) was deduced as described by the protocols of the PubMLST databases (http://pubmlst.org/paeruginosa/). Sequences of drug resistance genes, including β-lactamase-encoding genes at the Lahey Clinic website (www.lahey.org/studies), aminoglycoside, chloramphenicol, and fosfomycin resistance genes registered in GenBank (http://www.ncbi.nlm.nih.gov/nuccore/), and quinolone resistance genes (21), were determined using CLC Genomics Workbench version 8.0.
Escherichia coli transformants expressing blaIMP-7 and blaIMP-51 were produced, and the recombinant IMP-7 and IMP-51 were purified as previously described (22). During the purification process, β-lactamase activity was monitored using nitrocefin (Oxoid Ltd., Basingstoke, United Kingdom). The initial rate of hydrolysis in 50 mM Tris-HCl (pH 7.4), 0.3 M NaCl, and 10 μM Zn(NO3)2 at 37°C was determined by UV-visible spectrophotometry (V-530; Jasco, Tokyo, Japan), with the reaction initiated by the addition of substrate into spectrophotometer cells, and UV absorption measured during the initial phase of the reaction. Km, kcat, and the kcat/Km ratio were determined using a Hanes-Woolf plot. Wavelengths and extinction coefficients were used for the analysis of β-lactam substrates (23–25). Km and kcat were determined using triplicate analyses.
A DNA plug of NCGM3025, digested with I-CeuI, was prepared, separated by pulsed-field gel electrophoresis, and subjected to Southern hybridization (26) using 16S rRNA and blaIMP-51 probes (12, 27).
P. aeruginosa NCGM3025 was resistant to all antibiotics tested, except for amikacin, colistin, and tigecycline. The isolate was susceptible to amikacin and intermediate to colistin and tigecycline. The MICs of β-lactams in NCGM3025 are shown in Table 1; the MICs of other antibiotics were 16 μg/ml for arbekacin, 16 μg/ml for amikacin, 1 μg/ml for colistin, 64 μg/ml for gentamicin, 16 μg/ml for ciprofloxacin, >1,024 μg/ml for fosfomycin, and 4 μg/ml for tigecycline. NCGM3025 was positive for IMP-type MBLs and AAC(6′)-Ib. Whole-genome sequencing revealed that the isolate had a novel blaIMP variant, designated blaIMP-51. Its predicted amino acid sequence revealed that IMP-51 differed from IMP-7 by an amino acid substitution (Ser262Gly) and from IMP-43 by two amino acid substitutions (Phe67Val and Ser262Gly). A phylogenetic tree showed that IMP-51 belonged to an IMP-7-like clade (Fig. 1). In addition to blaIMP-51, NCGM3025 had several drug resistance genes, including aac(6′)-Ib-cr, aac(6′)-Ib, aph(3′)-IIb, blaPAO, blaOXA-246, blaOXA-50, cmlA1, catB7, and fosA. The isolate had a point mutation in the quinolone-resistance-determining region of gyrA with an amino acid substitution of Ser83Ile in GyrA. The MLST of NCGM3025 was sequence type 235 (ST235).
TABLE 1.
MICs of β-lactams for P. aeruginosa NCGM3025 and E. coli transformants containing blaIMP-7 and blaIMP-51
| Antibiotic(s)a | MIC (μg/ml) of antibiotic for: |
|||
|---|---|---|---|---|
| P. aeruginosa NCGM3025 | E. coli DH5α(pHSG398/IMP-7) | E. coli DH5α(pHSG398/IMP-51) | E. coli DH5α(pHSG398) | |
| Ampicillin | >1,024 | 128 | 32 | 8 |
| Ampicillin-sulbactam | 512 | 64 | 8 | 4 |
| Penicillin G | >1,024 | 128 | 32 | 32 |
| Aztreonam | 32 | 0.063 | 0.063 | 0.063 |
| Cefepime | 256 | 8 | 8 | 0.063 |
| Cefotaxime | 1,024 | 32 | 64 | 0.031 |
| Cefoxitin | >1,024 | 512 | >2,048 | 16 |
| Cefozopran | 256 | 16 | 8 | 0.125 |
| Cefpirome | 32 | 2 | 0.5 | ≤0.007 |
| Ceftazidime | 256 | 512 | 128 | 0.5 |
| Ceftriaxone | >1,024 | 64 | 128 | 0.031 |
| Cephradine | >1,024 | 512 | 64 | 16 |
| Doripenem | 256 | 2 | 4 | 0.031 |
| Imipenem | 16 | 0.25 | 0.25 | 0.031 |
| Meropenem | 512 | 1 | 4 | 0.015 |
| Panipenem | 16 | 0.25 | 0.25 | 0.063 |
| Moxalactam | >1,024 | 256 | 1,024 | 0.125 |
The ratio of ampicillin to sulbactam was 2:1.
FIG 1.
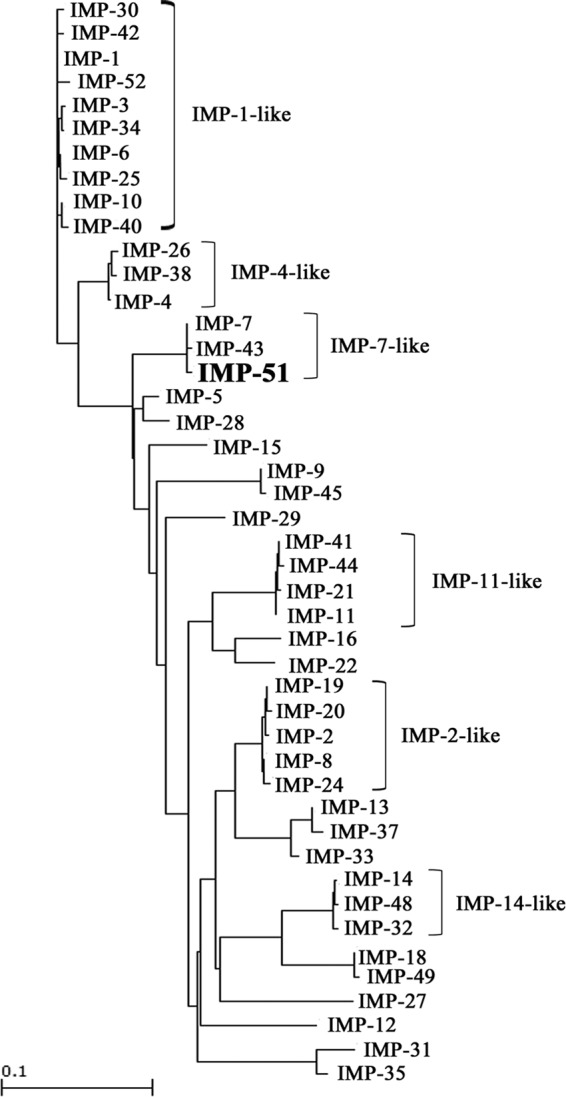
Dendrogram of 45 IMP-type MBLs for comparison with IMP-51. The dendrogram was calculated with the Clustal W2 program. Branch lengths correspond to the number of amino acid exchanges for IMP-type enzymes.
E. coli DH5α, expressing blaIMP-7 or blaIMP-51, showed a significant reduction in susceptibility to all tested β-lactams, except for aztreonam, compared with DH5α expressing a vector control (Table 1). E. coli DH5α expressing blaIMP-51 showed 4-fold higher MICs of cefoxitin, meropenem, and moxalactam, 4-fold lower MICs of ampicillin, ampicillin-sulbactam, penicillin G, cefpirome, and ceftazidime, and 8-fold lower MICs of cephradine than E. coli DH5α expressing blaIMP-7 (Table 1).
Recombinant IMP-7 and IMP-51 hydrolyzed all tested β-lactams, except for aztreonam (Table 2). IMP-51 showed markedly higher kcat/Km ratios for cefmetazole, cefotaxime, cefoxitin, doripenem, meropenem, and moxalactam and lower kcat/Km ratios for ampicillin, penicillin G, cefpirome, ceftazidime, cephradine, imipenem, and panipenem. In particular, the higher kcat values of IMP-51 than those of IMP-7 for doripenem and meropenem resulted in the higher kcat/Km ratios for IMP-51 (Table 2). The kcat/Km values of IMP-51 against cefepime were similar to those of IMP-7 (Table 2).
TABLE 2.
Kinetic parameters of IMP-7 and IMP-51 enzymesa
| β-Lactam | IMP-7 |
IMP-51 |
||||
|---|---|---|---|---|---|---|
| Km (μM)b | kcat (s−1)b | kcat/Km (μM−1 s−1) | Km (μM)b | kcat (s−1)b | kcat/Km (μM−1 s−1) | |
| Ampicillin | 116 ± 18 | 8.5 ± 1.3 | 0.02 | 872 ± 153 | 3.5 ± 0.6 | 0.004 |
| Penicillin G | 212 ± 18 | 17.5 ± 1.3 | 0.081 | 976 ± 188 | 4.6 ± 0.7 | 0.0048 |
| Aztreonam | NHc | NH | NH | NH | NH | NH |
| Cefepime | 58 ± 3 | 1.2 ± 0.1 | 0.020 | 56 ± 4 | 1.4 ± 0.1 | 0.025 |
| Cefmetazole | 47 ± 5 | 3.7 ± 0.1 | 0.078 | 1.8 ± 0.4 | 2.78 ± 0.01 | 1.5 |
| Cefotaxime | 12 ± 2 | 1.7 ± 0.1 | 0.15 | 5.7 ± 1.8 | 4.4 ± 0.2 | 0.93 |
| Cefoxitin | 120 ± 13 | 4.9 ± 0.2 | 0.041 | 2.2 ± 0.6 | 1.91 ± 0.02 | 0.88 |
| Cefpirome | 57 ± 5 | 2.0 ± 0.1 | 0.035 | 182 ± 25 | 3.4 ± 0.4 | 0.019 |
| Ceftazidime | 19 ± 3 | 0.34 ± 0.01 | 0.018 | 35 ± 4 | 0.03 ± 0.01 | 0.0085 |
| Cephradine | 55 ± 8 | 12 ± 1 | 0.22 | 75 ± 21 | 0.80 ± 0.08 | 0.011 |
| Doripenem | 46 ± 7 | 2.7 ± 0.2 | 0.059 | 61 ± 7 | 10.7 ± 0.4 | 0.18 |
| Imipenem | 104 ± 13 | 5.0 ± 0.2 | 0.048 | 312 ± 29 | 5.5 ± 0.3 | 0.018 |
| Meropenem | 59 ± 8 | 0.99 ± 0.07 | 0.017 | 51 ± 8 | 2.7 ± 0.1 | 0.053 |
| Panipenem | 40 ± 5 | 4.0 ± 0.2 | 0.099 | 230 ± 7 | 10.6 ± 0.2 | 0.046 |
| Moxalactam | 57 ± 6 | 4.6 ± 0.2 | 0.081 | 24 ± 3 | 5.0 ± 0.1 | 0.21 |
The proteins were initially modified by a His tag, which was removed after purification.
Km and kcat values represent the means ± standard deviations from three independent experiments.
NH, no hydrolysis was detected at substrate concentrations up to 1 mM and enzyme concentration up to 700 nM.
The differences of the kcat/Km values between IMP-7 and IMP-51 were well-correlated to those of the MICs of antibiotics between E. coli expressing blaIMP-7 and E. coli expressing blaIMP-51. Compared with IMP-7, IMP-51, which showed higher kcat/Km ratios for cefotaxime, cefoxitin, doripenem, meropenem, and moxalactam, conferred higher MICs for these antibiotics in E. coli, whereas IMP-51, which showed lower kcat/Km ratios for ampicillin, penicillin G, cefpirome, ceftazidime, and cephradine, conferred lower MICs for these antibiotics in E. coli (Table 1 and 2).
The sequence surrounding blaIMP-51 was determined to be tnpA-tnpR-intI1-blaIMP-51-aac(6′)-Ib-aac(6′)-cmlA1-blaOXA-246 (9,797 bp), which was obtained from a contig assembled by Genomic Workbench. The blaIMP-51 gene was located within a class I integron, of which the downstream region was not determined because it was not contained in the sequence of the contig. The genetic structure that included blaIMP-51 had a unique gene cassette array and was located on the chromosome by Southern hybridization (data not shown). In the structure, tnpA-tnpR (nucleotide 1 [nt 1] to nt 5,059) was identical to the sequence of the Tn1403-like transposon in a plasmid pOZ176 from P. aeruginosa PA96 isolated in China (28). The cmlA1-blaOXA-246 (nt 7,321 to nt 9,786) was similar to a part of the DK45-2 class 1 integron (nt 669 to nt 3,134) in P. aeruginosa DK45 isolated in South Korea (GenBank accession number GQ853420). The blaOXA-246 was first identified in a plasmid from P. aeruginosa pae943 isolated in China (GenBank accession number EU886980).
The Ser262Gly substitution in IMP-51 markedly affected the catalytic activities of the enzyme against β-lactams, especially against carbapenems. IMP-51 had higher kcat/Km ratios against doripenem and meropenem but lower kcat/Km ratios against imipenem and panipenem than those of IMP-7. These differences in catalytic activities may explain the high resistance of NCGM3025 against doripenem and meropenem (Table 1). Similarly, IMP-6 with a Ser262Gly substitution had higher activity against meropenem and panipenem than that of IMP-1 (29). Residue 262 is located near the Zn(II) binding site, which plays an important role in β-lactam turnover catalyzed by IMP-type MBLs (30). The Ser262Gly substitution in IMP-6 compared with that in IMP-1 (29) was found to stabilize the anionic intermediate of certain β-lactam substrates bound to IMP-6, enhancing catalysis (31).
In conclusion, a doripenem- and meropenem-resistant P. aeruginosa isolate producing IMP-51 has emerged in Vietnam. The Ser262Gly amino acid substitution in IMP-51 appeared to significantly increase its hydrolytic activity for doripenem and meropenem. This substitution may have arisen due to the selective pressure caused by the use of doripenem and meropenem.
Nucleotide sequence accession number.
The genomic environment surrounding blaIMP-51 was identified and deposited in GenBank under the accession number LC031883.
ACKNOWLEDGMENTS
This study was approved by the Bach Mai Hospital institutional review board (approval no. 38) and the Biosafety Committee at the National Center for Global Health and Medicine.
The study was supported by grants from International Health Cooperation Research (no. 27-S-1102 and 26-A-103) and a grant from the Research Program on Emerging and Reemerging Infectious Diseases from Japan Agency for Medical Research and Development (AMED).
REFERENCES
- 1.Bush K. 2001. New β-lactamases in Gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis 32:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- 2.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev 18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 5.Yong D, Toleman MA, Bell J, Ritchie B, Pratt R, Ryley H, Walsh TR. 2012. Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob Agents Chemother 56:6154–6159. doi: 10.1128/AAC.05654-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogalski TM, Gilbert MM, Devenport D, Norman KR, Moerman DG. 2003. DIM-1, a novel immunoglobulin superfamily protein in Caenorhabditis elegans, is necessary for maintaining bodywall muscle integrity. Genetics 163:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. 2013. FIM-1, a new acquired metallo-β-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob Agents Chemother 57:410–416. doi: 10.1128/AAC.01953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob Agents Chemother 48:4654–4661. doi: 10.1128/AAC.48.12.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother 38:71–78. doi: 10.1128/AAC.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekiguchi J, Morita K, Kitao T, Watanabe N, Okazaki M, Miyoshi-Akiyama T, Kanamori M, Kirikae T. 2008. KHM-1, a novel plasmid-mediated metallo-β-lactamase from a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother 52:4194–4197. doi: 10.1128/AAC.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-β-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob Agents Chemother 55:5143–5149. doi: 10.1128/AAC.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, Rossolini GM, Chong Y. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother 49:4485–4491. doi: 10.1128/AAC.49.11.4485-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavascki AP, Gaspareto PB, Martins AF, Goncalves AL, Barth AL. 2005. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing SPM-1 metallo-β-lactamase in a teaching hospital in southern Brazil. J Antimicrob Chemother 56:1148–1151. doi: 10.1093/jac/dki390. [DOI] [PubMed] [Google Scholar]
- 15.El Salabi A, Borra PS, Toleman MA, Samuelsen O, Walsh TR. 2012. Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob Agents Chemother 56:2241–2245. doi: 10.1128/AAC.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 43:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby GA, Munoz-Price LS. 2005. The new β-lactamases. N Engl J Med 352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Kitao T, Miyoshi-Akiyama T, Tanaka M, Narahara K, Shimojima M, Kirikae T. 2011. Development of an immunochromatographic assay for diagnosing the production of IMP-type metallo-β-lactamases that mediate carbapenem resistance in Pseudomonas. J Microbiol Methods 87:330–337. doi: 10.1016/j.mimet.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Tada T, Miyoshi-Akiyama T, Tanaka M, Narahara K, Shimojima M, Kitao T, Shimada K, Kirikae T. 2012. Development of an immunochromatographic assay for rapid detection of AAC(6′)-Ib-producing Pseudomonas aeruginosa. J Microbiol Methods 91:114–116. doi: 10.1016/j.mimet.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Kasai A, Mizuguchi Y, Araake M, Fujino T, Kikuchi H, Sasaki S, Watari H, Kojima T, Miki H, Kanemitsu K, Kunishima H, Kikuchi Y, Kaku M, Yoshikura H, Kuratsuji T, Kirikae T. 2007. Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J Clin Microbiol 45:979–989. doi: 10.1128/JCM.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tada T, Shrestha B, Miyoshi-Akiyama T, Shimada K, Ohara H, Kirikae T, Pokhrel BM. 2014. NDM-12, a novel New Delhi metallo-β-lactamase variant from a carbapenem-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother 58:6302–6305. doi: 10.1128/AAC.03355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boschi L, Mercuri PS, Riccio ML, Amicosante G, Galleni M, Frere JM, Rossolini GM. 2000. The Legionella (Fluoribacter) gormanii metallo-β-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 β-lactamases. Antimicrob Agents Chemother 44:1538–1543. doi: 10.1128/AAC.44.6.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. 1998. Overexpression, purification, and characterization of the cloned metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 42:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Queenan AM, Shang W, Flamm R, Bush K. 2010. Hydrolysis and inhibition profiles of β-lactamases from molecular classes A to D with doripenem, imipenem, and meropenem. Antimicrob Agents Chemother 54:565–569. doi: 10.1128/AAC.01004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A 90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. 2014. Biochemical analysis of the metallo-β-lactamase NDM-3 from a multidrug-resistant Escherichia coli strain isolated in Japan. Antimicrob Agents Chemother 58:3538–3540. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong J, Alexander DC, Ma JH, Deraspe M, Low DE, Jamieson FB, Roy PH. 2013. Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob Agents Chemother 57:3775–3782. doi: 10.1128/AAC.00423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pegg KM, Liu EM, George AC, LaCuran AE, Bethel CR, Bonomo RA, Oelschlaeger P. 2014. Understanding the determinants of substrate specificity in IMP family metallo-β-lactamases: the importance of residue 262. Protein Sci 23:1451–1460. doi: 10.1002/pro.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oelschlaeger P, Schmid RD, Pleiss J. 2003. Modeling domino effects in enzymes: molecular basis of the substrate specificity of the bacterial metallo-β-lactamases IMP-1 and IMP-6. Biochemistry 42:8945–8956. doi: 10.1021/bi0300332. [DOI] [PubMed] [Google Scholar]


