Abstract
Mutational acquired resistance is a major challenge in cancer therapy. Somatic tumours harbouring some oncogenic mutations are characterised by a high mortality rate. Surprisingly, preclinical evaluation methods do not show clearly resistance of mutated cancers to some drugs. Here, we implemented Raman spectral imaging to investigate the oncogenic mutation resistance to epidermal growth factor receptor targeting therapy. Colon cancer cells with and without oncogenic mutations such as KRAS and BRAF mutations were treated with erlotinib, an inhibitor of epidermal growth factor receptor, in order to detect the impact of these mutations on Raman spectra of the cells. Clinical studies suggested that oncogenic KRAS and BRAF mutations inhibit the response to erlotinib therapy in patients, but this effect is not observed in vitro. The Raman results indicate that erlotinib induces large spectral changes in SW-48 cells that harbour wild-type KRAS and BRAF. These spectral changes can be used as a marker of response to therapy. HT-29 cells (BRAF mutated) and SW-480 cells (KRAS mutated) display a smaller and no significant response, respectively. However, the erlotinib effect on these cells is not observed when phosphorylation of extracellular-signal-regulated kinase and AKT is monitored by Western blot, where this phosphorylation is the conventional in vitro test. Lipid droplets show a large response to erlotinib only in the case of cells harbouring wild-type KRAS and BRAF, as indicated by Raman difference spectra. This study shows the great potential of Raman spectral imaging as an in vitro tool for detecting mutational drug resistance.
Electronic supplementary material
The online version of this article (doi:10.1007/s00216-015-8875-z) contains supplementary material, which is available to authorized users.
Keywords: Raman imaging, Erlotinib, Oncogenic mutation, BRAF, KRAS, Colon cancer
Introduction
Cancer is one of the leading causes of death worldwide. Statistics from the World Health Organization indicate that 8.2 million cases of cancer death were registered in 2012 [1]. This high number of cancer deaths is due to the failure of many therapeutic strategies to suppress tumour progression. Some oncogenic mutations render tumours aggressive, which is accompanied by poor prognosis and a decreased response to therapy [2–4]. Therefore, researchers in drug development are competing to find new potent anticancer drugs that can overcome the mutational drug resistance. Hundreds of new anticancer drugs appear each year. To evaluate their potencies, they are studied in a series of in vitro and in vivo preclinical tests. The promising candidates are then proceeded to clinical trials, to be tested on human cancer patients [5]. However, preclinical methods do not predict the resistance of some oncogenic mutations to therapy, which is later detected in many clinical studies. For instance, inhibitors of epidermal growth factor receptor (EGFR) showed great therapeutic potential in solid tumour suppression [6]. However, some of them failed in patients with KRAS-mutated cancers [7].
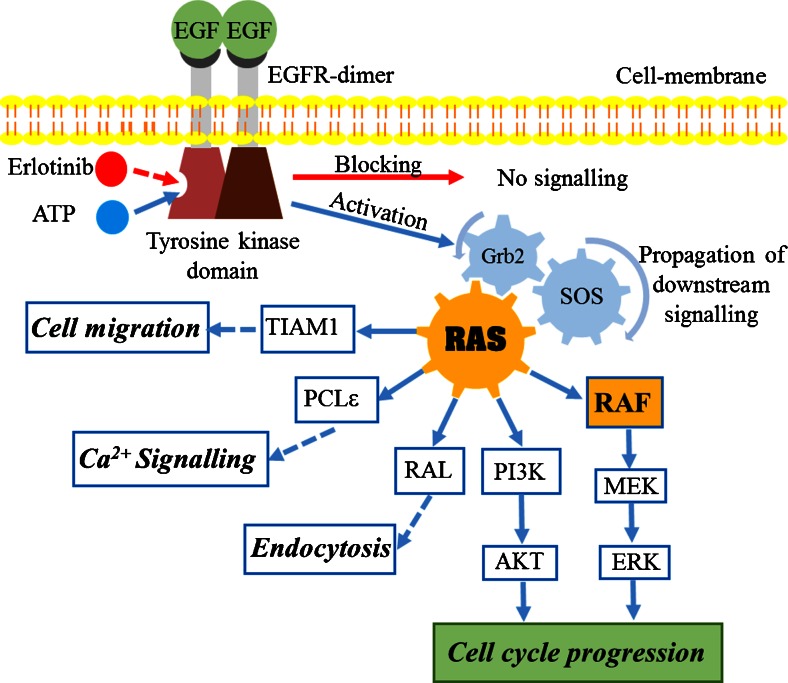
Owing to the overexpression of EGFR in many cancers, targeting EGFR is one of the effective strategies to suppress tumours in advanced stages [6,8]. EGFR is a transmembrane protein with extracellular, transcellular, and intracellular domains. It is a member of the ERBB family of receptors. Upon binding of a ligand such as epidermal growth factor (EGF), EGFR dimerises with other homologous members of the ERBB receptors, which promotes phosphorylation of the intracellular tyrosine kinase (TK) domain, which in turn stimulates cell cycle progression as shown in Fig. 1. Erlotinib (Tarceva) is a small molecule that inhibits the phosphorylation of the EGFR TK domain. It blocks downstream signalling transduction and tumour progression [9]. It is clinically approved by the US Food and Drug Administration and the European Medicines Agency for the treatment of advanced non-small-cell lung, and pancreatic cancers [10–15]. Erlotinib is still in clinical trials for the treatment of colon cancer [16].
Fig. 1.
Epidermal growth factor receptor (EGFR)–RAS downstream signalling cascade
RAS is a membrane-anchored guanosine triphosphatase protein [17]. It controls multiple downstream signalling pathways and acts as an on/off signalling switch (Fig. 1). Activated EGFR sets RAS to on mode, which in turn activates multiple effector proteins such as rapidly accelerated fibrosarcoma (RAF), phosphatidylinositiol 3-kinase (PI3K), TIAM, phospholipase Cε (PCLε), and RAL for cell proliferation and survival [17–19]. In the case of EGFR inhibition, RAS remains in off mode, and the cell cycle is arrested [20]. However, mutated RAS is chronically switched to on mode, and it becomes independent of EGFR activation. Therefore, RAS-mutated tumours are aggressive, invasive, and metastatic. Moreover, it is accompanied by a decreased response to EGFR-inhibition therapy [4,7,17,18,21–25]. KRAS mutation accounts for 86 % of all RAS mutations [26]. It is commonly found in the most deadly cancer types: lung (33 %), colon (40 %), and pancreatic (90 %) cancers [27,28]. KRAS mutation also is reported to be predictive for poor prognosis and low survival rate in cancer [22,25,29].
RAF is one of the well-identified RAS effector proteins, with serine/threonine kinase activity [17]. RAF is activated by binding with the active form of RAS, which subsequently stimulates the mitogen-activated protein kinase–extracellular-signal-regulated kinase (ERK) pathway proteins through a cascade of autophosphorylation events towards cell proliferation (Fig. 1) [30]. BRAF mutation occurred in up to 80 % of skin cancers and 5-10 % of colon cancers [31]. In addition, BRAF mutation is accompanied by an elevated kinase activity, which increases ERK phosphorylation [32,33]. Clinical studies showed that cancer patients with BRAF mutation have a relatively poor prognosis [25,34].
Oncogenic mutations are commonly detected by DNA sequencing and methods based on polymerase chain reaction [35,36]. In vitro assessment of drug effects is done separately by cytotoxicity assays [5]. Although, some in vivo methods such as genetically engineered cancer models revealed promising results in detecting drug resistance to mutations [37,38], they still have some drawbacks, such as requiring a long time and unpredictability of tumour formation [39]. To the best of our knowledge, no in vitro method has reported so far the impact of oncogenic mutations on response to EGFR molecularly targeted therapy.
Raman micro-spectroscopic imaging is an emerging technique in biomedical research. Raman spectroscopy can measure biological samples in an aqueous physiological environment. It is a label-free, non-invasive technique with high spectral/lateral resolution and great reproducibility [40–43]. Raman spectral imaging can classify cancerous human tissues [44,45]. It can be used for imaging of single cells and characterisation of subcellular components [46–50]. Furthermore, Raman imaging can be conducted to monitor drug uptake and its effect on single cells [51–57]. In our previous work we investigated the distribution and metabolism of erlotinib in SW-480 colon cancer cells using its unique –C ≡ C– band at 2100 cm−1, which is used as a marker band for erlotinib localisation [53].
Here, we implemented Raman imaging coupled with hierarchical cluster analysis (HCA) to monitor the response of colon cancer cells to erlotinib therapy. We report in vitro evidence that detects the effect of oncogenic KRAS and BRAF mutations on the cellular response to erlotinib. The Raman results show that colon cancer cells experience a large spectral response to erlotinib, but colon cancer cells expressing oncogenic BRAF or KRAS mutations experience small or no relevant effects, respectively. Furthermore, the largest effect is observed in lipid droplets of cancer cells harbouring wild-type BRAF and KRAS that were treated with erlotinib.
Material and methods
Cell culture
The colon cancer cell lines SW-48, HT-29, and SW-480 were purchased from American Type Culture Collection. Cells were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies, Darmstadt, Germany) supplemented with 10 % fetal bovine serum (Life Technologies, Darmstadt, Germany), 2 mM l-glutamine, and 5 % penicillin–streptomycin, and were incubated at 37 °C in a 10 % CO2 atmosphere. Cells were subcultured to 80 % confluence, detached by trypsin–EDTA (0.25 %) (Gibco trypsin solution, Life Technologies, Darmstadt, Germany), centrifuged at 1500 rpm for 3 min and diluted to 10 %, then seeded again in culture medium. Raman measurements were performed on cells grown on CaF2 windows (Korth Kristalle, Kiel, Germany) to avoid Raman scattering from regular glass slides. Cells were incubated with erlotinib (Tarceva; Roche, Switzerland) at 10 μg/ml at 37 °C in 10 % CO2 for 12 h. Subsequently, cells were fixed in 4 % paraformaldehyde (VWR International, Darmstadt, Germany) and then submerged in phosphate-buffered saline (Life Technologies, Darmstadt, Germany).
Confocal Raman microscopy
Cancer cells were measured with an alpha300 AR confocal Raman microscope (WITec, Ulm, Germany) as described previously [45,50,53]. Briefly, the setup excitation source was a frequency-doubled Nd:YAG laser of 532 nm (CrystaLaser, Reno, NV, USA) with an output power of around 40 mW. The laser radiation was coupled into a Zeiss microscope through a wavelength-specific single-mode optical fibre. An achromatic lens used as a collimator for the laser beam, and a holographic band-pass filter focused the beam on the sample through a Nikon NIR APO (×60/1.00 numerical aperture) water immersion objective. The sample was located on a piezoelectrically driven microscope scanning stage, which has an x,y resolution of 3 nm and a z resolution of 0.3 nm. Raman back-scattered light was collected through a microscopic objective and passed through a holographic edge filter into a multimode fibre (50-μm diameter) and into a 300-mm focal length monochromator, which incorporated a 600/mm grating blazed at 500 nm. A back-illuminated deep-depletion charge-coupled device camera operating at −60 °C was used to detect Raman spectra. We acquired Raman imaging measurements by raster scanning the laser beam over cells and collecting a full Raman spectrum at speed of 0.5 s per pixel, with a pixel resolution of 500 nm.
Multivariate analysis
HCA and principal component analysis (PCA) were performed on Raman spectra of cells. Raman hyperspectral results were exported to MATLAB 8.2 (The MathWorks, Natick, MA, USA). Scripts written in-house were used to perform data preprocessing and multivariate analyses. Raman spectra without a C–H band at 2850–3000 cm−1 were treated as the background and were deleted. An impulse noise filter was used to remove cosmic spikes, and the Raman spectra were interpolated to a reference wavenumber scale. All spectra were then baseline corrected with a third-order polynomial and were also vector normalised. HCA was performed on the regions from 700 to 1800 cm−1 and 2800 to 3050 cm−1 with use of Ward’s clustering in combination with the Pearson correlation distance. All Raman mean spectra obtained from HCA were normalised with the phenylalanine band near 1008 cm−1. We calculated Raman difference spectra of cells by subtracting the mean spectrum of erlotinib-treated cells from that of control cells for each component identified by HCA. PCA was performed on the mean spectra for all subcellular components identified by HCA of both control and erlotinib-treated cells, and the first three PCs generated denote the higher variances within the data set [58]. PC1 loadings of respective subcellular components of control and erlotinib-treated cells were paired for comparison.
Western blot
Phosphatase inhibitor/buffer mixture II (Sigma-Aldrich, Munich, Germany) and protease inhibitor cocktail (Roche) were used to harvest the cells. They were resolved by sodium dodecyl sulfate–10 % polyacrylamide gel electrophoresis, and transferred to Immobilon-P membranes (Millipore, Schwalbach am Taunus, Germany). The membranes were then incubated with antibodies to activate phosphorylated ERK1 and ERK2 (p-ERK; Cell Signaling Technology, Danvers, MA, USA), total ERK1 and ERK2 (Cell Signaling Technology, Danvers, MA, USA), phosphorylated AKT (p-AKT; Cell Signaling Technology, Danvers, USA), and total AKT (Cell Signaling Technology, Danvers, USA). Antibodies were detected with the appropriate anti-mouse horseradish peroxidase conjugated secondary antibody enhanced by chemiluminescence (Pierce, Life Technologies, Darmstadt, Germany), and images were captured with a Versa Doc 5000 imaging system (Bio-Rad, Munich, Germany).
Results and discussion
Downstream signalling and oncogenic mutations by Western blot
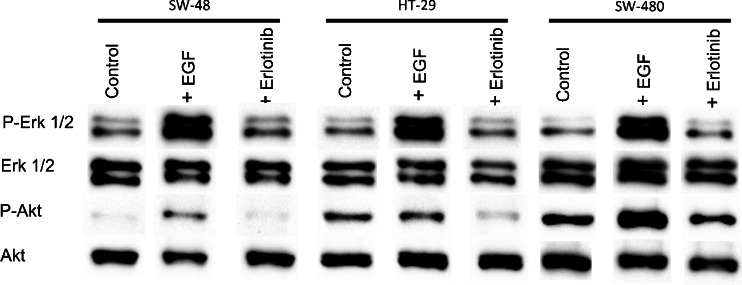
Binding of EGF to EGFR leads to dimerisation of EGFR, followed by phosphorylation of the EGFR TK domain. Subsequently, it leads to activation of the RAS machinery, which in turn activates multiple pathways towards cell proliferation (Fig. 1) [59,60]. Erlotinib blocks the receptor phosphorylation and consequently prevents RAS activation and inhibits cancer cell proliferation [10,11]. We performed Western blot experiments to detect the effect of erlotinib on the phosphorylation of two RAS downstream signalling pathways, RAF–ERK and PI3K–AKT, as shown in Fig. 2. It is expected that BRAF mutation in HT-29 cells and KRAS mutation in SW-480 cells can confer activation of p-ERK and p-AKT in control cells [61]. This activation is shown in Fig. 2. Although SW-48 cells harbour wild type KRAS and BRAF, they also show activation of p-ERK. This is because SW-48 cells carry a mutant EGFR allele [62]. EGF binding to EGFR increases the ERK and AKT phosphorylation level in these cancer cells.
Fig. 2.
Effect of EGF (50 ng/ml for 15 min) and erlotinib (10 μg/ml for 2 h) on ERK and AKT activation in SW-48, HT-29, and SW-480 colon cancer cells. Cell lysates were resolved by sodium dodecyl sulfate–Polyacrylamide gel electrophoresis and Western blot analysis with use of antibodies that recognise phosphorylated ERK1 and ERK2 (p-Erk 1/2) and phosphorylated AKT (p-Akt) or total ERK1 and ERK2 (Erk 1/2), and AKT (Akt)
Furthermore, no significant difference in p-ERK level was observed for erlotinib-treated SW-48, HT-29 and SW-480 cells compared with the control. This is perhaps due to the activating mutations of BRAF in HT-29 cells and KRAS in SW-480 cells as well as the EGFR mutation in SW-48 cells. Similar results were observed for the p-AKT level for erlotinib-treated SW-48 and SW-480 cells. On the other hand, the p-AKT level was reduced in erlotinib-treated HT-29 cells. This means that the Western blotting results did not significantly discriminate between the KRAS and BRAF status in these cells. These results are also in agreement with a study conducted to detect the effect of erlotinib on p-ERK and p-AKT in a series of pancreatic cancer cell lines with wild-type and mutated KRAS [63].
Effect of erlotinib on SW-48 cells by Raman spectral imaging
Erlotinib is a reversible competitive inhibitor of the TK domain of EGFR that binds to its adenosine 5′-triphosphate binding site. To detect the effect of erlotinib binding to the TK domain, several Raman measurements of SW-48 cells were performed. Cells were incubated with erlotinib at 10 μg/ml for 12 h. The concentration of erlotinib used in this study is similar to that detected in the plasma of cancer patients taking this drug. The oral dosage of erlotinib in clinics is 150 mg/day, and the erlotinib concentration in the plasma was found to be 2.5–29 μg/ml [64]. No contribution from erlotinib to the Raman spectra itself was expected because of its low concentration, which cannot be detected under our experimental conditions.
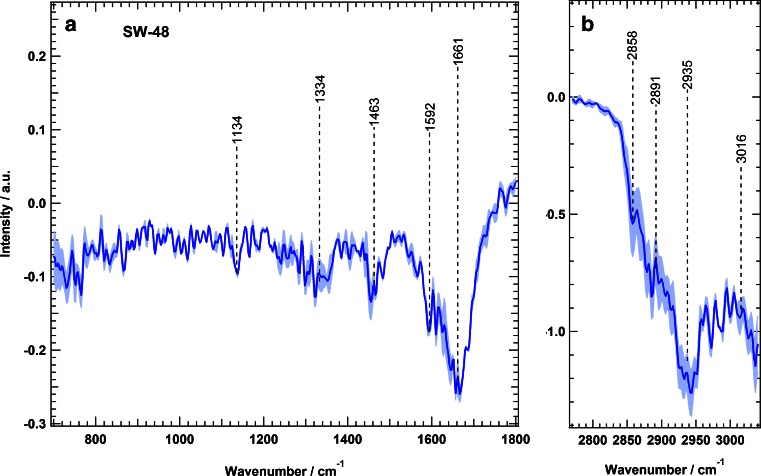
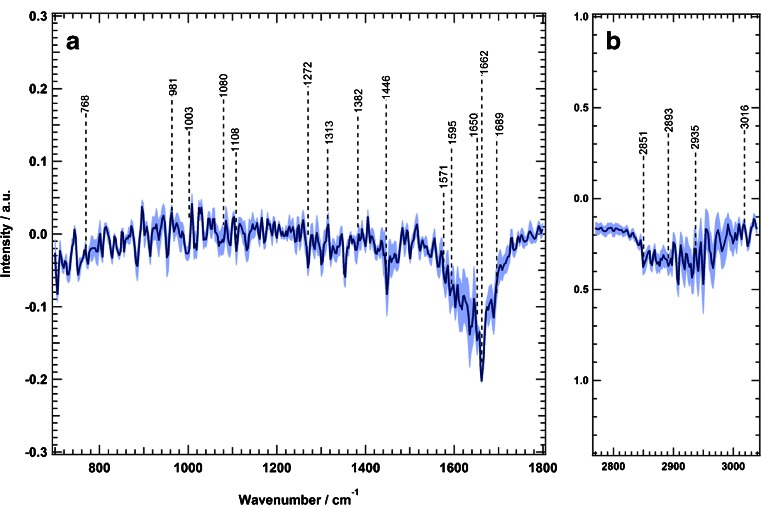
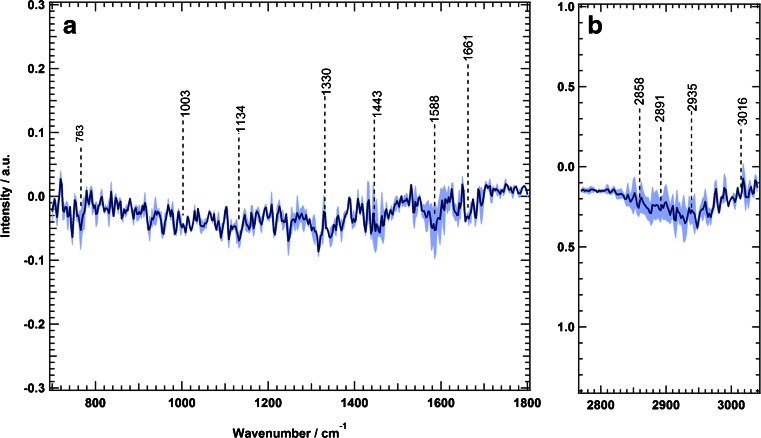
We calculated the Raman difference spectrum of SW-48 cells by subtracting the mean spectrum of erlotinib-treated cells from that of the control cells; the results are presented in Fig. 3. The difference spectrum shows large spectral changes, which are represented by negative peaks at 1134 cm−1 (C–C/C–N stretching), 1334 cm−1 (amide III), 1463 cm−1 (C–H and CH2 bending deformation), 1592 cm−1 (C = C bending), 1661 cm−1 (amide I), and 2858, 2891, and 2935 cm−1 (C–H stretching) [42,65,66]. These changes can be attributed to the changes in proteins, nucleic acids, lipids, and carbohydrates that are constituents of the cell resulting from erlotinib treatment. Erlotinib-driven biochemical changes of subcellular components would result in cell cycle arrest and eventually lead to apoptosis [6,67–69].
Fig. 3.
Raman difference spectra of SW-48 cells for control cells and erlotinib-treated cells in two spectral regions: a 700–1800 cm−1 and b 2800–3100 cm−1. The shading represents the standard deviation
These results encouraged us to improve the spatial resolution of the cell response to erlotinib. We intended to emphasise the changes of subcellular components in response to erlotinib treatment. Subcellular organelles were determined by Raman spectroscopy coupled with HCA. Figure 4 shows a Raman intensity image of SW-48 cells in the absence of erlotinib (control) based on the integrated Raman intensities of the C–H stretching vibration in the 2800–3100-cm−1 region. The size of the image is 45 × 45 μm2 (90 × 90 pixels).
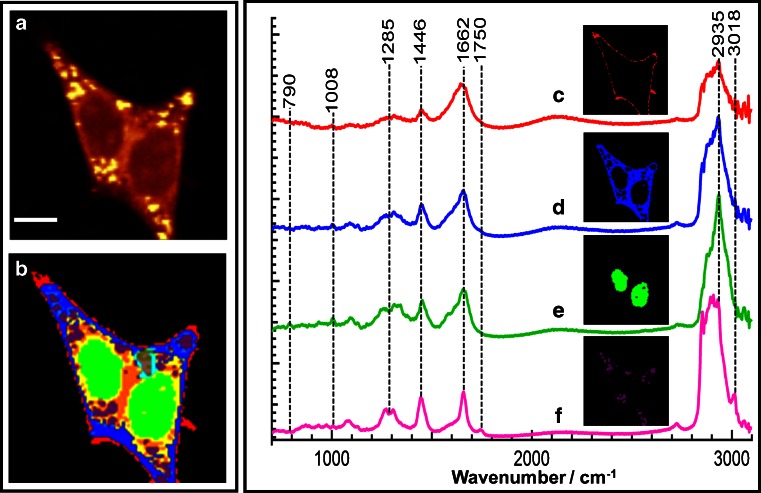
Fig. 4.
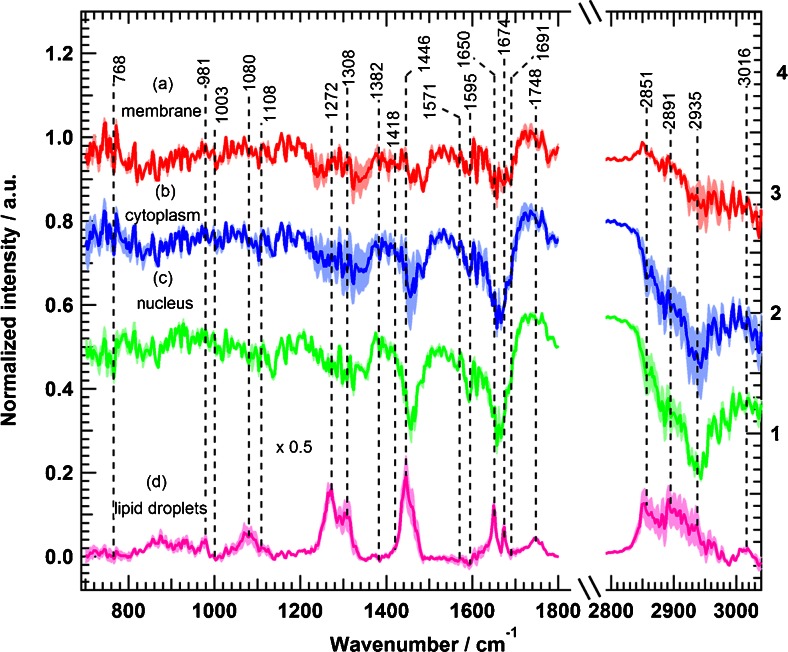
a Raman intensity image constructed in the C–H stretching region of SW-48 cells. b hierarchical cluster analysis (HCA) results based on the Raman data shown in a. Mean spectra and the respective spatial distribution of HCA clusters: c plasma membrane, d cytoplasm, e nucleus, and f lipid droplets. The scale bar in a is 9 μm
Next, HCA was performed in the spectral ranges from 700 to 1800 cm−1 and 2800 to 3100 cm−1 in order to construct a cluster map based on the Raman intensity image (Fig. 4, panel a); the result is shown in Fig. 4, panel b. The clusters were then merged to four major subcellular components: plasma membrane, cytoplasm, nucleus, and lipid droplets. This merging process was based on use of the spectral marker bands of each cell component. Afterwards, mean spectra of the four clusters were calculated (Fig. 4, spectra c–f). The outermost cluster (Fig. 4, spectrum c) is considered to contain the plasma membrane. However, the plasma membrane is smaller than the laser spot (less than 1 μm); therefore, the membrane cluster might contain contributions from the buffer and cytoplasm (Fig. 4, spectrum d). The mean spectrum of the nucleus (Fig. 4, spectrum e) contains characteristic Raman bands such as the stretching vibration of DNA phosphodiester at 790 cm−1. The spectrum of the cytoplasm (Fig. 4, spectrum d) and that of the nucleus (Fig. 4, spectrum e) both contain the Raman spectral band of the ring-breathing mode of phenylalanine at 1008 cm−1, which represents their protein constituent. However, cytoplasm spectra do not contain the DNA marker band at 790 cm−1 [55–57,70]. The Raman mean spectrum of lipid droplets (Fig. 4, spectrum f) shows characteristic bands of the C = O stretching vibration of the ester form of fatty acids at 1750 cm−1 and the = C―H stretching vibration of unsaturated fatty acids at 3018 cm−1. The spectra of the nucleus and lipid droplets are consistent with those reported previously [42,43,49,50].
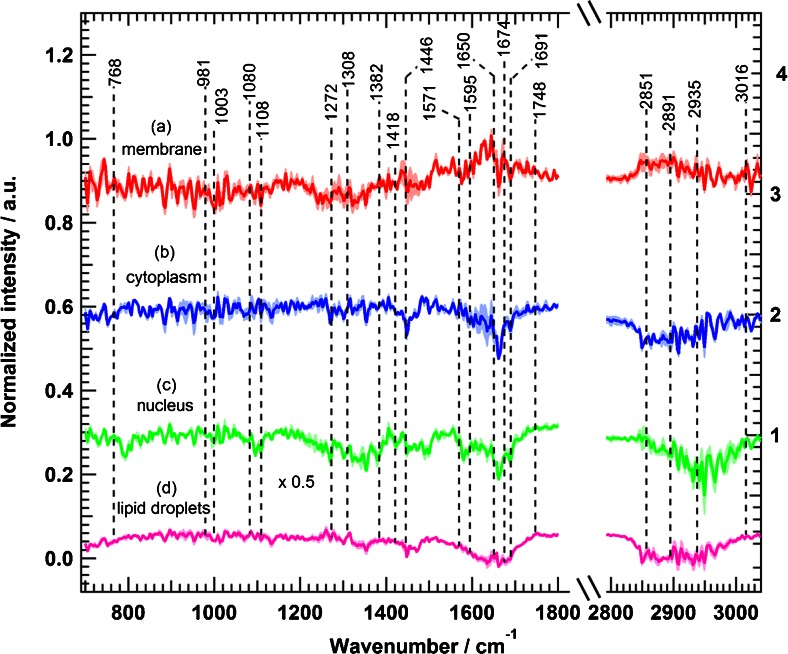
To monitor the effect of erlotinib on cells, several Raman measurements of control cells (SW-48) and cells treated with erlotinib were acquired. Data analysis and HCA were performed for all Raman measurements in ways similar to those for the data presented in Fig. 4. All Raman mean spectra obtained from HCA were normalised with the phenylalanine band near 1008 cm−1. We calculated Raman difference spectra of cellular components by subtracting the mean spectrum of erlotinib-treated cells from that of control cells for each component identified by HCA; the results are shown in Fig. 5. Obvious spectral changes of the four cellular components were detected. In addition, the pairwise loading spectra from PCA of each component displayed clear differences on erlotinib treatment (see Figs. S1–S4). For example, the Raman difference spectrum of the membrane (Fig. 5, spectrum a) indicates negative peaks at 1231–1259 cm−1 (amide III, and PO2−, phospholipid region), 1321 cm−1 (CH2 twisting), 1458–1483 cm−1 (CH deformation), 1642–1688 cm−1 (amide I), and 2851 and 2891 cm−1 (C–H stretching). These spectral differences refer to changes in the lipid and protein constituents of the plasma membrane. The cytoplasm difference spectrum (Fig. 5, spectrum b) displays larger changes than that of the membrane, and shows negative peaks at 1231–1259 cm−1 (amide III), 1321 cm−1 (CH2 twisting), 1455 cm−1 (C–H deformation), 1592 cm−1 (C = C bending), 1642–1688 cm−1 (amide I), and 2851–2950 cm−1 (C–H stretching). In the case of the nucleus, the Raman difference spectrum (Fig. 5, spectrum c) reveals clear changes at 1134 cm−1 (C–C/C–N stretching), 1231–1259 cm−1 (amide III), 1321 cm−1 (CH2 twisting), 1458 cm−1 (C–H deformation), 1592 cm−1 (C = C bending), 1657–1666 cm−1 (amide I), and 2851–2950 cm−1 (C–H stretching).
Fig. 5.
Raman difference spectra of SW-48 cells for control cells and erlotinib-treated cells for a plasma membrane, b cytoplasm, c nucleus, and d lipid droplets. The shading represents the standard deviation
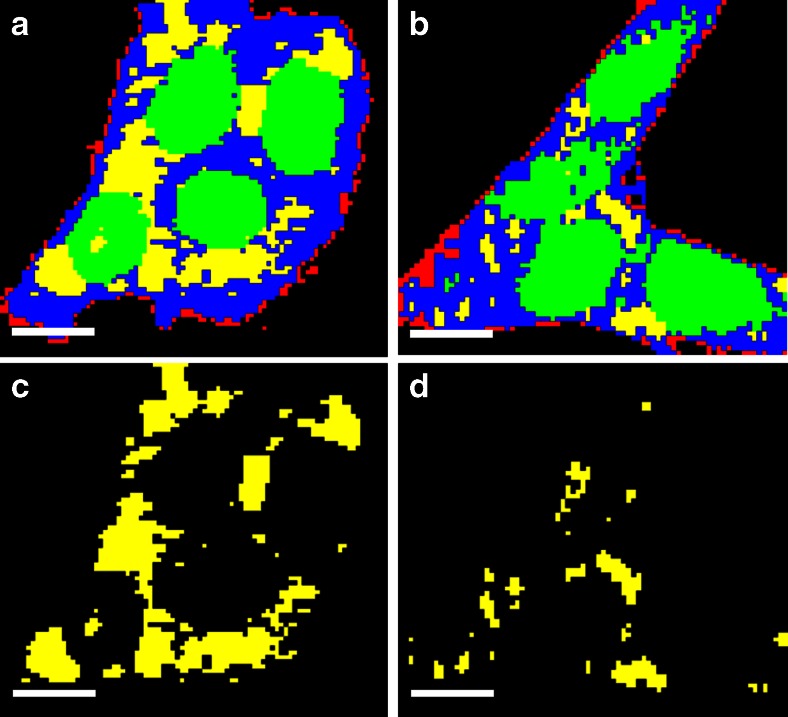
The largest changes of all the cell components were observed in the Raman difference spectrum of lipid droplets (Fig. 5, spectrum d). It exhibits intense positive peaks at 1272 cm−1 (CH3 twisting), 1308 cm−1 (CH2 twisting), 1446 cm−1 (C–H deformation), 1650–1674 cm−1 (C = C– stretching vibration of unsaturated fatty acids), 1744 cm−1 (=C–H stretching vibration of unsaturated fatty acids), 2851–2950 cm−1 (CH stretching), and 3016 cm−1 (=C–H stretching). These strong positive peaks refer to a decrease in the intensity of the mean spectrum of lipid droplets in erlotinib-treated cells. This might be produced as a result of a decrease in the number of lipid droplets in erlotinib-treated cells. In fact, lipid droplets play a major role in cell proliferation. It was believed that the only function of lipid droplets is to store and metabolise lipids. However, it is found that lipid droplets are also involved in protein storage, degradation, and trafficking processes [71]. Lipid droplets are also accumulated in cancer cells, and they induce proliferation in colon cancer [72,73]. In addition, lipid droplets decrease in number or disappear in cells treated with drugs, and the amount of lipid droplets might have some effect in drug resistance of tumour cells [74]. Even more, erlotinib treatment results in reducing accumulation of lipid droplets in human cardiomyocytes [75]. Thus, lipid droplets have become an important target in cancer therapy [72,73]. HCA images based on Raman measurements of SW-48 cells show that the number of lipid droplets decreases in cells treated with erlotinib (Fig. 6b, d) in comparison with control cells (Fig. 6a, c). Given these facts, we suggest that the positive peaks of lipid droplets in the difference spectrum (Fig. 5d) are produced as a result of reduction of lipid droplet numbers in erlotinib-treated cells when compared with control cells. In contrast to lipid droplets, the Raman difference spectra revealed negative peaks for the membrane, cytoplasm, and nucleus on erlotinib treatment, implying that the Raman intensity of these components is increased in erlotinib-treated cells. This is perhaps due to an increase in the expression level of proteins during apoptosis on erlotinib treatment [68,69,76].
Fig. 6.
HCA results based on the Raman intensity images of a SW-48 control cells and b SW-48 erlotinib-treated cells. The clusters are shown in red for plasma membrane, blue for cytoplasm, green for nuclei, and yellow for lipid droplets. c HCA cluster of lipid droplets from a. d HCA cluster of lipid droplets from b. The scale bar is 7 μm
Resistance of oncogenic mutations to erlotinib therapy by Raman spectral imaging
We applied the experimental conditions used for SW-48 cells to the other two colon cancer cell lines, HT-29, and SW-480, which have oncogenic mutations of BRAF (V600E), and KRAS (G12V), respectively [77]. Figure 7 displays the Raman difference spectrum of HT-29 cells, which shows relatively few spectral differences when compared with the Raman difference spectrum of SW-48 cells (Fig. 3). Spectral changes are observed (Fig. 7) at 1446, 1595–1690, 2851, 2893, and 2935 cm−1. Further clustering of four subcellular components of HT-29 cells was performed, and the Raman difference spectra detect some small spectral changes in the plasma membrane, cytoplasm, nucleus, and lipid droplets, as indicated in Fig. 8.
Fig. 7.
Raman difference spectra of HT-29 cells for control cells and erlotinib-treated cells in two spectral regions: a 700–1800 cm−1 and b 2800–3100 cm−1 . The shading represents the standard deviation
Fig. 8.
Raman difference spectra of HT-29 cells for control cells and erlotinib-treated cells for a plasma membrane, b cytoplasm, c nucleus, and d lipid droplets. The shading represents the standard deviation
In the case of the membrane (Fig. 7, spectrum a), the Raman difference spectrum shows changes at 1446, 1600–1660, and 2851–3935 cm−1. The cytoplasm difference spectrum (Fig. 7, spectrum b) displays negative spectral changes at 1448, 1650–1674, and 2851–2935 cm−1. In addition, the nucleus difference spectrum (Fig. 7, spectrum c) shows negative spectral changes at 1095, 1300–1360, 1583, 1650–1690, and 2891–2980 cm−1. Finally, the lipid droplet spectrum (Fig. 7, spectrum d) exhibits notable changes at 1272, 1595–1690, 1748, and 2851–2935 cm−1. To compare the observed spectral changes for both SW-48 cells (Fig. 3 or Fig. 5) and HT-29 cells (Fig. 7 or Fig. 8), the same intensity scale is used. HT-29 cells (Figs. 7 and 8), harbouring BRAF mutation, showed a small response to erlotinib treatment in comparison with SW-48 cells (Figs. 3, 5). Furthermore, Fig. 9 shows the calculated Raman difference spectrum of SW-480 cells. The spectrum reveals no significant spectral changes between control and erlotinib-treated cells. Thus, the Raman results show that HT-29 cells (BRAF mutated) and SW-480 cells (KRAS mutated) display a smaller and not significant change, respectively, on erlotinib treatment.
Fig. 9.
Raman difference spectra of SW-480 cells for control cells and erlotinib-treated cells in two spectral regions: a 700–1800 cm−1 and b 2800–3100 cm−1. The shading represents the standard deviation
It is worth mentioning that BRAF mutation affects mainly one signalling pathway for cell proliferation, whereas KRAS mutation affects multiple pathways (Fig. 1); therefore, the impact of BRAF mutation is less than that of KRAS mutation. In agreement with the present Raman results, clinical studies on human patients harbouring BRAF- or KRAS-mutated tumours have shown a limited response to TK inhibitors [22,25,29,34,78]. On the other hand, the Western blot results could not discriminate between erlotinib-treated cells harbouring wild-type KRAS and BRAF and those with KRAS and BRAF mutations through monitoring the ERK phosphorylation. In addition, the AKT phosphorylation is inhibited only in cells with BRAF mutations. In Western blot, antibodies are used to mark single proteins although cellular proteins are highly coupled in protein networks, and monitoring of a single protein may not reflect the in vivo cellular response to drugs such as erlotinib. This might explain the differences between the findings of in vitro drug studies and the drug effect observed in cancer patients. On the other hand, Raman microscopy reflects the overall chemical composition of the cell, and Raman difference spectra can detect the changes in the whole cell contents, which would be an efficient way to exhibit the overall cellular and subcellular component changes upon drug treatment.
Developing a new preclinical method that would predict the resistance of mutated cancers to therapy can reduce the cost, time, and effort spent in the new drug discovery process. It is one step further to increase the accuracy of in vitro drug evaluation, before these drugs proceed to clinical trials to be tested on human patients. In fact, clinical trials suffer from many ethical and economical limitations [5]. Consequently, increasing the accuracy of in vitro evaluation tests would have a significant impact on accelerating the process of new anticancer drug evaluation and final approval.
Conclusion
Raman microscopy coupled with clustering analysis such as HCA can be used to detect the subcellular changes induced by drug treatment. Raman difference spectra can also emphasise the oncogenic mutational drug resistance in EGFR-targeted therapy. Erlotinib resistance in BRAF- and KRAS-mutated colon cancers is clearly reflected in their Raman difference spectra. Cancer cells with wild-type KRAS and BRAF show clear changes upon erlotinib treatment, whereas BRAF- and KRAS-mutated cancer cells display a limited change or no significant changes, respectively. Consistently, clinical studies revealed that human cancer patients with BRAF- or KRAS-mutated tumours have the worst therapy prognosis. Accordingly, in vitro Raman imaging is a valuable addition that can enhance the sensitivity of the preclinical evaluation methods for new anticancer agents.
Electronic supplementary material
(PDF 1.86 mb)
Acknowledgments
We thank Carsten Kötting for helpful discussions. We also thank Dennis Petersen for his assistance in the PCA. This study was supported by the Protein Research Unit Ruhr Within Europe (PURE), the Ministry of Innovation, Science and Research (MIWF) of North Rhine–Westphalia, Germany, and the Center for Vibrational Microscopy, European Regional Development Fund, the European Union, and North Rhine–Westphalia, Germany, and the Deutsche Forschungsgemeinschaft under grant number: SFB 642. This study was also supported by a fellowship to H.K.Y. from the Konrad Adenauer Foundation.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.International Agency for Research on Cancer, World Health Organization . World cancer report 2014. Lyon: International Agency for Research on Cancer; 2014. [Google Scholar]
- 2.Fujimoto K. Transforming growth factor-β1 promotes Invasiveness after cellular transformation with activated Ras in Intestinal epithelial cells. Exp Cell Res. 2001;266:239–249. doi: 10.1006/excr.2000.5229. [DOI] [PubMed] [Google Scholar]
- 3.Sumimoto H, Miyagishi M, Miyoshi H, Yamagata S, Shimizu A, Taira K, Kawakami Y. Inhibition of growth and invasive ability of melanoma by inactivation of mutated BRAF with lentivirus-mediated RNA interference. Oncogene. 2004;23:6031–6039. doi: 10.1038/sj.onc.1207812. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, Duval A, Hamelin R, Machado JC, Schwartz S, Carneiro F, Seruca R. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26:158–163. doi: 10.1038/sj.onc.1209758. [DOI] [PubMed] [Google Scholar]
- 5.Zips D, Thames HD, Baumann M. New anticancer agents: in vitro and in vivo evaluation. In Vivo. 2005;19:1–7. [PubMed] [Google Scholar]
- 6.Goel S, Hidalgo M, Perez-Soler R. EGFR inhibitor-mediated apoptosis in solid tumors. J Exp Ther Oncol. 2007;6:305–320. [PubMed] [Google Scholar]
- 7.Raponi M, Winkler H, Dracopoli NC. KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol. 2008;8:413–418. doi: 10.1016/j.coph.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 9.Dowell J, Minna JD, Kirkpatrick P. Fresh from the pipeline: Erlotinib hydrochloride. Nat Rev Drug Discov. 2005;4:13–14. doi: 10.1038/nrd1612. [DOI] [PubMed] [Google Scholar]
- 10.Tang PA, Tsao M-S, Moore MJ. A review of erlotinib and its clinical use. Expert Opin Pharmacother. 2006;7:177–193. doi: 10.1517/14656566.7.2.177. [DOI] [PubMed] [Google Scholar]
- 11.Siegel-Lakhai WS. Current knowledge and future directions of the selective epidermal growth factor receptor inhibitors erlotinib (Tarceva®) and gefitinib (Iressa®) Oncologist. 2005;10:579–589. doi: 10.1634/theoncologist.10-8-579. [DOI] [PubMed] [Google Scholar]
- 12.Duckett DR, Cameron MD. Metabolism considerations for kinase inhibitors in cancer treatment. Expert Opin Drug Metab Toxicol. 2010;6:1175–1193. doi: 10.1517/17425255.2010.506873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchetti A, Milella M, Felicioni L, Cappuzzo F, Irtelli L, Del Grammastro M, Sciarrotta M, Malatesta S, Nuzzo C, Finocchiaro G, Perrucci B, Carlone D, Gelibter AJ, Ceribelli A, Mezzetti A, Iacobelli S, Cognetti F, Buttitta F. Clinical implications of KRAS mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia. 2009;11:1084–1092. doi: 10.1593/neo.09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brugger W, Triller N, Blasinska-Morawiec M, Curescu S, Sakalauskas R, Manikhas GM, Mazieres J, Whittom R, Ward C, Mayne K, Trunzer K, Cappuzzo F. Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:4113–4120. doi: 10.1200/JCO.2010.31.8162. [DOI] [PubMed] [Google Scholar]
- 15.Parsons BL, Myers MB. KRAS mutant tumor subpopulations can subvert durable responses to personalized cancer treatments. Pers Med. 2013;10:191–199. doi: 10.2217/pme.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townsley CA, Major P, Siu LL, Dancey J, Chen E, Pond GR, Nicklee T, Ho J, Hedley D, Tsao M, Moore MJ, Oza AM. Phase II study of erlotinib (OSI-774) in patients with metastatic colorectal cancer. Br J Cancer. 2006;94:1136–1143. doi: 10.1038/sj.bjc.6603055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 19.Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 21.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 22.Arrington AK, Heinrich EL, Lee W, Duldulao M, Patel S, Sanchez J, Garcia-Aguilar J, Kim J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int J Mol Sci. 2012;13:12153–12168. doi: 10.3390/ijms131012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 24.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 25.Eklöf V, Wikberg ML, Edin S, Dahlin AM, Jonsson B-A, Öberg Å, Rutegård J, Palmqvist R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108:2153–2163. doi: 10.1038/bjc.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 28.Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 29.Phipps AI, Buchanan DD, Makar KW, Win AK, Baron JA, Lindor NM, Potter JD, Newcomb PA. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer. 2013;108:1757–1764. doi: 10.1038/bjc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong H, Vikis HG, Guan K-L. Mechanisms of regulating the Raf kinase family. Cell Signal. 2003;15:463–469. doi: 10.1016/S0898-6568(02)00139-0. [DOI] [PubMed] [Google Scholar]
- 31.Borràs E, Jurado I, Hernan I, Gamundi M, Dias M, Martí I, Mañé B, Arcusa À, Agúndez JA, Blanca M, Carballo M. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer. 2011;11:406. doi: 10.1186/1471-2407-11-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zebisch A, Troppmair J. Back to the roots: the remarkable RAF oncogene story. Cell Mol Life Sci. 2006;63:1314–1330. doi: 10.1007/s00018-006-6005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leicht DT, Balan V, Kaplun A, Singh-Gupta V, Kaplun L, Dobson M, Tzivion G. Raf kinases: Function, regulation and role in human cancer. Biochim Biophys Acta. 2007;1773:1196–1212. doi: 10.1016/j.bbamcr.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalady MF, DeJulius KL, Sanchez JA, Jarrar A, Liu X, Manilich E, Skacel M, Church JM. BRAF Mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum. 2012;55:128–133. doi: 10.1097/DCR.0b013e31823c08b3. [DOI] [PubMed] [Google Scholar]
- 35.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VEH, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 36.Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P, Jackman DM, Janne PA. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free ülasma DNA. Clin Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh M, Johnson L. Using genetically engineered mouse models of cancer to aid drug development: an industry perspective. Clin Cancer Res. 2006;12:5312–5328. doi: 10.1158/1078-0432.CCR-06-0437. [DOI] [PubMed] [Google Scholar]
- 38.Kucherlapati R. Genetically modified mouse models for biomarker discovery and preclinical drug testing. Clin Cancer Res. 2012;18:625–630. doi: 10.1158/1078-0432.CCR-11-2021. [DOI] [PubMed] [Google Scholar]
- 39.Becher OJ, Holland EC, Sausville EA, Burger AM. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res. 2006;66:3355–3359. doi: 10.1158/0008-5472.CAN-05-3827. [DOI] [PubMed] [Google Scholar]
- 40.Notingher I, Hench LL. Raman microspectroscopy: a noninvasive tool for studies of individual living cells in vitro. Expert Rev Med Devices. 2006;3:215–234. doi: 10.1586/17434440.3.2.215. [DOI] [PubMed] [Google Scholar]
- 41.Popp J, Tuchin VV, Chiou A, Heinemann S (eds) (2011) Handbook of biophotonics. Volume 2: photonics for health care. Wiley-VCH, Weinheim
- 42.Matthäus C, Chernenko T, Newmark JA, Warner CM, Diem M. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophys J. 2007;93:668–673. doi: 10.1529/biophysj.106.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krafft C, Knetschke T, Funk RHW, Salzer R. Studies on stress-induced changes at the subcellular level by Raman microspectroscopic mapping. Anal Chem. 2006;78:4424–4429. doi: 10.1021/ac060205b. [DOI] [PubMed] [Google Scholar]
- 44.Krafft C, Codrich D, Pelizzo G, Sergo V. Raman and FTIR microscopic imaging of colon tissue: a comparative study. J Biophotonics. 2008;1:154–169. doi: 10.1002/jbio.200710005. [DOI] [PubMed] [Google Scholar]
- 45.Mavarani L, Petersen D, El-Mashtoly SF, Mosig A, Tannapfel A, Kötting C, Gerwert K. Spectral histopathology of colon cancer tissue sections by Raman imaging with 532 nm excitation provides label free annotation of lymphocytes, erythrocytes and proliferating nuclei of cancer cells. Analyst. 2013;138:4035–4039. doi: 10.1039/c3an00370a. [DOI] [PubMed] [Google Scholar]
- 46.Draux F, Gobinet C, Sulé-Suso J, Trussardi A, Manfait M, Jeannesson P, Sockalingum GD. Raman spectral imaging of single cancer cells: probing the impact of sample fixation methods. Anal Bioanal Chem. 2010;397:2727–2737. doi: 10.1007/s00216-010-3759-8. [DOI] [PubMed] [Google Scholar]
- 47.Klein K, Gigler AM, Aschenbrenner T, Monetti R, Bunk W, Jamitzky F, Morfill G, Stark RW, Schlegel J. Label-free live-cell imaging with confocal Raman microscopy. Biophys J. 2012;102:360–368. doi: 10.1016/j.bpj.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palonpon AF, Sodeoka M, Fujita K. Molecular imaging of live cells by Raman microscopy. Curr Opin Chem Biol. 2013;17:708–715. doi: 10.1016/j.cbpa.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 49.El-Mashtoly SF, Niedieker D, Petersen D, Krauss SD, Freier E, Maghnouj A, Mosig A, Hahn S, Kötting C, Gerwert K. Automated Identification of subcellular organelles by coherent anti-Stokes Raman scattering. Biophys J. 2014;106:1910–1920. doi: 10.1016/j.bpj.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krauß SD, Petersen D, Niedieker D, Fricke I, Freier E, El-Mashtoly SF, Gerwert K, Mosig A. Colocalization of fluorescence and Raman microscopic images for the identification of subcellular compartments: a validation study. Analyst. 2015 doi: 10.1039/c4an02153c. [DOI] [PubMed] [Google Scholar]
- 51.Dorney J, Bonnier F, Garcia A, Casey A, Chambers G, Byrne HJ. Identifying and localizing intracellular nanoparticles using Raman spectroscopy. Analyst. 2012;137:1111–1119. doi: 10.1039/c2an15977e. [DOI] [PubMed] [Google Scholar]
- 52.Ling J, Weitman SD, Miller MA, Moore RV, Bovik AC. Direct Raman imaging techniques for study of the subcellular distribution of a drug. Appl Opt. 2002;41:6006. doi: 10.1364/AO.41.006006. [DOI] [PubMed] [Google Scholar]
- 53.El-Mashtoly SF, Petersen D, Yosef HK, Mosig A, Reinacher-Schick A, Kötting C, Gerwert K. Label-free imaging of drug distribution and metabolism in colon cancer cells by Raman microscopy. Analyst. 2014;139:1155–1161. doi: 10.1039/c3an01993d. [DOI] [PubMed] [Google Scholar]
- 54.Meister K, Niesel J, Schatzschneider U, Metzler-Nolte N, Schmidt DA, Havenith M. Label-free imaging of metal-carbonyl complexes in live cells by Raman microspectroscopy. Angew Chem Int Ed. 2010;49:3310–3312. doi: 10.1002/anie.201000097. [DOI] [PubMed] [Google Scholar]
- 55.Nawaz H, Bonnier F, Knief P, Howe O, Lyng FM, Meade AD, Byrne HJ. Evaluation of the potential of Raman microspectroscopy for prediction of chemotherapeutic response to cisplatin in lung adenocarcinoma. Analyst. 2010;135:3070–3076. doi: 10.1039/c0an00541j. [DOI] [PubMed] [Google Scholar]
- 56.Nawaz H, Bonnier F, Meade AD, Lyng FM, Byrne HJ. Comparison of subcellular responses for the evaluation and prediction of the chemotherapeutic response to cisplatin in lung adenocarcinoma using Raman spectroscopy. Analyst. 2011;136:2450–2463. doi: 10.1039/c1an15104e. [DOI] [PubMed] [Google Scholar]
- 57.Nawaz H, Garcia A, Meade AD, Lyng FM, Byrne HJ. Raman micro spectroscopy study of the interaction of vincristine with A549 cells supported by expression analysis of bcl-2 protein. Analyst. 2013;138:6177. doi: 10.1039/c3an00975k. [DOI] [PubMed] [Google Scholar]
- 58.Bonnier F, Knief P, Lim B, Meade AD, Dorney J, Bhattacharya K, Lyng FM, Byrne HJ. Imaging live cells grown on a three dimensional collagen matrix using Raman microspectroscopy. Analyst. 2010;135:3169. doi: 10.1039/c0an00539h. [DOI] [PubMed] [Google Scholar]
- 59.Citri A, Yarden Y. EGF–ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 60.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeh JJ, Routh ED, Rubinas T, Peacock J, Martin TD, Shen XJ, Sandler RS, Kim HJ, Keku TO, Der CJ. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol Cancer Ther. 2009;8:834–843. doi: 10.1158/1535-7163.MCT-08-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vartanian S, Bentley C, Brauer MJ, Li L, Shirasawa S, Sasazuki T, Kim J-S, Haverty P, Stawiski E, Modrusan Z, Waldman T, Stokoe D. Identification of mutant K-Ras-dependent phenotypes using a panel of isogenic cell lines. J Biol Chem. 2013;288:2403–2413. doi: 10.1074/jbc.M112.394130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buck E. Inactivation of Akt by the epidermal growth factor receptor inhibitor erlotinib is mediated by HER-3 in pancreatic and colorectal tumor cell lines and contributes to erlotinib sensitivity. Mol Cancer Ther. 2006;5:2051–2059. doi: 10.1158/1535-7163.MCT-06-0007. [DOI] [PubMed] [Google Scholar]
- 64.Czejka M, Sahmanovic A, Buchner P, Steininger T, Dittrich C. Disposition of erlotinib and its metabolite OSI420 in a patient with high bilirubin levels. Case Rep Oncol. 2013;6:602–608. doi: 10.1159/000357211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salzer R, Siesler HW, editors. Infrared and Raman spectroscopic imaging. Weinheim: Wiley; 2009. [Google Scholar]
- 66.Diem M, Chalmers JM, Griffiths PR, editors. Vibrational spectroscopy for medical diagnosis. Chichester: Wiley; 2008. [Google Scholar]
- 67.Huether A, Höpfner M, Sutter AP, Schuppan D, Scherübl H. Erlotinib induces cell cycle arrest and apoptosis in hepatocellular cancer cells and enhances chemosensitivity towards cytostatics. J Hepatol. 2005;43:661–669. doi: 10.1016/j.jhep.2005.02.040. [DOI] [PubMed] [Google Scholar]
- 68.Ali S, Banerjee S, Ahmad A, El-Rayes BF, Philip PA, Sarkar FH. Apoptosis-inducing effect of erlotinib is potentiated by 3,3’-diindolylmethane in vitro and in vivo using an orthotopic model of pancreatic cancer. Mol Cancer Ther. 2008;7:1708–1719. doi: 10.1158/1535-7163.MCT-08-0354. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Ling Y-H, Lin R, Perez-Soler R. Erlotinib induces mitochondrial-mediated apoptosis in human H3255 non-small-cell lung cancer cells with epidermal growth factor receptorL858R mutation through mitochondrial oxidative phosphorylation-dependent activation of BAX and BAK. Mol Pharmacol. 2008;74:793–806. doi: 10.1124/mol.107.044396. [DOI] [PubMed] [Google Scholar]
- 70.Notingher I, Selvakumaran J, Hench LL. New detection system for toxic agents based on continuous spectroscopic monitoring of living cells. Biosens Bioelectron. 2004;20:780–789. doi: 10.1016/j.bios.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 71.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17:363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 72.Qi W, Fitchev PS, Cornwell ML, Greenberg J, Cabe M, Weber CR, Roy HK, Crawford SE, Savkovic SD. FOXO3 growth inhibition of colonic cells is dependent on intraepithelial lipid droplet density. J Biol Chem. 2013;288:16274–16281. doi: 10.1074/jbc.M113.470617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tirinato L, Liberale C, Di Franco S, Candeloro P, Benfante A, La Rocca R, Potze L, Marotta R, Ruffilli R, Rajamanickam VP, Malerba M, De Angelis F, Falqui A, Carbone E, Todaro M, Medema JP, Stassi G, Di Fabrizio E. Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells. 2014 doi: 10.1002/stem.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rak S, De Zan T, Stefulj J, Kosović M, Gamulin O, Osmak M. FTIR spectroscopy reveals lipid droplets in drug resistant laryngeal carcinoma cells through detection of increased ester vibrational bands intensity. Analyst. 2014;139:3407. doi: 10.1039/c4an00412d. [DOI] [PubMed] [Google Scholar]
- 75.Doherty KR, Wappel RL, Talbert DR, Trusk PB, Moran DM, Kramer JW, Brown AM, Shell SA, Bacus S. Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes. Toxicol Appl Pharmacol. 2013;272:245–255. doi: 10.1016/j.taap.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 76.Hülsmann HJ, Rolff J, Bender C, Jarahian M, Korf U, Herwig R, Fröhlich H, Thomas M, Merk J, Fichtner I, Sültmann H, Kuner R. Activation of AMP-activated protein kinase sensitizes lung cancer cells and H1299 xenografts to erlotinib. Lung Cancer. 2014;86:151–157. doi: 10.1016/j.lungcan.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2 doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ulivi P, Delmonte A, Chiadini E, Calistri D, Papi M, Mariotti M, Verlicchi A, Ragazzini A, Capelli L, Gamboni A, Puccetti M, Dubini A, Burgio M, Casanova C, Crinò L, Amadori D, Dazzi C. Gene mutation analysis in EGFR wild type NSCLC responsive to erlotinib: are there features to guide patient selection? Int J Mol Sci. 2014;16:747–757. doi: 10.3390/ijms16010747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1.86 mb)