Abstract
‘If G-quadruplexes form so readily in vitro, Nature will have found a way of using them in vivo’ (Statement by Aaron Klug over 30 years ago).
During the last decade, four-stranded helical structures called G-quadruplex (or G4) have emerged from being a structural curiosity observed in vitro, to being recognized as a possible nucleic acid based mechanism for regulating multiple biological processes in vivo. The sequencing of many genomes has revealed that they are rich in sequence motifs that have the potential to form G-quadruplexes and that their location is non-random, correlating with functionally important genomic regions. In this short review, we summarize recent evidence for the in vivo presence and function of DNA and RNA G-quadruplexes in various cellular pathways including DNA replication, gene expression and telomere maintenance. We also highlight remaining open questions that will have to be addressed in the future.
INTRODUCTION
The inherent propensity of guanines to self-associate forming four-stranded helical structures has been known since the early 1960s (1). Subsequently it was demonstrated that the conserved DNA sequence repeats of telomeres form G-quadruplex (or G4) structures in vitro (2,3). Since then numerous biochemical and structural analyses have established that nucleic acid sequences, both DNA and RNA, containing runs of guanines (G-tracts) separated by other bases spontaneously fold into G-quadruplex structures in vitro. The building blocks of G-quadruplexes are G-quartets that are formed through a cyclic Hoogsten hydrogen-bonding arrangement of four guanines with each other. The planar G-quartets stack on top of one another forming four-stranded helical structures. G-quadruplex formation is driven by monovalent cations such as Na+ and K+, and hence physiological buffer conditions favour their formation (Figure 1A).
Figure 1.
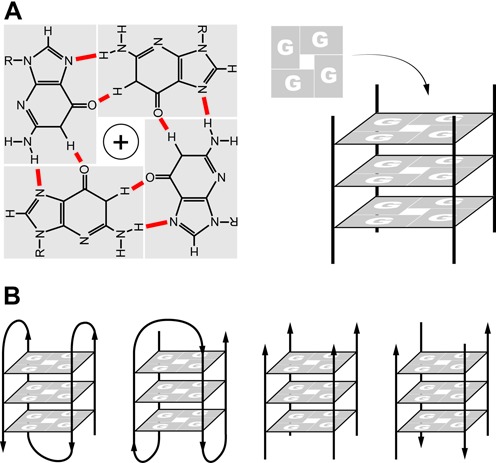
Structure of G-quadruplexes. G-quadruplexes form in vitro in DNA or RNA sequences containing tracts of three to four guanine. (A) The building blocks of G-quadruplexes are G-quartets that arise from the association of four guanines into a cyclic arrangement stabilized by Hoogsten hydrogen bonding (N1–N6 and N2–N7). The planar G-quartets stack on top of one another, forming four-stranded helical structures. G-quadruplex formation is driven by monovalent cations such as Na+ and K+. (B) G-quadruplex structures are polymorphic and can be sub-grouped into different families, as for example parallel or antiparallel according to the orientation of the strands and can be inter- or intramolecular folded. The type of structure depends on the number of G-tracts in a strand.
G-quadruplex structures are topologically very polymorphic and can arise from the intra- or inter- molecular folding of G-rich strands. Intra-molecular folding requires the presence of four or more G-tracts in one strand, whereas inter-molecular folding can arise from two or four strands giving rise to parallel or antiparallel structures depending on the orientation of the strands in a G-quadruplex (4,5) (Figure 1B). Knowledge of the precise 3D-structure (6) is important for the design of G-quadruplex stabilizing ligands used for probing the consequences of G-quadruplex stabilization on processes such as DNA replication and gene transcription, and as anticancer drugs to target G-quadruplexes in the promoters of oncogenes and at telomeres (7). The thermal stability of G-quadruplexes is dependent on features such as the number of G-quartets present in the structure and the length and composition of the loops formed by non-guanine bases (8,9). However, the thermal stability of a G-quadruplex in vitro may not correlate with its in vivo effect (11). In vitro many G-quadruplex DNA structures, once formed, are more thermodynamically stable than double-stranded DNA and importantly for biological function, their unfolding kinetics are much slower than those of DNA or RNA hairpin structures (10). Overall therefore, G-quadruplex structures are likely to obstruct DNA and RNA metabolism and hence their formation must be regulated. Over the last several years, the increasingly direct evidence for the presence of G-quadruplexes in vivo, and the identification of proteins that specifically regulate G-quadruplex folding and unfolding have started to provide insights into G-quadruplex occurrence and function.
Potential G-quadruplexes in the human genome
Computational analyses of the human genome searching with the consensus sequence (G3+N1–7G3+N1–7G3+N1–7G3+) revealed that it contains over 300 000 sequences that have the potential to form G-quadruplexes (pG4 or PQS) (12). This likely is an oversimplification as non-consensus sequences may form G-quadruplexes (13), as well as an underestimation since long runs of repeated DNA sequences are missing from the available sequence database (S. Schuster, personal communication). Significantly, it was found that the localization of pG4s is non-random: pG4 colocalize with functional regions of the genome and furthermore, are highly conserved between different species (8) indicating a selection pressure to retain such sequences at specific genomic sites. This conservation is highest among mammalian species and decreases in non-mammalian species and lower organisms (14). Finally, pG4s are also present in bacteria (15) and human RNA and DNA viruses (16–18).
The highest abundance of pG4s is at telomeres, which in humans consist of 5 to 10 000 bp of the tandemly arranged TTAGGG repeat. They are also highly enriched in gene promoters, at the border between introns and exons and target immunoglobulin gene class switch recombination (9). Most interestingly, recently it has emerged that 90% of human DNA replication origins contain pG4 motifs, and in higher densities near origins that are used frequently (19–21) (Figure 2A–C). Genome-wide analysis of DNA breakpoints in different cancer types show a significant enrichment in pG4s in the vicinity of somatic copy number alterations (22), as well as telomeres being favoured targets of persistent DNA damage response in aging (23). It has also been found that in about 3000 human genes, pG4s are present in the region specifying the 5′-UTR of the encoded mRNAs and may repress translation (Figure 2D) (24,25). The long G-rich RNA transcripts of telomeric DNA, TERRA, are also rich in pG4s (26). All these observations of the conserved presence of pG4s in important genomic regions suggest that they provide a regulatory role through their ability to form G-quadruplex structures. Whether all, or a subset of pG4s present in genomes have an in vivo function remains to be established.
Figure 2.
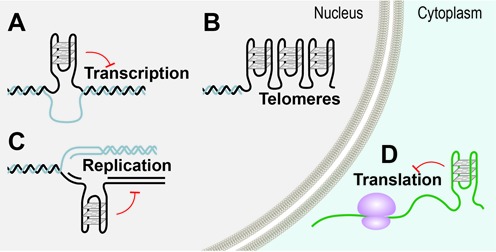
Possible locations of G-quadruplex structures in cells. Genome wide searches have revealed the location of G-rich regions with G-quadruplex forming potential (pG4). pG4s are non-randomly distributed in the genome and promoters and telomeres are particularly enriched in these sequences. In the nucleus, G-quadruplex formation can occur in double stranded G-rich regions when DNA becomes transiently single stranded, during (A) transcription and (C) replication and (B) at the single stranded telomeric G-rich overhangs. Outside the nucleus, G-quadruplexes can also form in mRNA and (D) are involved in translational control. Red T-bars indicate impediments to transcription, replication and translation.
For biology, the important question is if, where and when the mapped pG4s form G-quadruplex structures in vivo. Genomic DNA is primarily double stranded and stabilized through being packaged into chromatin. Two exceptions are the very tips of the linear eukaryotic chromosomes that have the conserved feature of ending in a single-stranded G-overhang and in mRNA. In double stranded DNA, the opportunity for forming G-quadruplexes arises during DNA replication, transcription and repair when DNA is rendered transiently single stranded through the breaking of Watson-Crick base pairing, which would permit the alternative Hoogsteen base pairing present in G-quadruplexes to take place (27,28) (Figure 2). Indeed, emerging in vivo observations are consistent with G-quadruplex formation and resolution providing a regulatory role in these pathways (see below). In addition, it can be envisaged that G-quadruplex formation could be favoured by superhelical stress, molecular crowding (29) as well as specific G-quadruplex binding proteins (30). Furthermore, contrary to the Watson-Crick base pairing dogma, it has been found that Hoogsteen base pairs transiently form in canonical double stranded DNA (31), suggesting that G-quadruplex formation may not necessarily require the prior melting of the DNA double helix through, for instance, DNA replication.
A breakthrough in establishing the presence and location of G-quadruplexes in vivo came over a decade ago from the development of specific antibodies directed against telomric DNA G-quadruplexes. This permitted the first direct visualization by in situ immuno staining in the micronuclei of the ciliate Stylonychia lemnae (32). Later, a G-quadruplex antibody was used to map the location of such structures in human genomic DNA using immuno-precipitation followed by deep sequencing of the selected DNA fragments (33). This study revealed their presence at multiple genomic sites: in gene promoters and both 5′ and 3′ untranslated regions (UTRs), suggesting roles in both transcriptional initiation and termination, within introns and in subtelomeric regions. It should be noted that the immuno-precipitation of the G-quadruplex structures was carried out with isolated and fragmented DNA, so it cannot be excluded that G-quadruplex formation occurred during the analysis. Another approach to identify G-quadruplexes genome wide was by using the G-quadruplex interacting drug pyridostatin, which leads to replication and transcription dependent DNA damage. Chromatin immuno precipitation with an antibody directed against the DNA damage marker histone gamma H2AX identified genes enriched in pG4s (34). Since these observations are consistent with G-quadruplexes being involved in several biological processes, the expectation is that G-quadruplex formation is dynamic and regulated, and hence their distribution may differ in variously differentiated cells. Therefore, it would be of considerable interest to map the occurrence of G-quadruplexes in cells with different transcriptional repertoires.
Regulation of G-quadruplex formation
An essential consideration for the possible participation of G-quadruplexes in biology is the kinetics of formation (folding) and resolution (unfolding). As might have been anticipated, experimental evidence is accumulating for the role of protein chaperones and helicases in the regulation of folding and unfolding, respectively. The folding kinetics of G-quadruplexes forming sequences is sequence dependent and ranges from ms to minutes. For sequences that fold very fast such as human telomeric repeats, the time scale of folding is in the range of that of DNA replication (35). For others, the kinetics of G-quadruplex formation can be increased dramatically by protein chaperones (36). Much of the available evidence for the role of chaperones comes from in vitro studies and most of the identified proteins function at telomeres: the S. cerevisiae telomere double strand binding protein Rap1, was shown two decades ago to promote G-quadruplex formation (37): as was the regulatory subunit of yeast telomerase Est1 (38): and the human telomeric binding protein TRF2 has been implicated in both DNA and RNA (TERRA) G-quadruplex binding (39). Convincing in vivo evidence has started to emerge: the in vitro observation that ciliate telomere end-binding protein TEBPβ enhances the formation of G-quadruplexes by 105–6 fold (36) has been confirmed by in vivo experiments that demonstrate that TEBPβ regulates G-quadruplex formation at telomeres in a cell cycle dependent manner (40). The human DNA mismatch recognizing factor MutSα binds to G-quadruplexes and promotes synapsis of transcriptionally activated immunoglobulin switch regions (41). Of particular interest, recent studies suggest that nucleophosmin (NPM1) interacts with several G-quadruplex regions in ribosomal DNA, both in vitro and in vivo. NPM1 is an abundant nucleolar protein implicated in ribosome maturation and export and is the most frequently mutated gene in acute myeloid leukaemia (42). Significantly nucleolin, (NCL) an essential nucleolar phosphoprotein for which there is old in vitro evidence that it binds DNA G-quadruplexes with high affinity (43), was very recently shown to be sequestered specifically in a conformation dependent manner by G-quadruplexes present in aborted RNA transcripts arising from the expansion of the hexanucleotide repeat (GGGGCC)n (Figure 4B). This led to nucleolar stress and perturbations in RNA processing, initiating a molecular cascade that resulted in a type of a neurodegenerative disorder (44).
Figure 4.
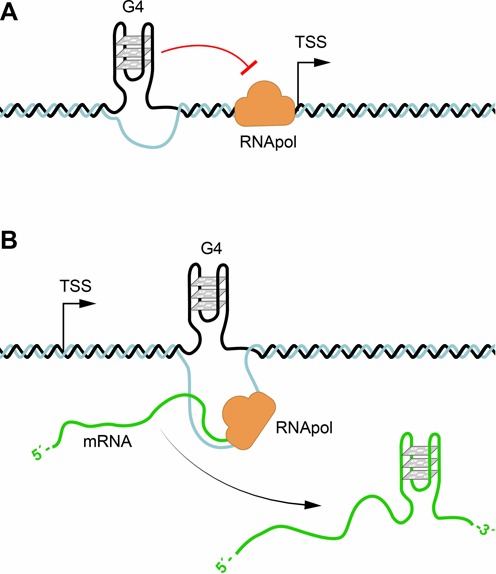
G-quadruplexes in transcription and translation. (A) pG4 sequences are present in about 50% of human genes promoters. G-quadruplex formation could impair initiation of transcription by the RNA polymerase, or if present in the antisense strand inhibit transcription. (B) The presence of G-quadruplexes formed in the 5′ UTR of mRNAs can regulate translation as well as lead to aborted RNA transcripts in hexanucleotide repeat expansion diseases.
Evidence implicating specialized helicases in the unwinding of DNA or RNA G-quadruplexes is now accumulating. Helicases are a large family of ATP-dependent nucleic acid unwinding enzymes that have a major role in genome maintenance. Importantly, loss-of-function mutations in a distinct family of DNA helicases linked to various cancers and genetic disorders have provided a direct link between pG4 sequences and genome instability. It is emerging that G-quadruplex helicases play important roles in DNA replication and telomere maintenance (see below) (45). The human helicases WRN and BLM and S. cerevisiae Sgs1 are involved in telomere maintenance and have G-quadruplex-unwinding activity in vitro and contain a conserved RQC domain that binds G-quadruplexes with high affinity (30). Mutations in WRN cause the Werner syndrome and mutations in BLM the Blooms syndrome, that give rise to premature ageing (adult progeria) and increased risk of cancer, respectively. The integrity of the C. elegans DOG-1gene (deletion of guanine rich DNA) is crucial for the stability of G-tracts in the genome (46). Furthermore, the introduction of a DNA sequence that forms G-quadruplexes in vitro was highly mutagenic and was removed from genomes lacking DOG-1 (47). Studies on the mammalian FANCJ helicase, the orthologue of the C. elegans DOG-1 helicase, reinforce these findings. The FANCJ helicase is associated with the heritable cancer susceptibility disorder Fanconi anaemia. Cell lines from patients lacking FANCJ accumulate large genomic DNA deletions that map to pG4s (48). The demonstration that FANCJ preferentially unwinds G-quadruplexes over other DNA substrates in vitro suggested that the FANCJ helicase, like DOG-1, functions in resolving potential replication impediments caused by DNA G-quadruplexes (48). Mutations in the mammalian DNA helicase regulator of telomere length RETL1 confers increased susceptibility to certain types of cancers (49). Because of its similarity to human FANCJ and the C. elegans DOG-1 helicases, and because of the observed increase in telomere fragility by treatment with the G-quadruplex stabilizing compound TMPyP4, RETL1 was implicated in G-quadruplex unwinding (50). However, direct biochemical evidence for this activity is lacking. Studies on the PIF1 DNA helicase family, which is conserved from bacteria to man, provide strong evidence for a potent G-quadruplex unwinding activity in vitro (51). A genome wide chromatin immuno-precipitation of the S. cerevisiae Pif1 revealed pG4s at a major subset of Pif1 binding sites and that such sites in Pif1 deficient cells are prone to DNA double strand breaks (52–54). The human PIF1 also appears to act on pG4s (34). It is also emerging that in the absence of G-quadruplex helicases, a number of nucleases act to process G-quadruplexes leading to G-tract deletions. Nucleases such as the yeast Kem1 (55) and human FEN1, EXO1 and DNA2 are known to cleave G-quadruplexes in vitro (56). EXO1 and FEN1 play a role in DNA replication and are involved in telomere maintenance. Depletion of these nucleases causes telomere disfunction (57,58). In addition, the single-strand binding replication protein RPA that plays an important role in telomere maintenance, has been shown to aid the unfolding of G-quadruplexes by shifting the equilibrium from a folded to an unfolded state (59).
For RNA G-quadruplexes, convincing evidence for their processing into single stranded RNA comes from the identification and characterization of the ATP dependent human RNA helicase RHAU (60,61). RHAU is a DEAH-box helicase that exhibits G4-RNA (and G4-DNA) binding and resolving activity. RNA immunoprecipitation (RIP)-chip analysis identified approximately 100 RNAs associated with RHAU in vivo and the majority contained pG4s sequences. One target is the human telomerase RNA TER and binding of RHAU depends on the presence of a stable G4 structure in the 5′-region of TER, both in vivo and in vitro (62). The functional consequences of knockdown of RHAU are impaired telomerase assembly and changes in telomere length (63). These data provide strong evidence that there are proteins that directly regulate G-quadruplexes resolution, or removal altogether, to prevent replication fork stalling and DNA breakage, and RNA folding.
The positive role of DNA and RNA G-quadruplex structures in biology is less well documented. In addition to their role in specifying metazoan DNA replication origins and at telomeres (discussed below), it is emerging that G-quadruplexes might be exploited as DNA recombination sites. Studies in a gram-negative bacterium have shown that a G-quadruplex is necessary to serve as the recombination initiation site in vivo and is required for pilin antigenic variability (64,65).
G-quadruplexes at telomeres
The tandem organization of G-rich telomeric DNA repeats is almost universally conserved in eukaryotes and such sequences are well known for forming G-quadruplexes in vitro. Direct in vivo evidence for the presence of G-quadruplexs at telomeres first came over 10 years ago from studies using very specific antibodies directed against G-quadruplexes (32,66). These studies showed that an antiparallel intermolecular G-quadruplex structure is formed at the macronuclear telomeres of Stylonychia, and importantly that the G-quadruplex structure is resolved during DNA replication, suggesting that G-quadruplexes might act as a telomeric capping structure (Figure 3A and B). The observation that G-quadruplex unfolding is regulated by a cell cycle dependent phosphorylation of the telomere end binding protein TEBPβ and a telomerase associated RecQ-like helicase (64,67,68,30), provided an important mechanistic insight into the spatial and temporal regulation of G-quadruplex folding and unfolding to permit telomere synthesis by the telomerase when required.
Figure 3.
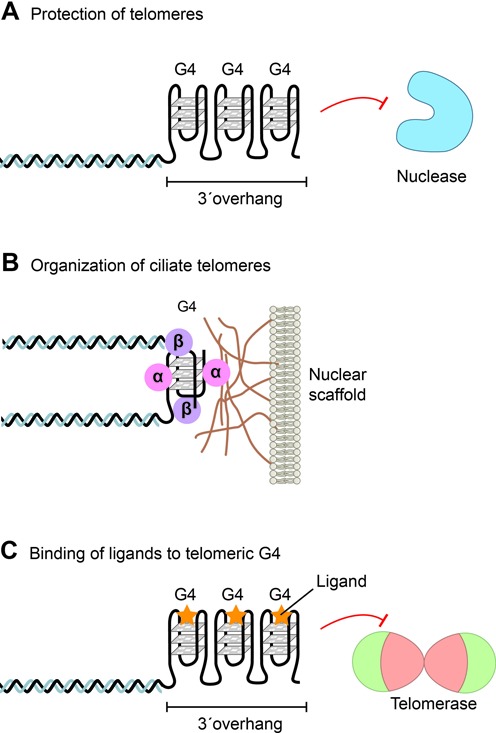
G-quadruplexes at telomeres. The G-rich overhang of telomeres can form G-quadruplex structures involved in telomere end protection and telomeric DNA metabolism. (A) The long human G-rich overhang can form strings intramolecularly folded G-quadruplexes that may offer end protection against nucleases or regulate telomerase activity. (B) Ciliate telomeres form intermolecular G-quadruplex structures involving two telomeres promoted by the telomere-end binding protein TEBPβ. Telomeres are attached to a sub-nuclear structure (the nuclear matrix or scaffold) via an interaction of the telomere-end binding protein TEBPα. (C) Stabilizing of G-quadruplexes by G-quadruplex binding ligands (yellow stars) impairs telomere repeat synthesis by the telomerase enzyme and lead to telomere shortening (modified after (80)).
For human telomeres, the first indication that G-quadruplexes may be present came from the observation that G-quadruplex stabilizing ligands impaired telomere metabolism and lead to telomere shortening (Figure 3C) (69–71). This approach was based on the observation that telomeric G-quadruplexes inhibit telomerase activity (72). A number of G-quadruplex stabilizing ligands are now available and it has become evident that many do not target the telomerase enzyme, but the telomere itself (73–75). Other approaches to identify G-quadruplexes at human telomeres have included the binding of a radiolabelled G-quadruplex ligand to metaphase chromosomes (76), the use of a fluorescent cyanine dye (77) and, more recently, the use of an engineered, structure-specific antibody against human G-quadruplexes (78,79). In all cases, signals were detected at the ends of chromosomes, albeit not all ends. The resolution of light microscopy is insufficiently high to decipher whether binding occurred at the very end of the chromosome or at subtelomeric regions, also known to adopt G-quadruplex structures in vitro (80). For these reasons, whether G-quadruplexes are present at human telomeres remains to be established. However, the finding that a number of helicases that are known to unwind G-quadruplexs in vitro (such as the RecQ helicases WRN and BLM), localize to telomeres and are required for telomere integrity in vivo, provide strong circumstantial evidence for the existence of such structures at mammalian telomeres (81,82,30,45,83,84).
The primary function of WRN and BLM helicases appears to be to ensure appropriate replication of telomeric DNA by aiding the unwinding G-quadruplex impediments (Figure 5). The WRN helicase is necessary for preventing dramatic telomere loss during lagging strand replication of the G-rich strand and the consequent accumulation of chromosome aberrations such as chromosome fusions (85). WRN colocalizes and physically interacts with the critical telomere binding proteins TRF2 and POT1 (86) and both WRN and BLM bind with high affinity to POT1 (87), suggesting that DNA binding telomeric proteins may function in helicase recruitment. RETL1 deficient cells exhibit telomere fragility (50,81), which is enhanced by the G-quadruplex stabilizing agent TMPyP4. This again suggests that G-quadruplexes form at telomeres and if not resolved result in DNA damage. However, the role of RETL1 appears to be more general in aiding replication genome wide as it interacts directly with the replication clamp PCNA (49).
Figure 5.
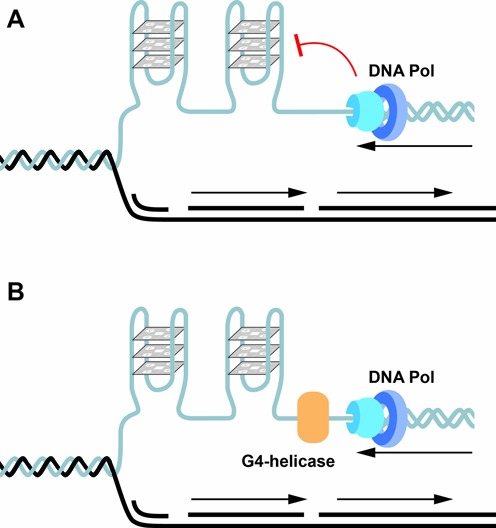
G-quadruplexes and replication. G-quadruplexes formed during replication when the DNA is transiently single stranded impede replication and have to be resolved to permit the replication machinery including DNA polymerase to proceed for both leading and lagging strand DNA synthesis. G-quadruplexes are known to be resolved by G-quadruplex unwinding helicases such as FANCJ that has a 5′–3′ directionality. Other helicases such as BLM and WRN have a 3′–5′ directionality. Other proteins or enzymes such as polymeras also function in the successful bypass of G-quadruplexes.
G-quadruplexes in transcription and translation
The finding that about 50% of human genes contain pG4s near their promoter regions suggested a role for G-quadruplexes in regulating gene expression (Figure 4A). Interestingly, pG4s are more frequent in oncogenes or regulatory genes than in house keeping, or tumour suppressor genes (88,89), (for review (9,84)). The first evidence that pG4s at promoters (Figure 4A) have an effect on gene expression came from studies on the oncogene c-MYC, for which it was shown that mutations of the pG4, or the addition of a G-quadruplex stabilizing ligand affected transcription in vivo (90,91). More convincingly, genome wide gene expression studies in yeast and human cells show changes in numerous genes after addition of the G-quadruplex binding ligand TMPyP4 and significantly, the genes whose expression was affected were enriched in pG4s (Figure 4A) (92,93). Studies using a G-quadruplex specific single chain antibody also showed alterations in gene expression of genes containing pG4s and the effect was not only limited to promoters but also on pG4s at the ends of genes, suggesting a possible involvement of G-quadruplexes in both transcriptional initiation and termination (94,95). Furthermore, genome wide analysis of human cells has revealed that the binding sites of the transcription associated helicases XPB and XPD significantly overlap with pG4s. Since these helicases bind and unfold G-quadruplexes they are likely recruited to G-quadruplexes in the genome to aid transcription (96).
Genome wide studies also revealed that pG4s are highly enriched at sites DNaseI hypersensitivity (97) and correlate with low nucleosome occupancy (93). Consequently, pG4s could both affect the deposition of regulatory proteins and/or alter chromatin structure and stability. The link between G-quadruplexes and chromatin is highlighted by the chromatin remodeller ATRX (alpha-thalassaemia/mental retardation X-linked) which together with its interacting partner DAXX functions as a histone-chaperone complex in the deposition of the histone variant H3.3, thus stabilizing chromatin (98). ATRX localizes to pericentric heterochromatin and telomeres, and genome-wide analyses in mouse and human cells revealed that it binds to GC-rich sequences of which a significant fraction are pG4s. In vitro ATRX was shown to bind G-quadruplexes (99). Disease-causing mutations in ATRX affect a variety of fundamental nuclear processes, including transcription. The ATRX syndrome is associated with thalassemia, which is attributed to the down-regulation of α-globin through the binding of ATRX to pG4s repeats upstream of the gene. Interestingly, both the proximity and the length of the variable number tandem repeats affected the severity of gene silencing, suggesting a direct cause by pG4s. Alternatively, based on observations from replication studies in avian DT40 cells (100,101), it was concluded that transcriptional silencing occured via G-quadruplex impeded DNA replication (Figure 5) affecting transcription through the inappropriate inheritance of epigenetic histone marks. Interestingly, epigenetic instability can arise from a G-quadruplex located at a distance of up to 3500 base pairs from the transcription start-site (13). Although these observations provide compelling evidence for the involvement of pG4s in the regulation of gene expression in vivo, whether the mechanism for transcriptional regulation is directly through G-quadruplex formation remains to be proven. If G-quadruplex formation is the mechanism, then their occurrence (and regulation) should differ between various differentiated cells.
Computational predictions have revealed that RNA pG4s are also present in the 5′ untranslated region (UTR) of many genes and the involvement of G-quadruplexes in the regulation of translation has been demonstrated by in vitro and in vivo studies (Figure 2D) (24). Over a decade ago, pioneering studies on fragile X mental retardation, arising from (CGG)n repeat expansion, first suggested that G-quadruplexes in mRNA were involved in neuronal function (102) and were the target of the mental retardation protein FMRP. FMRP binds specifically to its own mRNA through a G-quadruplex present in its 5′ UTR, which impedes translation giving rise to altered brain mRNA translational profiles (103). A recent investigation on the role of the RNA helicase eIF4A, using ribosome footprinting to provide snapshots of translation across the transcriptome, has revealed that the hallmark of eIF4A dependent transcripts is a 12-nucleotide pG4 signature (CGG)4 that can form RNA G-quadruplex structures (104). eIF4A promotes T-cell acute lymphoblastic leukaemia by aiding the translation of mRNAs with long and complex UTRs. Hence, these results implicate RNA G-quadruplexes in the regulation of translation of a number of oncogenes (Figure 4B) (104). In addition, pG4 in the 3′ UTR of some mRNAs have been shown to be involved in alternative polyadenylation and the shortening of the transcripts (25).
Direct evidence for proteins that bind and resolve RNA G-quadruplexes is scant, with the exception of recent studies on the RNA helicase RHAU. This ATP-dependent resolvase was shown to bind with a high affinity and specificity many RNAs containing pG4s including the telomerase RNA TER, and resolves G-quadruplexes in vitro (60,62). Mutations in the pG4 in TER give rise to a telomeric phenotype (105). In addition, as mentioned above, G-quadruplexes that form in m-RNA arising from hexanucleotide repeat expansion (GGGGCC)n lead to aborted RNA transcripts and perturbation of RNA processing (44).
The long G-rich telomere repeat containing RNA (TERRA) that result from the transcription of human and yeast telomeric DNA (106,107) unsurprisingly adopt G-quadruplex structures in vitro (26,108). In vitro studies have shown that the mammalian telomere binding protein TRF2 can interact with TERRA G-quadruplexes, which may suggest a role in telomere organization (39). Given the great structural repertoire of RNAs, it is likely that additional roles of RNA G-quadruplexes (and other higher order structures) in regulatory processes will emerge.
G-quadruplexes in replication and genome instability
DNA G-quadruplexes appear to have a dual role in the regulation of DNA replication: as impediments to replication that in mutant backgrounds lead to genome instability, and as components of metazoan replication origins (Figures 5 and 6).
Figure 6.
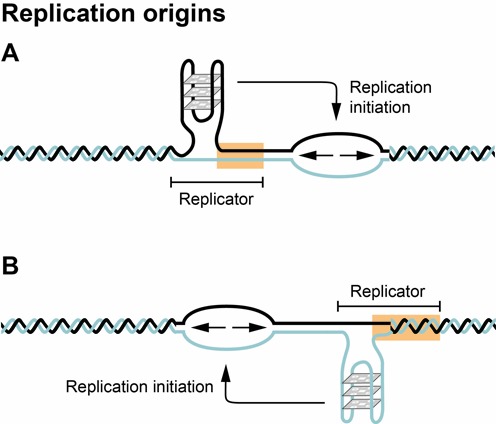
G-quadruplexes and the initiation of DNA replication. Origins of replication in mice and humans are GC-rich and contain pG4s. G-quadruplex formation is required for the initiation of DNA replication and the localization of the G-quadruplex determines the site of initiation (modified after (117)).
There is mounting evidence that both specialized DNA polymerases and helicases function in the replication of G-quadruplexes to prevent genetic and epigenetic instability (Figure 5) (45,109). Insights into the role of G-quadruplex helicases in DNA replication have emerged from studies on genome instability syndromes that are caused by loss of function mutations in one or other of such helicases (Figure 5A). In C. elegans containing a loss of function mutation in the DOG-1 gene, deletions accumulate in genomic regions containing pG4s (46). Studies on other helicases, as for example FANCJ, WRN, BLM and Pif1, support and extend these findings. While the WRN helicase seems to be primarily required for replication of telomeric DNA, consistent with Werner syndrome patients (who have mutations in the WRN helicase) showing premature senescence and accelerated telomere shortening. FANCJ as well as Pif1 or BLM helicase also act at internal genomic regions and are required for genome integrity (45,109). Mutations in the FANCJ helicase leads to large deletions that map to pG4s in the genome (48). The demonstration that FANCJ preferentially unwinds G-quadruplexes in vitro suggested that the FANCJ helicase like DOG1, functions by resolving G-quadruplexes formed during replication (Figure 5B). In vivo evidence for replication fork stalling caused by pG4s came from an elegant study in an avian DT40 cell line deficient in the translesion polymerase REV1 (100). In this study, the sites of stalled forks were mapped to pG4s and further the authors demonstrated that stalling was due to the presence of a G-quadruplex forming sequence, rather than a G-rich sequence. Significantly, it was found that failure to maintain processive DNA replication leads to the uncoupling of DNA synthesis and histone recycling, and further that pG4-associated epigenetic instability is due to mutations in three helicases implicated in the unwinding of G-quadruplex structures (FANCJ, WRN and BLM) (100,101). Recent observations also provide direct evidence for ATRX playing a role in aiding replication. ATRX dysfunction induced replication defects and increased the number of DNA damage response mechanisms at telomeres in mouse embryonic stem cells (110). This observation taken together with the knowledge that ATRX targets pG4s (97), suggest that in this case also G-quadruplexes could also be the cause of replication fork stalling. Mutations in human ATRX have recently been identified in tumours that maintain their telomeres by a telomerase independent pathway (the alternative lengthening of telomeres (ALT)) involving homologous recombination, implicating replication defects at telomeres in ALT cells (111). Evidence for genome instability arising from defective replication has also come from studies in which the human subtelomeric minisatellite CBE1, which forms G-quadruplex structure in vitro (80), was inserted into the genome of Pif1 deficient yeast (52,54). It was further shown that replication slowed down in the vicinity of pG4s and that DNA breakage occurred at these sites in Pif1 deficient S. cerevisiae. Convincingly for the role of Pif1 in resolving G-quadruplexes, G-quadruplex associated DNA breakage could be suppressed by the ectopic expression of Pif1 (51,112). Similarly, in the study using antibodies directed against G-quadruplexes (78) it was shown that the number of DNA damage foci significantly increased during S-phase, suggesting that the foci represent G-quadruplexes formed during replication. While this might be true, co-localization of these foci with newly synthesized DNA would strengthen the conclusion (78).
The large size of metazoan genomes, where replication occurs from about 30 000–50 000 replication origins in human or mouse cells (and possibly contain at least 10 times more potential origins), prevented their identification for many years. However, improved genome wide analyses of active sites of initiation of DNA replication have now been able to address the sequence specificity of replication origins. Contrary to the AT rich consensus sequences for origins in S. cerevisiae, 80–90% of all origins in mouse and human cells contain GC-rich regions forming G-rich repeated elements (Figure 6) (19,21,113). This GC rich feature is conserved in the replication origins of plants (114). These sequence elements have the characteristic features of G-quadruplex forming sequences, and such structures might be more important than sequence specificity for marking the initiation of DNA replication from either DNA strand (Figure 6) (115). Recently it was argued that the high occurrence of pG4s at origins might be an overestimation due to the inefficient digestion of G-quadruplex structure by the exonuclease used in the mapping (116). Nevertheless, convincing experimental data show that a G-quadruplex is indeed required for the initiation of replication in vivo (117). However, how potential replication origins within a replicon are selected for activation in different cells types remains to be understood and may involve the crosstalk between genome, chromatin organization and replication and transcription (115,118). These data are not only important for our understanding of the control of DNA replication in higher eukaryotes, but may have an impact on applied science by allowing the construction of autonomously replicating vectors to be used in gene and stem cell therapy.
CONCLUSIONS
Research on the role of G-quadruplexes in cells has developed into an exciting field of modern biology. Mapping their occurrence in vivo has been difficult, but increasingly direct evidence is accumulating for their formation in both DNA and RNA, suggesting that beyond the primary sequence, DNA and RNA higher order structures provide a nucleic acid based control mechanism involved in regulating multiple biological pathways such as transcription, replication, translation and telomere structure.
Evidence suggests that G-quadruplex formation can serve both beneficial and regulatory roles in cells such as forming the capping structure of telomeres, the specification of DNA replication origins in vertebrates, and deletorious effects as they can impede the progression of replicative DNA polymerases. Evidence is also accomulating for the regulation of G-quadruplex structure formation by specific protein chaperones that bind or promote G-quadruplexes and helicases that can resolve them to promote faithful copying of the genome and prevent DNA damage. It remains to be determined whether all, or a subset of potential G4 motifs function in cells. However, since mechanisms have evolved to regulate their formation, there would be no selection pressure to remove these structures from where they could pose a potential problem.
For appropriate biological function, both the formation and resolution of G-quadruplexes need to be regulated in a tight spatiotemporal manner and should be specific for each cell type. Pioneering studies already indicate that this presumption is correct. The regulation of G-quadruplex formation through the cell cycle has been shown for ciliates and human cells (66,78), but it would be most insightful to know their occurrence and location during cellular differentiation to understand the regulation of G-quadruplex formation and the functional consequences. There is a need to obtain a better understanding of how G-quadruplex stabilizing proteins or G-quadruplex resolving helicases are recruited to specific sites in the genome and at the appropriate time. Also, since DNA is packaged into chromatin, it will be essential to establish how chromatin structure influences G-quadruplex formation and vice versa.
Acknowledgments
Hans J. Lipps and Daniela Rhodes thank their colleagues for many stimulating discussions and Mattias Carlene for the illustrations.
FUNDING
Hans J. Lipps was funded by Deutsche Forschungsgemeinschaft and Daniela Rhodes by Nanyang Technological University, a Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001]. Funding for open access charge: Ministry of Education (MOE) AcRF [MOE2012-T3-1-001].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipps H.J., Gruissem W., Prescott D.M. Higher order DNA structure in macronuclear chromatin of the hypotrichous ciliate Oxytricha nova. Proc. Natl. Acad. Sci. U.S.A. 1982;79:2495–2499. doi: 10.1073/pnas.79.8.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 4.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson G.N., Lee M.P., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 7.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug. Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.König S.L.B., Evans A.C., Huppert J.L. Seven essential questions on G-quadruplex. Biomol. Concepts. 2010;1:197–213. doi: 10.1515/bmc.2010.011. [DOI] [PubMed] [Google Scholar]
- 9.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M.P., Phan A.T., Nicolas A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiavone D., Guilbaud G., Murat P., Papadopoulou C., Sarkies P., Prioleau M.N., Balasubramanian S., Sale J.E. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J. 2014;33:2507–2520. doi: 10.15252/embj.201488398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frees S., Menendez C., Crum M., Bagga P.S. QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs. Human Genomics. 2014;8:8. doi: 10.1186/1479-7364-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaume N., Pathak R., Yadav V.K., Kota S., Misra H.S., Gautam H.K., Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41:76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundquist W.I., Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norseen J., Johnson F.B., Lieberman P.M. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009;83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuesuwan B., Kern J.T., Thomas P.W., Rodriguez M., Li J., David W.M., Kerwin S.M. Simian virus 40 large T-antigen G-quadruplex DNA helicase inhibition by G-quadruplex DNA-interactive agents. Biochemistry. 2008;47:1896–1909. doi: 10.1021/bi701747d. [DOI] [PubMed] [Google Scholar]
- 19.Cayrou C., Gregoire D., Coulombe P., Danis E., Mechali M. Genome-scale identification of active DNA replication origins. Methods. 2012;57:158–164. doi: 10.1016/j.ymeth.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Cayrou C., Coulombe P., Vigneron A., Stanojcic S., Ganier O., Peiffer I., Rivals E., Puy A., Laurent-Chabalier S., Desprat R., et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21:1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.M., Lemaitre J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 22.De S., Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewitt G., Jurk D., Marques F.D., Correia-Melo C., Hardy T., Gackowska A., Anderson R., Taschuk M., Mann J., Passos J.F. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 2012;3:708–716. doi: 10.1038/ncomms1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bugaut A., Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaudoin J.D., Perreault J.P. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013;41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., Suzuki Y., Ito K., Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardin C.C., Watson T., Corregan M., Bailey C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG) Biochemistry. 1992;31:833–841. doi: 10.1021/bi00118a028. [DOI] [PubMed] [Google Scholar]
- 28.Li W., Wu P., Ohmichi T., Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi D., Karimata H., Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- 30.Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Nikolova E.N., Kim E., Wise A.A., O'Brien P.J., Andricioaei I., Al-Hashimi H.M. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam E.Y., Beraldi D., Tannahill D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796–1780. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez R., Miller K.M., Forment J.V., Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang A.Y., Balasubramanian S. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012;134:19297–19308. doi: 10.1021/ja309851t. [DOI] [PubMed] [Google Scholar]
- 36.Fang G., Cech T.R. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 37.Rhodes D., Giraldo R. Telomere structure and function. Curr. Opin. Struct. Biol. 1995;5:311–322. doi: 10.1016/0959-440x(95)80092-1. [DOI] [PubMed] [Google Scholar]
- 38.Li Q.J., Tong X.J., Duan Y.M., Zhou J.Q. Characterization of the intramolecular G-quadruplex promoting activity of Est1. FEBS Lett. 2013;587:659–665. doi: 10.1016/j.febslet.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 39.Biffi G., Tannahill D., Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paeschke K., Juranek S., Rhodes D., Lipps H.J. Cell cycle-dependent regulation of telomere tethering in the nucleus. Chromosome Res. 2008;16:721–728. doi: 10.1007/s10577-008-1222-x. [DOI] [PubMed] [Google Scholar]
- 41.Larson E.D., Duquette M.L., Cummings W.J., Streiff R.J., Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 2005;15:470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 42.Chiarella S., De Cola A., Scaglione G.L., Carletti E., Graziano V., Barcaroli D., Lo Sterzo C., Di Matteo A., Di Ilio C., Falini B., et al. Nucleophosmin mutations alter its nucleolar localization by impairing G-quadruplex binding at ribosomal DNA. Nucleic Acids Res. 2013;41:3228–3239. doi: 10.1093/nar/gkt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dempsey L.A., Sun H., Hanakahi L.A., Maizels N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 44.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosh R.M. Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung I., Schertzer M., Rose A., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 47.Kruisselbrink E., Guryev V., Brouwer K., Pontier D.B., Cuppen E., Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 48.London T.B., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vannier J.B., Sandhu S., Petalcorin M.I., Wu X., Nabi Z., Ding H., Boulton S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 50.Vannier J.B., Pavicic-Kaltenbrunner V., Petalcorin M.I., Ding H., Boulton S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 51.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribeyre C., Lopes J., Boule J.B., Piazza A., Guedin A., Zakian V.A., Mergny J.L., Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z., Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 56.Vallur A.C., Maizels N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16508–16512. doi: 10.1073/pnas.0805327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saharia A., Guittat L., Crocker S., Lim A., Steffen M., Kulkarni S., Stewart S.A. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leon-Ortiz A.M., Svendsen J., Boulton S.J. Metabolism of DNA secondary structures at the eukaryotic replication fork. DNA Repair. 2014;19:152–162. doi: 10.1016/j.dnarep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 59.Safa L., Delagoutte E., Petruseva I., Alberti P., Lavrik O., Riou J.F., Saintome C. Binding polarity of RPA to telomeric sequences and influence of G-quadruplex stability. Biochimie. 2014;103:80–88. doi: 10.1016/j.biochi.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 60.Lattmann S., Giri B., Vaughn J.P., Akman S.A., Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giri B., Smaldino P.J., Thys R.G., Creacy S.D., Routh E.D., Hantgan R.R., Lattmann S., Nagamine Y., Akman S.A., Vaughn J.P. G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011;39:7161–7178. doi: 10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lattmann S., Stadler M.B., Vaughn J.P., Akman S.A., Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–9404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Booy E.P., Meier M., Okun N., Novakowski S.K., Xiong S., Stetefeld J., McKenna S.A. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012;40:4110–4124. doi: 10.1093/nar/gkr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cahoon L.A., Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walia R., Chaconas G. Suggested role for G4 DNA in recombinational switching at the antigenic variation locus of the Lyme disease spirochete. PLoS One. 2013;8:e57792. doi: 10.1371/journal.pone.0057792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 67.Paeschke K., Juranek S., Simonsson T., Hempel A., Rhodes D., Lipps H.J. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 68.Postberg J., Tsytlonok M., Sparvoli D., Rhodes D., Lipps H.J. A telomerase-associated RecQ protein-like helicase resolves telomeric G-quadruplex structures during replication. Gene. 2012;497:147–154. doi: 10.1016/j.gene.2012.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun D., Thompson B., Cathers B.E., Salazar M., Kerwin S.M., Trent J.O., Jenkins T.C., Neidle S., Hurley L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 70.Read M., Harrison R.J., Romagnoli B., Tanious F.A., Gowan S.H., Reszka A.P., Wilson W.D., Kelland L.R., Neidle S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 72.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 73.Di Antonio M., Rodriguez R., Balasubramanian S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods. 2012;57:84–92. doi: 10.1016/j.ymeth.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vy Thi Le T., Han S., Chae J., Park H.J. G-quadruplex binding ligands: from naturally occurring to rationally designed molecules. Curr. Pharm. Des. 2012;18:1948–1972. doi: 10.2174/138161212799958431. [DOI] [PubMed] [Google Scholar]
- 75.Rizzo A., Salvati E., Biroccio A. Methods of studying telomere damage induced by quadruplex-ligand complexes. Methods. 2012;57:93–99. doi: 10.1016/j.ymeth.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 76.Granotier C., Pennarun G., Riou L., Hoffschir F., Gauthier L.R., De Cian A., Gomez D., Mandine E., Riou J.F., Mergny J.L., et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Q., Xiang J., Yang S., Zhou Q., Li Q., Tang Y., Xu G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. recognizing mixed G-quadruplex in human telomeres. Chem. Commun. (Camb.) 2009:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- 78.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr, Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2013;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piazza A., Serero A., Boule J.B., Legoix-Ne P., Lopes J., Nicolas A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 2012;8:e1003033. doi: 10.1371/journal.pgen.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ding H., Schertzer M., Wu X., Gertsenstein M., Selig S., Kammori M., Pourvali R., Poon S., Vulto I., Chavez E., et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 82.Lin W., Sampathi S., Dai H., Liu C., Zhou M., Hu J., Huang Q., Campbell J., Shin-Ya K., Zheng L., et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013;32:1425–1439. doi: 10.1038/emboj.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paeschke K., McDonald K.R., Zakian V.A. Telomeres: structures in need of unwinding. FEBS Lett. 2010;584:3760–3772. doi: 10.1016/j.febslet.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 86.Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 87.Opresko P.L., Mason P.A., Podell E.R., Lei M., Hickson I.D., Cech T.R., Bohr V.A. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 88.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 93.Hershman S.G., Chen Q., Lee J.Y., Kozak M.L., Yue P., Wang L.S., Johnson F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huppert J.L., Bugaut A., Kumari S., Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fernando H., Sewitz S., Darot J., Tavare S., Huppert J.L., Balasubramanian S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gray L.T., Vallur A.C., Eddy J., Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hegyi H. Enhancer-promoter interaction facilitated by transiently forming G-quadruplexes. Sci. Rep. 2015;5:9165–9170. doi: 10.1038/srep09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clynes D., Higgs D.R., Gibbons R.J. The chromatin remodeller ATRX: a repeat offender in human disease. Trends Biochem. Sci. 2013;38:461–466. doi: 10.1016/j.tibs.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 99.Law M.J., Lower K.M., Voon H.P., Hughes J.R., Garrick D., Viprakasit V., Mitson M., De Gobbi M., Marra M., Morris A., et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 100.Sarkies P., Reams C., Simpson L.J., Sale J.E. Epigenetic instability due to defective replication of structured DNA. Mol. Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., Sale J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 103.Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 104.Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J., et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sexton A.N., Collins K. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011;31:736–743. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 107.Schoeftner S., Blasco M.A. Chromatin regulation and non-coding RNAs at mammalian telomeres. Semin. Cell. Dev. Biol. 2010;21:186–193. doi: 10.1016/j.semcdb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 108.Martadinata H., Heddi B., Lim K.W., Phan A.T. Structure of long human telomeric RNA (TERRA): G-quadruplexes formed by four and eight UUAGGG repeats are stable building blocks. Biochemistry. 2011;50:6455–6461. doi: 10.1021/bi200569f. [DOI] [PubMed] [Google Scholar]
- 109.Wickramasinghe C.M., Arzouk H., Frey A., Maiter A., Sale J.E. Contributions of the specialised DNA polymerases to replication of structured DNA. DNA Repair. 2015;29:83–90. doi: 10.1016/j.dnarep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 110.Clynes D., Jelinska C., Xella B., Ayyub H., Taylor S., Mitson M., Bachrati C.Z., Higgs D.R., Gibbons R.J. ATRX dysfunction induces replication defects in primary mouse cells. PLoS One. 2014;9:e92915. doi: 10.1371/journal.pone.0092915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shay J.W., Reddel R.R., Wright W.E. Cancer. Cancer and telomeres–an ALTernative to telomerase. Science. 2012;336:1388–1390. doi: 10.1126/science.1222394. [DOI] [PubMed] [Google Scholar]
- 112.Boule J.B., Vega L.R., Zakian V.A. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 113.Cadoret J.C., Meisch F., Hassan-Zadeh V., Luyten I., Guillet C., Duret L., Quesneville H., Prioleau M.N. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Costas C., Sanchez Mde L., Sequeira-Mendes J., Gutierrez C. Progress in understanding DNA replication control. Plant Sci. 2011;181:203–209. doi: 10.1016/j.plantsci.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 115.Leonard A.C., Mechali M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013;5:a010116. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Foulk M.S., Urban J.M., Casella C., Gerbi S.A. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25:725–735. doi: 10.1101/gr.183848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Valton A.L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintome C., Riou J.F., Prioleau M.N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gilbert D.M. Replication origins run (ultra) deep. Nat. Struct. Mol. Biol. 2012;19:740–742. doi: 10.1038/nsmb.2352. [DOI] [PubMed] [Google Scholar]


