Abstract
Ribosomes are large complexes of RNA and protein that perform the essential task of protein synthesis in the cell. Ribosomes also serve as the initiation point for several translation-associated functions. To perform these tasks efficiently, ribosomes interact with a myriad of non-ribosomal proteins and RNAs. Given that most of these interactions are transient, purification of the interacting factors in complex with the ribosome can be a challenging undertaking. Here, we review methods commonly used to isolate ribosomes and study ribosome associated factors. We also discuss crucial parameters for designing and executing ribosome association studies. Finally, we present a detailed protocol for reporter based enrichment assays that are employed to selectively isolate ribosomes translating a particular message of interest. These protocols can be used to study a wide range of ribosome-associated functions.
Keywords: tmRNA, SmpB, trans-translation, ribosome, translation, sucrose gradient
1. INTRODUCTION
The bacterial 70S ribosome is a 2.7 MDa particle composed of two subunits, the large or 50S subunit and the small or 30S subunit. Basic techniques to isolate bacterial ribosomes were developed in the 1960s and 1970s (1–4), and are still widely used with relatively minor modifications. Most of these techniques require differential or density gradient ultra-centrifugation of cell lysates to yield ribosomes or ribosomal subunits of varying purity (Fig. 1). Crude ribosomes can be obtained by ultracentrifugation of clarified cell lysate at 100,000 × g. Tight-coupled ribosomes are compact and highly active ribosomal particles that are prepared by sucrose cushion centrifugation (5, 6). Linear sucrose gradients are the predominant option for obtaining ribosome profiles, separating polyribosomes and intact 70S ribosomes from the 30S and 50S subunits. Selection of the optimal method depends on the application, the protein or RNA of interest, and the specific requirements of the experiment.
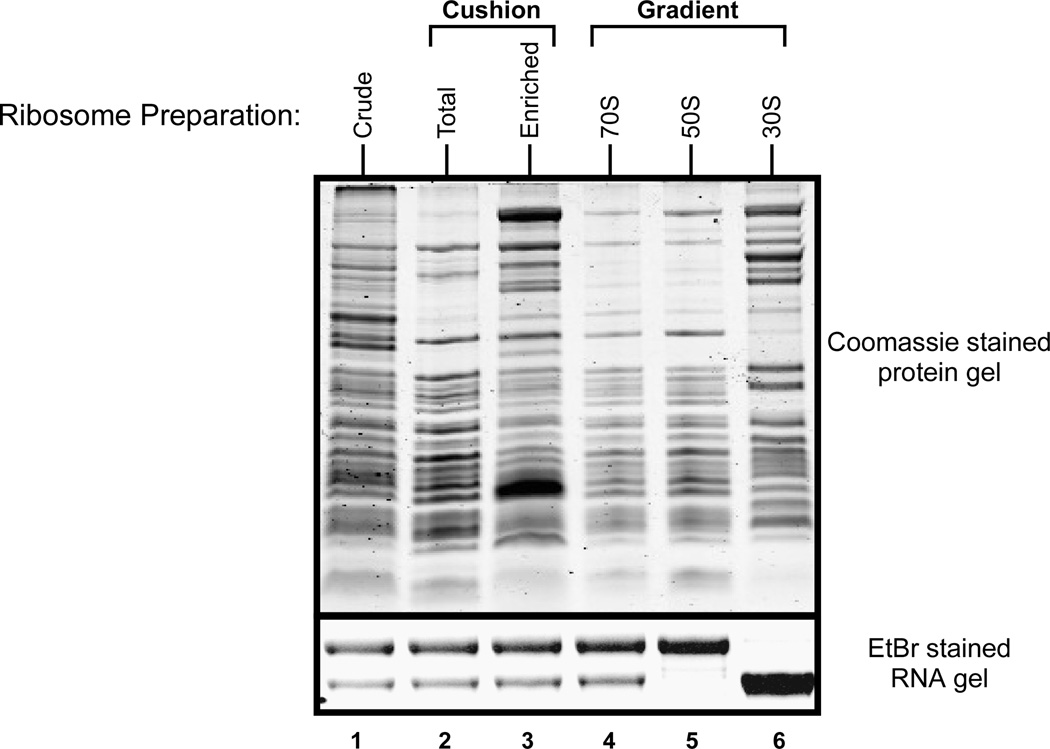
Figure 1. Protein and RNA profile of ribosomes purified by various isolation techniques.
From left to right, Lane 1: Crude Ribosomes, Lane 2: Tight-coupled ribosomes from sucrose cushion, Lane 3: Ribosomes enriched post sucrose cushion, Lanes 4, 5, and 6 are 70S, 50S, and 30S ribosomal profiles, respectively, from an analytical sucrose gradient. Top: Coomassie stained protein gel. Bottom: Ethidium Bromide (EtBr) stained RNA gel showing the 23S and 16S rRNA bands.
In addition to decoding genetic information for protein synthesis, ribosomes also serve as a platform for a number of co-translational processing events such as protein folding (7, 8), enzymatic processing of the nascent polypeptide chain (9), and degradation of defective or nonstop mRNAs (10, 11). These processes require the ribosome to associate with a number of non-ribosomal proteins and RNAs during the course of its function. Furthermore, ribosomes are known to associate with several regulatory and translation quality control factors. These include the SmpB-tmRNA ribosome rescue complex (12) and stress/starvation sensors such as RelA (13) and RelE (14). Non-ribosomal factors can interact with actively translating ribosomes or with one of the two ribosomal subunits. Knowing where these ribosome-binding factors lie on the functional ribosome landscape can provide significant information about their cellular roles. For instance, ribosome maturation factors usually associate with individual subunits and may not be present on mature 70S particles (15). Enzymes involved in post-translational modifications of the nascent polypeptide generally associate with the large subunit of the intact ribosome. Initial information on the binding site and subunit preference of a protein or RNA of interest can be obtained by western or northern blot analysis of a linear sucrose gradient profile of ribosomes. More directed experiments should then be designed to further characterize these interactions.
Factors that interact with functional ribosomes, particularly those that interact transiently or in a stage-specific manner with translating ribosomes, are best studied by isolating translationally active ribosomes. Such active ribosomes can be obtained either by polyribosome preparations, which can be arduous, or by specially designed affinity purification protocols that enable the enrichment of actively translating ribosomes using marked nascent polypeptide or RNA tags (16–18). Reporter based ribosome enrichment provides a powerful method for isolating ribosomes actively translating a particular mRNA. A protocol for enriching ribosomes translating a nonstop mRNA that lacks in-frame stop codons was developed in our laboratory to study trans-translation factors. This protocol can be easily tailored to study other translation-associated functions. In this approach, a reporter mRNA encoding an N-terminally His6-tagged protein is expressed in the desired strain. Total ribosomes are isolated and passed over a Ni2+-NTA column to specifically capture/enrich ribosomes translating the reporter transcript. Eluted ribosomes can then be used for western or northern blot analysis to determine if the factor of interest associates with the captured ribosomes (Fig. 2A). We typically use an N-terminally His6-tagged λ-cI-N nonstop reporter mRNA to study trans-translation factors ((16, 19, 20) and Fig. 2B). Quantitative estimation of the differential enrichment of trans-translation factors (SmpB, tmRNA, or RNase R) on ribosomes can be obtained by using a nonstop reporter and a control “normal” reporter that contains an in-frame stop codon (Fig. 3, and (19)). Modifications of this method can afford a simple and powerful means of isolating active ribosomes, which can then be analyzed for the relevant interactions. Any affinity tag or epitope can be used to capture the protein, RNA, ribosome, and/or a combination of these factors. However, care must be exercised to ensure that the placement of the epitope does not interfere with the biological function of these factors. For instance, we have demonstrated that the SmpB protein is essential for recognition of stalled ribosomes by tmRNA (12), and that the C-terminal domain of SmpB plays a critical role in accommodation of tmRNA into the ribosomal A-site (21). Therefore, appending a His6 epitope, or any other epitope, to the C-terminus of SmpB would be a poor choice, as it severely affects the biological function of the protein. In contrast, appending an N-terminal His6 tag to SmpB does not interfere with its biological activity and has been successfully used to identify SmpB interacting partners (22).
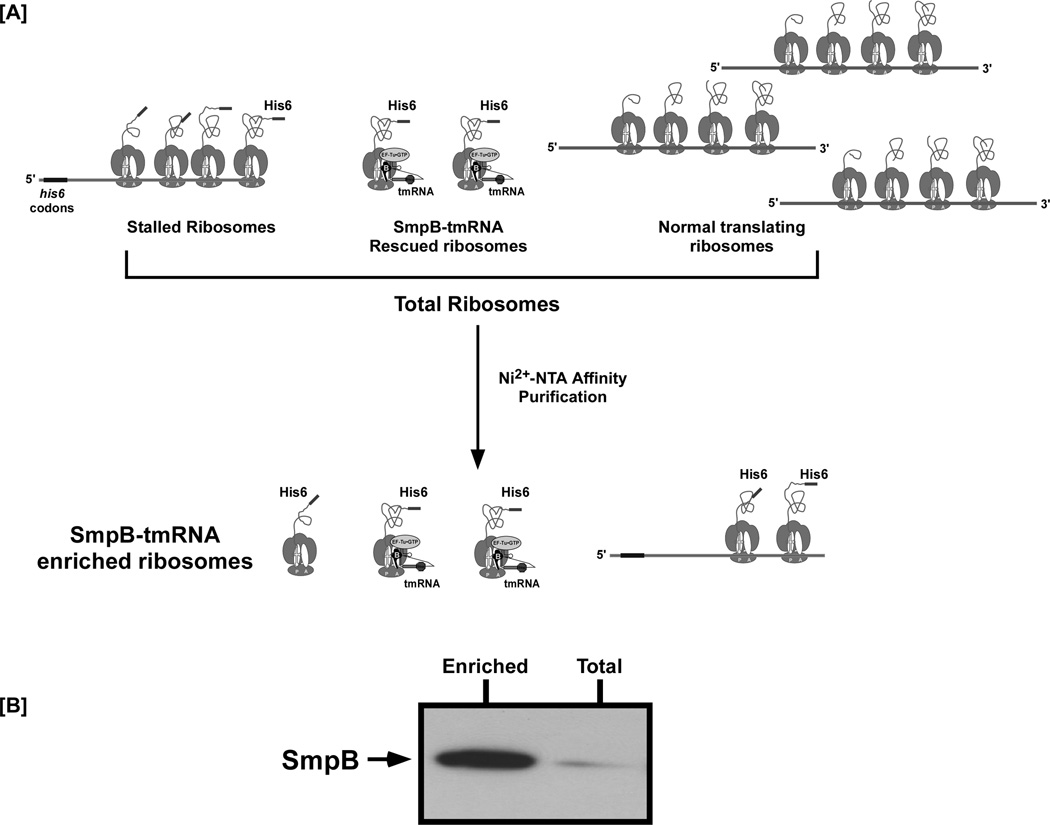
Figure 2. Reporter based enrichment assay for ribosome associated trans-translation factors.
[A] Schematic representation of a ribosome enrichment experiment. Total ribosomes can be obtained via any of the methods described in the main text. Isolated total ribosomes comprise a mixture of those translating normal cellular mRNA, or the reporter nonstop mRNAs encoding a His6 epitope tag. Stalled and rescued trans-translating ribosomes can be separated from the normal ribosome pool by using a suitable affinity column. The figure depicts enrichment of ribosomes translating a nonstop mRNA encoding an N-terminal His6-tagged reporter protein using a Ni2+-NTA affinity column. Enriched ribosomes are subjected to western blot analysis, using antibodies specific to the protein of interest. [B] Representative western blot showing enrichment of SmpB on ribosomes translating λ-cI-nonstop mRNA. Cushion purified total ribosomes were segregated into normal and stalled or trans-translating ribosomes using Ni2+-NTA column chromatography. Total and eluted enriched ribosomes were normalized by A260 and resolved by electrophoresis on a 10% SDS-acrylamide gel. The gel was used for electrophoretic transfer and western blot analysis using anti-SmpB antibodies. SmpB was enriched on captured ribosomes translating the reporter nonstop mRNA.
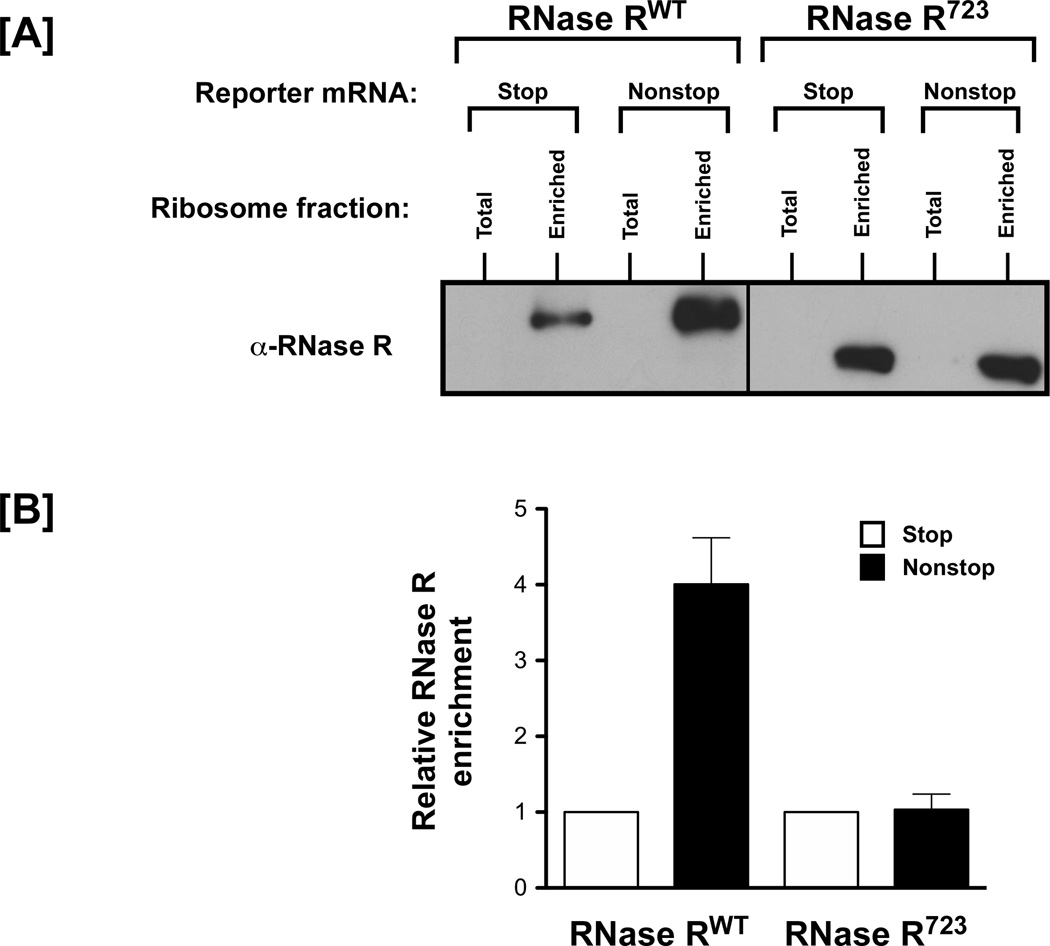
Figure 3. Selective enrichment of RNase R on ribosomes, translating a nonstop mRNA, is dependent on its C-terminal tail.
[A] Ribosomes were enriched for fractions translating the λ-cI nonstop and stop reporter mRNAs and tested for presence of RNase R using western blot analysis. A C-terminal truncation variant of RNase R (RNase R723) was also tested in this assay. [B] Quantitation of the fold-increase in levels of RNase R on ribosomes translating nonstop message compared to the control stop message. The level of RNase R is higher on enriched ribosomes that are stalled on the defective nonstop reporter mRNA. RNase RWT enriches more on stalled ribosomes than the C-terminally truncated RNase R723. This result illustrates that the ribosome enrichment assay is specific and quantitative.
While designing co-purification experiments, careful consideration should be given to buffer composition. Protein-ribosome interactions exhibit a range of salt sensitivities. For example, ribosomal protein S1 and trigger factor are at the two extremes of the salt sensitivity spectrum. Interactions of S1 with the ribosome are known to be highly salt sensitive, with S1 falling off the ribosome at as low as 100 mM NH4Cl (23). In contrast, interactions of trigger factor with the ribosome are highly resistant to salt washes, with trigger factor remaining bound to the ribosome at >500 mM NH4Cl (24). In general, proteins with binding sites within the interior of intact 70S ribosome are more resistant to dissociation as compared to surface bound proteins.
Most ribosome binding proteins will pellet along with crude ribosomes under low stringency salt conditions (<100 mM NH4Cl). A caveat to be mindful of is that use of low stringency condition could result in higher background signal, due largely to association of proteins that do not normally bind ribosomes or have any translation related function. Therefore, it is preferable to isolate ribosomes using either higher stringency conditions or the sucrose cushion approach (Fig. 1). However, it should be kept in mind that the higher stringency approaches might also cause the dissociation of some ribosome-surface associated factors. Consequently, the optimal method for one’s favorite ribosome associated factor often needs to be empirically determined. Use of non-physiological salt concentration can result in artifactual interactions and erroneous conclusions. For instance, SmpB and tmRNA are essential components of the ribosome rescue system that function as a complex during all stages of the trans-translation process (20, 25, 26). SmpB had been suggested to interact with the ribosome in the absence of tmRNA. However, some of these interactions were most likely due to nonspecific and off-pathway binding of SmpB to stalled ribosomes. Indeed, work done by Sundermeier et al. (20) convincingly demonstrated that the reported SmpB-ribosome interactions in the absence of tmRNA were non-physiological and attributable to the use of low stringency salt conditions during ribosome purification and binding studies.
Another important aspect while designing ribosome association studies is the scale of the experiment. At any given time, only a fraction of ribosomes in the cell may be associated with the protein or RNA of interest. These interactions can also be very transient and labile. This makes detection of these factors in small-scale cultures very difficult. The amount of starting material should depend on the sensitivity of the detection method and the nature of the interaction. Plasmid-borne expression of the desired protein or RNA is sometimes used to improve the signal, as it shifts the equilibrium towards greater interaction. However, over-expression might lead to increased non-specific interactions that can be misleading. It is therefore important to keep the expression of the factor of interest as close to its endogenous level as possible.
As discussed above, isolation of auxiliary factors in context of the ribosome needs robust and physiologically relevant ribosome isolation techniques. Here, we provide a compilation of methods routinely used in our laboratory to obtain ribosomes, with special emphasis on ribosome-protein interactions. Finally, we provide details of specialized protocols developed in our laboratory to study interactions of ribosomes with RNase R and other trans-translation factors. This protocol can be readily adapted to studying specialized factors that transiently associate with ribosomes in other functional contexts. Particular care has been taken to maintain, as much as possible, physiologically relevant buffer and salt conditions during the entire processing and purification process.
2. MATERIALS
All solutions should be prepared using ultrapure Milli-Q or DEPC treated water (see Note 1 and Note 2). All plasticware and glassware should be new or treated with RNase ZAP (Ambion) (see Note 3). All reagents are analytical grade or higher. All buffers should be filter sterilized and stored at 4°C (see Note 4). Change gloves frequently to minimize RNase contamination (see Note 1).
2.1. Stocks
2 M Tris pH 7.5: Weigh 242.27 g of Tris and transfer to a glass beaker. Add 750 mL of Milli-Q water. Mix using a magnetic stir-bar and adjust the pH to 7.5 using HCl (see Note 5). Adjust the volume to 1 L with Milli-Q water.
1 M MgCl2: Add 95.21 g of MgCl2 to 700 mL of water. Adjust the volume to 1 L with Milli-Q water. Use caution while mixing MgCl2 in water, as the reaction is exothermic.
500 mM EDTA: Add 186.12 g of EDTA to 800 mL of water. Adjust the pH to 8.0 with NaOH. Adjust the volume to 1 L with Milli-Q water, filter, and store at room temperature.
100 mM PMSF: Dissolve 0.87 g of PMSF (phenylmethylsulfonyl fluoride) in 50 mL of ethanol and store at −20°C (see Note 6).
2.2. Buffers
Buffer A: 20 mM Tris (pH 7.5), 300 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 6 mM β-mercaptoethanol (β-ME) (see Note 7), 10 units/mL SuperASE-In (Ambion).
Buffer B: 20 mM Tris (pH 7.5), 300 mM NH4Cl, 10 mM MgCl2, 2 mM β-ME.
Buffer C: 20 mM Tris (pH 7.5), 300 mM NH4Cl, 10 mM MgCl2, 2 mM β-ME, 250 mM Imidazole. Store away from light at 4°C.
Storage Buffer: Buffer A containing 10% glycerol.
Wash Buffer: 20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2.
2.3. Cell growth and lysis
LB broth: Dissolve 25 g of LB broth Miller mixture per 1 L of water. Sterilize by autoclaving.
Wash Buffer (see Section 2.2)
2.4. French Press
Pre-chilled 32 mL French press cell.
Buffer A (see Section 2.2).
PMSF, and DNase I (RNase free).
2.5. Chemical lysis
Lysozyme.
Bacterial protein extraction reagent: B-PER (Pierce # 78243).
DNase I (RNase free).
2.6. Crude ribosome preparation
Buffer A (see Section 2.2).
Beckman Ultra-Clear centrifuge tubes (# 344058).
Beckman SW28 swinging bucket rotor and ultracentrifuge (see Note 8).
2.7. Tight coupled ribosomes
Buffer A (see Section 2.2).
Storage Buffer (see Section 2.2).
32% sucrose solution in Buffer A.
Beckman polycarbonate bottles (# 355649).
Beckman 50.2 Ti rotor and ultracentrifuge.
2.8. Gradient purified ribosomes
Buffer A (see Section 2.2).
10% and 40% sucrose solutions in Buffer A.
Storage buffer (see Section 2.2).
20 mL syringes.
Stainless steel blunt-end large bore needle.
18 gauge or 22 gauge needles.
Beckman Ultra-Clear centrifuge tubes (# 344058).
Beckman SW28 swinging-bucket rotor and ultracentrifuge.
BioComp Gradient Master 107ip.
UV transparent 96 well plates.
Plate reader (Molecular devices SpectraMax M5E, or equivalent).
2.9. Reporter based enrichments assays
Buffer B (see Section 2.2). Depending on the Ni2+-NTA slurry used for purification, Buffer B can contain 10 mM Imidazole.
Ni2+-NTA resin (GE Healthcare # 17-5318-02 or Sigma Aldrich # P6611).
Buffer C (see Section 2.2).
Bio-Rad micro-Biospin columns.
Collection tubes.
Suitable antibodies for detection of protein of interest.
Materials for SDS-PAGE and western blot analysis.
3. METHODS
3.1. Cell culture and preparation of S30 extracts
Grow cells to mid-log phase, Optical Density at 600 nm of ≈ 0.5 (OD600 ≈ 0.5), in LB using baffled flasks at 37°C with vigorous shaking (250 RPM). Culture volumes and growth conditions can be varied depending on the nature of the experiment (see Note 9). Generally, the N-terminally His6-tagged reporter of interest is induced at OD600 ≈ 0.5 for 1 hour.
Harvest and wash cells with Wash Buffer (see Section 2.2). The cell pellet may be stored at −80°C. Perform all subsequent manipulations at 4°C.
-
Although a myriad of cell lysis methods exist, we routinely use either the French press or chemical lysis method. For isolating intact and functional ribosomes, we use the French press method for cell lysis. The French press cell should be pre-chilled at 4°C. Resuspend cell pellets in Buffer A (15 mL Buffer A containing 0.1 mM PMSF/liter of culture) and transfer to a pre-chilled French press cell (see Note 10). Lyse cells by passage through a French press at 10,000 psi. Collect the lysed cells at a rate of ~15 drops per minute. Add DNase I to a final concentration of 5 U/mL and incubate for 15 minutes at 4°C. Spin the cell lysate at 30,000 × g for 30 minutes. To avoid transferring unwanted cellular debris, recover the top 75–85% of the supernatant and spin again at 30,000 × g for 30 minutes. The supernatant obtained in this step is referred to as the S30 fraction.
For analytical gradients and culture volumes of less than 200 mL, we use the chemical lysis approach. Resuspend cell pellets in Buffer A (100 µL/100 mL of culture) containing 0.4 µg/µL of lysozyme. Incubate at RT for 1 minute. Freeze cells at −80°C. Add 500 µL of B-PER cell lysis reagent containing 10 U/mL of DNase I and 10 mM MgCl2. Incubate on ice for 5 minutes. Subject cells to two additional cycles of freezing at −80°C and thawing on ice (see Note 11). Adjust the volume up to 1 mL with Buffer A and spin at 30,000 × g for 30 minutes at 4°C. To avoid transferring unwanted cellular debris, use the top 75–85% of the supernatant for ribosome preparation.
3.2. Crude ribosome preparations
This method separates ribosomes from the majority of other lower molecular weight cellular components.
Spin the S30 fraction from a 750 mL culture at 100,000 × g for 1 hour in Ultra-Clear centrifuge tubes, using a Beckman SW28 rotor. Culture volumes can vary depending on the experiment.
Rinse the resulting crude ribosome pellet with Buffer A, with a gentle swirling motion. It is important to note that a salt wash procedure will remove some ribosomal proteins thus generating a heterogeneous ribosome population (see Note 12).
The ribosome pellet can be resuspended in any buffer of choice for further analysis. If intact ribosomes are required for downstream processing, use a buffer containing 10 mM MgCl2. For subunit preparations, resuspend the ribosome pellet in low MgCl2 Buffer A (containing 1 mM MgCl2) before layering on a sucrose gradient. The quality of the ribosome preparation should be checked at this stage (see Note 13).
3.3. Tight-coupled ribosomes
Tight-coupled ribosomes refer to compact and highly active ribosomal particles purified via a sucrose cushion.
Prepare 32% sucrose solution in Buffer A.
Transfer 12.5 mL of 32% sucrose solution to an ultracentrifuge tube and carefully layer an equal volume of the S30 supernatant on top by pipetting along the walls of the tube (see Note 14). The tube must be filled to the brim to prevent cracking during ultra-centrifugation. The exact volumes can vary depending on the tube used.
Normalize weight across the tubes using Buffer A (see Section 2.2).
Spin in a Beckman 50.2 Ti rotor at 100,000 × g for 16 hours at 4°C.
Discard the supernatant. The resulting ribosomes form a clear pellet. Occasionally, the clear ribosome pellet may be covered with a brown film. Wash the pellet in Buffer A to remove the brownish material. Resuspend the tight-coupled ribosomes in a buffer of choice for downstream processing. Ribosomes can be flash frozen in storage buffer and kept at −80°C for long-term storage.
3.4. Gradient purified ribosomes
Sucrose gradients are used to separate intact ribosomes from the individual subunits. Analytical gradients are used to determine the profile of ribosomes and subunits in cells. Gradients can be used to separate polyribosomes and 70S ribosomes from the 50S and 30S subunits based on their density. Fractions from analytical gradients can be used to determine the binding sites of non-ribosomal proteins on the ribosome. Small cultures (100–200 mL) are sufficient for this analysis. For this purpose, S30 fractions from chemically lysed cells (maximum 1 mL, 100–200 A260 units) can be directly layered on a gradient (see below). Preparative gradients from larger cultures may be needed to observe more transient interactions. For preparative gradients, crude ribosomes (see Section 3.2), or tight-coupled ribosomes (see Section 3.3) are completely resuspended in Buffer A and used. For ribosomal subunit preparations, it is important to limit the magnesium concentration in the buffer to 1.0 mM. Below, we provide a detailed protocol for preparative gradients:
Resuspend tight-coupled ribosomes (see Section 3.3), or crude ribosomes (see Section 3.2), in 1 mL Buffer A for isolating 70S ribosomes and in low MgCl2 Buffer A (containing 1 mM MgCl2) for subunit preparations. Typically, we use ribosomes isolated from 750 mL cultures.
Prepare a 10–40% linear sucrose gradient. Linear sucrose gradients can be prepared in several different ways. We generate linear sucrose gradients using the BioComp Gradient Master 107ip (see Note 15).
Load resuspended ribosomes (250–500 µL volume) onto a 40 mL 10–40% sucrose gradient. The percent range of the gradient can be varied when better resolution of the 70S ribosomes, or one of the subunits, is required. Overloading the gradient can impair the ability of the gradient to sufficiently resolve ribosomal subunits.
Spin the gradients at 82,705 × g for 16 hours in a Beckman SW28 rotor.
Fractionate the gradient either by using a fraction collector (see Note 16) or as follows: Mount the gradient tube onto a ring stand. Carefully pierce the bottom of the tube with an 18- or 22-gauge needle and remove the needle such that the flow is drop-wise (see Note 17). Fast flow rates can reduce resolution and make it more difficult to collect precise fractions. Collect the fractions in a 96 well plate, approximately 300 µL per well.
Determine the A260 values for each fraction by using a plate reader. Fractions might need to be diluted to obtain readings within the accuracy range of the spectrophotometer (see Note 18). Remember to use UV-transparent plates and mix the fractions well before measurement.
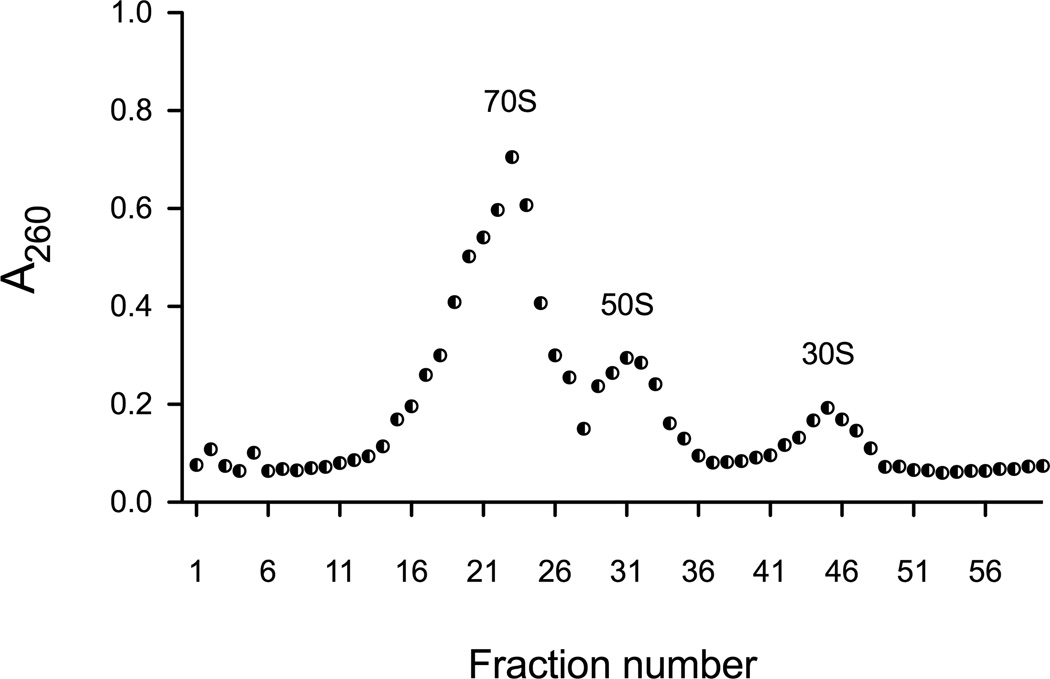
Plot the A260 values against the fraction number to determine the profile of the 70S, 50S and 30S fractions (Fig. 4).
The concentration of intact 70S ribosome and its subunits can be calculated based on the observation that 1 A260 unit of 50S is equivalent to 36 pmol/mL, 1 A260 unit of 30S is equivalent to 72 pmol/mL, and 1 A260 unit of 70S is equivalent to 24 pmol/mL (27).
Pool and flash freeze the required fractions in liquid nitrogen and store at −80°C. Glycerol can be added to a final concentration of 10% to serve as a cryo-protectant.
Figure 4. A260 profile of S30 extract separated on a sucrose gradient.
S30 extract from 100 mL of cell culture was layered onto a 10–40% sucrose gradient and spun at 82,705 × g for 16 hours in a Beckman SW28 rotor. Fractions were collected in a UV transparent 96 well plate and normalized for volume. A260 readings were obtained using a plate reader and plotted versus fraction number.
3.5. Reporter based ribosome enrichment
Crude ribosome pellets (see Section 3.2) or tight-coupled ribosomes prepared by the sucrose cushion method (see Section 3.3) can be used to perform ribosome enrichment assays. Generally, a 750 mL starting culture is required per enrichment experiment.
Resuspend ribosome pellets in 5 mL of Buffer B (enrichment buffer) by gentle rocking at 4°C. The buffer normally contains 10 mM Imidazole, unless a different source of Ni2+-NTA is used for downstream processing (see Note 19).
Centrifuge the resuspended sample at 30,000 × g for 30 minutes. Carefully transfer the supernatant to a fresh tube.
Add 40 µL of Buffer B equilibrated Ni2+-NTA resin (GE Healthcare) and incubate for 2 hours at 4°C.
Mount a 1 mL Bio-Rad spin column onto a collection tube. Load the ribosome-Ni2+-NTA slurry into the column. Allow the liquid to drain by gravity flow. The column should not be spun at this stage, and care must be exercised to prevent the resin from drying out.
Wash the column by addition of 1 mL of Buffer B. Allow the resin to settle for 1 minute before opening the stopcock to permit even flow of the buffer through the settled resin. Repeat this procedure 4-times.
To elute bound ribosomes, wipe the bottom of the column with Kimwipe paper and transfer it to a collection tube. Add 200 µL of Buffer C, allow it to stand for 1 minutes, and then spin in a microcentrifuge at 6000 × g for 1minute. Repeat the elution step by addition of another 200 µL of Buffer C.
Quantify the eluted samples by measuring A260 values. Assess enrichment of the factor of interest using western or northern blot analysis (see Note 20, and Fig. 5).
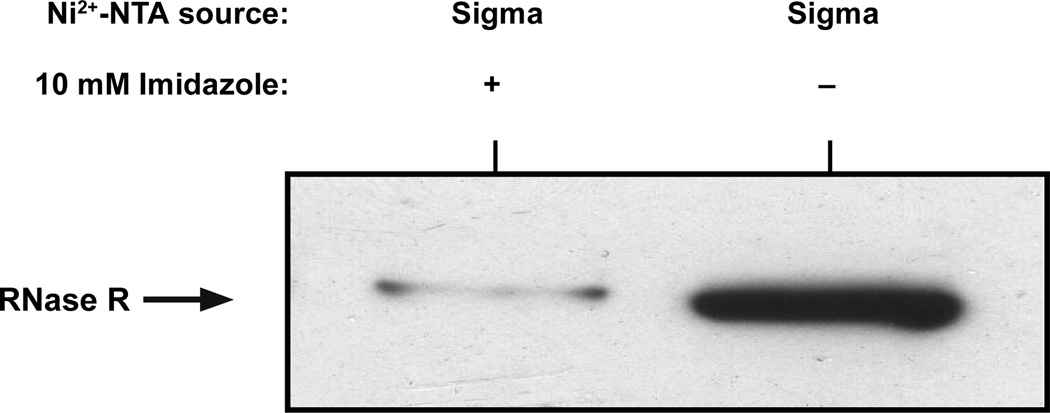
Figure 5. Ribosome enrichment and the effect of Imidazole on the Ni2+-NTA resin.
Ni2+-NTA resins from various manufacturers have different binding capacities and exhibit distinct sensitivities to the presence of Imidazole in enrichment buffer (Buffer B). We typically use Ni2+-NTA resin from GE-Healthcare with enrichment Buffer B that contains 10 mM Imidazole. We have also used Ni2+-NTA slurry from Sigma-Aldrich. However, Ni2+-NTA slurry from Sigma-Aldrich has lower binding capacity and does not work well with buffers containing Imidazole. To illustrate this point, we chose Ni2+-NTA slurry from Sigma-Aldrich and performed ribosome enrichment assays in the presence or absence of 10 mM Imidazole in the Buffer B. Shown is a western blot of enriched ribosomes probed with anti-RNase R antibodies.
Footnotes
Ribosome isolation protocols are generally robust and, with a little care, can yield reproducible and clean preparations. Certain important considerations and precautions are listed below:
- Use RNase-free water.
- Wear disposable gloves to prevent contamination by ribonucleases present on your hands.
- Use pre-wrapped disposable plasticware when possible.
- Use reagents of the highest quality available.
- Filter-sterilize all buffers for long-term storage.
- Add DEPC to a final concentration of 0.1% to water.
- Incubate at 37°C overnight.
- Autoclave to degrade DEPC.
Glassware can be made RNase free by incubating in 0.1% DEPC overnight at 37°C followed by autoclaving for 30 minutes. Alternatively, use commercially available products such as RNase ZAP (Ambion).
All manipulations must be carried out at 4°C. We recommend that all solutions be prepared 1 day in advance to permit sufficient time for cooling.
The pH adjustment of Tris-containing solutions should be done at the final working temperature.
PMSF is toxic and should be handled with care. A stock solution of PMSF can be prepared in isopropanol, methanol, or ethanol at a concentration of 100 mM. It can be stored at −20°C. However, long-term storage is not recommended. PMSF should be added to solutions and buffers immediately before use.
Reducing agents (β-ME/DTT) should always be added to buffers immediately before use.
To help minimize potential RNase activity, we recommend pre-chilling rotors and centrifuges.
Broad specificity periplasmic RNases can be avoided by using an RNase I deficient strains, such as MRE600 or the Keio RNase I knockout strain. E. coli K12 strains A19, D10, or CAN20-19E can also be used. For functional studies, it is preferable to use RNase I deficient strains. However, with a little care, intact and fully functional ribosomes can easily be isolated from wild type E. coli strains.
It is advisable to maintain low concentrations of EDTA in buffers during the initial stages of cell lysis and S30 preparation, even in the presence of MgCl2, to prevent metal-induced RNA cleavage. For isolation of intact 70S ribosomes the MgCl2 concentration should be much higher (10-fold or more) than the EDTA concentration.
Additional freeze-thaw cycles can be used if required. If the lysate is very viscous add an additional 2 U/mL of DNase I and incubate for 1–5 minutes at room temperature
It is important to keep in mind that a salt wash procedure on any ribosome pellet will partially remove some ribosomal proteins, and can thus generate a heterogeneous ribosome population. A number of ribosomal proteins have been observed to dissociate with a buffer containing 0.5 M NH4Cl (28).
The quality of a ribosome preparation should be determined by standard RNA gel analysis. The integrity of 16S, 23S and 5S rRNA can be directly visualized using 1.0–1.5% formaldehyde agarose gels (see Fig. 1).
Care must be taken while layering the cleared cellular lysate on top of 32% sucrose solution. Two distinct layers should be observed in order to achieve consistency between experiments.
- Prepare a 10% and a 40% sucrose solution in Buffer A. For subunit preparation, use Buffer A containing 1 mM MgCl2.
- To layer 10% sucrose solution on top of the 40% sucrose solution, we use a BioComp marker block to designate the half-full point in the centrifuge tube. This point can vary depending on the type of tube-cap that is used. The long cap is designed to leave a 10 mm gap above the finished gradient and a short cap is designed to leave a 4 mm gap above the finished gradient. We prefer using the long cap. Using the marker block, make a half-full mark on the tube with a fine-tip permanent marker. Gently pipette the 10% sucrose solution to the half-full mark. Place an equivalent volume of the 40% sucrose solution in a syringe equipped with a blunt-end stainless steel needle. The stainless steel needle must be sufficiently long to span the length of the centrifuge tube. Insert the tip of the needle to the bottom of the tube and slowly dispense the 40% sucrose solution to upwardly displace the 10% solution. It is important to maintain the distinct interface between the two solutions.
- Add approximately 1 mL of the 10% sucrose solution to the top, gently close the tube with the long BioComp cap, and place the tubes in the BioComp Gradient Master. When inserting the cap, it is important to ensure that there are no air bubbles trapped inside the tube. The presence of any air bubbles can interfere with gradient formation. Select the parameters of the desired gradient (10–40% w/v sucrose in this case) according to the manufacturer's instructions. Once the gradient is ready, carefully remove the cap and load the desired volume (250–500 µL) of your ribosome sample.
A density gradient fractionator available from Teledyne Isco can be used when fractioning the sucrose gradient instead of the described method. This system allows the A260 of a fractionated sample to be monitored throughout the fractionation process.
Use caution when piercing the bottom of the tube with a needle. Do not push the needle all the way into the gradient. Holding the centrifuge tube firmly in one hand and the needle in the other, carefully push the needle into the bottom of the tube with a slight twisting motion. The needle should be barely visible on the interior of the tube. Once the tube is pierced the needle should be removed before collection. Alternatively, a generic flow regulator (two-way stopcock) can be attached to the needle prior to piercing the tube to ensure an even flow through the needle and stopcock.
For preparative gradients, we usually dilute the sample 1:20 in water. It is important to mix the samples in the original plate before taking aliquots for dilution.
Not all Ni2+-NTA agarose resins work similarly in enrichment experiments. Our enrichment protocol is optimized with the GE-Healthcare Ni2+-NTA agarose resin (#17-5138-02). We have also used Ni2+-NTA resin from Sigma Aldrich (# P6611) with good success. It is essential to be cognizant of the differences in the protein binding capacity (mg protein/ml of resin) and sensitivity to Imidazole of the various Ni2+-NTA resins. Choosing the optimal buffer conditions, during the binding and washing steps, are critical for obtaining valid and reproducible ribosome enrichment results. As an example, we have examined the difference in enrichment of RNase R on ribosomes using the Sigma Aldrich slurry under two different buffer conditions. We performed enrichment assays to test the efficiency of the Sigma slurry in Buffer B with and without 10 mM Imidazole. The samples were eluted in enrichment Buffer C. Normalized samples were evaluated by western blot analysis, using anti-RNase R antibodies, to probe for the presence of RNase R (Fig. 5).
To obtain an accurate quantification of enrichment of a factor of interest on the captured ribosomes, it is important to normalize samples based on their A260 value.
REFERENCES
- 1.Britten RJ, Roberts RB. High-Resolution Density Gradient Sedimentation Analysis. Science. 1960;131:32–33. doi: 10.1126/science.131.3392.32. [DOI] [PubMed] [Google Scholar]
- 2.McQuillen K, Roberts RB, Britten RJ. Synthesis of Nascent Protein by Ribosomes in Escherichia Coli. Proc Natl Acad Sci U S A. 1959;45:1437–1447. doi: 10.1073/pnas.45.9.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nomura M, Tissières A, Lengyel P. Ribosomes. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 4.Ron EZ, Kohler RE, Davis BD. Magnesium ion dependence of free and polysomal ribosomes from Escherichia coli. J Mol Biol. 1968;36:83–89. doi: 10.1016/0022-2836(68)90221-0. [DOI] [PubMed] [Google Scholar]
- 5.Bonincontro A, Nierhaus KH, Onori G, Risuleo G. Intrinsic structural differences between "tight couples" and Kaltschmidt-Wittmann ribosomes evidenced by dielectric spectroscopy and scanning microcalorimetry. FEBS Lett. 2001;490:93–96. doi: 10.1016/s0014-5793(00)02415-7. [DOI] [PubMed] [Google Scholar]
- 6.Risuleo G, Gualerzi C, Pon C. Specificity and properties of the destabilization, induced by initiation factor IF-3, of ternary complexes of the 30-S ribosomal subunit, aminoacyl-tRNA and polynucleotides. Eur J Biochem. 1976;67:603–613. doi: 10.1111/j.1432-1033.1976.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 7.Kramer G, et al. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 8.Wegrzyn RD, Deuerling E. Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell Mol Life Sci. 2005;62:2727–2738. doi: 10.1007/s00018-005-5292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bingel-Erlenmeyer R, et al. A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature. 2008;452:108–111. doi: 10.1038/nature06683. [DOI] [PubMed] [Google Scholar]
- 10.Ge Z, Karzai AW. Co-evolution of multipartite interactions between an extended tmRNA tag and a robust Lon protease in Mycoplasma. Mol Microbiol. 2009;74:1083–1099. doi: 10.1111/j.1365-2958.2009.06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- 12.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) Embo J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Block R, Haseltine AW. Purification and properties of stringent factor. J Biol Chem. 1975;250:1212–1217. [PubMed] [Google Scholar]
- 14.Hurley JM, Cruz JW, Ouyang M, Woychik NA. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5' end of coding regions in vivo. J Biol Chem. 2011;286:14770–14778. doi: 10.1074/jbc.M110.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shajani Z, Sykes MT, Williamson JR. Assembly of Bacterial Ribosomes. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 16.Sundermeier T, et al. Studying tmRNA-mediated surveillance and nonstop mRNA decay. Methods Enzymol. 2008;447:329–358. doi: 10.1016/S0076-6879(08)02217-9. [DOI] [PubMed] [Google Scholar]
- 17.Youngman EM, Green R. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z, Reed R. Purification of functional RNA-protein complexes using MS2-MBP. Curr Protoc Mol Biol. 2003;Chapter 27(Unit 27):23. doi: 10.1002/0471142727.mb2703s63. [DOI] [PubMed] [Google Scholar]
- 19.Ge Z, Mehta P, Richards J, Karzai AW. Non-stop mRNA decay initiates at the ribosome. Mol Microbiol. 2010;78:1159–1170. doi: 10.1111/j.1365-2958.2010.07396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundermeier TR, Karzai AW. Functional SmpB-ribosome interactions require tmRNA. J Biol Chem. 2007;282:34779–34786. doi: 10.1074/jbc.M707256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci U S A. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karzai AW, Sauer RT. Protein factors associated with the SsrA.SmpB tagging and ribosome rescue complex. Proc Natl Acad Sci U S A. 2001;98:3040–3044. doi: 10.1073/pnas.051628298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szer W, Hermoso JM, Leffler S. Ribosomal protein S1 and polypeptide chain initiation in bacteria. Proc Natl Acad Sci U S A. 1975;72:2325–2329. doi: 10.1073/pnas.72.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesterkamp T, Hauser S, Lutcke H, Bukau B. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci U S A. 1996;93:4437–4441. doi: 10.1073/pnas.93.9.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dulebohn D, et al. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- 26.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 27.Christodoulou J, et al. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 2004;101:10949–10954. doi: 10.1073/pnas.0400928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnirke A, Geigenmuller U, Rheinberger HJ, Nierhaus LH. The allosteric three-site model for the ribosomal elongation cycle. Analysis with a heteropolymeric mRNA. J Biol Chem. 1989;264:7291–7301. [PubMed] [Google Scholar]
- 29.Luthe DS. A simple technique for the preparation and storage of sucrose gradients. Anal Biochem. 1983;135:230–232. doi: 10.1016/0003-2697(83)90755-8. [DOI] [PubMed] [Google Scholar]