Abstract
Mannosylphospho dolichol synthase (DPMS) is a critical enzyme in the biosynthesis of lipid-linked oligosaccharide (LLO; Glc3Man9GlcNAc2-PP-Dol), a pre-requisite for asparagine-linked (N-linked) protein glycosylation. We have shown earlier that DPMS is important for angiogenesis, i.e., endothelial cell proliferation. This is true when cAMP is used for intracellular signaling. During cAMP signaling, DPMS is activated and ER stress is reduced. To understand the activation of DPMS at the molecular level we have isolated a cDNA clone for the DPMS gene (bDPMS) from the capillary endothelial cells of bovine adrenal medulla. DNA sequencing and the deduced amino acid sequence have established that bDPMS has a motif to be phosphorylated by cAMP-dependent protein kinase (PKA). Based on the sequence information Serine 165 has been found to be the phosphorylation target in bDPMS. Hydropathy Index when plotted against amino acid number indicates the presence of a hydrophobic region around the amino acid residues 120–160, supporting that bDPMS has one membrane spanning region. The recombinant bDPMS has now been purified as His-tag protein with an apparent molecular weight of Mr 33 kDa. Additionally, we show here that overexpression of DPMS is indeed angiogenic. The capillary endothelial cells proliferate at a higher rate carrying the DPMS overexpression plasmid over the parental cells or the vector.
Keywords: Angiogenesis, Mannosylphospho dolichol synthase EC 2.41.83, cAMP, Lipid-linked oligosaccharide, N-linked glycoprotein, Breast cancer
Introduction
Mannosylphospho dolichol synthase (DPMS; EC 2.4.1.83) catalyzes the transfer reaction GDP-mannose+Dol-P<==003E;Dol-P-Man+GDP in the rough endoplasmic reticulum (ER) [1–7]. The synthesis occurs at the cytoplasmic face of the ER membrane [8–10]. Dol-P-Man serves as a ‘key’ mannosyl donor in a number of biochemical processes, such as (1) in the assembly of the precursor oligosaccharide-lipid Glc3Man9GlcNAc2-PP-Dol (LLO) in N-glycosylation of proteins, (2) in the synthesis of glycosylphosphatidylinositol (GPI) anchors, (3) in O-glycosylation of proteins in yeast, and (4) in C-mannosylation of Trp-7 in human ribonuclease 2 (RNase 2). In addition, Dol-P-Man has been recognized as an activator of N-acetylglucosaminyl 1-phosphate (GlcNAc-1P) transferase [11].
DPMS, an Mr 31 kDa protein and is distributed throughout the nature. Dpm1 is a structural gene in Saccharomyces cerevisiae and it is essential for their viability, because disruption of the Dpm1 gene is lethal and does not allow the organism to grow at nonpermissive temperature [12]. Similarly, a partial deficiency of DPMS has been found to be associated with some variants of congenital disorder of glycosylation (CDG) [13, 14]. These patients exhibit developmental delay, seizures, hyptonia, and dysmorphic function [15]. Mutation of DPMS has also been reported in human lymphoma Thy-1-class E cells and are defective in Dol-P-Man biosynthesis [16].
The gene for Dpm1 has now been cloned from a protozoan parasite to human [17–19] and based on the available sequence the Dpm1 has been grouped as Type I (Saccharomyces cerevisiae, Ustilago maydis, Trypanosoma brucei, and Leishmania mexicana) [19, 20], and Type II (human, Schizosaccharomyces pombe and Caenorhabditis briggsiae) [19]. Type I is a single-component enzyme represented by Saccharomyces cerevisiae Dpm1 [12] and the Type II enzyme contains a catalytic subunit DPM1 and two accessory proteins, DPM2 and DPM3 [21–23]. Members of the Type I share 50% to 60% amino acid identity and have a transmembrane domain near the C-terminus [19, 20], whereas those of the Type II members have only 30% amino acid identity to the former group members and lack the transmembrane domain [19, 21]. DPM2, a very hydrophobic protein of 84 amino acids, associates with the N-terminal hydrophobic protein (~60 amino acids) of DPM3. DPM3, via its C-terminal hydrophilic portion (~30 amino acids), associates with DPM1 (260 amino acids). The C-terminal 24 amino acids of DPM1 are necessary for association with DPM3, and DPM2 associates with DPM3 using its first transmembrane domain, which includes Phe-21 and Tyr-23 [22, 23]. DPM1 stabilized with DPM3 in the absence of DPM2 and expressing the DPMS activity indicates that DPM2 is not essential for the enzymatic reaction. On the other hand, the specific activity of the enzyme was 10 times higher when DPM2 was present. Therefore, DPM2 plays a role in the enzymatic reaction and its mutation is responsible for defective biosynthesis of Dol-P-Man in CHO Lec 15 mutant cells [21].
We have been studying the regulation of N-linked protein glycosylation and its relationship to angiogenesis. Angiogenesis is the formation of new blood vessels, an essential physiological event during growth and development. It is also important for tumor progression and metastasis. In our study, we have used isoproterenol, a β-receptor agonist which enhances intracellular cAMP as a stimulant to study the molecular details of angiogenesis. β-Adrenoreceptor is a trimeric G-protein coupled receptor and when activated the rate of LLO biosynthesis and turnover are increased. As a result the N-glycosylation of Factor VIIIc and other cellular proteins is enhanced [24, 25]. Factor VIIIc in capillary endothelial cells (i.e., eFVIIIc) has ~18% N-linked glycan and its expression precedes the cell proliferation [26, 27]. Dol-P-Man synthesis when evaluated either by metabolic labeling of cells with 2-[3H]mannose or by measuring the synthase activity in isolated microsomes following isoproterenol treatment, the level of the enzyme activity was high. Propranolol (a β-antagonist), but not actinomycin D (an antibiotic which binds to DNA duplexes and interferes with the action of enzymes engaged in replication and transcription) abolished such activation [28]. Thus, suggesting phosphorylation modification of the protein and not its increased gene expression was associated with enhanced DPMS activity in the stimulated cells.
We report here successful cDNA cloning of the DPMS gene from a non-transformed capillary endothelial cell line. The protein sequence deduced from the DNA sequence indicated the presence of a cAMP-dependent protein phopshorylation motif. Expression and purification of the protein indicated that the recombinant his-tagDPMS is an Mr 33 kDa protein. In addition, overexpression of DPMS accelerates angiogenesis.
Materials and methods
The capillary endothelial cells used in this study were from the laboratory stock. Minimal essential medium with Earle’s salt (EMEM), glutamine, antibiotic mixture (penicillin– streptomycin–fungizone), phosphate-buffer saline (PBS), pH 7.2 and 7.4 and trypsin–versine were obtained from BioSource, Camarillo, CA. Fetal bovine serum was a product of HyClone Laboratories, Logan, UT. Dimethyl-sulfoxide, Sepharose CL-2B, nystatin, bovine serum albumin (crystallized) and 8Br-cAMP were obtained from Sigma Aldrich, St. Louis, MO. mRNA isolation kit, FreeStyle, 293 expression system, pcDNA3.1/His vector, OptiMEM, 293fectin were purchased from Invitrogen, Carlsbad, CA. ZAP-cDNA synthesis kit was from Stratagene, La Jolla, CA. NP-40, Kaleidscope protein molecular weight markers, DC protein assay reagents, and other electrophoresis reagents were obtained from Bio-Rad Laboratories, Hercules, CA. Rhodamine-conjugated goat anti-rabbit IgG and Hoechst 33342 were from Molecular Probes, Eugene, OR. Cell culture supplies were from Sarstedt, Newton, NC. [α-32P]-dATP (800 Ci/mol) was from GE Healthcare, Piscataway, NJ. Restriction enzymes were from New England BioLabs, Beverly, MA.
Culturing of capillary endothelial cells
The stock of capillary endothelial cells was maintained in EMEM containing 10% fetal bovine serum (heat-inactivated), glutamine (2 mM), penicillin (50 units/ml), streptomycin (50 µg/ml), fungizone (2.5 µg/ml), and nystatin (1,000 units/ml) at 37°C in a humidified incubator (5% CO2–95% air) in tissue culture flasks or dishes without collagen underlay or other extracellular matrix components as described earlier [29]. Cells were routinely monitored by light microscopy and subcultured once a week. The viability of cells was determined by trypan blue (0.4%) exclusion.
Isolation of Total RNA
A total of 20 µg mRNA was obtained from 2 mg total RNA isolated from about 2 × 109 cells of bovine endothelial cells according to the standard method [30] using mRNA isolation kit from Invitrogen, CA.
Construction of a cDNA library and the isolation of a cDNA clone for bovine DPMS (bDPMS)
Ten micrograms of mRNA was used to prepare double-stranded cDNA using ZAPcDNA synthesis kit (Stratagene, CA) as it produced vector-ready directional cDNA that can be used in conjunction with the Uni-ZAP XR. During the first-strand synthesis 0.5 µl of [α-32P]-dATP (800 Ci/mole) was used to follow the cDNA synthesis as well as to detect [32P]-labeled double-stranded cDNA during the size fractionation using Sepharose CL-2B gel filtration Drip column provided in the kit. The primer design of the kit was GAGAGAGA GAGAGAGAGAGAACTAGTCTCGAG (A)18 3′. cDNA (>400 bp) were purified by phenol-chloroform and ethanol precipitation, ligated to the EcoRI adopter, digested with EcoRI/XhoI and then it was cloned into predigested UniZAP XR vector EcoRI and XhoRI, CIAP treated according to the manufacturer’s protocol. After the ligation, 1 µl of each ligation reaction including the control was used with Gigapack III Gold packaging extract according to the manufacturer’s protocol. The efficiency of packaging was determined by titrating the phage found to be about 1 × 106 plaque/µg of vector using XL1-Blue MRF’ and VCS257 cells on LB agar plates. The cDNA library was amplified and the titer was 1010 pfu/ml.
Screening and isolation of a cDNA clone for bDPMS
Screening of the cDNA library was done using [32P]-labeled probe hybridization method [30]. To prepare the probe, RT-PCR method was used with mRNA as the template. The primers were designed based on the homologous sequences of known Dpm1 (forward primer 5′TTCCTCCCAACTTG GATTCACC3′; and reverse primer 5′ATCATAGATGATG GAAGCCCAG3′) using PCR kit of Stratagene, CA. The RT-PCR product was gel purified, and was used as [32P]-labeled probe for screening the cDNA library by the standard method [30]. We had 5 positive, but incomplete clones (based on the homologous known sequences). Three of the clones had 5′-end of the homologous genes, and 2 of them had 3′-end. We have used these DNAs as the template to isolate the bDpm1 gene by PCR amplification using 5′ BamHI and 3′XhoI primers (forward primer 5′AAT GGATCCACCATGGCTGCCGAGGAAGCAAG3′ and reverse primer 5′CGCCTCGAGTTATGTAGTAGCAAAGA GAGTC3′) suitable to clone into bluescript vector. The PCR product was digested with BamHI and XhoI, gel purified, cloned into the vector for transformation into DH5α cells, screened, and sequenced to obtain full-length bDPMS gene. The clone KB-3 was used for confirmation studies.
Expression of recombinant bDPMS protein
(1) Selection of expression system: The FreeStyle™ 293 expression system was chosen to allow large-scale transfection of 293 human embryonic kidney cells in suspension culture in a defined, serum-free medium. The advantage of this system is that a denser culture could be obtained. The vector pcDNA3.1/His was designed for a high-level expression and purification of recombinant proteins in mammalian hosts, and suitable for the FreeStyle™ 293 expression system. We chose to clone the bDPMS gene at the KpnI and XhoI sites so that N-terminal His-tag DPM1 could be expressed containing the Xpress™ Epitope with the provision to digest the expressed protein with Enterokinase (EK) digestion to remove the tag, if necessary. The bDPMS (KB-3) gene was cloned into the pcDNA3.1/His A vector at the KpnI and XhoI site and transformed DH5α cells. The transformants were screened using the PCR colony screening method and the primers as mentioned above. Based on the DNA sequence, clone bDPMS-pcDNA-7 was chosen for transformation studies. DPMS-pcDNA-7 plasmid was used to transform FreeStyle™ 293 cells according to the manufacturer’s protocol. (2) Transfection: FreeStyle™ 293 cells were seeded at a density of 3 × 105 viable cells/ml and grown in 40 ml of FreeStyle™ medium in 250 ml of sterile polycarbonate disposable Erlenmeyer flasks shaken in an orbital shaker at 37°C humidified incubator with 8% CO2 for 3–5 days. The doubling time of the cell was about 20–25 h. Cells were counted manually using a hemacytometer and their viability was examined by Trypan blue exclusion. 3×107 cells were collected in 50 ml sterile centrifuge tube by centrifuging at 1,000 rpm for 5 min at room temperature. The pellet was suspended into 28 ml of pre-warmed FreeStyle™ media in a 250 ml to a density of 1 × 106 viable cells/ml and incubated in a 37°C orbital shaker incubator. To prepare the lipid–DNA complex for transfection, 30 µg of bDPMS-pcDNA-7 plasmid was diluted with Opti-MEM to a total volume of 1.0 ml. Separately, 40 µl of 293fectin was also diluted into Opti-MEM to a total volume of 1.0 ml, and incubated at room temperature for 5 min. The diluted plasmid DNA was then added to the diluted 293fectin solution and gently mixed, and incubated for 30 min at room temperature to allow the DNA-293fectin complexes to form. During this incubation period, the cell suspension was removed and an aliquot of 28 ml cell suspension was placed into sterile, disposable 125 ml Erlenmeyer flasks. 2 ml of DNA-293fectin complex was then added into each flask containing the cells and incubated at 37°C. For a negative control, 2 ml of Opti- MEM1 instead of DNA-293fectin complex was added. Cells were incubated as above for 24, 48, 72, and 96 h. (3) SDS-PAGE analysis: 5 ml of cell suspension was centrifuged, and suspended in 100 µl of SDS-PAGE sample buffer, sonicated, heated at 95°C for 3 min, centrifuged and 20 µl was analyzed on a 10% SDS-PAGE. 72 h post-transfection exhibited optimal expression of the recombinant bDPMS protein, and a large scale expression using 1 l of media was carried out using the same experimental condition.
Purification of expressed His-tagbDPMS
Cells were pelleted by centrifugation at 3,000 rpm for 10 min at 4°C and washed with PBS, pH 7.4. To isolate the microsomal membranes, the cells were homogenized in 0.1 M Tris–HCl (pH 7.0) containing 0.25 M sucrose and 1 mM EDTA using a dounce homogenizer. The microsomal membranes were prepared following a procedure described earlier by Banerjee et al. [31]. Briefly, the homogenate was centrifuged at 600×g for 10 min and the resulting supernatant at 9,000×g for 10 min. The 9,000×g supernatant was then centrifuged at 39,000×g for 15 min and the resulting particulate fraction was suspended in the homogenization buffer and stored frozen at −20°C in multiple aliquots until used. The microsomal membrane proteins (10 mg) were solubilized by incubating in ice for 10 min in 1.25 ml of Tris–HCl, pH 7.0 containing 60 mM sucrose, 25 µM EDTA, 2.5% NP-40 and 0.4% β-mercaptoethanol. It was then diluted with 15 ml of a mixture of water (10.5 ml), glycerol (4 ml) and β-mercaptoethanol (50 µl). The mixture was vortexed for 30 s and centrifuged at 100,000×g for 1 h at 4°C. The supernatant was used as the source for DPMS. The His-tag fusion protein was purified using ProBond (a nickel-chelating resin) purification system (Invitrogen, CA). The cell extract was added to 4 ml of ProBond resin in a 50 ml centrifuge tube, mixed gently at 4°C for 60 min. The mixture was loaded into an empty column and the flow-through (FT) was collected. The FT was re-applied to the column to minimize losses of the target proteins. Subsequently, the resin was washed with the washing buffer (20 mM imidazole, 0.3% N-lauroyl sarcosine, 50 mM CAPS buffer and 0.3 NaCl, pH 8) until the OD280 reading reached or fell below the base line. The 6× His-tagged fusion protein was eluted using the Elution buffer (wash buffer containing 300 mM imidazole). One milliliter fractions were collected and analyzed on 10% SDS-PAGE gel followed by Coomassie staining.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE)
The purity of protein was verified by the 10% SDS-PAGE using Laemmli’s procedure [32].
Quantification of protein
Protein was quantified by DC Protein Assay kit of Pierce using bovine serum albumin as the standard.
Generation of DPMS overexpression plasmid and isolation of stable transfectants
Cellular RNA was isolated with TRIzol reagent and treated with DNase to remove any possible genomic DNA contamination. Total RNA was quantified in a NanoDrop Bioanalyzer. First strand cDNA was synthesized using the iScript™ cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. The primers for amplifying full DPMS gene were: 5′-ccgctcgagATGGCTGCCGAGGAAGCAAGTC-3′ (forward) and 5′-ccgggatccCTCCAGAAAACAGACTTGCCT TACCT-3′ (reverse), which provided with the Xho I and BamH I site (underlined). PCR reactions were carried out as follows: initial denaturation at 94°C for 4 min, 30 cycles at 94°C for 50 s, 64°C for 50 s, 72°C for 50 s, and a final extension for 10 min at 72°C in 50 µl containing 2 µl each of cDNA, 0.2 µM each primer, 0.2 mM dNTP, and 2 units of Taq DNA polymerase. After amplification, 5 µl of each reaction mixture was detected by 1.5% agarose gel electrophoresis followed by ethidium bromide staining. PCR products were purified and then cloned into Xho I and BamH I sites of pEGFP-N1 to obtain pEGFP-N1-DPMS overexpression plasmid. The plasmids were confirmed by Xho I / BamH I double digestion and DNA sequencing. Transfection of the capillary endothelial cells with pEGFP-N1-DPMS overexpression plasmid and pEGFP-N1 vector was performed using Lipofectamine™ Reagent and Plus™ Reagent (Invitrogen) according to the manufacturer’s protocol. After transfection, the cells were cultured in regular EMEM containing 0.55 mg/ml of G418 for 4 weeks, and cells overexpressing DPMS and pEGFP-N1 vector alone were collected.
Characterization of DPMS overexpressing clones
The stable DPMS-transfected capillary endothelial cells were maintained in EMEM containing 10% fetal bovine serum (heat-inactivated), glutamine (2 mM), penicillin (50 units/ml), streptomycin(50 µg/ml), nystatin (1,000 units/ml) and G418 (0.55 µg/ml) at 37°C in an humidified incubator (5% CO2–95% air) in tissue culture flasks. The presence of DPMS was monitored by western blotting, immunofluorescence microscopy, RT-PCR, restriction double digest, and DNA sequencing.
The vector has a GFP construct. Therefore, the protein expression was monitored following immunofluorescence GFP.
For DNA sequencing plasmid was isolated and 20 µl sequencing reaction was designed according to the BigDye terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) containing 3.2 pmol primer (Forward Primer: 5′-TT TCCAAAATGTCGTAACAACTCCG-3′; ReversePrimer:5′-TGAACTTGTGGCCGTTTACGT-3′) and 300 ng plasmid. The reaction was carried out as follows: initial denaturation at 96°C for 1 min, 25 cycles at 96°C for 10 s, 50°C for 50 s, 60°C for 4 min, and rapid thermal ramp to 4°C. This was followed using BigDye XTerminator™ Purification Kit (Applied Biosystems), and then electrophoresis on the ABI PRISM 310 Genetic Analyze.
Immunofluorescence microscopy
Capillary endothelial cells and their DPMS overexpressing clones were subcultured on cover glasses (15 mm diameter, Warner Instruments, Hamden, CT) for 24 h under standard conditions. The cells were washed three times with PBS, pH 7.4 and fixed for 5 min in ice-cold methanol. The cells were washed three times with PBS, pH 7.4 and incubated with DPMS antibody (rabbit polyclonal 1:500 dilution) for 1 h at room temperature. After washing three times with PBS, pH 7.4, the cells were incubated with rhodamine-conjugated goat anti-rabbit IgG (1:300 dilution) and Hoechst 33342 (1:10,000 dilution) for 30 min at room temperature. After washing three times with PBS, pH 7.4, the cover glasses were mounted and examined in a Zeiss AxiocamMRc (Carl Zeiss, Germany) fluorescent microscope.
Cell proliferation assay
DPMS overexpressing cells were seeded into 24-well plates (2 × 104 cells/well) in normal medium containing 10% serum and 0.55 mg/ml G418. After 24 h the medium was removed, cells were washed three-times with PBS (pH7.4) and incubated in a serum-free medium for 24 h. At the end the medium was replaced with normal medium containing 10% serum and 0.55 mg/ml G418. The cell numbers were counted after every 24 h for seven days.
Results
Identification of bDPMS clone
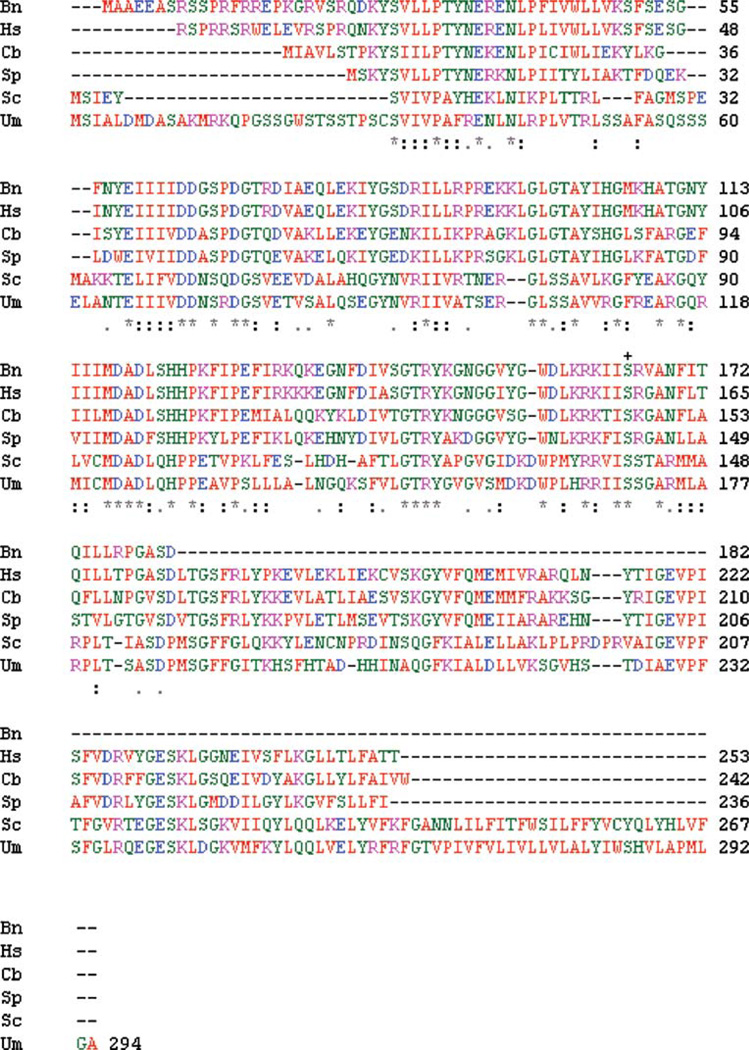
DNA of the positively identified clone carrying the bDPMS gene was sequenced by the dideoxy method of Sanger [33]. Figure 1(a) shows the DNA sequence of the bDPMS gene. Amino acid sequence of the bovine capillary endothelial cell DPMS was then deduced from the above DNA sequence of bDPMS and is shown in Fig. 1(b). The cloned bDPMS may have an incomplete gene, but it codes for a protein of 182 amino acid residues, and carries at serine-165 in a consensus sequence RKIIS165, a motif for phsphorylation by cAMP-dependent protein kinase (PKA) (Fig. 2). Furthermore, the homology search for bDPMS indicated that the bDPMS has 85% homology with human DPM1, 26% homology with Saccharomyces cerevisiae, 55% homology with Caenorhabditis briggsiae, 56% homology with Schizosaccharomyces pombe, and 26% homology with Ustilago maydis, respectively.
Fig 1.
Nucleotide sequence of the bDPMS gene and its deduced amino acid sequence. a Nucleotide sequence of bovine capillary endothelial cell DPMS. DNA sequencing of the DPMS gene was carried out with chain-terminating inhibitors method of Sanger [33] as described in “Materials and methods”. b Deduced amino acid sequence exhibited the presence of a PKA phosphorylation motif “RKIIS”
Fig 2.
DPMS sequence alignment. DPMS protein sequence from mammalian and nonmammalian sources was aligned using the CLUSTALW program (http://www.ebi.ac.uk/Tools/clustalw). (+) indicates the PKA phosphorylation site
Hydropathy Profile of DPMS
Determination of the threedimensional structure of a membrane protein, i.e., its topology—is generally much more difficult that determining its amino acid sequence, either directly or by gene sequencing. The presence of unbroken sequences of more than 20 hydrophobic residues in a membrane protein is commonly taken as evidence that these sequences traverse the lipid bilayer, acting as hydrophobic anchors or forming transmembrane channels. Virtually all integral proteins have at least one such sequence.
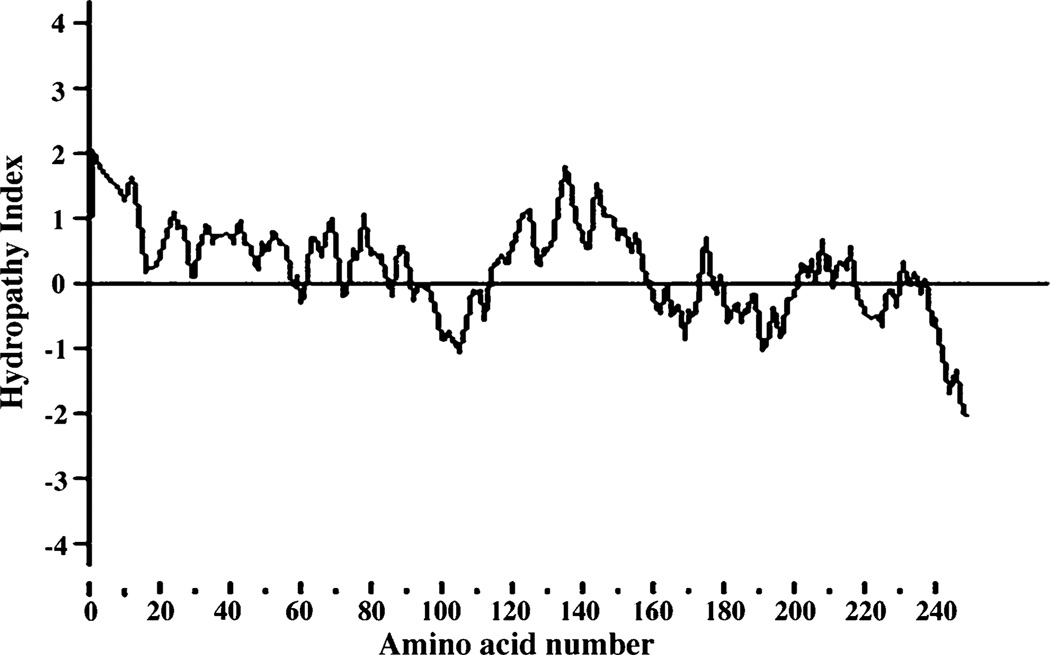
What can we predict about the secondary structure of the membrane-spanning portions of an integral protein, DPMS? An α-helical sequence of 20 to 25 residues is just long enough to span the thickness (30 Å) of the lipid bilayer [the length of an α-helix is 1.5 Å (0.15 nm) per amino acid residue]. A polypeptide chain surrounded by lipids, having no water molecule with which to hydrogen-bond, will tend to form α-helices or β-sheets, in which intrachain hydrogen bonding is maximized. If the side chains of all amino acids in a helix are nonpolar, hydrophobic interactions with the surrounding lipids further stabilize the helix. Several simple methods of analyzing amino acid sequences yield reasonably accurate predictions of secondary structure for transmembrane proteins. The relative polarity of each amino acid has been determined experimentally by measuring the free energy change accompanying the movement of that amino acid side chain from a hydrophobic solvent into water. This free energy of transfer, which can be expressed as a hydropathy index, ranges from very endergonic for amino acids with aromatic or aliphatic hydrocarbon side chains. The overall hydropathy index (hydrophobicity) of a sequence of amino acids is estimated by summing the free energies of transfer for the residues in the sequence. To scan a polypeptide sequence of DPMS for potential membrane-spanning segments, we have calculated the hydropathy index for successive segments (called windows) of a given size from 7 to 20 residues. Hydropathy Index when plotted against amino acid number it indicates the presence of a hydrophobic region around the amino acid residues 120–160 supporting that bDPMS has one membrane spanning region. Hydropathy analysis of bovine DPMS drawn according to Kyte and Doolittle [34] predicts a single hydrophobic helix for DPMS near the C-terminus (Fig. 3). Positive hydropathy index between the amino acid residues strongly support the presence of a transmembrane region in bDPMS.
Fig 3.
Hydropathy profile of bovine capillary endothelial cell DPMS. The hydropathy character of DPMS was displayed according to Kyte and Doolittle [34]
Purification of the expressed His-tagDPMS
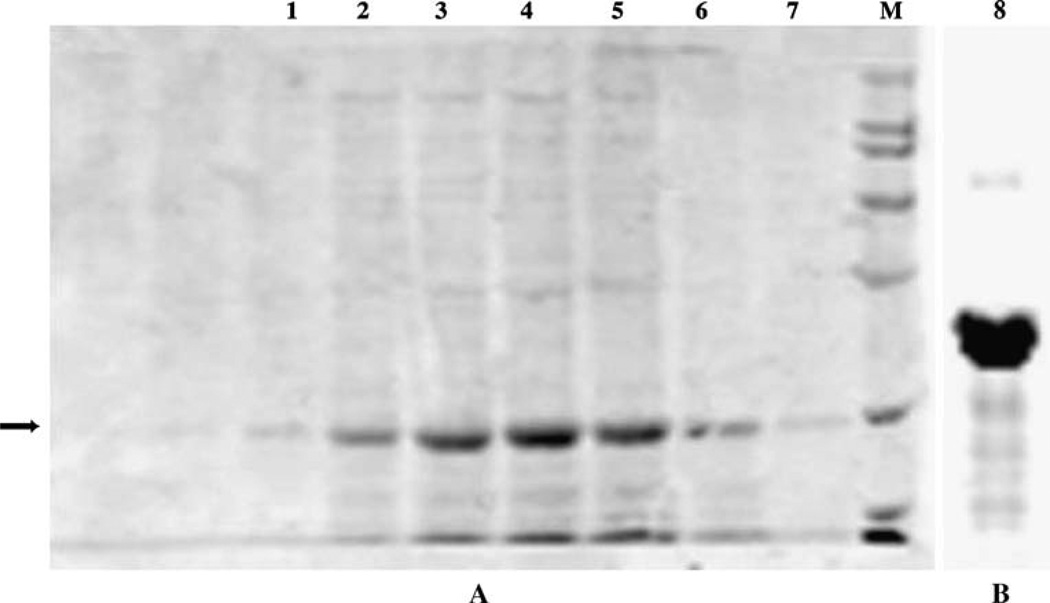
Clone bDPMS-pcDNA-7 was used for the expression studies. The cells were pelleted by centrifugation at 3,000 rpm for 10 min at 4°C. The microsomal membranes were isolated and the proteins were solubilized as mentioned under “Materials and methods”. The His-tag fusion protein was purified using ProBond purification system (Invitrogen, CA) and the fractions were analyzed by SDS-PAGE. Figure 4 shows that most of the Mr 33 kDa protein was eluted between fractions 3–7. The fractions were pooled, and re-chromatographed over another 2 ml ProBond column by the same method. The purified His-tagDPMS (Mr 33 kDa) was more than 97% pure based on the SDS-PAGE analysis. Using this technique we have purified approximately 50 µg bDPMS protein from one liter cell suspension.
Fig 4.
SDS-PAGE analysis of ProBond column fractions. DPMS was solubilized and purified as a His-tag protein using ProBond (a nickel-chelating resin) as described in “Materials and methods”. The fractions were separated on a 10% SDS-PAGE and the protein was stained with Coomassie blue. a First ProBond fractions. Lanes 1–7 are fraction numbers 1 to 8; Lane M: Broad Mol Wt. Marker containing Mr 6.5, 14, 21, 31.5, 45, 66, 97 and 200 kDa protein bands (Bio-Rad). b Second ProBond column profile of pooled fractions (20 µg protein; separate gel). Arrow indicates the position of an Mr 33 kDa protein band
Characterization of DPMS overexpressing clones
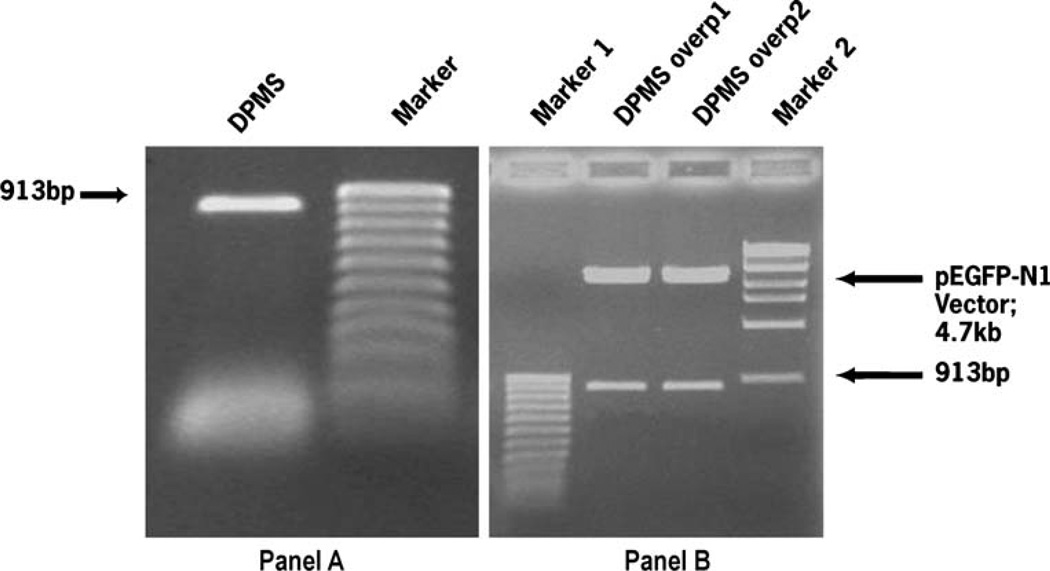
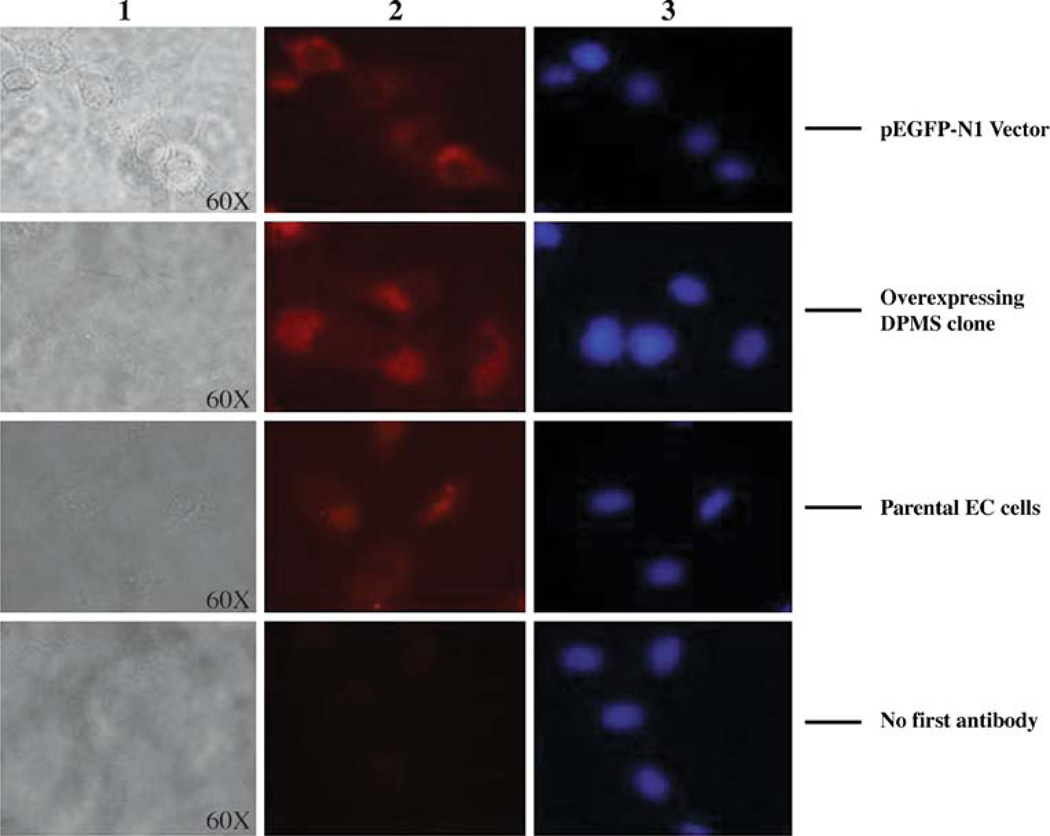
Overexpressing DPMS gene from capillary endothelial cell was cloned by RT-PCR and confirmed by digestion with restriction enzymes BamHI and XhoI. Both RT-PCR (panel A, Fig. 5) and the double restriction digest (panel B, Fig. 5) identified a 913 bp fragment on agarose gels. DPMS protein expression was also monitored by fluorescence microscopy and western blot (data not shown). Capillary endothelial cells harboring DPMS overexpressing plasmid expressed DPMS higher than pEGFP-N1 vector and the parental cells (Fig. 6).
Fig 5.
Cloning and confirmation of overexpressing DPMS gene. Overexpressing DPMS gene from capillary endothelial cells has been isolated and confirmed as mentioned in“Materials and methods”. The DPMS gene was cloned by RT-PCR and confirmed by restriction enzyme digestion followed by agarose gel electrophoresis and ethidium bromide staining. Panel a, aga-rose gel profile of the RT-PCR product; Panel b, agarose gel profile of the restriction digest of clone-1 and clone-2
Fig 6.
DPMS Immunofluorescence microscopy. The capillary endothelial cells (parental line, cells transfected with vector alone and the cells overexpressing DPMS) were cultured as mentioned in “Materials and methods”. The cells were processed for immunological detection of DPMS by fluorescence microscopy. Column 1 bright field; column 2 DPMS staining; column 3 = Hoechst 33342 staining for nucleus
Relationship between DPMS and angiogenesis
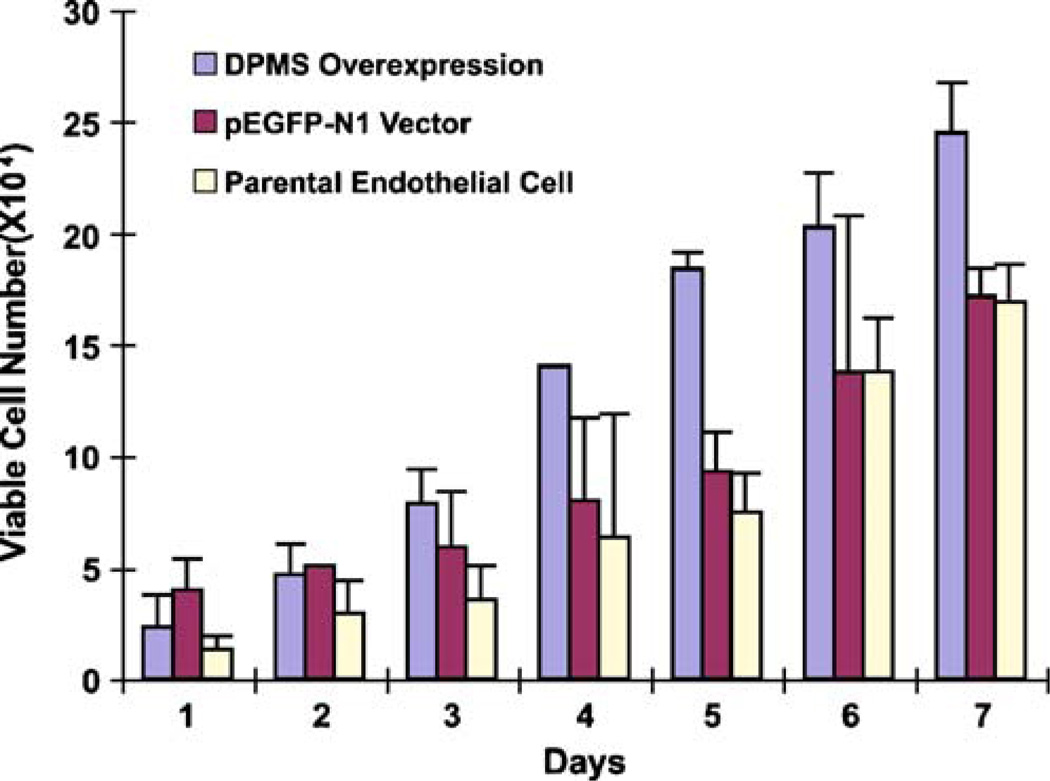
It has been proposed earlier that phosphorylation activation of DPMS accelerates the lipid-linked oligosaccharide biosynthesis and turnover in capillary endothelial cells. This consequently resulted in increased cellular proliferation. To establish further that DPMS plays a significant role in angiogenesis, the rate of cellular proliferation was analyzed in a capillary endothelial cell line overexpressing DPMS. The results in Fig. 7 clearly demonstrate a higher rate of cellular proliferation in DPMS overexpressing cells compared to the pEGFP-N1 vector and the parental cells.
Fig 7.
Proliferation of capillary endothelial cells overexpressing DPMS. 2×104 cells/well were plated in 24-well clusters and synchronized for 24 h in serum-free media. The cells were then cultured in a medium containing 10% fetal bovine serum and counted in a hemocytometer after every 24 h for seven days. blue-colored square overexpressing DPMS cell clone, purple-colored square pEGFP-N1 Vector; yellow-colored square parental cell line
Discussion
Mannosylphospho dolichol synthase (DPMS), the only glycosyltransferase of the protein N-glycosylation pathway is currently known to undergo regulation by PKA-mediated-phosphorylation signal [31, 35]. This post-translational modification alters the DPMS activity to interact adequately with the cellular microenvironment. The signals could be either a neurotransmitter, or a hormone (β-adrenergic or peptidergic), or it may be a cytokine or a chemokine that generates intracellular cAMP [36]. Phosphorylation of DPMS irrespective of its remain bound to the ER membrane or a purified recombinant protein enhances the catalytic activity of the enzyme. Vmax of the enzyme is increased by two to six folds without appreciably altering the Km for GDP-mannose [31, 35]. In addition, the phosphorylated DPMS has exhibited higher enzyme turnover (kcat) and enzyme efficiency (kcat/Km). SDS-PAGE followed by autoradiography of 32P-labeled DPMS has detected a Mr 32 kDa phosphoprotein. Active 32P-labeled enzyme when analyzed by anti-DPMS antibody affinity column chromatography both radioactivity and the enzyme activity have co-migrated [37]. Dephosphorylation with alkaline phosphatase at pH 7.0 did reduce its activity by ~87% [31] and immunoblotting with anti-phosphoserine antibody also identified a phosphoserine residue in in vitro phosphorylated recombinant DPMS from Saccharomyces cerevisiae [35]. Replacing the serine-141 at the PKA phosphorylation site of the recombinant DPMS from Saccharomyces cerevisiae by alanine (i.e., S141A) by site-directed mutagenesis has reduced the phosphorylation activation of the enzyme by ~50%. [35].
Protein kinase type I-deficient Chinese hamster ovary (CHO) cells have also exhibited reduced DPMS activity with a Km for GDP-mannose 160–400% higher than that of the wild type. The kcat for the DPMS has reduced two to four folds in the mutant cells and exogenously added Dol-P has failed to rescue the Km for GDP-mannose [38]. DPMS is a ‘key’ step in the LLO biosynthesis, a pre-requisite for protein N-glycosylation. Therefore, it is expected that up- or down-regulation of the DPMS activity should reflect appropriately to the level of LLO and the glycoproteins. In fact, phosphorylation up-regulation of DPMS has increased the LLO biosynthesis and turnover [39]. These are significantly reduced in PKA-deficient CHO cells [38]. Protein glycosylation of many cellular glycoproteins in these mutants is also impaired [38]. This deficiency however, has been restored by rescuing the mutation of protein kinase type I by somatic cell genetics i.e., in a revertant. The LLO biosynthesis as well as the protein N-glycosylation also has reached almost to the normal level [40].
To test that phosphorylation activation of DPMS has an impact on cellular proliferation, we have cultured capillary endothelial cells in the presence of 2 mM 8BrcAMP. The results have indicated that under the experimental condition (1) the cell proliferation is increased; (2) the cell doubling time is reduced; and (3) 47% cells had entered in the S phase. Expression of pro-angiogenic molecule Bcl-2 and the apoptotic molecule caspase-3 remained unaffected [24]. Decreased expression of the ER chaperone GRP-78/Bip also an “ER stress response identifier” with a concomitant increase in the cytoplasmic chaperone HSP-70 expression has also been observed. These have resulted in increased DPMS activity with an up-regulation of LLO biosynthesis and its rapid turnover and increased glycosylation of endothelial cell factor VIIIc. Therefore, identification of a PKA phosphorylation motif in the isolated capillary endothelial cell bDPMS has supported our hypothesis that upregulation of DPMS activity by phosphorylation is fundamental and may involve in angiogenesis for breast and other solid tumor growth and metastasis. This in fact has been supported in an analogous experiment in which the DPMS has been overexpressed in capillary endothelial cells. The results presented here (Fig. 6) has unequivocally supported that overexpression of DPMS is indeed increased the rate of capillary endothelial cell proliferation, i.e., angiogenesis. Therefore, we conclude that DPMS is a potential “angiogenic switch”.
Acknowledgement
The editorial assistance of Ms. Laura M. Bretaña is greatly appreciated. The work was partly supported by the funds from the University of Puerto Rico Medical sciences Campus and the grants from the Department of Defense DAMD17-03-1-0754, NIH U54-CA096297and the Susan G. Komen for the cure BCTR58206 (DKB) and the NIH/NCRR/RCMI grant G12-RR03035 (KB).
Abbreviations
- EMEM
minimal essential medium with Earle’s salt
- PBS
phosphate-buffer-saline
- NP-40
Nonident P-40
- cAMP
adenosine 3′,5′-cyclic monophosphate
- RNA
ribonucleic acid
- mRNA
messenger ribonucleic acid
- DNA
deoxyriboynucleic acid
- PKA
cAMP-dependent protein kianse
- ER
endoplasmic reticulum
- DPMS
mannosylphospho dolichol synthase
- PCR
polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
- EDTA
ethylenediamine tetraacetic acid
- Dol-P-Man
dolichol-P-mannose
- GDP
guanosine diphosphate
- DTT
dithiothretol
- Dol-P
dolichyl monophosphate
- DMSO
dimethyl sulfoxide
- SDS
sodium dodecylsulfate
- PAGE
polyacrylamide gel electrophoresis
- LLO
lipid-linked oligosaccharide Glc3Man9GlcNAc2-PP-Dol
Contributor Information
Krishna Baksi, Department of Anatomy and Cell Biology, School of Medicine, Universidad Central del Caribe, Bayamón, PR 00930-3100, USA.
Zhenbo Zhang, Department of Biochemistry, School of Medicine, University of Puerto Rico, Medical Sciences Campus, San Juan, PR 00936-5067, USA.
Aditi Banerjee, Department of Biochemistry, School of Medicine, University of Puerto Rico, Medical Sciences Campus, San Juan, PR 00936-5067, USA.
Dipak K. Banerjee, Email: dbanerjee@rcm.upr.edu, Department of Biochemistry, School of Medicine, University of Puerto Rico, Medical Sciences Campus, San Juan, PR 00936-5067, USA.
References
- 1.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Tannner W, Lehle L. Protein glycosylation in yeast. Biochim. Biophys. Acta. 1987;906:81–99. doi: 10.1016/0304-4157(87)90006-2. [DOI] [PubMed] [Google Scholar]
- 3.Orlean P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:5796–5806. doi: 10.1128/mcb.10.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menon AK, Mayor S, Schwarz RT. Biosynthesis of glycosylphosphatidylinositol lipids in Trypanosoma brucei: involvement of mannosylphosphoryldolichol as the mannose donor. EMBO J. 1990;9:4249–4258. doi: 10.1002/j.1460-2075.1990.tb07873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Englund PT. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu. Rev. Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 6.Herscovics A, Orlean P. Glycoprotein biosynthesis in yeast. FASEB. J. 1993;7:540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- 7.Doucey MA, Hess D, Cacan R, Hofsteenge J. Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell. 1998;9:291–300. doi: 10.1091/mbc.9.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke BL, Naylor C, Lennarz WJ. Comparative studies on mannosylphosphoryl dolichol and glucosylphosphoryl dolichol synthases. Chem. Phys. Lipid. 1989;51:239–247. doi: 10.1016/0009-3084(89)90011-x. [DOI] [PubMed] [Google Scholar]
- 9.Hirschberg CB, Snider MD. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 1987;56:63–87. doi: 10.1146/annurev.bi.56.070187.000431. [DOI] [PubMed] [Google Scholar]
- 10.Abeijon C, Hirschberg CB. Topography of glycosylation reactions in the endoplasmic reticulum. Trends Biol. Sci. 1992;17:32–36. doi: 10.1016/0968-0004(92)90424-8. [DOI] [PubMed] [Google Scholar]
- 11.Kean EL, Rush JS, Waechter CJ. Activation of GlcNAc-P-P-dolichol synthesis by mannosylphosphoryldolichol is stereospecific and requires a saturated alpha-isoprene unit. Biochemistry. 1994;33:10508–10512. doi: 10.1021/bi00200a036. [DOI] [PubMed] [Google Scholar]
- 12.Orlean P, Albright C, Robbins PW. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 1988;263:17499–17507. [PubMed] [Google Scholar]
- 13.Marquardt T, Denecke J. Congenital disorders of glycosylation: review of their molecular bases, clinical presentations and specific therapies. Eur. J. Pediatr. 2003;162:359–379. doi: 10.1007/s00431-002-1136-0. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Westphal V, Srikrishna G, Mehta DP, Peterson S, Filiano J, Karnes PS, Patterson MC, Freeze HH. Dolichol phosphate mannose synthase (DPM1) mutations define congenital disorder of glycosylation Ie (CDG-Ie) J. Clin. Invest. 2000;105:191–198. doi: 10.1172/JCI7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Developmental abnormalities of glycosylphosphatidylinositol-anchor deficient embryos revealed by Cre/loxP system. Lab. Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- 16.Chapman A, Trowbridge IS, Hyman R, Kornfeld S. Structure of the lipid-linked oligosaccharides that accumulate in class E Thy-1-negative mutant lymphomas. Cell. 1979;17:509–515. doi: 10.1016/0092-8674(79)90259-9. [DOI] [PubMed] [Google Scholar]
- 17.Mazhar-Tabrizi R, Eckert V, Blank M, Muller R, Mumberg D, Funk M, Schwarz RT. Cloning and functional expression of glycosyltransferases from parasitic protozoans by heterologous complementation in yeast: the dolichol phosphate mannose synthase from Trypanosoma brucei brucei. Biochem. J. 1996;316:853–858. doi: 10.1042/bj3160853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman JW, Specht CA, Ceganes BX, Robbins PW. The isolation of a Dol-P-Man synthase from Ustilago maydis that functions in Saccharomyces cerevisiae. Yeast. 1996;12:765–771. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C765::AID-YEA974%3E3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 19.Colussi PA, Taron CH, Mack JC, Orlean P. Human and Saccharomyces cerevisiae dolichol phosphate mannose synthases represent two classes of the enzyme, but both function in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 1997;94:7873–7878. doi: 10.1073/pnas.94.15.7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilgoutz SC, Zawadzki JL, Ralton JE, McConville MJ. Evidence that free GPI glycolipids are essential for growth of Leishmania mexicana. EMBO J. 1999;18:2746–2755. doi: 10.1093/emboj/18.10.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomita S, Inoue N, Maeda Y, Ohishi K, Takeda J, Kinoshita T. A homologue of Saccharomyces cerevisiae Dpm1p is not sufficient for synthesis of dolichol-phosphate-mannose in mammalian cells. J. Biol. Chem. 1998;273:9249–9254. doi: 10.1074/jbc.273.15.9249. [DOI] [PubMed] [Google Scholar]
- 22.Maeda Y, Tomita S, Watanabe R, Ohishi K, Kinoshita T. DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1, and binding of dolichol phosphate. EMBO J. 1998;17:4920–4929. doi: 10.1093/emboj/17.17.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda Y, Tanaka S, Hino J, Kanagawa K, Kinoshita T. Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 2000;19:2475–2482. doi: 10.1093/emboj/19.11.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee DK, Martínez JA, Baksi K. Significance of protein N-glycosylation in breast tumor angiogenesis. In: Maragoudakis ME, Papadimitriou E, editors. Angiogenesis: Basic Science and Clinical Applications. Trivandrum, Kerala, India: Transworld Research Network; 2007. pp. 287–308. [Google Scholar]
- 25.Banerjee DK, Oliveira CM, Tavárez JJ, Katiyar VN, Saha S, Martínez JA, Banerjee A, Sánchez A, Baksi K. Importance of a Factor VIIIc-like Glycoprotein Expressed in Capillary Endothelial Cells (eFactor VIIIc) in Angiogenesis. In: Wu Albert., editor. Molecular Immunology of Complex Carbohydrates III. 2009. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee DK, Tavárez JJ, Oliveira CM. Expression of blood clotting factor VIII: C gene in capillary endothelial cells. FEBS Lett. 1992;306:33–37. doi: 10.1016/0014-5793(92)80831-z. [DOI] [PubMed] [Google Scholar]
- 27.Martínez JA, Torres-Negrón I, Amigó LA, Banerjee DK. Expression of Glc3Man9GlcNAc2-PP-Dol is a prerequisite for capillary endothelial cell proliferation. Cell Mol Biol (Noisy-legrand) 1999;45:137–152. [PubMed] [Google Scholar]
- 28.Baksi K, Tavárez-Pagán JJ, Martínez JA, Banerjee DK. Unique structural motif supports mannosylphospho dolichol synthase:) an important angiogenesis regulator. Current Drug Targets. 2008;9:262–271. doi: 10.2174/138945008783954916. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee DK, Ornberg RL, Youdim MB, Heldman E, Pollard HB. Endothelial cells from bovine adrenal medulla develop capillary-like growth patterns in culture. Proc. Natl. Acad. Sci. USA. 1985;82:4702–4706. doi: 10.1073/pnas.82.14.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch EF, Sambrook J. In molecular cloning: a laboratory manual, Cold Spring Harbor laboratory. New York: Cold Spring Harbor; 1982. [Google Scholar]
- 31.Banerjee DK, Kousvelari EE, Baum BJ. cAMP-mediated protein phosphorylation of microsomal membranes increases mannosylphosphodolichol synthase activity. Proc. Natl. Acad. Sci. USA. 1987;84:6389–6393. doi: 10.1073/pnas.84.18.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee DK, Carrasquillo EA, Hughey P, Schutzbach JS, Martínez JA, Baksi K. In vitro phosphorylation by cAMP-dependent protein kinase up-regulates recombinant Saccharomyces cerevisiae mannosylphosphodolichol synthase. J. Biol. Chem. 2005;280:4174–4181. doi: 10.1074/jbc.M406962200. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee DK, Vendrell-Ramos M. Is asparagine-linked protein glycosylation and obligatory requirement for angiogenesis? Indian J. Biochem. Biophys. 1993;30:389–394. [PubMed] [Google Scholar]
- 37.Banerjee DK, DaSilva JJ, Bigio B. Mannosylphosphodolichol synthase activity is associated with a 32 kDa phosphoprotein. Bioscience Report. 1999;19:169–177. doi: 10.1023/a:1020221602373. [DOI] [PubMed] [Google Scholar]
- 38.Banerjee DK, Aponte E, DaSilva JJ. Low expression of lipid-linked oligosaccharide due to a functionally altered Dol-P-Man synthase reduces protein glycosylation in cAMP-dependent protein kinase deficient Chinese hamster ovary cells. Glycoconj. J. 2004;21:479–486. doi: 10.1007/s10719-004-5538-2. [DOI] [PubMed] [Google Scholar]
- 39.Martínez JA, Tavárez JJ, Oliveira CM, Banerjee DK. Potentiation of angiogenic switch in capillary endothelial cells by cAMP: a crosstalk between up-regulated LLO biosynthesis and the HSP-70 expression. Glycoconj. J. 2006;21:209–220. doi: 10.1007/s10719-006-7926-2. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee DK. Requirement of protein kinase type I for cAMP-mediated up-regulation of lipid-linked oligosaccharide for asparagine-linked protein glycosylation. Cell. Mol. Biol. (Noisy-le-grand) 2007;53:55–63. [PubMed] [Google Scholar]