Abstract
Development and improvement of quality control tests for live attenuated vaccines are a high priority because of safety concerns. Live attenuated influenza vaccine (LAIV) viruses are 6:2 reassortants containing the hemagglutinin (HA) and neuraminidase (NA) gene segments from circulating influenza viruses to induce protective immune responses, and the six internal gene segments from a cold-adapted Master Donor Virus (MDV). LAIV candidate viruses for the 2012–2013 seasons, A/Victoria/361/2011-CDC-LV1 (LV1) and B/Texas/06/2011-CDC-LV2B (LV2B), were created by classical reassortment of A/Victoria/361/2011 and MDV-A A/Leningrad/134/17/57 (H2N2) or B/Texas/06/2011 and MDV-B B/USSR/60/69. In an attempt to provide better identity and stability testing for quality control of LV1 and LV2B, sensitive real-time RT-PCR assays (rRT-PCR) were developed to detect the presence of undesired gene segments (HA and NA from MDV and the six internal genes from the seasonal influenza viruses). The sensitivity of rRT-PCR assays designed for each gene segment ranged from 0.08 to 0.8 EID50 (50% of Egg Infectious Dose) per reaction for the detection of undesired genes in LV1 and from 0.1 to 1 EID50 per reaction for the detection of undesired genes in LV2B. No undesired genes were detected either before or after five passages of LV1 or LV2B in eggs. The complete genome sequencing of LV1 and LV2B confirmed the results of rRT-PCR, demonstrating the utility of the new rRT-PCR assays to provide the evidence for the homogeneity of the prepared vaccine candidate.
Keywords: Influenza virus, Reassortants, Live vaccine, Real time RT-PCR, Homogeneity test
1. Introduction
Influenza A and B viruses are responsible for annual epidemics and significant morbidity and mortality in the human population worldwide (Thompson et al., 2003). Vaccination is an important intervention for preventing influenza and reducing the public health impact of epidemics and pandemics (Ambrose et al., 2011; Osterholm et al., 2012). Global Pandemic Influenza Action Plan to increase vaccine supply was initiated by the World Health Organization (WHO) in November of 2006 and refined further in 2011. One of the objectives was to expand vaccine production capacity by building new production facilities to strengthen pandemic influenza preparedness and response. The Institute of Experimental Medicine (IEM, St Petersburg, Russia) contributed to the Global Pandemic Influenza Action Plan by establishing an agreement with the WHO to supply Russian LAIV reassortant viruses to manufacturers in developing countries who could then provide influenza vaccines to the public (Rudenko et al., 2011). The increased international demand of Russian LAIV reassortant viruses prompted the WHO and IEM to establish a back-up facility at the Centers of Disease Control and Prevention (CDC), Influenza Division to prepare and incorporate quality assessment of LAIV reassortants for international use.
Seasonal influenza vaccines contain two types A viruses (H1N1 and H3N2) and one type B virus to elicit immunity to currently circulating influenza viruses (Fiore et al., 2009). Two major types of influenza vaccines are licensed for human use: trivalent inactivated influenza vaccine (TIV) and LAIV. LAIV is administered intranasally which mimics natural infection and protects against the disease caused by the influenza viruses (Aleksandrova, 1977; Cox et al., 2004; Maassab and DeBorde, 1985).
The development of LAIV became possible with the generation of cold-adapted (ca) type A and B Master Donor Viruses (MDVs). Both MDVs have three phenotypes: (1) allow efficient viral replication at 25 °C and 33 °C (ca), (2) restrict replication at temperatures above 39 °C (temperature-sensitive, ts), and (3) do not cause classical influenza-like illness and are restricted in replication in the lower respiratory tract (attenuated, att) (Maassab and Bryant, 1999; Murphy and Coelingh, 2002; Rudenko et al., 1996). LAIV are 6:2 reassortant viruses that contain the HA and NA gene segments from circulating influenza viruses to induce protective immune responses and the six internal gene segments (PB1, PB2, PA, NP, M, and NS) from MDVs provide ca, ts and att phenotypes of LAIV. In addition to conferring the ts and ca phenotypes, MDV also allows the reassortant viruses to replicate efficiently in embryonated chicken eggs as all licensed LAIVs are currently produced in embryonated chicken eggs.
Due to constant antigenic drift of circulating influenza viruses, the individual strains of the vaccine are updated annually based on WHO recommendations. A/Victoria/361/2011 (H3N2)-like and B/Wisconsin/1/2010-like viruses were recommended by the WHO for vaccine use in the 2012–2013 influenza season. Classical reassortment methods were used to produce vaccine candidate viruses by co-infection of A/Victoria/361/2011 and MDV-A A/Leningrad/134/17/57 (H2N2) or B/Texas/06/2011 and MDV-B B/USSR/60/69 followed by genotyping a number of virus progeny for selection of the 6:2 reassortants. To enhance the genetic homogeneity, virus cloning by serial limiting dilution in eggs was performed. Sensitive control of reassortant genetic composition is necessary to provide evidence that there are no contaminating wild type (wt) viruses present that may replicate efficiently in patients and cause illness and to assure that the vaccine strain does not contain HA and NA from MDVs. A challenge for the limiting dilution cloning in eggs method is the possibility of obtaining an impure vaccine candidate. According to WHO guidelines for generation and characterization of LAIV candidates, biological materials for vaccine production, such as seed viruses and intermediates should be fully and up-to-date characterized to assure the quality, safety, and efficacy of LAIV for intranasal administration (WHO, 2009). The safety concerns necessitate accurate, sensitive and reliable methods to characterize the quality of LAIV seed candidates generated by classical reassortment. The current techniques for identity/homogeneity testing of reassortant influenza vaccines are the ones used for genotyping – analysis of restriction fragment length polymorphism of viral genes amplified by RT-PCR (Fulvini et al., 2011; Klimov and Cox, 1995; Sakamoto et al., 1996), multiplex RT-PCR (Ha et al., 2006; Lee et al., 2010) and Sanger sequencing – all are based on conventional RT-PCR method. In an attempt to provide better quality control of LAIV candidate viruses and to ensure the uniform composition of the A/Victoria/361/2011(H3N2)-CDC-LV1 (LV1) and B/Texas/06/2011-CDC-LV2B (LV2B) reassortants (designated as Master Virus Seed, MVS), real-time RT-PCR assays were developed to confirm the lack of any genetic material of undesired origin in the reassortant viruses.
2. Materials and methods
2.1. Viruses
Egg adapted wt influenza virus strains A/Victoria/361/2011 (passage history in eggs – E3/E2, where E#/means number of egg passages in submission laboratory and/E# means number of egg passages in CDC) and B/Texas/06/2011 (E4) were obtained from CDC repository. The clones of cold-adapted MDV-A A/Leningrad/134/17/57(H2N2), and MDV-B B/USSR/60/69 were obtained at the IEM, St Petersburg, Russia. MDV stocks were prepared in 10-day-old SPF eggs (Charles River Laboratories Inc., Wilmington, MA) and stored at −80 °C. Infectious titer of wt and MDVs were measured by 50% egg infectious dose per milliliter (EID50/ml) as previously described (Huprikar and Rabinowitz, 1980). Briefly, 10-fold serial dilutions of allantoic fluid were made in PBS (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH of 7.4) and 0.2 ml of each dilution was inoculated into 10-day-old SPF eggs. Five eggs were infected with each virus dilution and incubated at 32 °C for 48 h for influenza A virus and 72 h for influenza B virus. Harvested allantoic fluids were tested for HA activity using 0.5% turkey red blood cells (tRBC). The virus titer was calculated using Reed and Muench method (Reed and Muench, 1938).
2.2. Reassortment of ca donor and wt influenza viruses
Reassortant influenza viruses that possess the internal genes of MDV-A or MDV-B and the surface genes of A/Victoria/361/2011 or B/Texas/06/2011, respectively, were prepared according to a method developed by the IEM, St Petersburg, Russia (Aleksandrova, 1977; Wareing et al., 2002). Briefly, equal doses (~106 EID50) of the donor and wt strains were inoculated into 10-day-old SPF eggs and incubated at 32 °C for 2 (influenza A) or 3 (influenza B) days. HA-positive allantoic fluids were combined and diluted 1:10 with antiserum prepared against the MDV-A in ferrets and against MDV-B in rabbits (provided by IEM, St Petersburg, Russia, through WHO). The virus–serum mixtures were incubated overnight at 4 °C and then passaged once in SPF eggs at 25 °C for 6 days. If needed, a blind passage at 32 °C was performed. All HA-positive allantoic fluids were combined. A second selective passage of HA-positive allantoic fluids was then carried out in the presence of antiserum at 25 °C followed by cloning procedures (Wareing et al., 2002).
2.3. Isolation of viral RNA
RNA was isolated from influenza virus-containing allantoic fluids and purified on the MagnaPure LC (Roche, San Francisco, CA) using the MagNA Pure Total Nucleic Acid Kit (Roche, San Francisco, CA) following the manufacturer’s instructions. Clarified allantoic fluid of infected eggs (200 μL) was used for RNA isolation. RNAs were eluted in a final volume of 50 μL of RNase-free water.
2.4. Genotyping of reassortants
Genome composition of reassortant influenza viruses was assessed by standard hemagglutination inhibition assay (for HA) and pyrosequencing analysis (for NA and the other six genes) (Deng et al., 2011).
2.5. Real time RT-PCR (rRT-PCR)
Strain-specific and segment-specific primers/probe sets for rRT-PCR were designed for genes of A/Leningrad/134/17/57 (Table 1), A/Victoria/361/2011 (Table 2), B/Texas/06/2011 (Table 3) and B/USSR/60/69 using GenScript Real-time PCR TaqMan Primer Design (GenScript, Piscataway, NJ) and PrimerExpress 3.0 Software (Applied Biosystems, Foster City, CA). Primers/probe sets for B/USSR/60/69 are not provided for the reasons of intellectual property rights associated with this master donor virus. Primers/probe sets for universal detection of H3 and H2 subtypes, and specific detection of PB1 gene of LAIV-A (FluMist) were provided by Diagnostic Development Team, VSDB, Influenza Division, CDC (primer and probes available upon request). The rRT-PCR was performed using SuperScript® III Platinum®One-Step quantitative RT-PCR Kit (Invitrogen, Carlsbad, CA) on Mx3000P thermal cycler (Agilent Technologies, Santa Clara, CA). The reaction was conducted in a total volume of 25 μl containing 0.8 μM of each primer and 0.2 μM of probe and 5 μl of viral RNA. Reaction conditions were as follows: one cycle of 50 °C for 15 min, followed by 2 min at 95 °C, and 45 cycles of 15 s at 95 °C and 1 min at 60 °C. The data were analyzed using MxPro QPCR Software.
Table 1.
Primers and probes used for real-time RT-PCR amplification of A/Leningrad/134/17/57 (H2N2).
| Primers/probesa | Gene | Nucleotides | Sequence 5′→3′ |
|---|---|---|---|
| NSLenF150 | NS | 150–169 | CGGTCTGAACATCGAAACAG |
| NSLenR230 | NS | 210–230 | AGTGCCTCATCGGATTCTTCC |
| NSLenP197r | NS | 173–197 | FAM-TCCACTATCTGCTTTCCAACACGGG-BHQ1 |
| MLenF427 | M | 427–445 | GCCTTGGGCCTGGTATGTG |
| MLenR488 | M | 465–488 | CTATGAGACCTATGCTGGGAGTCA |
| MlenP447 | M | 447–463 | FAM-AACCTGTGAACAGATTG-BHQ1 |
| NALenF946 | NA | 946–965 | TATGTGTGCTCAGGGCTTGT |
| NALenR1041 | NA | 1021–1041 | TGGATTCCCTCTCTCATTGTT |
| NALenP967 | NA | 967–986 | FAM-GGCGACACACCCAGGAACGA-BHQ1 |
| NPLenF743 | NP | 743–764 | CAGGAAATGCTGAGATCGAAGA |
| NPLenR813 | NP | 789–813 | AGCAACTGACCCTCTCAATATGAGT |
| NPLenP769 | NP | 769–785 | FAM-ATCTTTCTGGCACGGTC-BHQ1 |
| PALenF1222 | PA | 1222–1242 | CAGAATGAGTTCAACAAGGCA |
| PALenR1301r | PA | 1282–1301 | GGAGCCACATCTTCTCCAAT |
| PALenP1243 | PA | 1243–1265 | FAM-TGCGAGCTGACCGATTCAATCTG-BHQ1 |
| PB2LenF976 | PB2 | 976–995 | GGCGGGTTCACATTTAAGAG |
| PB2LenR1053 | PB2 | 1034–1053 | TGTTTGAAGATTGCCCGTAA |
| PB2LenP1026r | PB2 | 1001–1026 | FAM-TTCCTCTCTCTTGACTGATGATCCGC-BHQ1 |
F and R in the primer name indicate forward and reverse direction, respectively. P indicates probe, and r indicates probe in reverse direction.
Table 2.
Primers and probes used for real-time RT-PCR amplification of A/Victoria/361/2011 (H3N2).
| Primers/probesa | Gene | Nucleotides | Sequence 5′→3′ |
|---|---|---|---|
| NSVicF149 | NS | 149–169 | TCGGTCTAGACATCAAAGCAG |
| NSVicR234 | NS | 211–234 | TTTAAGTGCCTCATCAGATTCTTC |
| NSVicP170 | NS | 170–193 | FAM-CCACCCATGTTGGAAAGCAAATTG-BHQ1 |
| MVicF809 | M | 809–833 | TGGATTCTTGATCGTCTTTTTTTCA |
| MVicR875 | M | 854–874 | GCCTCTTTTAAGGCCGTGTTT |
| MVicP834 | M | 835–851 | FAM-ATGCGTCTATCGACTCT-BHQ1 |
| NAVicF1106 | NA | 1106–1125 | CGTCACGCTTAGGGTATGAA |
| NAVicR1215 | NA | 1197–1216 | AACCGGACCTATCACCTCTG |
| NAVicP1136 | NA | 1136–1158 | FAM-TCATTGAAGGCTGGTCCAACCCT-BHQ1 |
| NPVicF838 | NP | 838–858 | GCGTATGGACCTGCAGTATCC |
| NPVicR908 | NP | 888–908 | GGGTCTATTCCCACCAAGGAA |
| NPVicP861 | NP | 861–878 | FAM-TGGGTACGACTTCGAAAA-BHQ1 |
| PAVicF1280 | PA | 1280–1299 | AAATTGGAGAGGACGTAGCC |
| PAVicR1360 | PA | 1341–1360 | TACAATGGGACACCTCTGCT |
| PAVicP1300 | PA | 1300–1323 | FAM-CCAATTGAGCACATTGCAAGCATG-BHQ1 |
| PB1VicF659 | PB1 | 659–679 | GAGCTTTGACATTGAACACGA |
| PB1VicR731 | PB1 | 712–731 | GGTGTTGCAATAGCCCTTCT |
| PB1VicP704r | PB1 | 680–704 | FAM-TTGCCTCTCTCTGCATCTTTGGTCA-BHQ1 |
| PB2VicF999 | PB2 | 999–1017 | AAGCGGGTCATCAGTCAAA |
| PB2VicR1076 | PB2 | 1052–1076 | CCCTCATGTACTCTTATTCTCAATG |
| PB2-VicP1050r | PB2 | 1024–1050 | FAM-TTGGAGATTGCCTGTAAGCACCTCTTC-BHQ1 |
F and R in the primer name indicate forward and reverse direction, respectively. P indicates probe, and r indicates probe in reverse direction.
Table 3.
Primers and probes used for real-time RT-PCR amplification of B/Texas/06/2011.
| Primers/probesa | Gene | Nucleotides | Sequence 5′ →3′ |
|---|---|---|---|
| HATex_F226 | HA | 226–245 | ACAGATCTGGATGTGGCCTT |
| HATex_R287 | HA | 267–287 | GCTTTAGCAGAAGGTGTGGTC |
| HATex_P266r | HA | 247–266 | FAM-CCCACACACATTGGCCTGCC-BHQ1 |
| NATex_F718 | NA | 718–737 | ATAACTGATGGCCCAGCTTC |
| NATex_R837 | NA | 817–837 | GCATGTGCATTCCTCAGTATG |
| NATex_P779r | NA | 753–779 | FAM-CGGCCTTCTCGAATCTTAAGGAATCTG-BHQ1 |
| MTex_F256 | M | 256–275 | ATGGGAACAACAGCAACAAA |
| MTex_R358 | M | 339–358 | CATGGCCTTCTGCTATTTCA |
| MTex_P304r | M | 282–304 | FAM-TTCTCTCAGCCAGAATCAGGCCC-BHQ1 |
| NSTex_F377 | NS | 377–396 | GGAGGTGCCTTGATGACATA |
| NSTex_R461 | NS | 438–461 | TCTTTGTTGTTCATGTCCCTTAAT |
| NSTex_P426r | NS | 403–426 | FAM-TGGGCCATCAACATCATCTGGTTC-BHQ1 |
| NPTex_F159 | NP | 159–179 | AGCAACCACAAGCAGTGAAGA |
| NPTex_R298 | NP | 279–298 | TGAGTCCAGCTTTGACCATC |
| NPTex_P263r | NP | 243–263 | FAM-TCGCCCAGTTTCACCACCATG-BHQ1 |
| PATex_F260 | PA | 260–279 | TAGCATGGATGGTCCAAAGA |
| PATex_R338 | PA | 316–338 | TAATCAAACAAATCAGCCAGATA |
| PATex_P314r | PA | 288–314 | FAM-TTGGGAGTCTCTATCCCATGCTCTTGA-BHQ1 |
| PB1Tex_F141 | PB1 | 141–166 | TGAGTACTCGAACAAAGGAAAACAGT |
| PB1Tex_R215 | PB1 | 194–215 | GGCCCATTTGTTGGATCTATCA |
| PB1TexP168 | PB1 | 168–187 | FAM-TGTTTCTGACATCACAGGAT-BHQ1 |
| PB2Tex_F587 | PB2 | 587–607 | AAGGAACGATGATAACTCCCA |
| PB2Tex_R657 | PB2 | 637–657 | GAACCTTCTCCTGGCAACTAA |
| PB2Tex_P636r | PB2 | 614–636 | FAM-TTCCCTCTCGAGCATGTATGCCA-BHQ1 |
F and R in the primer name indicate forward and reverse direction, respectively. P indicates probe, and r indicates probe in reverse direction.
RNAs were extracted from viruses in allantoic fluid with known infectious titer. Serial 10-fold dilutions were prepared in RNase-free water. Standard curves were generated with duplicate samples over a template dilution range of 6 logs by using obtained cycle threshold (Ct) with a difference between replicates of < 0.5 Ct values. The resulting Ct values for each dilution were plotted versus virus EID50. The linear regression analysis was applied to determine the slopes. By using obtained Ct values the amplification efficiencies were calculated as E = (10(−1/slope) − 1) × 100.
2.6. Reassortant virus passaged in eggs for stability analysis
Reassortant MVS (Master Virus Seed) stocks were tested for genome stability and the absence of contamination by wt viruses after five consecutive passages in SPF eggs (An et al., 2009). Briefly, vaccine candidate viruses were diluted with PBS to the final virus concentration of 5 × 105 EID50/ml and three 10-day-old SPF eggs were inoculated with 0.2 ml of the diluted virus. The eggs were incubated for 48 h (influenza A) or 72 h (influenza B) at 32 °C, then were chilled (4 °C 3 h to overnight) and allantoic fluids from each egg were harvested and pooled together. The virus titer was estimated in HA assay using 0.5% tRBC. The average ratio between number of influenza virus particles and HA units has been reported to be 107 particles/HA per mL with the deviation about 8%, depending of virus strains and type of RBC (Desselberger, 1975; Donald and Isaacs, 1954; Isaacs, 1957; Tyrrell and Valentine, 1957). Thus, allantoic fluid with 500 HA titer is estimated to contain about 5 × 109 virus particles per mL. Approximately ten virus particles correspond to one EID50/mL (Donald and Isaacs, 1954). The infectivity of allantoic fluid with 500 HA unit would correspond to about 5 × 108 EID50/mL. To keep the same level of multiplicity of infection in the following passages, the allantoic fluid with HA titer of 500 were diluted 1000 times with PBS to final 5 × 105 EID50/mL and SPF eggs were inoculated with 0.2 mL/egg. After the 5th passage in eggs, allantoic fluids were collected and clarified by low speed centrifugation at 300 × g for 10 min and used for further analysis.
2.7. Genomic sequence analysis
The complete viral cDNAs for each segment (PB2, PB1, PA, HA, NP, NA, M and NS) of the cloned viruses were synthesized from purified viral RNA using AccessQuick RT-PCR system (Promega, Madison, USA). Viral genome fragments (eight for PB2, PB1 and PA, six for HA, five for NP and NA, and three for M and NS) were amplified using overlapping M13 tagged gene segment specific primers. RT-PCR products were resolved by the 2% E-Gel 96 agarose electrophoresis system (Invitrogen, Carlsbad, CA) and were purified by ExoSAP-IT system (Affymetrix/USB, Cleveland, OH). Sanger sequencing of the cDNA was performed with the M13 primers using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA). The sequencing extension products were purified using the Agencourt CleanSEQ (Agencourt/Beckman Coulter, Brea, CA) and analyzed using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) and 3730 Data Collection v3.0 software (Applied Biosystems, Foster City, CA). Trace files were assembled in Sequencer (Gene Codes Corporation, Ann Arbor, MI). BioEdit Sequence Alignment Editor (Thomas Hall/Ibis Biosciences, Carlsbad, CA) software was used to align consensus sequences of gene segments with the corresponding sequences of MDV-A and wt A/Victoria/361/2011 or MDV-B and wt B/Texas/06/2011 viruses.
3. Results
3.1. Specificity and sensitivity of the designed rRT-PCR assays
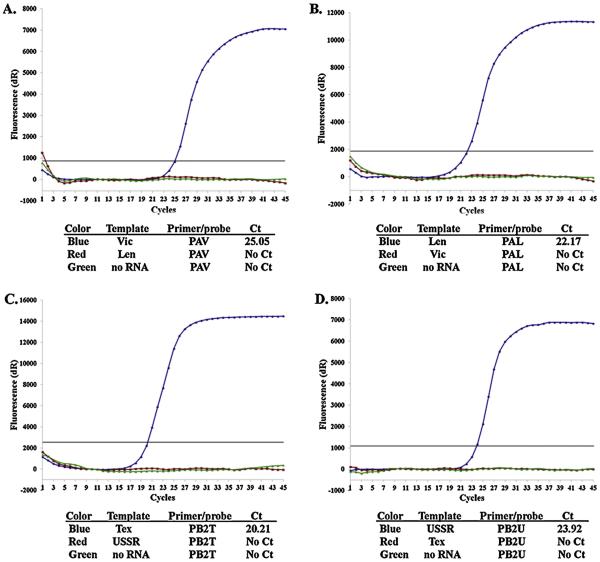
For each wt-MDV pair used to create reassortants, primers/probe sets for rRT-PCR were designed to match the genome of only one of the virus strain of the pair (Tables 1–3). To verify the specificity of the primer sets, viral RNAs were isolated and purified from 107 EID50/mL wt and MDV viruses, and used for rRT-PCR reaction. Each virus RNA (MDV and wt) was tested by rRT-PCR with all primers/probe sets designed for that pair. These primers amplified only a specific product. None of the cross-templates resulted in a positive signal, confirming that the designed primers/probe sets were strain and segment specific. The examples of test amplification curves are shown in Fig. 1.
Fig. 1.
Fluorescence amplification curves of rRT-PCR assays using RNAs purified from A/Victoria/361/11 (Vic) or MDV-A (Len) (A and B) or B/Texas/06/2011 (Tex) or MDV-B (USSR) (C and D). Primer/probe sets for Vic PA (A), Len PA (B), Tex PB2 (C) or USSR PB2 (D) were used for the reactions.
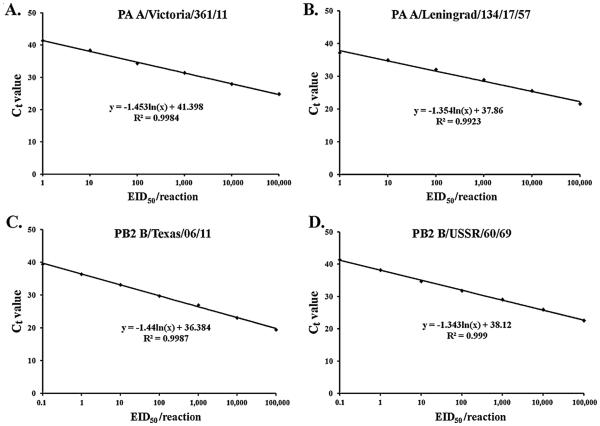
The sensitivity was determined for each rRT-PCR primers/probe set for the detection of the undesired genetic materials in the prepared clonal MVS stocks. The amplification efficiency between 90% and 110% means doubling of the amplicon at each cycle. This corresponds to a slope of standard curve −3.1 to −3.6. Standard curves obtained showed that designed assays had amplification efficiency in 90–110% range and R2 values were 0.98–0.99 (Fig. 2).
Fig. 2.
Standard curves demonstrating the limit of detection studies of rRT-PCR assays for PA gene of A/Victoria/361/11 (A), PA gene of MDV-A (B), PB2 gene of B/Texas/06/2011 (C) and PB2 gene of MDV-B (D). Ten-fold dilutions of viral RNA were plotted against the threshold cycle. The coefficient of determination (R2 ) and the equation of the regression curve (y) calculated are shown.
The limit of detection was determined as the lowest concentration at which positive signal was obtained (Table 4). The limit of detection of rRT-PCR using primer/probe sets for A/Victoria/361/2011 PA and PB2 was at 0.08 EID50 per reaction, followed by 0.8 EID50 for PB1, NP, M and NS, and 1.3 EID50 for MDV-A HA and NA. In the case of influenza B strains, B/Texas/06/2011 PB2, PB1, PA, and NS primer/probe sets were most sensitive at 0.1 EID50/reaction followed by 1 EID50 for NP and M. LoD for MDV-B HA NA genes were 1 EID50/per reaction.
Table 4.
Detection limits of each rRT-PCR assay.
| Virus strain | Primers/probe | Limit of detection (EID50/rxna) | Virus strain | Primers/probe | Limit of detection (EID50/rxn) |
|---|---|---|---|---|---|
| A/Victoria/361/2011 | PB2V | 0.08 | B/Texas/06/2011 | PB2T | 0.1 |
| PB1V | 0.8 | PB1T | 0.1 | ||
| PAV | 0.8 | PAT | 0.1 | ||
| NPV | 0.8 | NPT | 1 | ||
| MV | 0.8 | MT | 1 | ||
| NSV | 0.8 | NST | 0.1 | ||
| MDV-A | H2 | 1.3 | MDV-B | HAU | 1 |
| NAL | 1.3 | NAU | 1 |
rxn, rRT-PCR reaction containing 5 μl of RNA.
3.2. Genetic homogeneity test
Using the highly specific and sensitive rRT-PCR protocol as described above, the genetic homogeneity of reassortant viruses, LV1 and LV2B, were analyzed using RNA corresponding to 105 EID50/per reaction. MDV and wt viruses were also tested as controls. For both reassortants, the positive signals were detected only with primers/probe specific for internal genes of MDV and HA and NA genes of wt (A/Victoria/361/2011 for LV1 and B/Texas/06/2011 for LV2B). No undesired genes were detected in the reassortant viruses with a high sensitivity level of detection (Table 5). The data showed that the A/Victoria/361/2011(H3N2)-CDC-LV1 reassortant does not contain genetic material from PB2, PB1, PA, NP, M and NS genes of A/Victoria/361/2011 or HA and NA genes of originating from A/Leningrad/134/17/57. Similarly, the B/Texas/06/2011-CDC-LV2B reassortant does not contain any genetic material of B/Texas/06/2011 genes PB2, PB1, PA, NP, M, NS or HA and NA genes of B/USSR/60/69 origin, confirming of genetic homogeneity of the reassortant viruses.
Table 5.
Cycle threshold (Ct values of RNA from LV1 and LV2B corresponding to 105 EID50/per reaction.
| Gene | A/Victoria/361/2011(H3N2)-CDC-LV1 Assays specific for |
B/Texas/06/2011-CDC-LV2B Assays specific for |
||
|---|---|---|---|---|
| A/Leningrad/134/17/57 (MDV-A) | wt A/Victoria/361/2011 | B/USSR/60/69 (MDV-B) | wt B/Texas/06/2011 | |
| PB2 | 22.23 | No Cta | 22.81 | No Ct |
| PB1 | 21.45 | No Ct | 23.08 | No Ct |
| PA | 21.14 | No Ct | 22.35 | No Ct |
| NP | 22.27 | No Ct | 22.91 | No Ct |
| M | 22.39 | No Ct | 20.73 | No Ct |
| NS | 21.31 | No Ct | 23.11 | No Ct |
| HA | No Ct | 22.89 | No Ct | 22.07 |
| NA | No Ct | 21.76 | No Ct | 20.46 |
No signal was detected after 40 cycles of amplification.
The genome sequence analysis of LV1 and LV2B was completed by alignment of the six internal protein gene segments (PB2, PB1, PA, NP, M, and NS) with those of corresponding MDVs and the two surface antigen segments, HA and NA, with the corresponding RNA segments of the wt A/Victoria/361/2011 or B/Texas/06/2011. The rRT-PCR analysis was consistent with the full genome sequence analysis. The alignment of sequences for the six internal gene segments of LV1 detected no nucleotide changes. LV2B contained only one nucleotide change in the NP gene segment (T66C) that did not result in amino acid substitution. No other mutations in the remaining internal gene segments were identified. The HA and NA sequences of LV1 were identical to one of the egg adapted A/Victoria/361/2011. The HA sequence of wt B/Texas/06/2011, E4 isolate, which was used for reassortment, was shown to have a mix at position 596 (A or C) compared to E3 isolate which had 596A. The A596C change led to amino acid Q200P substitution, since this mutation appeared during passage of virus in eggs it constitutes an egg adapted change. LV2B HA gene had 596C indicating the presence of 200P. NA sequence was identical to the NA of wt virus.
3.3. Genetic stability test
To test the genetic stability, LAIV reassortant viruses, LV1 and LV2B were subjected to five passages in SPF eggs. The infectious titers of original LAIV reassortants were 109.3 and 109.2 EID50/mL for LV1 and LV2B, respectively. Passages were done with 0.2 ml of a 10−3 dilution of the virus which corresponds to approximately 105 EID50 of virus per egg. RNA purified from five time passaged viruses was subjected to rRT-PCR procedure to confirm the homogeneity of the virus. No signal was detected in the reactions with RNA from the passaged LV1 reassortant when Victoria PB2, PB1, PA, NP, M, NS, or MDV-A NA and HA primers/probe sets were used. Similarly, no signal was detected with LV2B RNA when Texas internal genes and MDV-B NA and HA primers/probe sets were used, confirming again that the obtained reassortants were genetically homogeneous and did not contained genetic material of undesired origin.
The LV1 and LV2B viruses passaged five times in eggs were also characterized by complete full genome sequencing. Sequence alignments of all gene segments of the passaged virus with corresponding sequences of the original LV1 or LV2B did not reveal any nucleotide differences between them, indicating the genetic stability of the LAIV reassortant genome.
4. Discussion
Tests to characterize the quality of live attenuated influenza vaccine viruses are being continuously developed and improved (Buonagurio et al., 2006; Yeolekar and Dhere, 2012). The WHO recently updated recommendations to provide vaccine manufacturers and national regulatory authorities with guidance in developing specific processes to assure the quality, safety, and efficacy of live attenuated influenza vaccines for intranasal administration (WHO, 2009). The testing guidelines for the infectivity (potency), identity, sterility and stability of vaccine were outlined in this document. According to the guideline, LAIV candidates should be characterized by an identity test during their preparation. The identity test should include appropriate methods to identify the HA and NA antigens and to obtain phenotypic and genetic information for the vaccine viruses (WHO, 2009). Genotypic characterization of a vaccine virus should include its complete sequence, and may include analysis of viral subpopulations and its genetic stability. The stability of the genotype and phenotype should also be demonstrated following viral passages beyond the level used in vaccine production (WHO, 2009). In this study, the real-time RT-PCR assays were developed for both type A and type B LAIV reassortants, and its sensitivity was tested in an attempt to provide better identity and stability testing for quality control of LAIV candidate viruses.
LAIV candidates for the 2012–2013 seasons, A/Victoria/361/2011-CDC-LV1 (LV1) and B/Texas/06/2011-CDC-LV2B (LV2B) were created by the classical viral reassortment method. The genetic identity of vaccine candidate viruses was demonstrated by pyrosequencing assay at the stage of vaccine candidate selection. The pyrosequencing approach as well as most of the other current techniques used for genotyping and screening of reassortant influenza viruses are based on conventional RT-PCR technique (Fulvini et al., 2011; Ha et al., 2006; Kiseleva et al., 2011; Klimov and Cox, 1995; Lee et al., 2010; Sakamoto et al., 1996). The sensitivity of the RT-PCR was shown to be at least log10 lower compared to rRT-PCR technique in a number of reports (Chen et al., 2007; Emery et al., 2004; Kaida et al., 2008; Rodrigues et al., 2011; Zhao et al., 2007). For example, the rRT-PCR H5 gene assay designed for avian influenza reproducibly determined lowest amount of viral RNA corresponding 0.05 EID50 per reaction in contrast to detection limit of 3 EID50 in cnRT-PCR (Chen et al., 2007). Real time RT-PCR is currently recognized as the most sensitive and reliable technique for detection and identification of viral subpopulations in diagnostics. The highest sensitivity of rRT-PCR assays was demonstrated for the detection of influenza viruses of different origin and composition (Chen et al., 2007; Monne et al., 2008; Nakauchi et al., 2011; Shu et al., 2011). The rRT-PCR assays reported for detection of subtype H5, H7, and H9 avian influenza viruses had sensitivity from 0.5 to 2.74 EID50/per reaction (Monne et al., 2008). The sensitivity of rRT-PCR assays developed in the present study was shown to range from 0.08 to 0.8 EID50 per reaction for the detection of undesired genes in LV1 and from 0.1 to 1 EID50/reaction for the detection of undesired genes in LV2B. Therefore, primer/probe sets used in the current study allowed genome detection as sensitive as those reported in other studies using rRT-PCR assays (Chen et al., 2007; Monne et al., 2008; Spackman et al., 2002; Wise et al., 2004). However, it should be noted that although the designed primers and probes for MDVs are expected to work for the next MDV–wt reassortment pairs, the primer and probes sequences for next selected for vaccine wt virus must be checked and updated regularly and the limit of detection should be identified using that specific wt RNA.
During the vaccine manufacturing process, MVS undergoes three to four additional passages before blending the monovalent pool to formulate LAIV (Buonagurio et al., 2006; WHO, 2009). It is important to determine that no other genetic material is present in the MVS which could be assured by rRT-PCR homogeneity test. In the present study, the homogeneity of the original and passaged LV1 and LV2B viruses were analyzed by rRT-PCR and genetic stability was confirmed by complete sequencing of all gene segments. The rRT-PCR data demonstrate that neither of the reassortant viruses contained genetic material from internal genes of wt parental viruses or HA and NA genes of MDV origin with a high sensitivity levels of assays (Table 5). The rRT-PCR results were confirmed by sequencing analysis. No nucleotide change was found in sequences of passaged five times LV1 or LV2B viruses indicating the high level of genetic stability. The data obtained in the present study are in good agreement with the previous study on genetic stability of FluMist/CAIV-T vaccine, which demonstrated a remarkable sequence identity for all vaccine intermediates throughout the manufacturing process (Buonagurio et al., 2006)
The fact that no undesired genes were detected after five passages of MVS by rRT-PCR proves that the developed rRT-PCR assays were adequate and sufficient to provide the evidence for the homogeneity of the prepared vaccine candidate. In conclusion, both rRT-PCR and sequencing results provide conclusive safety controls for further manufacturing processing of MVS. The real-time RT-PCR assays developed in the present study are in compliance with WHO recommendations to provide further quality control steps to ensure safe use of LAIV.
Acknowledgments
We thank Larisa Rudenko (IEM, St Petersburg, Russia) for providing MDVs, serum to MDVs and protocols for generation and characterization of reassortants. We thank Xiyan Xu for providing wt viruses, Amanda Balish for technical assistance on the project, Jan Mabry for help in preparation of serum to reassortant virus, Angie Foust for help in lyophilization of reassortant, Stephen Lindstrom for providing primers/probe sets for universal detection of H3 and H2 subtypes and detection of PB1 gene of LAIV-A virus (FluMist). We also thank Nancy Cox and Julie Villanueva for the support of project development at CDC. The work described in this report was supported by the World Health Organization (WHO) and the U.S. Department of Health and Human Services Biomedical Advanced Research and Development Authority (HHS BARDA).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
References
- Aleksandrova GI. Use of the genetic recombination method for obtaining vaccinal strains of the influenza virus. Voprosy Virusologii. 1977;4:387–395. [PubMed] [Google Scholar]
- Ambrose CS, Levin MJ, Belshe RB. The relative efficacy of trivalent live attenuated and inactivated influenza vaccines in children and adults. influenza and Other Respiratory Viruses. 2011;5:67–75. doi: 10.1111/j.1750-2659.2010.00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An WQ, Yang PH, Duan YQ, Luo DY, Tang C, Jia WH, Xing L, Shi XF, Zhang YJ, Liu XF, Wang XL. Generation and characterization of a cold-adapted attenuated live H3N2 subtype influenza virus vaccine candidate. Chinese Medical Journal. 2009;122:2880–2885. [PubMed] [Google Scholar]
- Buonagurio DA, Bechert TM, Yang CF, Shutyak L, D’Arco GA, Kazachkov Y, Wang HP, Rojas EA, O’Neill RE, Spaete RR, Coelingh KL, Zamb TJ, Sidhu MS, Udem SA. Genetic stability of live, cold-adapted influenza virus components of the FluMist/CAIV-T vaccine throughout the manufacturing process. Vaccine. 2006;24:2151–2160. doi: 10.1016/j.vaccine.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Chen W, He B, Li C, Zhang X, Wu W, Yin X, Fan B, Fan X, Wang J. Real-time RT-PCR for H5N1 avian influenza A virus detection. Journal of Medical Microbiology. 2007;56:603–607. doi: 10.1099/jmm.0.47014-0. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Brokstad KA, Ogra P. influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scandinavian Journal of Immunology. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- Deng YM, Caldwell N, Barr IG. Rapid detection and subtyping of human influenza A viruses and reassortants by pyrosequencing. PLoS ONE. 2011;6:e23400. doi: 10.1371/journal.pone.0023400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U. Relation of virus particle counts to the hemagglutinating activity of influenza virus suspensions measured by the HA pattern test and by use of the photometric HCU method. Archives of Virology. 1975;49:365–372. doi: 10.1007/BF01318246. [DOI] [PubMed] [Google Scholar]
- Donald HB, Isaacs A. Some properties of influenza virus filaments shown by electron microscopic particle counts. Journal of General Microbiology. 1954;11:325–331. doi: 10.1099/00221287-11-2-325. [DOI] [PubMed] [Google Scholar]
- Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, Meyer RF, Tong S, Cook BT, Holloway BP, McCaustland KA, Rota PA, Bankamp B, Lowe LE, Ksiazek TG, Bellini WJ, Anderson LJ. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerging Infectious Diseases. 2004;10:311–316. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore AE, Bridges CB, Cox NJ. Seasonal influenza vaccines. Current Topics in Microbiology and Immunology. 2009;333:43–82. doi: 10.1007/978-3-540-92165-3_3. [DOI] [PubMed] [Google Scholar]
- Fulvini AA, Ramanunninair M, Le J, Pokorny BA, Arroyo JM, Silverman J, Devis R, Bucher D. Gene constellation of influenza A virus reassortants with high growth phenotype prepared as seed candidates for vaccine production. PLoS ONE. 2011;6:e20823. doi: 10.1371/journal.pone.0020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SH, Kim HA, Kim YH, Kim JS, Lee KH, Park SY, Park WJ, Seong BL. A multiplex RT-PCR method for screening of reassortant live influenza vaccine virus strains. Journal of Virological Methods. 2006;134:154–163. doi: 10.1016/j.jviromet.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Huprikar J, Rabinowitz S. A simplified plaque assay for influenza viruses in Madin–Darby kidney (MDCK) cells. Journal of Virological Methods. 1980;1:117–120. doi: 10.1016/0166-0934(80)90020-8. [DOI] [PubMed] [Google Scholar]
- Isaacs A. Particle counts and infectivity titrations for animal viruses. Advances in Virus Research. 1957;4:111–158. doi: 10.1016/s0065-3527(08)60597-7. [DOI] [PubMed] [Google Scholar]
- Kaida A, Kubo H, Shiomi M, Kohdera U, Iritani N. Evaluation of real-time RT-PCR compared with conventional RT-PCR for detecting human metapneumovirus RNA from clinical specimens. Japanese Journal of Infectious Diseases. 2008;61:461–464. [PubMed] [Google Scholar]
- Kiseleva IV, Larionova NV, Teley LC, Rudenko LG. [Restriction analysis of genome composition of live influenza vaccine] Voprosy Virusologii. 2011;56:28–32. [PubMed] [Google Scholar]
- Klimov AI, Cox NJ. PCR restriction analysis of genome composition and stability of cold-adapted reassortant live influenza vaccines. Journal of Virological Methods. 1995;52:41–49. doi: 10.1016/0166-0934(94)00133-2. [DOI] [PubMed] [Google Scholar]
- Lee EY, Lee KH, Jung EJ, Jang YH, Seo SU, Kim HA, Seong BL. Genotyping and screening of reassortant live-attenuated influenza B vaccine strain. Journal of Virological Methods. 2010;165:133–138. doi: 10.1016/j.jviromet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Maassab HF, Bryant ML. The development of live attenuated cold-adapted influenza virus vaccine for humans. Reviews in Medical Virology. 1999;9:237–244. doi: 10.1002/(sici)1099-1654(199910/12)9:4<237::aid-rmv252>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Maassab HF, DeBorde DC. Development and characterization of cold-adapted viruses for use as live virus vaccines. Vaccine. 1985;3:355–369. doi: 10.1016/0264-410x(85)90124-0. [DOI] [PubMed] [Google Scholar]
- Monne I, Ormelli S, Salviato A, De Battisti C, Bettini F, Salomoni A, Drago A, Zecchin B, Capua I, Cattoli G. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. Journal of clinical microbiology. 2008;46:1769–1773. doi: 10.1128/JCM.02204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Coelingh K. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunology. 2002;15:295–323. doi: 10.1089/08828240260066242. [DOI] [PubMed] [Google Scholar]
- Nakauchi M, Yasui Y, Miyoshi T, Minagawa H, Tanaka T, Tashiro M, Kageyama T. One-step real-time reverse transcription-PCR assays for detecting and subtyping pandemic influenza A/H1N1 2009, seasonal influenza A/H1N1, and seasonal influenza A/H3N2 viruses. Journal of Virological Methods. 2011;171:156–162. doi: 10.1016/j.jviromet.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infectious Diseases. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method for estimating fifty percent endpoints. American Journal of Hygiene. 1938;27:5. [Google Scholar]
- Rodrigues R, Telles JN, Essere K, Ducournau C, Roqueplo C, Levieuge A, Davoust B, Parola P, Paranhos-Baccala G, Peyrefitte CN. Development of a one step real time RT-PCR assay to detect and quantify Dugbe virus. Journal of Virological Methods. 2011;176:74–77. doi: 10.1016/j.jviromet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko L, van den Bosch H, Kiseleva I, Mironov A, Naikhin A, Larionova N, Bushmenkov D. Live attenuated pandemic influenza vaccine: clinical studies on A/17/California/2009/38 (H1N1) and licensing of the Russian-developed technology to WHO for pandemic influenza preparedness in developing countries. Vaccine. 2011;29(Suppl. 1):A40–A44. doi: 10.1016/j.vaccine.2011.04.122. [DOI] [PubMed] [Google Scholar]
- Rudenko LG, Lonskaya NI, Klimov AI, Vasilieva RI, Ramirez A. Clinical and epidemiological evaluation of a live, cold-adapted influenza vaccine for 3-14-year-olds. Bulletin of the World Health Organization. 1996;74:77–84. [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Kino Y, Oka T, Herlocher ML, Maassab F. Gene analysis of reassortant influenza virus by RT-PCR followed by restriction enzyme digestion. Journal of Virological Methods. 1996;56:161–171. doi: 10.1016/0166-0934(95)01909-x. [DOI] [PubMed] [Google Scholar]
- Shu B, Wu KH, Emery S, Villanueva J, Johnson R, Guthrie E, Berman L, Warnes C, Barnes N, Klimov A, Lindstrom S. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. Journal of Clinical Microbiology. 2011;49:2614–2619. doi: 10.1128/JCM.02636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of Clinical Microbiology. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA: Journal of the American Medical Association. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Tyrrell DA, Valentine RC. The assay of influenza virus particles by haemagglutination and electron microscopy. Journal of General Microbiology. 1957;16:668–675. doi: 10.1099/00221287-16-3-668. [DOI] [PubMed] [Google Scholar]
- Wareing MD, Marsh GA, Tannock GA. Preparation and characterisation of attenuated cold-adapted influenza A reassortants derived from the A/Leningrad/134/17/57 donor strain. Vaccine. 2002;20:2082–2090. doi: 10.1016/s0264-410x(02)00056-7. [DOI] [PubMed] [Google Scholar]
- WHO . WHO Recommendations to Assure the Quality, Safety, and Efficacy of influenza Vaccines (Human, Live Attenuated) for Intranasal Administration. WHO; Geneva: 2009. [Google Scholar]
- Wise MG, Suarez DL, Seal BS, Pedersen JC, Senne DA, King DJ, Kapczynski DR, Spackman E. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. Journal of Clinical Microbiology. 2004;42:329–338. doi: 10.1128/JCM.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeolekar LR, Dhere RM. Development and validation of an egg-based potency assay for a trivalent live attenuated influenza vaccine. Biologicals: Journal of the International Association of Biological Standardization. 2012;40:146–150. doi: 10.1016/j.biologicals.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Zhao C, Li Z, Yan B, Harrison TJ, Guo X, Zhang F, Yin J, Yan Y, Wang Y. Comparison of real-time fluorescent RT-PCR and conventional RT-PCR for the detection of hepatitis E virus genotypes prevalent in China. Journal of Medical Virology. 2007;79:1966–1973. doi: 10.1002/jmv.21040. [DOI] [PubMed] [Google Scholar]