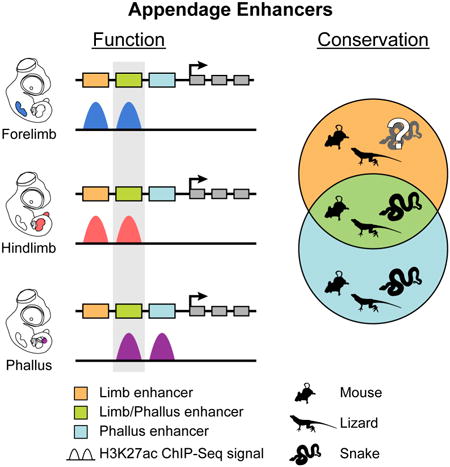
Summary
The amniote phallus and limbs differ dramatically in their morphologies but share patterns of signaling and gene expression in early development. Thus far, the extent to which genital and limb transcriptional networks also share cis-regulatory elements has remained unexplored. We show that many limb enhancers are retained in snake genomes, suggesting that these elements may function in non-limb tissues. Consistent with this, our analysis of cis-regulatory activity in mice and Anolis lizards reveals that patterns of enhancer activity in embryonic limbs and genitalia overlap heavily. In mice, deletion of HLEB, an enhancer of Tbx4, produces defects in hindlimbs and genitalia, establishing the importance of this limb-genital enhancer for development of these different appendages. Further analyses demonstrate the HLEB of snakes has lost hindlimb enhancer function while retaining genital activity. Our findings identify roles for Tbx4 in genital development and highlight deep similarities in cis-regulatory activity between limbs and genitalia.
Graphical abstract
Introduction
The phallus is a feature present in many amniotes and likely evolved in a common ancestor more than 310 million years ago (Gredler et al., 2014; Hedges et al., 2004). Although the phallus differs from limbs in both form and function, the development of the phallus and limbs share certain features. Both limbs and external genitalia form through the establishment of tissue outgrowths from the main body axis during embryogenesis, and both possess regional signaling centers that direct growth and patterning. The limb buds and genital tubercle (GT), the outgrowth from which the penis and clitoris develop, also express many of the same transcription factors and signaling molecules at equivalent stages of outgrowth (Cohn, 2011; Tschopp et al., 2014). For instance, Tbx4, a transcription factor required for murine hindlimb development, is also expressed in the GT (Chapman et al., 1996; Naiche and Papaioannou, 2003). Furthermore, signaling pathways essential for limb development are also critical for the formation of mammalian external genitalia (Lin et al., 2009; 2013; Miyagawa et al., 2009; Seifert et al., 2010; Suzuki et al., 2003).
Important functional links between the transcriptional networks that control formation of genitalia and limbs have been found through studies of human syndromes, particularly those involving mutations in HOX transcription factors (Goodman et al., 1997; Mortlock and Innis, 1997). These findings, in conjunction with animal studies, led to the proposal that the similar developmental features and gene expression patterns of the limbs and phallus stem from the use of common regulatory cassettes and that the phallus may have evolved through co-option of a preexisting digit or limb developmental program (Kondo et al., 1998).
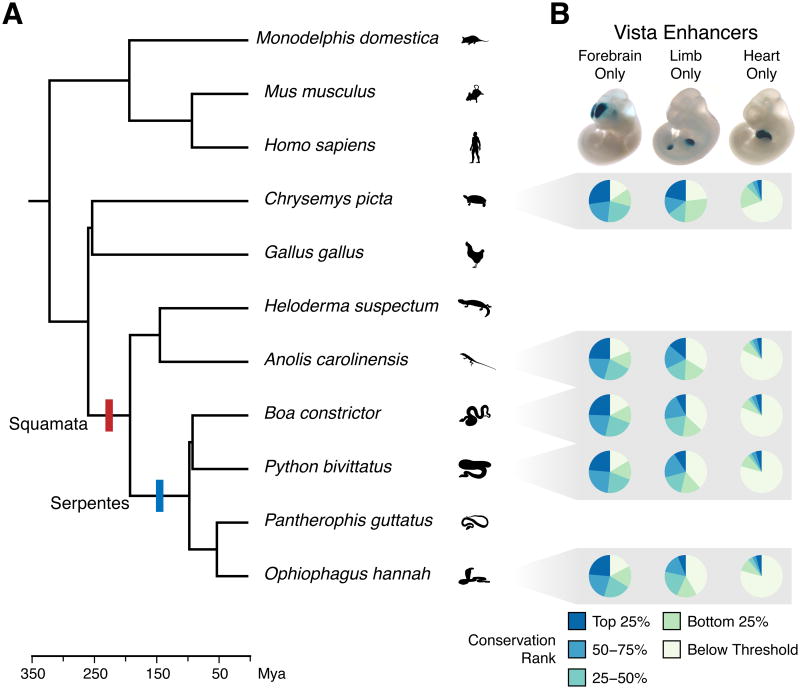
Lonfat and colleagues reported that the HoxA and HoxD gene clusters display similar regulatory topologies in the digits and the genitalia, with a combination of shared and tissue-specific enhancer-promoter interactions (Lonfat et al., 2014). Still, whether gene expression in limbs and external genitalia is primarily controlled by shared or distinct cis-regulatory elements remains unknown. If the latter, then a species that evolved limb loss would be expected to also lose limb enhancers over time. Snakes represent one of the most successful and ancient limb-reduced lineages (Vitt and Caldwell, 2009). Fossil evidence indicates that complete loss of forelimbs in snakes evolved more than 100 million years ago (Figure 1A; Pyron and Burbrink, 2012; Tchernov et al., 2000; Zaher et al., 2009). Some older lineages of living snake species, such as members of the Booidea (e.g., boas and pythons), retain a highly reduced hindlimb, but most other modern snakes lack evidence of any forelimb or hindlimb development. Thus, the analysis of snake genomes provides an opportunity to gain insights into the functional constraints on limb enhancer evolution.
Figure 1.
Enhancer elements display similar patterns of conservation across reptiles. (A) Chronogram of the major amniote lineages highlighting the divergence of the Squamata and the Serpentes. Phylogeny and dates adapted from Pyron (2010). (B) A comparison of the degree of conservation of VISTA enhancers positive for forebrain-only, limb-only, or heart-only enhancer activity (Visel et al. 2007) in the genomes of Chrysemys picta (painted turtle), Anolis carolinensis (green anole lizard), Boa constrictor (boa), Python bivittatus (Burmese python), and Ophiophagus hannah (king cobra). Pie charts indicate the relative proportion of enhancers in each similarity score category, with forebrain-only enhancers being most conserved, and heart-only enhancers being least conserved. See also Figure S1 and Table S1.
Here we use a combination of comparative genomics, chromatin profiling, and functional analyses to investigate the cis-regulatory elements of the developing limbs and phallus. We find extensive overlap in the patterns of cis-regulatory activity in these different appendage types in mice, and show that many of these elements are present in snake genomes. Functional tests of one such enhancer, the HLEB element from the Tbx4 locus, demonstrate an important role for this cis-regulatory element during hindlimb and urogenital development in mice. We provide evidence that the limb and genital activity of HLEB is conserved between mice and Anolis lizards, but has functionally diverged in snakes.
Results
Conservation of Limb Enhancers in Snakes
Genome assemblies for Boa constrictor (boa), Python bivittatus (Burmese python), and Ophiophagus hannah (king cobra) have recently become available (Bradnam et al., 2013; Castoe et al., 2013; Vonk et al., 2013). We reasoned that the evolution of limb loss in snakes might lead to substantial sequence divergence or complete loss of genomic regions that function exclusively in the limbs, such as limb-specific cis-regulatory elements. To examine limb regulatory elements in snakes we used the VISTA enhancer database (Visel et al., 2007), which contains thousands of human and mouse sequences that have been tested for enhancer activity in mouse embryos at embryonic day 11.5 (E11.5).
We searched the VISTA enhancer database for functionally validated limb enhancers. Of 208 VISTA enhancers with any limb activity, we selected a subset of 65 with highly restricted, limb-specific patterns of enhancer activity. As controls we selected comparable sets of forebrain and heart specific VISTA enhancers (Table S1). Starting with mouse enhancer sequences, we examined the conservation of these tissue-specific regulatory elements in the genomes of boa, Burmese python, and king cobra, as well as the genomes of a limbed squamate, Anolis carolinensis (green anole lizard), and Chrysemys picta (painted turtle). Tissue-specific enhancers exhibited different overall levels of conservation: forebrain enhancers had the highest degree of conservation and heart enhancers the lowest among the genomes analyzed (Figure 1B).
Despite the ancient limb reduction that occurred in the snake lineage, we found that snake genomes retain many sequences orthologous to mammalian limb enhancers. Levels of conservation were comparable across all three snake genomes, and the overall pattern of conservation of individual enhancers in snakes was quite similar to that observed in Anolis (Figure S1). Of note, similarity scores generated in our analyses likely underestimate actual conservation levels, because of errors in genome assemblies.
Activity of Mammalian Limb Enhancers in the Phallus
The apparent conservation of many “limb” enhancers in snakes suggests they might function in other, non-limb tissues. This could be a derived feature of snakes or a more ancient feature common to both limbed and limb-reduced animals. Therefore, we reassessed the tissue specificity of limb enhancers in mice. Because many genes are co-expressed in limbs and genitalia, cis-regulatory elements might be shared between limb and genital appendages (Tschopp et al., 2014). To determine whether shared expression patterns between limbs and genitalia reflect the action of shared or appendage-specific enhancers, we examined patterns of enhancer activity in embryonic mouse limbs and genitalia using ChIP-Seq against H3K27ac, a histone modification that marks active enhancers and promoters (Creyghton et al., 2010).
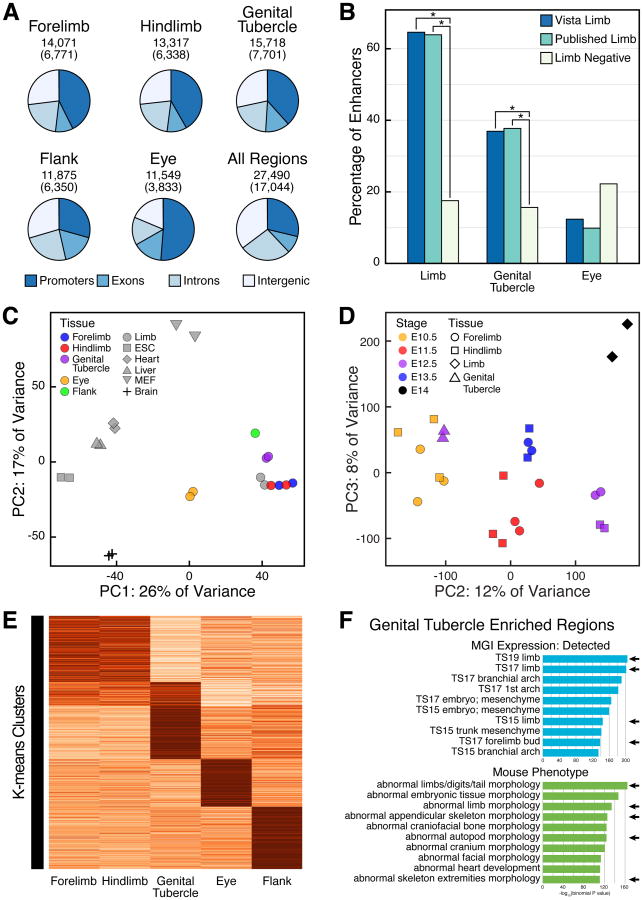
Outgrowth of the mouse forelimb is initiated at E9.0, and that of the hindlimb at E9.5. By comparison, the paired swellings that merge to form the GT do not develop until E10.5. To compare enhancer activity at similar stages of development, we performed H3K27ac ChIP-Seq on mouse appendages approximately two days after outgrowth is initiated: E11.5 for forelimb, E11.5 for hindlimb, and E12.5 for GT. As controls, we also performed ChIP-Seq on E11.5 flank and E10.5 eye tissues. Across all ChIP samples, we identified 27,490 genomic regions enriched for H3K27ac (Figure 2A; Table S2). We examined enrichment of H3K27ac at the 65 VISTA limb enhancers used in our comparative genomic analysis, another 61 limb enhancers previously reported in the literature (Table S1), and the VISTA forebrain and heart enhancers. In embryonic limbs, we found that 65% (42 of 65, Fisher's exact test p=1.74 × 10-15) of the VISTA limb enhancers and 64% (39 of 61, p=2.38 × 10-14) of the published limb enhancers had significant H3K27ac enrichment compared to non-limb VISTA enhancers (Figure 2B), demonstrating that many, but not all, embryonic limb enhancers are marked by H3K27ac at E11.5. We note that limb enhancers that are active in a small subset of limb cells may not appear to be enriched in our whole-limb H3K27ac data sets.
Figure 2.
Genome-wide analysis of H3K27ac signal reveals cis-regulatory overlap between the limbs and the genital tubercle. (A) Annotation of H3K27ac-enriched regions in the mouse E11.5 forelimb, E11.5 hindlimb, E12.5 GT, E11.5 flank, E10.5 eye, and all regions combined. For each category the total number of enriched regions, the number of putative enhancers in parentheses, and the relative proportions of regions that overlap exons, promoters, introns, and intergenic regions are indicated. (B) Percentage of validated enhancers that overlap H3K27ac-enriched regions from the limb, GT, and eye. Both H3K27ac limb regions and GT regions are significantly enriched at VISTA limb enhancers and published limb enhancers. Asterisks indicate p < 0.001 (Fisher's exact test). (C) Principal component analysis (PCA) of H3K27ac signal at shared regions from mouse tissues reveals similar signal profiles between limb-derived and GT tissues. (D) PCA of limb and GT H3K27ac signal at putative enhancers shows E12.5 GT associates with E10.5 limb signal. (E) K-means clustering of H3K27ac activity at putative enhancer regions identified from mouse E11.5 forelimb, E11.5 hindlimb, E12.5 GT, E10.5 eye, and E11.5 flank tissues identifies a set of elements (K-means Cluster #2) active in the limbs and GT. (E) The top ten MGI Expression and Mouse Phenotype enriched terms for GT H3K27ac-enriched regions generated using GREAT (McLean et al., 2010). Appendage-associated terms are indicated with arrows. See also Figure S2 and Tables S2, S3, and S4.
In the GT, we found that 37% (24 of 65, p=5.75 × 10-5) of VISTA limb enhancers and 38% (23 of 61, p=2.45 × 10-4) of the published limb enhancers are significantly enriched for H3K27ac compared to non-limb VISTA enhancers (Figure 2B). Of the 81 VISTA and published limb enhancers that are enriched for H3K27ac in limbs, 57% (46 of 81) are also enriched for H3K27ac in the GT. Much lower levels of overlap were observed with VISTA forebrain and heart enhancers. To expand our analysis to additional tissue types and developmental stages, we searched for enrichment of H3K27ac at limb enhancers using H3K27ac ChIP-Seq data from embryonic tissues analyzed as part of the mouse ENCODE project (Shen et al., 2012). We found much lower levels of H3K27ac overlap with the limb enhancers in non-limb embryonic tissues than in our limb and GT H3K27ac ChIP-Seq data (Figure S2).
Global Patterns of cis-Regulatory Activity in Limb and Genital Appendages
To broadly compare patterns of enhancer activity between the limbs and the GT, we analyzed all putative enhancer elements identified by H3K27ac ChIP-Seq in our mouse forelimb, hindlimb, GT, flank, and eye datasets. From the H3K27ac-enriched regions identified in each tissue, we excluded those that overlap gene promoters or exons. This identified 6,771 regions in the forelimb, 6,338 regions in the hindlimb, 7,701 regions in the GT, 6,350 regions in the flank, and 3,833 regions in the eye as putative enhancers (Figure 2A; Table S3). We analyzed these regions for associated genes using GREAT (McLean et al., 2010). Putative enhancers identified in the GT were enriched for terms associated with limb development (Figure 2F). Similar limb terms were enriched in both the forelimb and hindlimb putative enhancers lists, while putative eye enhancers were associated with eye and neural development, and flank enhancers were associated with skeletal and mesenchymal terms (Figure S2 and data not shown). For forelimb enhancers, 67.0% overlapped a putative hindlimb enhancer, while 45.9% overlapped a GT enhancer. For hindlimb enhancers, 70.0% overlapped a forelimb enhancer and 46.3% overlapped a GT enhancer, indicating extensive sharing of H3K27ac activated regions between the forelimbs, hindlimbs, and GT.
To compare variation in H3K27ac signal at all putative enhancers across a large number of tissues, we performed a principal component analysis (PCA) of H3K27ac signal intensity at enhancer regions. Included in the PCA were H3K27ac ChIP-Seq data from either 16 mouse tissues from the mouse ENCODE project, or a subset of 6 embryonic ENCODE tissues. These analyses showed H3K27ac signatures in E11.5 forelimbs, E11.5 hindlimbs, and E12.5 GT clustering with each other, and with mouse ENCODE E14.5 limb data (Figures 2C and S2). To evaluate similarities in cis-regulatory activity between E12.5 GT and limbs of different developmental stages, we performed an additional PCA using our own E11.5 limb and E12.5 GT H3K27ac datasets as well as 9 published H3K27ac datasets collected from a range of limb stages (Cotney et al., 2012; Cotney et al., 2013; Shen et al., 2012). In this appendage PCA, principal component 1 (PC1) associated with technical variation between datasets (Figure S2), while plotting PC2 against PC3 demonstrated that E12.5 GT clustered together with E10.5 forelimb and E10.5 hindlimbs (Figure 2D). Together these analyses revealed extensive sharing of appendage-specific cis-regulatory activity between embryonic limbs and GT.
We investigated patterns of cis-regulatory activity in embryonic limbs and genitalia further by grouping relative H3K27ac signal at enhancer regions using k-means clustering. However, instead of H3K27ac signal organizing into five categories based on tissue type, we found a set of 4,458 elements most strongly marked by H3K27ac in forelimbs and hindlimbs that formed a limb cluster, a set of 3,599 regions that formed a GT cluster, and a set of 1,585 regions that formed a distinct limb-GT cluster of elements that are strongly marked in both the limbs and GT (Figure 2E; Table S4). This limb-GT category represents a sizable proportion of all enhancer elements that have activity in both limb and genital appendage types. The remaining enriched regions formed eye specific and flank-specific clusters.
Properties of Limb, Limb-GT, and GT Enhancers
We examined whether the putative enhancers in our mouse limb, limb-GT, and GT clusters are significantly enriched near particular gene categories. To increase specificity, we excluded from this analysis any elements in the three appendage clusters that are marked by H3K27ac in any non-appendage tissues. This left 1,927 limb-specific elements, 1,467 GT-specific elements, and 700 elements that are limb-GT specific (Table S4). All three enhancer categories were strongly enriched for limb-associated terms (Figure 3A). Moreover, the GT and limb-GT elements are also enriched near genes associated with external genitalia and urogenital tissues. These include genes that are expressed and function within the developing genitalia and urogenital system.
Figure 3.

Properties of limb, limb-GT, and GT specific enhancers. (A) The top ten MGI Expression and Mouse Phenotype terms associated with limb, limb-GT, and GT specific enhancers using GREAT are enriched predominately with limb and skeletal development terms. Terms associated with urogenital and reproductive systems appear in the limb-GT and GT groups (arrows). (B) The top three de novo motifs found from the limb, limb-GT, and GT specific enhancers and their best matches to known motifs using HOMER (Heinz et al., 2010). (C) Percent of enhancers that liftOver to other tetrapod and fish genomes. Limb and limb-GT specific enhancers are more conserved in sarcopterygian genomes (Anolis, Xenopus, coelacanth). See also Figure S3 and Table S5.
We also compared the enriched motifs present in each enhancer category using HOMER (Heinz et al., 2010). Overall, the limb, limb-GT, and GT enhancers each show enrichment of a number of transcription factor binding motifs associated with limb development (Figure 3B; Table S5). The limb enhancers in particular showed strong enrichment for a homeobox-containing motif identified from a limb P300 ChIP-Seq dataset. For the GT enhancers, one of the top motifs is associated with Isl1, a transcription factor important for early hindlimb development and a marker for genital development (Kawakami et al., 2011; Tschopp et al., 2014).
Our initial examination of limb enhancer conservation in reptiles relied on enhancers in the VISTA database. We revisited the conservation of appendage enhancers using the mouse elements we identified via H3K27ac enrichment. We compared the level of conservation between the enhancer classes by examining their conservation in other tetrapod and fish genomes (Figure 3C). The limb and limb-GT enhancers display the highest degree of conservation with 16.8% and 14.9% of regions detectable in Anolis, respectively. In contrast, only 11.2% of GT enhancers were detected in Anolis. Similar trends were seen in the Xenopus and coelacanth genomes, with fewer GT enhancers present relative to limb or limb-GT enhancers. For all three enhancer categories the rate of successful liftOver to the fish genomes was under 3%. We also compared conservation of the mouse Limb, limb-GT, and GT enhancers in Anolis and snake genomes using LASTZ to calculate similarity scores. We found similar levels of conservation for all enhancers in the Anolis to boa and Anolis to Burmese python comparisons (Figure S3). In contrast, while the similarity scores of GT enhancers in Anolis and King cobra were comparable, we found the scores for limb and limb-GT enhancers trended lower in cobra relative to Anolis (p=0.0018 and p=0.032, respectively). Hence, we find evidence that limb and limb-GT enhancers exhibit greater sequence divergence in king cobra than in Anolis.
Functional Conservation of Appendage Enhancers
Since sequence conservation does not necessarily predict conservation of enhancer function, we performed H3K27ac ChIP-Seq on forelimb, hindlimb, and external genitalia (hemiphallus) tissue collected from Anolis carolinensis embryos. This allowed us to evaluate the conservation of enhancer activity between mouse and lizard appendages. We compared H3K27ac read coverage across the subset of mouse limb, limb-GT, and GT enhancers that exhibit sequence conservation in Anolis (Figures 4A, 4B, and S4). This revealed clear evidence of limb enhancer activity for Anolis orthologs of the mouse limb and limb-GT elements. For the limb enhancer orthologs, 37.3% and 44.8% overlap regions with significant enrichment in the Anolis forelimb and hindlimb H3K27ac data, respectively (Figure 4C; Table S6). Similarly, for the limb-GT enhancers, 40.2% and 46.7% are marked in the Anolis forelimb and hindlimb data. In contrast, we found that only 4.2% of limb and 16.8% of limb-GT enhancers were marked by significant H3K27ac enrichment in the hemiphalluses of Anolis embryos. Additionally, only 16.7% of GT enhancer orthologs overlapped regions significantly enriched in Anolis hemiphalluses, a ratio that is similar to the overlap with forelimb and hindlimb enriched regions (14.4% and 20.7%, respectively). Thus, at the developmental stages we analyzed cis-regulatory activity of orthologous enhancers is more often conserved between mouse and lizard limbs than mouse and lizard genitalia.
Figure 4.
H3K27ac enrichment at orthologous mouse and lizard enhancers. (A) H3K27ac coverage across limb, limb-GT, and GT specific enhancers in mouse E11.5 forelimb (blue), E11.5 hindlimb (red), and E12.5 GT (purple). H3K27ac signal was averaged across all mouse limb, limb-GT, and GT enhancers with a conserved ortholog in Anolis carolinensis. (B) H3K27ac coverage across A. carolinensis orthologs of mouse limb, limb-GT, and GT specific enhancers. H3K27ac signal analyzed in A. carolinensis embryonic tissues from stage 7-8 forelimb, stage 7-8 hindlimb, and stage 9-11 hemiphallus. (C) Fraction of A. carolinensis orthologs of mouse limb, limb-GT, and GT specific enhancers that overlap H3K27ac-enriched regions in A. carolinensis embryonic tissues. See also Figure S4 and Table S6.
Conservation of Limb and Genital cis-Regulatory Activity at the Tbx4 Locus
In order to investigate the function of limb-GT cis-regulatory elements in finer detail we chose to focus on the Tbx4 locus. The Tbx4 gene, which encodes a T-box transcription factor, is expressed during formation of the hindlimb and is essential for its development (Naiche and Papaioannou, 2003). Although studies of Tbx4 function in the GT have not been reported, Tbx4 is also expressed in the developing genitalia (Chapman et al., 1996). Previously, we identified two Tbx4 hindlimb enhancers, HLEA and HLEB (Menke et al., 2008). In our mouse H3K27ac k means clustering analysis, HLEA appeared in the limb cluster and HLEB in the limb-GT cluster (Table S4). Transgenic mouse lines carrying HLEA and HLEB Hsp68LacZ transgenes drive reporter gene expression in patterns that closely match their H3K27ac profiles, with both enhancers driving hindlimb expression and only HLEB driving expression in the GT (Figures 5A and S5).
Figure 5.
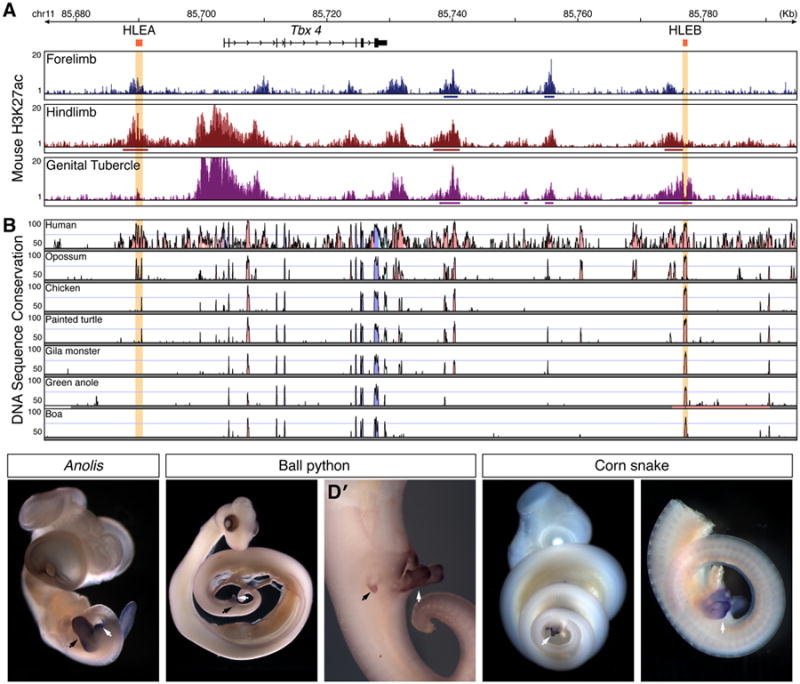
Conservation of Tbx4 enhancers and gene expression. (A) H3k27ac enrichment at the Tbx4 promoter demonstrates activity in the mouse hindlimb and GT, but not the forelimb. Similarly, the HLEB element of Tbx4 is active in the hindlimb and GT, while the HLEA element displays significant H3K27ac enrichment in the hindlimb only. H3K27ac signal from mouse E11.5 forelimb, E11.5 hindlimb, and E12.5 GT replicates are displayed as overlays of fold-enrichment compared to input; regions of significant enrichment are indicated by solid lines underneath signal profiles. (B) VISTA plot of sequence conservation using pairwise comparisons between mouse and other amniote species shows conservation of HLEB but not HLEA from human to boa. The Tbx4 gene is indicated in black. Red peaks correspond to conserved non-coding regions and blue peaks to conserved coding regions. The Tbx4 hindlimb enhancers HLEA and HLEB are indicated in orange (Menke et al., 2008). (C-E′) Tbx4 in situ hybridization on squamate embryos. Scale bars indicate 500 μm. (C) Anolis lizard embryos express Tbx4 in the hindlimbs and the developing hemiphalluses. (D) Ball python (Python regius) embryos display Tbx4 expression in the rudimentary hindlimbs and the hemiphalluses (D′). (E) Corn snake embryos (Pantherophis guttatus) show Tbx4 expression in the hemiphalluses (E′). Black arrows indicate location of hindlimbs; white arrows indicate hemiphalluses. See also Figure S5.
Sequence alignments of the mouse Tbx4 locus with orthologous loci from other amniotes revealed few noncoding regions with deep sequence conservation (Figure 5B). We uncovered no evidence of HLEA conservation in snakes, Anolis, or in a second lizard species, the Gila monster. In contrast, HLEB is conserved in all amniote species we examined, including lizards and snakes (Figures 5B and 6B). Despite the relative paucity of highly conserved non-coding regions at the Tbx4 locus, Tbx4 expression in hind appendages is a deeply conserved trait among vertebrates (Tanaka et al., 2002). We found that Tbx4 expression is readily detected in the hindlimbs and hemiphalluses of Anolis embryos (Figure 5C). We also examined Tbx4 expression in two species of snakes: Pantherophis guttatus (corn snake) and Python regius (ball python). Like king cobras, corn snakes are members of the Colubroidea and completely lack hindlimb buds. Ball pythons, on the other hand, develop rudimentary hindlimb buds. We observed intense Tbx4 expression throughout the hemiphalluses of corn snake and ball python embryos (Figures 5D-5E′; also see Leal and Cohn, 2015). Tbx4 expression in ball python hindlimbs was also apparent although staining was less intense than that observed in the external genitalia (Figure 5D′).
Figure 6.
Enhancer activity of HLEB elements from limbed and limb-reduced animals. (A) H3K27ac signal across A. carolinensis HLEB reveals significant enrichment in embryonic Anolis hindlimb and hemiphallus chromatin. (B) VISTA plot of pairwise sequence comparison of Anolis HLEB against multiple amniote species. Regions in red indicate 70% identity over a window size of at least 100 base pairs. (C-F′) Mouse embryos carrying HLEB Hsp68LacZ transgenes from mouse (C and C′), Anolis lizard (D and D′), Burmese python (E and E′), and king cobra (F and F′). Blue stain indicates the expression of LacZ. Scale bars indicate 500 μm. See also Figure S6.
Given that HLEB is among the most conserved non-coding regions near Tbx4, we investigated whether the enhancer activity of this element is conserved in limbed and limb-reduced species. We found that HLEB displays H3K27ac enrichment in the hindlimbs and hemiphalluses of Anolis embryos, suggesting that HLEB is active in both of these tissues (Figure 6A). Further tests of an Anolis HLEB Hsp68LacZ transgene showed that Anolis HLEB is sufficient to drive robust reporter gene expression in the hindlimbs and GT of E12.5 mouse embryos in patterns similar to that of mouse HLEB (Figures 6C-D′, and S6). In contrast, king cobra HLEB and Burmese python HLEB transgenes did not drive expression in mouse hindlimbs (Figures 6E, 6F, and S6). Moreover, unlike Anolis HLEB, which drove expression throughout the mouse GT, king cobra HLEB produced LacZ staining in a subset of cells located in the ventral half of the GT, and Burmese python HLEB staining was restricted to the base of the GT (Figures 6E′ and 6F′). Therefore, both the hindlimb and the genital specificity of HLEB are conserved between Anolis and mouse, but the HLEB of snakes has lost hindlimb enhancer activity while retaining subdomains of GT activity.
Reduced Tbx4 Expression After Deletion of a Limb-GT Enhancer
To determine the relative importance of HLEB in promoting Tbx4 expression, we used homologous targeting in ES cells to delete a 732 bp region that spans all squamate-mammal conserved sequences found within the mouse HLEB region (Figure S7). After germline transmission and subsequent breeding to a Cre deleter strain, the HLEB-deleted Tbx4 allele (Tbx4DelB) contained a single LoxP site and an FRT site at the location of the deleted enhancer. Whole mount in situ hybridization revealed subtle reductions in Tbx4 expression in the hindlimb buds of DelB/DelB embryos at E10.5 (Figure S7). By E11.5, Tbx4 levels in the GT were also lower in DelB/DelB embryos than in wild type (Figure 7A).
Figure 7.
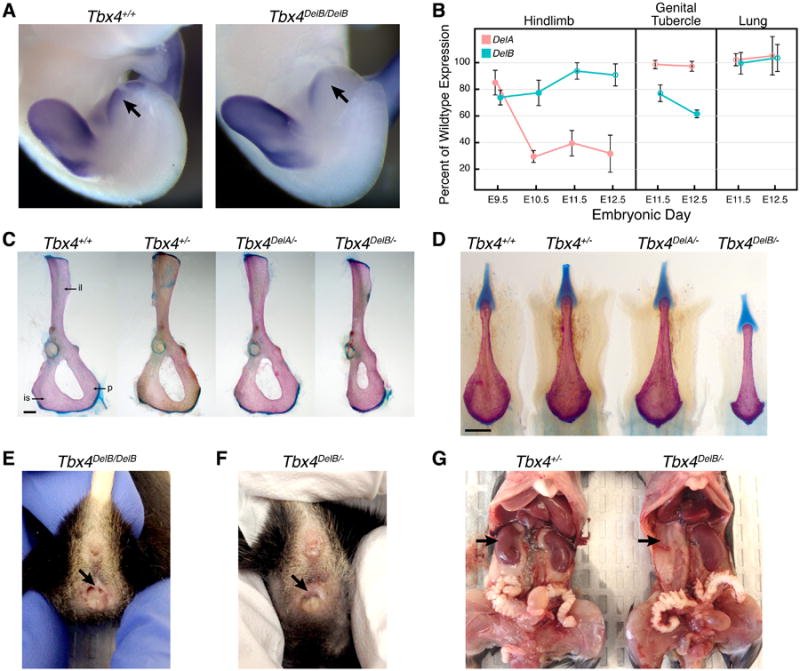
Deletion of hindlimb enhancer B (HLEB) affects Tbx4 expression and results in pelvic and urogenital-specific defects in adult mice. (A) E11.5 embryos homozygous for the HLEB deletion (DelB/DelB) show decreased expression of Tbx4 in the GT (arrows) by in situ hybridization. (B) Relative amounts of Tbx4 transcript produced from DelA and DelB alleles in embryonic hindlimb, GT, and lung as measured by an allele-specific expression assay. Error bars mark one standard deviation; filled points indicate values significantly different from wild-type expression (p < .05, t-test). (C) The deletion of HLEB affects pelvic bone morphology. In Tbx4DelB/- adult mice the posterior bones of the pelvis are reduced compared to Tbx4+/+, Tbx4+/-, and Tbx4DelA/- mice. Abbreviations: ilium (il), ischium (is), pubis (p). (D) The deletion of HLEB affects the baculum of adult male mice. Male Tbx4DelB/- mice have a significantly shortened baculum. (E-G) Additional examples of urogenital defects in HLEB knockout mice. Tbx4DelB/DelB adult female mice have a higher prevalence of vaginal septa (E) compared to wild-type littermates. In addition to vaginal septa, Tbx4DelB/- females occasionally have imperforate vagina (F). Adult HLEB knockout mice also display other urogenital defects, such as kidney agenesis (G). Scale bars indicate 500 μm. See also Figures S7 and Table S7.
In previous work, we showed deletion of HLEA results in a significant decrease in Tbx4 hindlimb expression (Menke et al., 2008). To compare the relative effect of deleting HLEA (DelA allele) or HLEB, we quantified Tbx4 expression levels across early and late stages of hindlimb bud development. We found that both the DelA and DelB alleles show reduced Tbx4 expression (Figure 7B), but the relative effect of deleting HLEA or HLEB varied by stage. At E9.5 the DelB allele shows a greater reduction in Tbx4 expression than the DelA allele (74% and 85% of wild-type, respectively). By E10.5, after emergence of hindlimb buds, the DelA allele expresses Tbx4 at only 30% of wild-type and remains at this reduced level through E12.5, whereas Tbx4 expression from the DelB allele recovers to near wild-type levels by E11.5.
The H3K27ac profiles of HLEA and HLEB indicated that only HLEB is active during GT development. Consistent with this, the DelB allele, but not the DelA allele, has significantly reduced Tbx4 transcription in this appendage (Figure 7B). In addition, loss of neither HLEA nor HLEB had a detectable effect on Tbx4 expression in the lungs, a tissue with prominent Tbx4 expression (Chapman et al., 1996).
Altered Hindlimb Morphology in HLEB Knockout Mice
Matings between DelB/+ heterozygotes yielded DelB/DelB offspring at frequencies that did not differ significantly from Mendelian expectations (20 +/+, 63 DelB/+, 22 DelB/DelB). Examination of adult animals revealed alterations to the proximal hindlimb skeleton. Specifically, the widths of the ischium and pubis bones of DelB/DelB animals were reduced by 13% (p=0.001) and 8% (p=0.002), respectively. In addition, the total dorsal ventral span of the ischium and pubis was reduced by 7% (p=3 × 10-5). In contrast, the ilium and the leg bones were relatively unaffected by the deletion of HLEB. We bred the DelB allele against a Tbx4 null allele (Naiche and Papaioannou, 2003) and found that DelB/- mice were viable but had severe defects in pelvic morphology (Figure 7C). As in DelB/DelB animals, abnormalities in the pelvis of DelB/- mice were most apparent in the ischium and pubis, both of which were dramatically reduced in size. In addition, the ilium (8.5%, p=0.02), femur (4.8%, p=4 × 10-5), and patella (5.8%, p=5 × 10-5) were significantly smaller in DelB/- animals than in +/- littermates.
In contrast to DelB/- mice, DelA/- mice exhibited comparatively normal pelvic morphology (Figure 7C). Furthermore, the distal hindlimb bones of DelA/- mice displayed a variety of defects not found in DelB/- animals, including malformed and fused anklebones, and fused first and second metatarsals (Figure S7). These defects are similar to those we have previously observed in DelA/DelA mice (Menke et al., 2008), although the severity of these malformations is enhanced in DelA/- animals.
Urogenital Defects in HLEB Knockout Mice
Given the reduced Tbx4 expression observed in the GT of DelB/DelB embryos, we inspected the genitalia of DelB/DelB adults for structural defects. External examination of DelB/DelB males revealed no gross abnormalities. Therefore, we examined the baculum bones of adult males for alterations in size or shape. The baculum, or penis bone, is present within the phallus of many mammals, including mice, and baculum size correlates with reproductive success in mice (Stockley et al., 2013). Measurement of bacula uncovered significant decreases in length (3.5%, p=0.007) and width (3.7%, p=0.029) in DelB/DelB adult males, compared with wild-type animals. Moreover, striking reductions in baculum sizes of DelB/- males were apparent by visual inspection (Figure 7D). By comparison, we detected no significant alterations in baculum size in DelA/DelA or DelA/- males (Table S7).
Although we did not observe major abnormalities in the external genitalia of DelB/DelB males, we found that approximately half of adult DelB/DelB females presented with a vaginal septum. The vaginal septum in these females spanned the vagina in a dorsal-ventral orientation and bisected the vaginal canal, resulting in two separate vaginal openings (Figure 7E). The presence of vaginal septa has been shown to severely impair female fertility and has been reported to occur spontaneously at frequencies as high as 11% in C57BL/6J mice (Gearhart et al., 2004), the same genetic background onto which we back crossed the DelB allele. Nevertheless, in our mouse colony we detected much higher rates of this vaginal defect in DelB/DelB females than in wild-type littermates (+/+ = 0/16, DelB/+ = 2/19, DelB/DelB = 7/15). Moreover, DelB/- females presented with either a vaginal septum (8 of 10 DelB/- females) or imperforate vagina, the complete absence of a vaginal opening (2 out of 10; Figure 7F).
Further examination of DelB/- animals revealed additional defects of the urogenital system. DelB/- males exhibited enlarged bulbourethral glands and severe malformations of the seminal vesicles, which appeared to be overgrown with excessive branching (Figure S7). Female DelB/- mice with imperforate vagina had distended uterine horns caused by the accumulation fluids that normally drain through the vagina. Finally, we observed a range of kidney malformations, including unilateral kidney agenesis, hyperplasia, and hydronephrosis (Figures 7G and S7).
Discussion
Primitive snakes retain a highly reduced hindlimb that includes pelvic bones and a femur (Cohn and Tickle, 1999). As such, the maintenance of some aspects of the limb developmental program might, in part, explain the retention of limb cis-regulatory elements in boa and python that we have uncovered. In contrast, advanced snakes (Colubroidea), which originated approximately 85 million years ago and include the majority of extant snake species (Pyron and Burbrink, 2012), completely lack forelimbs or hindlimbs. Because advanced snakes likely evolved complete limb loss in the late Cretaceous, there has been substantial time for limb-specific regulatory elements to diverge. However, the presence of limb enhancer elements in king cobra, a member of the Colubroidea, suggests that there is selective pressure to retain these elements in limbless species.
Although many amniote limb enhancers are still found in the genomes of snakes, there are numerous snake-specific mutations in these elements and substantial variation in the degree of conservation of individual enhancers (Figures S1 and S3). In the absence of limbs, one might expect cis-regulatory elements to lose limb-specific activities, while retaining activities in other tissues or evolving entirely new functions. Our analyses of HLEB orthologs suggest that this element has functionally diverged along such a path with snake HLEBs losing hindlimb function but retaining activity within subdomains of the external genitalia. The Burmese python HLEB also displays enhancer activity in the nasal region of transgenic mouse embryos, a site of activity not observed with mouse, Anolis, or cobra orthologs (Figures 6 and S6). Thus python HLEB may have evolved a new function, a hypothesis that will require further study to verify.
Our H3K27ac ChIP-Seq profiles of limb and genital appendages in mice demonstrate that these different appendage types exhibit many shared limb-GT enhancers. We believe, however, that the persistence of limb enhancers in snake genomes is unlikely to be solely attributable to a role for these elements in external genitalia. For instance, deletion of HLEB from mice not only affects development of the hindlimb and the GT, but also impacts formation of other reproductive and urogenital tissues. Furthermore, we find that of the mouse limb-GT enhancers with orthologs in Anolis, a relatively small proportion exhibit conserved enhancer activity in the hemiphallus as compared to the limbs of Anolis embryos (16.8% in the hemiphallus vs. 40.2% in the forelimb and 46.7% in the hindlimb; Figure 4). The modest number of limb-GT enhancers with conserved genital activity might reflect the striking anatomical differences between the genitalia of these species (Gredler et al., 2014). In most amniotes, including mammals, the external genitalia form as paired swellings, which then merge to form a single phallus. In contrast, the paired genital swellings of squamates do not fuse and instead form a pair of hemiphalluses. Moreover, unlike other amniotes, squamates do not incorporate endodermal derivatives into their external genitalia. Nevertheless, it remains possible that at earlier stages of development hemiphalluses might exhibit greater evidence of enhancer activity conservation than we detected in our samples.
It is well established that the transcription of individual genes is often driven by multiple enhancers with similar or overlapping activities (Hong et al., 2008). We find that in mouse limbs and genitalia there is an additional layer of complexity with appendage genes often displaying a combination of limb, limb-GT, and GT enhancer elements (Table S4). Deletion of the HLEA or HLEB enhancers from the Tbx4 locus demonstrates the effect of developmental context on the relative importance of these enhancer elements. Removal of HLEA results in mild reductions in Tbx4 expression during early stages of hindlimb development, but dramatic reductions at later stages. By comparison, the function of HLEB is more important during early phases of hindlimb formation. Differences in the relative importance of HLEA and HLEB in driving early and late phases of Tbx4 expression are reflected in the hindlimb phenotypes that result from their deletion. When HLEB is removed, the pelvis, which is patterned early during hindlimb development, is altered. Whereas when HLEA is deleted, the distal leg and foot, which are patterned later, are strongly impacted. Therefore, the function of these enhancers in the hindlimb is not equivalent and their activity is not completely redundant.
While both HLEA and HLEB are active during hindlimb development, only HLEB functions in the external genitalia. Since deletion of HLEB does not eliminate Tbx4 expression in the GT, we can infer that HLEB works in conjunction with other limb-GT or GT enhancers to induce Tbx4 transcription. Nonetheless, deletion of HLEB is sufficient to induce alterations in the genitalia of male and female mice. Thus, shared cis-regulatory elements of the limbs and GT can play important roles in both structures, and the deletion of HLEB has revealed additional roles for Tbx4 in vagina, phallus, and urogenital development.
Though the limbs and external genitalia display similar patterns of gene expression and rely on many of the same signaling pathways, their development is not completely analogous (Cohn, 2011). However, the extensive sharing of enhancers that we have detected between embryonic limbs and external genitalia indicates that their similarities are not superficial. Indeed, similarities in gene regulation extend down to the level of the cis-regulatory elements that control gene transcription. Presently, it is unclear whether genital and limb-specific morphologies in amniotes are shaped primarily through modulation of the common appendage enhancers, the appendage-specific enhancers, or both. Mechanistic studies of how these different enhancer classes interact with and are regulated by transcription factors during limb and genital development will be required to fully elucidate the behavior of these regulatory elements in different developmental contexts.
Experimental Procedures
Mouse Strains
Tbx4 null and Tbx4 DelA alleles have been previously reported (Naiche et al., 2003; Menke et al., 2008). The Tbx4 DelB allele was produced through homologous targeting in mouse ES cells. See the Supplemental Experimental Procedures for additional details. Transgenic mouse strains carrying mouse HLEA and HLEB Hsp68LacZ transgenes were previously described (Menke et al., 2008; Infante et al., 2013), and Anolis HLEB, king cobra HLEB, and Burmese python HLEB Hsp68LacZ transgenic mouse embryos were produced by Cyagen Biosciences (Santa Clara, CA) through pronuclear injections. See the Supplemental Experimental Procedures for further details. For linear measurements of morphology, significant differences between means were determined using Student's t-test. All mouse work was reviewed by the University of Georgia Institutional Animal Care and Use Committee and was performed under an approved Animal Use Protocol.
Enhancer conservation
The mouse genomic coordinates (mm9) of functionally validated limb enhancers were downloaded from the VISTA enhancer database (http://enhancer.lbl.gov, download date 24 Jan 2014). Forebrain Only, Limb Only, and Heart Only enhancer categories were determined by selecting only enhancers with annotated activity in each target tissue and excluding any that had annotated activity in any other tissues. Additional functionally validated limb enhancers were taken from the literature and where necessary the genomic coordinates were translated to the mouse. Genome assemblies for painted turtle (chrPic1, UCSC Genome Browser), anole lizard (anoCar2, UCSC Genome Browser), Burmese python (version 5.0.2, NCBI), king cobra (OphHan1.0, NCBI), boa constrictor (assembly version 6C, Assemblathon2, http://gigadb.org) were subdivided and queried with enhancer sequences in parallel using the LASTZ aligner (Harris, 2007) with the HoxD55 scoring matrix and the parameters “--gap=400,30 --seed=12 of 19 --transition --hspthresh=3000 --entropy --ydrop=3400 --gappedthresh=6000 --inner=2000”. LASTZ alignments were compiled into a sqlite database for each genome. For each enhancer the highest scoring alignment was used to calculate a similarity score as a metric of conservation by scaling the alignment score by the length of the enhancer sequence. Multiple non-redundant hits for the same enhancer that were within 2 kb on the same scaffold or contig were combined. All calculations and database functions were performed in R (http://cran.r-project.org).
Chromatin Immunoprecipitation
ChIP-Seq was performed as previously described (Infante et al., 2013) on embryonic mouse tissues collected from E10.5 (eye), E11.5 (forelimb, hindlimb, and flank), and E12.5 (GT). Anolis ChIP-Seq was performed on tissues collected from stages 7 to 8 (forelimbs and hindlimbs) and on stages 9 to 11 (hemiphallus). Anolis embryos were staged according to Sanger (Sanger et al., 2007). An Anti-H3K27ac monoclonal antibody (Millipore, #05-1334) was used for all ChIP-Seq experiments. ChIP-Seq reads were aligned to the mouse genome (mm9) or Anolis carolinensis genome (anoCar2) using bowtie (1.1.0; Langmead et al., 2009) with the options “--best --sam -t -l 28 -n 3 -m1”. Aligned ChIP-Seq datasets were assessed for quality of enrichment using ENCODE statistics calculated using phantompeakqualtools (https://code.google.com/p/phantompeakqualtools/). Enriched regions were determined using macs2 (2.0.10.20130501; Zhang et al., 2008) with the options “--bdg --SPMR --nomodel --qvalue=0.05”. Extension size for macs2 was determined for each ChIP-Seq replicate based the cross-correlation profile calculated by phantompeakqualtools. For each sample, enriched regions within 1 kb were combined into a single regions using BEDTools (version 2.19.0; Quinlan and Hall, 2010); merged regions with a minimum overlap of 1 bp from each replicate were then combined to create a dataset of reproducible enriched regions. Reproducibly enriched regions that did not overlap within 1 kb of a transcription start site or any exons of UCSC known genes (Kent et al., 2002) were designated potential enhancers.
Mouse ENCODE and Additional Appendage ChIP-Seq Data
Aligned reads from H3K27ac ChIP-Seq experiments in 16 mouse tissues and cell lines were downloaded from the ENCODE database (https://www.encodeproject.org; Shen et al., 2012). Enriched regions for two replicates each of adult heart (ahe), adult liver (ali), adult brown adipose tissue (bat), adult bone marrow (bon), adult cortical plate (cor), embryonic E14.5 brain (ebr), embryonic E14.5 heart (ehe), embryonic E14.5 liver (eli), embryonic stem cell (esc), adult kidney (kid), embryonic E14.5 limb (lim), mouse embryonic fibroblast (mef), adult olfactory bulb (olf), adult small intestine (sma), adult spleen (spl), and adult thymus (thy) were determined and processed as described above. Additional H3K27ac aligned read datasets for mouse E10.5 forelimb, E10.5 hindlimb, E11.5 forelimb, E11.5 hindlimb, E12.5 forelimb, E12.5 hindlimb, E13.5 forelimb, and E13.5 hindlimb (Cotney et al., 2012, Cotney et al., 2013) were downloaded from the GEO database (series GSE30641 and GSE42413) and enriched regions were determined and processed as described above.
Principal Component Analysis
Potential enhancer regions from either 11 tissue types (E11.5 forelimb, E11.5 hindlimb, E12.5 genital tubercle, E10.5 eye, E11.5 flank, and ENCODE embryonic limb, embryonic brain, embryonic heart, embryonic liver, embryonic stem cell, and embryonic fibroblast), 21 tissue types (E11.5 forelimb, E11.5 hindlimb, E12.5 genital tubercle, E10.5 eye, E11.5 flank, and all 16 additional mouse ENCODE tissues), or 12 appendage tissue datasets (E11.5 forelimb, E11.5 hindlimb, E12.5 genital tubercle from this study; E10.5 forelimb, E10.5 hindlimb, E11.5 forelimb, E11.5 hindlimb, E12.5 forelimb, E12.5 hindlimb, E13.5 forelimb, and E13.5 hindlimb from Cotney et al., 2012 and Cotney et al., 2013; E14.5 limbs from Shen et al., 2012) were intersected with BEDTools and shared regions were compiled into a matrix using R. Reads overlapping shared regions were counted using Rsubread (Liao et al., 2013). The counts were then extracted into a DGEList and converted into RPKM values using EdgeR (Robinson et al., 2010). Principal components analysis was performed on the resulting RPKM matrix using the prcomp function in R. Principal components were plotted using the R package ggplot2 (Wickham, 2009).
K-means Clustering
All potential enhancer regions from H3K27ac ChIP-Seq experiments on mouse E11.5 forelimb, E11.5 hindlimb, E12.5 genital tubercle, E10.5 eye, and E11.5 flank were combined into a single matrix. Reads overlapping any regions in the combined matrix were counted using Rsubread. The counts were then extracted into a DGEList and converted into RPKM values using EdgeR. RPKM values for a given regions were normalized and by subtracting the mean value across tissues and dividing by the standard deviation (Cotney et al., 2013). K-means clustering was performed using the kmeans function in R.
H3K27ac Coverage at Matched Mouse and Anolis Enhancers
Mouse enhancer coordinates (mm9) were translated to the Anolis carolinensis genome (anoCar2) using the UCSC liftOver tool. Only mouse enhancer regions that lifted over successfully to the Anolis genome were used to compare H3K27ac signal at orthologous enhancers between species. To normalize the H3K27ac reads across tissue samples withinin each species, scaling factors for each dataset were calculated using CHANCE (Diaz et al., 2012). H3K27ac read coverage was then scored in 25 bp windows +/- 2 kb from the center point of orthologous enhancers from normalized bigWig files using deepTools package (Ramírez et al., 2014). The deepTools computeMatrix script was used to calculate matrix of coverage values. This matrix was then processed in R to produce signal plots of average enrichment, or processed using the deepTools heatmapper script to produce heatmaps of coverage over all orthologous enhancer regions.
Motif Finding
Enriched motifs in mouse Limb, limb-GT, and GT enhancers were determined using HOMER (Heinz et al., 2010). For each enhancer category, the central 500 bp of each enhancer regions was used for de novo and known motif searches. A background sequence dataset was created by combining all putative enhancer regions from mouse E11.5 forelimb, E11.5 hindlimb, E12.5 genital tubercle, E10.5 eye, and E11.5 flank datasets, omitting any regions that were present in the input sequences.
In Situ Hybridization and LacZ Staining
Adult Anolis sagrei were collected from the University of South Florida Botanical Gardens (Tampa, FL, USA), housed at the University of Georgia, and bred to produce embryos in accordance with an approved Animal Use Protocol. Corn snake (Pantherophis guttatus) embryos were a gift from David W. Hall. Ball python eggs were purchased from J. Kobylka Reptiles (Toccoa, GA). Whole-mount mRNA in situ hybridizations were performed as described for other species (Wilkinson, 1992). Templates for mouse, Anolis, and corn snake tbx4 riboprobes were generated by PCR amplification from embryonic mouse, Anolis, and corn snake cDNAs. Template for python tbx4 probe was produced through gene synthesis of the predicted Burmese python mRNA by Life Technologies (Grand Island, NY). LacZ staining of transgenic mouse embryos was performed as described (DiLeone et al., 1998).
Allele-Specific Gene Expression Analysis
Quantification of Tbx4 expression from wild-type, DelA, and DelB alleles was accomplished through the use of a naturally occurring SNP within the Tbx4 transcript that differs between the strains used to create our targeted Tbx4 alleles and the DBA/2J mouse strain (Menke et al., 2008). See the Supplemental Experimental Procedures for further details.
Supplementary Material
Acknowledgments
We thank David W. Hall for corn snake eggs and Laurie Walker for permission and assistance collecting lizards in Tampa, FL. We also thank V.E. Papaioannou for providing Tbx4 knockout mice and Anna Lau for editorial assistance. This work was supported by grants awarded to D.B.M from NSF (#1149453), NIH (HD081034), and by start-up funds from the University of Georgia. Additional support to J.D.L. was provided by the Sharon Stewart Aniridia Trust. K.K.J. was supported by a graduate fellowship from the Southern Regional Education Board Doctoral Scholars Program. This study was supported in part by resources from the Georgia Advanced Computing Resource Center.
Footnotes
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables.
Author Contributions: C.R.I. and D.B.M designed the study. C.R.I. performed all comparative genomics and bioinformatic analyses of ChIP-Seq data. J.D.L. directed the design and collection of mouse eye ChIP-Seq data with S.P. and K.K.J. All other mouse and Anolis ChIP-Seq data was generated by A.G.M., C.R.I., and S.P. Transgenic and knockout mice were designed by D.B.M. and generated by S.P. Analysis of mouse knockouts was performed by A.G.M., S.P., and J.S.W. C.R.I. and D.B.M wrote the manuscript.
Accession Numbers: H3K27ac ChIP-Seq data generated for this work has been deposited in the Gene Expression Omnibus (GEO Accession Number GSE64055).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert SB, Chapman JA, Chapuis G, Chikhi R, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2:1–1. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, de Koning APJ, Hall KT, Card DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu W, et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci USA. 2013;110:20645–20650. doi: 10.1073/pnas.1314475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399:474–479. doi: 10.1038/20944. [DOI] [PubMed] [Google Scholar]
- Cohn MJ. Development of the external genitalia: Conserved and divergent mechanisms of appendage patterning. Dev Dyn. 2011;240:1108–1115. doi: 10.1002/dvdy.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Nellore A, Song JS. CHANCE: comprehensive software for quality control and validation of ChIP-seq data. Genome Biol. 2012;13:R98. doi: 10.1186/gb-2012-13-10-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone RJ, Russell LB, Kingsley DM. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart S, Kalishman J, Melikyan H, Mason C, Kohn DF. Increased incidence of vaginal septum in C57BL/6J mice since 1976. Comp Med. 2004;54:418–421. [PubMed] [Google Scholar]
- Goodman FR, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli ML, Lapi E, Majewski F, McGaughran J, McKeown C, Reardon W, et al. Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci USA. 1997;94:7458–7463. doi: 10.1073/pnas.94.14.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredler ML, Larkins CE, Leal F, Lewis AK, Herrera AM, Perriton CL, Sanger TJ, Cohn MJ. Evolution of External Genitalia: Insights from Reptilian Development. Sex Dev. 2014;8:311–326. doi: 10.1159/000365771. [DOI] [PubMed] [Google Scholar]
- Harris RS. Ph D Thesis. The Pennsylvania State University; 2007. Improved pairwise alignment of genomic DNA. [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow Enhancers as a Source of Evolutionary Novelty. Science. 2008;321:1314–1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante CR, Park S, Mihala AG, Kingsley DM, Menke DB. Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev Biol. 2013;374:234–244. doi: 10.1016/j.ydbio.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Marti M, Kawakami H, Itou J, Quach T, Johnson A, Sahara S, O'Leary DD, Nakagawa Y, Lewandoski M, Pfaff S, Evans SM, Izpisua Belmonte JC. Islet1-mediated activation of the β-catenin pathway is necessary for hindlimb initiation in mice. Development. 2011;138:4465–4473. doi: 10.1242/dev.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Zákány J, Innis JW, Duboule D. Of fingers, toes and penises. Nature. 1998;390:1–1. doi: 10.1038/36234. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal F, Cohn MJ. Development of Hemipenes in the Ball Python Snake Python regius. Sex Dev. 2015;9:6–20. doi: 10.1159/000363758. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucl Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yin Y, Veith GM, Fisher AV, Long F, Ma L. Temporal and spatial dissection of Shh signaling in genital tubercle development. Development. 2009;136:3959–3967. doi: 10.1242/dev.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yin Y, Bell SM, Veith GM, Chen H, Huh SH, Ornitz DM, Ma L. Delineating a Conserved Genetic Cassette Promoting Outgrowth of Body Appendages. PLoS Genet. 2013;9:e1003231. doi: 10.1371/journal.pgen.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science. 2014;346:1004–1006. doi: 10.1126/science.1257493. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- Miyagawa S, Moon A, Haraguchi R, Inoue C, Harada M, Nakahara C, Suzuki K, Matsumaru D, Kaneko T, Matsuo I, et al. Dosage-dependent hedgehog signals integrated with Wnt/beta-catenin signaling regulate external genitalia formation as an appendicular program. Development. 2009;136:3969–3978. doi: 10.1242/dev.039438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock DP, Innis JW. Mutation of HOXA13 in had-foot-genital syndrome. Nat Genet. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- Pyron RA. A likelihood method for assessing molecular divergence time estimates and the placement of fossil calibrations. Syst Biol. 2010;59:185–194. doi: 10.1093/sysbio/syp090. [DOI] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT. Extinction, ecological opportunity, and the origins of global snake diversity. Evolution. 2012;66:163–178. doi: 10.1111/j.1558-5646.2011.01437.x. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucl Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger TJ, Losos JB, Gibson-Brown JJ. A developmental staging series for the lizard genus Anolis: A new system for the integration of evolution, development, and ecology. J Morphol. 2008;269:129–137. doi: 10.1002/jmor.10563. [DOI] [PubMed] [Google Scholar]
- Seifert AW, Zheng Z, Ormerod BK, Cohn MJ. Sonic hedgehog controls growth of external genitalia by regulating cell cycle kinetics. Nat Commun. 2010;1:1–9. doi: 10.1038/ncomms1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockley P, Ramm SA, Sherborne AL, Thom MDF, Paterson S, Hurst JL. Baculum morphology predicts reproductive success of male house mice under sexual selection. BMC Biol. 2013;11:1–1. doi: 10.1186/1741-7007-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Bachiller D, Chen YP, Kamikawa M, Ogi H, Haraguchi R, Ogino Y, Minami Y, Mishina Y, Ahn K, et al. Regulation of outgrowth and apoptosis for the terminal appendage: external genitalia development by concerted actions of BMP signaling. Development. 2003;130:6209–6220. doi: 10.1242/dev.00846. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Münsterberg A, Anderson WG, Prescott AR, Hazon N, Tickle C. Fin development in a cartilaginous fish and the origin of vertebrate limbs. Nature. 2002;416:527–531. doi: 10.1038/416527a. [DOI] [PubMed] [Google Scholar]
- Tchernov E, Rieppel O, Zaher H, Polcyn MJ, Jacobs LL. A fossil snake with limbs. Science. 2000;287:2010–2012. doi: 10.1126/science.287.5460.2010. [DOI] [PubMed] [Google Scholar]
- Tschopp P, Sherratt E, Sanger TJ, Groner AC, Aspiras AC, Hu JK, Pourquié O, Gros J, Tabin CJ. A relative shift in cloacal location repositions external genitalia in amniote evolution. Nature. 2014;18:391–394. doi: 10.1038/nature13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitt LJ, Caldwell JP. Herpetology. Burlington, MA: Elsevier; 2009. [Google Scholar]
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJR, Kerkkamp HME, Vos RA, Guerreiro I, Calvete JJ, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- Wilkinson DG. In situ hybridization: a practical approach. Oxford University Press; 1992. [Google Scholar]
- Zaher H, Apesteguía S, Scanferla CA. The anatomy of the upper cretaceous snake Najash rionegrina Apesteguía & Zaher, 2006, and the evolution of limblessness in snakes. Zool J Linnean Soc. 2009;156:801–826. [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.