Abstract
Affinity reagents recognizing biomarkers specifically are essential components of clinical diagnostics and target therapeutics. However, conventional methods for screening of these reagents often have drawbacks such as large reagent consumption, the labor-intensive or time-consuming procedures, and the involvement of bulky or expensive equipment. Alternatively, microfluidic platforms could potentially automate the screening process within a shorter period of time and reduce reagent and sample consumption dramatically. It has been demonstrated recently that a subpopulation of tumor cells known as cancer stem cells possess high drug resistance and proliferation potential and are regarded as the main cause of metastasis. Therefore, a peptide that recognizes cancer stem cells and differentiates them from other cancer cells will be extremely useful in early diagnosis and target therapy. This study utilized M13 phage display technology to identify peptides that bind, respectively, to colon cancer cells and colon cancer stem cells using an integrated microfluidic system. In addition to positive selection, a negative selection process was integrated on the chip to achieve the selection of peptides of high affinity and specificity. We successfully screened three peptides specific to colon cancer cells and colon cancer stem cells, namely, HOLC-1, HOLC-2, and COLC-1, respectively, and their specificity was measured by the capture rate between target, control, and other cell lines. The capture rates are 43.40 ± 7.23%, 45.16 ± 7.12%, and 49.79 ± 5.34% for colon cancer cells and colon cancer stem cells, respectively, showing a higher specificity on target cells than on control and other cell lines. The developed technique may be promising for early diagnosis of cancer cells and target therapeutics.
I. INTRODUCTION
Cancers, with a hallmark of uncontrolled cell division, are the leading cause of death in developed countries, where people could live long enough to suffer the non-communicable diseases. According to the World Health Organization statistics, cancers caused about 8.2 × 106 deaths or 14.6% of all human deaths.1 Colorectal cancer (CRC) is now the most commonly diagnosed cancer in the USA and its mortality is the second among all cancers. The five-year survival rate of patients with early-stage CRC can reach 90%, reflecting the importance of early diagnosis of the disease.2,3
However, two commonly used CRC biomarkers—carcinoembryonic antigen (CEA) and carbohydrate antigen (CA) 19-9, suffer from low specificity. Because both CEA and CA19-9 could be also detected with significant values in other kinds of cancers such as pancreatic cancer and lung cancer.4 Therefore, a more specific biomarker for CRC is essential for diagnosis of CRC. For CRC stage IV (metastasis) patients, although there are now six drugs used for treatment of metastatic CRC, there is no improvement of survival rates for metastatic CRC patients. They only have a five-year survival rate less than 10%.5 More research for more effective therapeutics of CRC may help in the increase of patients' survival rates.
Recently, a subpopulation of tumor cells that initiate the differentiation of cells to cancer cells is considered as cancer stem cells (CSCs). CSCs are a rare population that can reconstitute a new tumor with a similar composition and phenotype to the tumor of origin. They have the ability to initiate tumor cells. Also, the metastasis of cancer is caused by migratory CSCs. CSCs are defined as having three main properties including the capacity of self-renewing indefinitely, the ability to recreate the full repertoire of cancer cells of the parent tumor, and the expression of a distinctive set of surface biomarkers.6 Furthermore, CSCs also have a slow cell-cycle and exhibit multiple-drug resistances.
CSCs are commonly obtained using selection of, and enrichment for cells with prospectively identified cell surface markers.7 A set of surface markers including a cluster of differentiation 24 (CD24), CD44, CD133, CD166, and epithelial cell adhesion module (Ep-CAMhigh) were employed for identification and isolation of colorectal CSCs (CRSCs).1 Among them, CD24, CD44, and CD133 are the most commonly used. Unfortunately, these surface biomarkers are not specific enough, and it is also found that these markers are presented in a portion of normal stem cells, progenitor cells, or even some portions of normal tissue of colon. Furthermore, not only they are used to isolate CRSCs but also they are used for isolation of other cancer's CSCs.8 Therefore, biomarkers that may detect CRC specifically and distinguish CRC and CRSC are indeed of urgent need in clinical diagnosis and target therapeutics.
Among the methods of screening cell-specific affinity reagents, the phage display library has been demonstrated to be a promising tool.9 Phage display technology is based on the ability to express polypeptides as fusions to capsid of phages, which was first discover in 1985.10 Surface display of peptides could be achieved by inserting a random peptide encoding DNA sequence into the gene for capsid of phages and was first successfully displayed by using filamentous phages (M13 phages). M13 phages are approximately 0.9–1.0 μm in length and 5–10 nm in diameter. The displayed peptides generally target biologically relevant sites on the surface of target proteins.
Under a process called bio-panning or in-vitro selection, affinity reagents with a high specificity to target proteins could be screened by using the phage display library.11 However, the traditional method of peptide screening using the phage display library is relatively time-consuming and labor-intensive, which involves a 5–6 days process for one panning.12 Briefly, the target molecules or cells need to be first immobilized on the substrate for peptide selection and take at least one day. Second, the panning process is consisted of incubation between the phage display library and target cells (30 min to several hours), washing (several hours), and amplification (overnight). Third, polyethylene glycol (PEG) precipitation of the amplified phages is performed, which needs another day. Fourth, the titer of the amplified phages must be measured quantitatively such that the amplified phages reach a cut-off concentration of at least 109 pfu/ml for the next round of the panning process.
In addition to time-consuming and labor-intensive issues, traditional phage display library methods for affinity reagents screening have another serious issue that the screened peptide candidates may be up to several hundred after the entire screening process, indicating that a tedious validation process of these candidates is inevitable. Also, the traditional methods lack precise stringency control in each step of the panning process, especially in the washing step. In recent years, with the advancement of the microfluidic technology, several groups have performed the phage display screening assay on microfluidic systems. For instance, a micro-device for phage selection stringency study has been reported to increase the specificity of selected targets by varying magnetic and fluidic conditions.13 Alternatively, a simple micro-channel was used for adherent cells to isolate specific peptides with improved binding affinity and specificity.14 Furthermore, a microfluidic device with elastomeric and multiplexed features was reported to identify high-affinity peptides.15 However, the target binding, washing, and phage amplification have not yet been integrated into a single microfluidic system to perform the entire screening process by using the phage display screening assay. Recently, our group has demonstrated an integrated microfluidic system, which could perform the entire screening process for ovarian cancer cell-specific peptides automatically.16 However, this microfluidic system could be only applied for a single bio-panning process. Furthermore, only one negative selection was included, and it could not provide the washing stringency during the screening process. We therefore aim to propose a new strategy in this work for control of stringency in the washing step such that the peptide candidates with weak affinities could be washed away. Moreover, we would like to improve the specificity of screened peptides to target cells during the selection experiment. Therefore, a positive selection process for the target cells and a negative selection process for the control cells are incorporated at each step. Finally, six bio-panning rounds would be automated in this new integrated microfluidic system.
In order to tackle the above-mentioned issues, a magnetic-bead based microfluidic platform has been proposed in this work for the screening of peptides using the phage display library. This combination has several advantages than the traditional methods. First, the time for incubating the phage display library and the target cells (or the control cells) is greatly shortened, since an efficient micromixer is used. Therefore, the total time of the entire on-chip panning is reduced to only 6 h. Second, the washing stringency (for instance, shear force) can be precisely controlled such that the final number of peptide candidates could be greatly reduced. Third, the phage amplification chambers are implemented on the chip such that a whole phage display bio-panning can be performed on this microfluidic platform. More importantly, the traditional method suffers from a limitation that the amount of cells needed for the screening experiment is relatively large (105–107 cell/run). When the target is harvested from organs or tissues, it is nearly impossible to get enough cells for the screening experiments.17 With the microfluidic approach, the amount of the cells could be greatly reduced. Furthermore, the entire screening process could be automated with less human intervention on an integrated microfluidic system. In summary, both the new features of micro-devices and simplification of a more robust experimental protocol provide the improved results and successful on-chip phage display assay in this study.
II. MATERIALS AND METHODS
A. Preparation of CRCs and CRSCs
All cell culture medium and reagents used in this study were purchased from Life Technologies, USA unless otherwise indicated. The CRC cell line (HCT-8) was cultured in Roswell Park Memorial Institute (RPMI) 1640 medium. The CRSC was sorted from HCT-8 by using a flow cytometric approach based on the cells presenting CD44 surface protein. CRSCs were suspension-cultured in serum-free Dulbecco's Modified Eagle Medium (DMEM) supplemented with nutrient mixture F-12, GlutaMax-l, 1% of nutrient mixture N2, 2% of nutrient mixture B27, 20 ng/ml human fibroblast growth factor 2 (Sigma-Aldrich, MO), and 50 ng/ml Epidermal growth factor (Sigma-Aldrich, MO) was used.14 Collection of cells was performed prior to the screening process. In this study, an external magnetic field was used for collection of cells. Epithelial Enrich magnetic beads (Epi-Enrich, Dynabeads®, diameter = 4.5 μm, Invitrogen™, USA) surface-coated with a monoclonal antibody towards the human EpCAM antibody was used for capturing the CRCs and CRSCs when the cell-bead complexes were collected with an external magnet. Cell viability was measured before and after the flow binding assay by trypan blue exclusion assay, and the cell viability ranges from 85% to 99%.
B. Experimental process of on-chip phage display
A screening process using the phage display library was performed on an integrated microfluidic chip for screening cell-specific peptides. Phage display library is based on a combinatorial library of random dodecapeptides fused to a minor coat protein (g3p) of M13 phage. The displayed peptide, in this case, 12-mer is expressed at the N-terminus of g3p. The phage display peptide library and E. coli K12 ER2738 were from Ph.D.™-12 Phage Display Peptide Library Kit (New England Biolabs, USA). Note that negative and positive selection processes were performed sequentially to screen peptides with high affinity and specificity for CRCs and CRSCs. First, 10 μl of M13 phage display peptide library (∼1011 pfu/ml, prepared in tris buffer saline (TBS)) was pre-mixed with 200-μl control cells (HCT-8 or CRSC/105 cells/ml, prepared in phosphate-buffered saline buffer (PBS, Merck Ltd., Darmstadt, Germany)), which were pre-isolated by Epithelial Enrich magnetic beads (4 108/ml). They were then incubated for 10 min by activating a micromixer. A magnet was then used to collect captured control cells (HCT-8 or CRSC)/peptide complexes as a negative selection process. The supernatant containing the phage after the negative selection which was then transported to a positive selection chamber to perform the subsequent positive selection. At the same time, target cells (CRSC or HCT-8) pre-isolated by Epithelial Enrich magnetic beads were added to the supernatant from the previous step for 10-min incubation. After the positive selection, a magnet was used to collect target cells (CRSC or HCT-8)/peptide complexes. Two hundred microliters PBS was then used to wash away the unbounded or weak-binding phages for 5 times. Note that the shear force was increased each round to increase the washing stringency. The applied gauge pressures of the micropump for washing were −13.5 kPa, −27 kPa, −40 kPa, −53.5 kPa, −65 kPa, and −75 kPa, respectively, to provide the required flow velocity and shear force. Finally, the magnetically collected target cells/bead/phage complexes were transported to a phage amplification chamber, which was pre-loaded with Escherichia coli ER2738 for amplification of screened phages.18 Note that the working process about the optimization of phage amplification time is shown in supplementary material.29
Experimental procedure performed on the integrated microfluidic chip for screening of cancer cell-specific peptides is schematically shown in Figure 1. First, the control cells (200 μl, 5 × 104/ml) (for negative selection) pre-isolated with magnetic beads (10 μl, 4 × 105/ml) and the phage display library (10 μl, 1011 pfu/ml) were loaded into the inlet chamber at a total volume of 210 μl. At the same time, the target cells (200 μl, 5 × 104/ml) (for positive selection) pre-isolated with magnetic beads (10 μl, 4 × 105/ml) and E. coli (500 μl, 106 cells/ml) were loaded into the target cell inlet and the amplification chamber, respectively. Second, the control cells coated with magnetic beads were incubated with the phage display library for 10 min by activating the micromixer. Then, a magnet was used to collect the phages captured by the control cells. Third, the supernatant was then transported to the positive selection chamber by activating the micropump to perform the subsequent positive selection by incubating the supernatant and the target cells for 10 min. Later, a magnet was again used to collect the target-cell captured phages and then was washed for 5 times with 200 μl 1 × PBS. Note that the applied gauge pressure for activating the pumping varies with each different rounds of panning processes. Finally, the target cells/phage/beads complexes were transported to the phage amplification chamber (pre-loaded with E. coli) for 5-h amplification. Finally, the temperature was raised to 65 °C for 15 min to kill E. coli. Note that all the steps of incubation, transportation, and washing could be automated in the proposed microfluidic system. However, the loading of the buffer, phage library (phages for next panning round), and cell-bead complexes were performed by manual operation. Furthermore, the experimental process of polymerase chain reaction (PCR) and the total time of the entire on-chip panning process could be found in supplementary material.29 After 6 round of on-chip screening, the sixth round PCR products were used for TA cloning. The DNA was cloned into pCR™2.1-TOPO® (Invitrogen, USA), total 33 colonies for HCT-8 specific peptide and 55 colonies for CRSC specific peptides were selected and checked by using plasmid extraction mini kit (FavorPrepTM Plasmid Extraction Kit/Favorgen, Taiwan) and PCR to ensure that each individual colony has DNA segment of interest.16,24
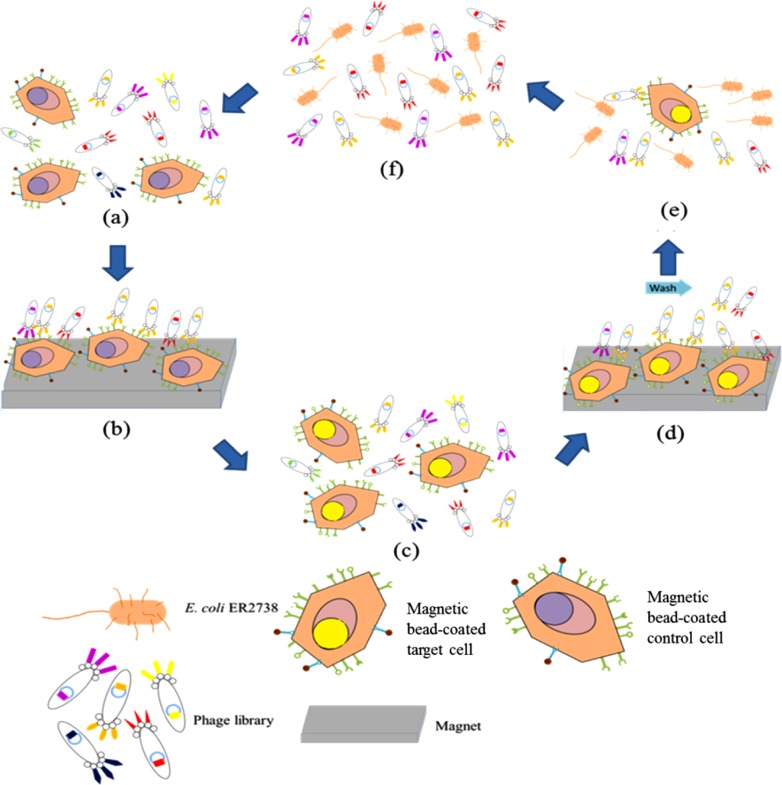
FIG. 1.
Schematic illustration of the experimental procedure performed on the integrated microfluidic chip for screening of cancer cell-specific peptides, including positive selection, negative selection, and phage amplification steps. (a) Negative selection using magnetic bead-coated control cells and a phage display library. (b) Removal of unwanted phages by applying a magnet and transporting the phages of interest to the positive selection chamber. (c) Positive selection using magnetic bead-coated target cells and the phage display library from the step (b). (d) Collection of phages of interest by using a magnet and the washing of un-bound or weakly bound phages. (e) Amplification of phage binding on target cells by propagating on E. coli.
C. Chip design and fabrication
A microfluidic chip was designed and fabricated to perform the above-mentioned process. It was made of two polydimethylsiloxane (PDMS, Sylgard 184A and Sylgard 184B, Sil-More, USA) layers—a thick PDMS layer was used for air control and a thin PDMS layer for liquid channel, on a glass substrate (G-Tech Optoelectronics Corp., Taiwan). The thick PDMS layer contained a fluid transport module, and the thin PDMS layer contained fluid channel. Therefore, this microfluidic chip was developed for transporting and incubating of samples/reagents in the screening process.
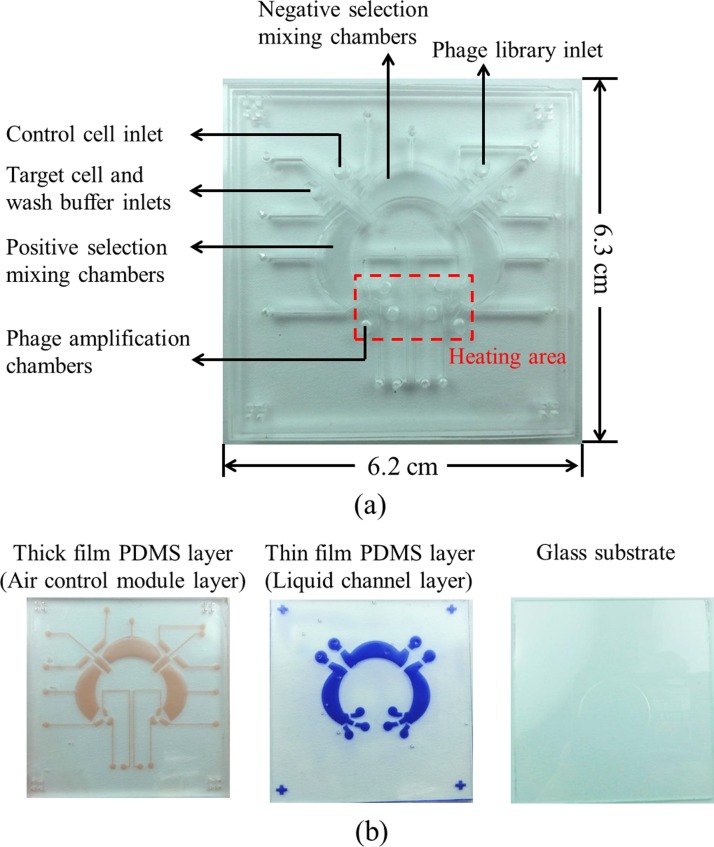
Furthermore, Figure 2(a) was a photograph of the integrated microfluidic chip and a the top-view of the chip, which contains closed-chamber micromixers/micropumps,19 normally closed microvalves,20 reservoirs, and a temperature control module. The dimensions of the chip were measured to be 6.2 cm × 6.3 cm. Figure 2(b) showed separated layers of the chip, containing an air control module layer, a fluid channel layer, and a glass substrate. The reason for using the closed-chamber micropumps/micromixers and normally closed microvalve was to prevent cross contamination of samples and reagents. During the screening step, avoiding environmental bacteria is important due to the fact that phages may infect the contaminated bacteria. As a result, we may lose potential phage candidates. The normally closed valve only opens when needed. Thus, no contamination of target and control cells, or E. coli during the positive and negative selections could occur. In addition, a temperature control module using a thermoelectric cooler for controlling the temperature of the phage amplification process and the removal of E. coli residue process was adopted.
FIG. 2.
(a) A photograph of the integrated microfluidic chip for phage display bio-panning process, which contains closed-chamber micropumps/micromixers, normally closed microvalves, chambers, and a temperature control module underneath six phage amplification chambers. Note that the heating area is used for E. coli culture in 37 °C. (b) The layers marked in red and blue represent the air control module layer and the fluid channel layer, respectively. Separated layers of the integrated microfluidic chip are composed of an air control module layer, a fluid channel layer, and a glass substrate.
The fabrication of the microfluidic chip was performed by using a standard PDMS casting process.21 The microfabrication of the master molds on polymethylmethacrylate (PMMA) plates was performed using a computer-numerical-control machining process with a 0.5-mm drill bit.22 The rotational speed and feed rate of the drill bit were set to be 26 000 rpm and 15 mm/min, respectively, followed by a PDMS replication process to obtain the inverse microstructures of the microchannels, the liquid microchambers, and the air chambers. Briefly, the silicone elastomer and the elastomer curing agent of PDMS were first mixed in a mass ratio of 10:1 and then poured on the PMMA master molds. Next, the mixture of PDMS underwent vacuuming to remove the bubbles formed during mixing and therefore air pockets in the replicas would be prevented. The PDMS on the master molds was then cured at 80 °C in an oven for 1 h. Finally, the PDMS with microstructures was mechanically peeled from the master molds. To combine two layers of PDMS and glass, an oxygen plasma treatment was performed to bind them together.21 Note that the process for measurement of the pumping rate and the specific dimensions of the integrated microfluidic chip could be found in supplementary material.29 Furthermore, it is important to note that every microfluidic chip was autoclaved before the on-chip phage display process. The screening of cell-specific peptides was performed based on the processes shown in Figure 1. Note that the whole process was repeated 6 times, and each time the stringency (shear force) was increased gradually to obtain peptides with higher affinity and specificity.
D. Measurement of shear force
In order to improve the affinity and specificity of the screened peptides, the stringency of the washing step, i.e., shear force, was increased gradually in each panning process. In this study, the shear force was controlled by applying different gauge pressures to cause different flow velocities of the micropump. The flow velocity was measured by using plastic beads and a high-speed charge-coupled-device camera (Mikrotron/MC1311, Germany). Note that the flow measurement was made at the mid-height of the chamber. The flow velocity was then calculated by an imaging-acquisition method. The shear force was further calculated using the equation as follows:
where τ is shear stress, μ is dynamic viscosity (μPBS, 20 °C = 1.05 × 10−3 N s/m2), u is the flow velocity, y is the mid-height of the chamber, and A is the area of the cross-section (A = 0.05712 cm2).
E. Specificity test and dissociation constant (Kd) measurement
The specificity of the screened peptide was tested by measuring the capture rate of the cell-specific peptide on other cell lines. The screened peptide was first conjugated on carboxylic functionalized magnetic beads. Then, the beads were incubated with different cell lines (A549, MCF7, HepG2, BxPC3, and NIH3T3) with a cell number of 2 × 105. Afterwards, a magnetic rack was used to pellet the cells and 1 × PBS (200 μl) was used to wash away the unbounded cells and debris for 5 times. Finally, a hemocytometer was used to calculate the cells captured by the specific peptide conjugated beads. The capture rate was calculated as the captured cell number divided by the total cell number added to perform the cell capture experiments.
The Kd value of the peptide was measured using flow cytometry (C6 Cytometer—BD Biosciences, BD Accuri™, USA).23,24 The cell number of the target cells was fixed to be 2 × 105 and the concentration of the specific peptide used varied with 2-fold serial dilutions from 1 μM to 10 nM. Note that the binding site of the specific peptide was assumed to be only one. Therefore, the equilibrium Kd value of the fluorescent ligand was obtained by fitting a plot of the specific binding intensity versus (Y) the peptide concentration (X) to the equation Y = BmaxX/(Kd + X) using GraphPad Prism 6.0 (La Jolla, CA, USA), where Bmax represents the maximum specific binding.
III. RESULTS AND DISCUSSION
A. Chip characterization
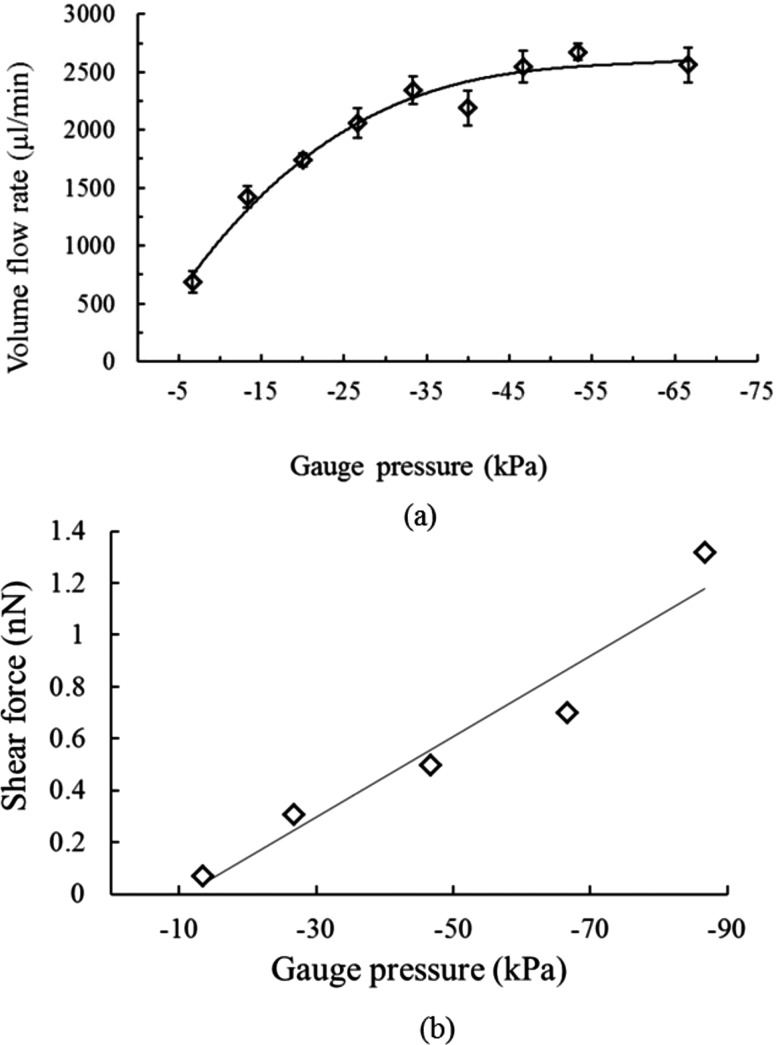
The performance of the micropump was first characterized, and the relationship between the volume flow rate and the applied gauge pressure was shown in Figure 3(a). Note that there was no sag of the PDMS membranes observed during the pumping process. The experimental results showed that the volume flow rate increased with the applied gauge pressure. At a gauge pressure of −48.5 kPa, the volume flow rate has a maximum value of 2674 μl/min, which is 16 times superior to the one in our previous work.16 Note that we have a capability to gradually increase the shear force during the screening runs by varying the pumping rate with the new designed micropump. The pumping rate of the micropump was observed to be saturated with a gauge pressure of −48.5 kPa and higher. This may be caused by the fact that the deformed PDMS membrane has occupied all space for membrane deformation at this pressure. Therefore, even with a higher gauge pressure, the pumping rate did not increase.
FIG. 3.
(a) The relationship between the volume flow rate and the applied gauge pressure of the micropump when operated at different gauge pressures. Note that three consecutive experiments were repeated. The volume flow rate was measured to be 2674 μl/min when actuated at a gauge pressure of −33.5 kPa. (b) The relationship between the shear force and the applied gauge pressure. Note that three consecutive experiments were repeated with a deviation less than 1%. Shear force was increased in each panning experiment. The shear force of each round was 0.07 nN, 0.14 nN, 0.21 nN, 0.28 nN, 0.35 nN, and 0.42 nN, respectively. Note that the flow measurement was made at the mid-height of the channel.
Note that the micropump could also work as a micromixer.25 A mixing frequency of 0.2 Hz was used to perform the incubation process between the cell-bead complexes and the phage display library because a gentle mixing during incubation was considered to allow more phages binding on target cells and having a greater opportunity to obtain potential candidates.
In order to identify peptides with higher affinity and specificity, shear forces of the washing process at each panning round were increased to provide the required selection stringency. The shear force applied to the micropump was carefully adjusted not to higher than the regular protein-protein interacting force typically in the range of several nanonewtons.26 Therefore, the applied shear force of the microfluidic chip was set within a range of 0−2.1 nN. As shown in Figure 3(b), the applied gauge pressures of each panning round were −13.5 kPa, −27 kPa, −40 kPa, −53.5 kPa, −65 kPa, and −75 kPa, respectively, resulting in the maximum shear force to be around 1.32 nN, which is within the range of specific protein-protein interactions. Note that the shear force was fine-tuned to be 0.07 nN, 0.14 nN, 0.21 nN, 0.28 nN, 0.35 nN, and 0.42 nN, respectively, during the washing step of each panning round.
As mentioned above, the shear force was adjusted to control the washing stringency during the washing process. However, in order to avoid influence caused by the shear force during other steps, the incubation step, a low gauge pressure was chosen during sample/reagent transportation. Although the micropump has a maximum pumping rate at −48.5 kPa, a gauge pressure of −13.5 kPa was used for sample/reagent transportation. Pressure lower than this value was not chosen because it took too long to complete the sample and reagent transportation. Note that the variation of the applied gauge pressure and the resulting shear force was lower than 1%. With this approach, the increase of the shear force at the washing step of each panning round could wash away weak-binding phages, allowing the identification of peptides with better affinity and specificity since only phages with relatively higher affinity could be retained and amplified subsequently. The obtained candidates should be dramatically less than the ones from to traditional methods, and expected to have a higher Kd value.
B. Optimization of the phage amplification time
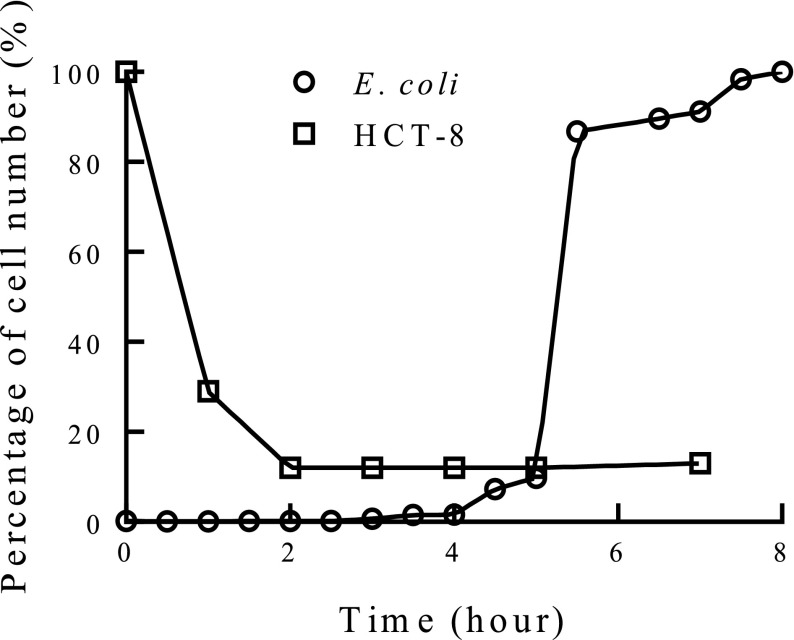
Figure 4 showed the HCT-8 cell survival rate and the E. coli growth curve in Lysogeny broth (LB broth). After 3 h, the survival rates of HCT-8 cells reached a saturated value and nearly did not change. Therefore, it was assumed that almost all phages were released from the HCT-8 cells after 3 h of incubation. Furthermore, the E. coli growth is also an important factor for amplification of captured phages. The mid-log phase of the E. coli growth curve is expected to have the fastest phage propagation in E. coli. Hence, the cross-over point of the cell survival rate and the E. coli growth curve in LB broth was chosen to be the optimal time for phage amplification, which was 5 h in this study. The survival rate of CRSC in LB broth is similar to HCT-8, and the detailed data could be found in supplementary material.29
FIG. 4.
Optimization of the amplification time of phages. The crossover point of E. coli ER2738 growth curve and the survival rate of HCT-8 cells in LB broth was chosen to be the optimal time for phage amplification using E. coli ER2738, and the optimal amplification time of phages was measured to be 5 h. Note that three consecutive experiments were performed and the error was less than 2%. (100% E. coli represents the highest cell number after 8-h culture. Percentage of E. coli were defined as the cell number at a specific time divided by the highest cell number.)
C. Screening of cell-specific peptides
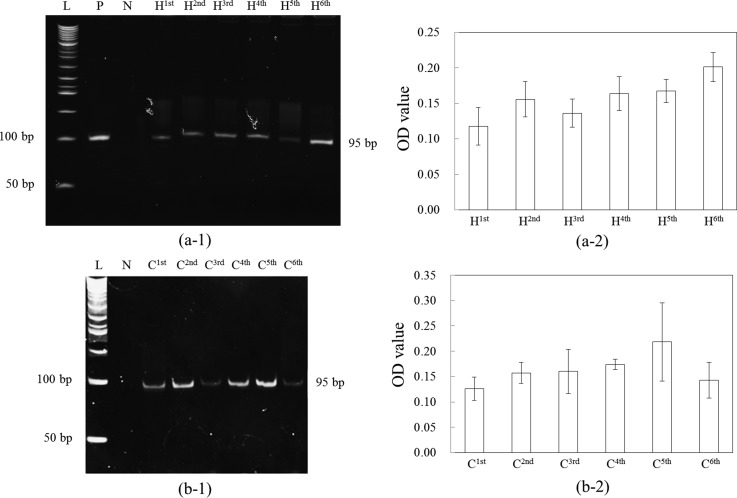
In the screening process of the phage display library, negative and positive selections were sequentially performed to obtain cell-specific binding peptides with high affinity and specificity. Negative selection using magnetic beads to capture M13 phage display peptides that were not of interest was first performed such that peptides binding on non-target cells (control cells) could be removed. Then, the supernatant was collected to perform the subsequent positive selection. As mentioned previously, the target cells surfaced-coated with magnetic beads were incubated with the supernatant to perform the positive selection. Those collected cells captured with M13 phage display peptides of interest were then transported to the phage amplification chamber containing E. coli for phage amplification. These steps were automatically performed for six times to get highly specific and high-affinity phages. PCR and polyacrylamide gel electrophoresis (PAGE) were further used to confirm whether each round of selection is successful. Figure 5 shows the results for six rounds of screening using (a-1) HCT-8 as target cells and CRSC as control cells and (b-1)28 CRSC as target cells and HCT-8 as control cells. Note that phage library was used as a positive control and double-distilled water (ddH2O) was used as a negative control for PCR confirmation. The figure shows that successful screening of phage display library was achieved by using the microfluidic chip. Figures (a-2) and (b-2) showed that the optical density (OD) values analysis with several PAGE results (n = 5), which indicated that the amount of PCR products was observed to increase and become saturated from H1st to H6th and from C1st to C6th. It also indicates that the increase of stringency, i.e., shear force, did not wash away all the phage bound on the target cells. The E. coli infection process could amplify phages even though the shear force was increased. In this work, two peptides (HOLC-1 and HOLC-2) specific to HCT-8 and one peptide specific to CRSC (COLC-1) were further used for measurements of Kd and cell capture rates.
FIG. 5.
Polyacrylamide gel electrophoresis analysis of screened peptides used (a-1) HCT-8 (H) as target cells and CRSC (C) as control cells, and (a-2) presented the OD values analysis with several PAGE results (n = 5), indicating that the amount of PCR product was observed to increase and become saturated from H1st to H6th; (b-1) CRSC as target cells and HCT-8 as control cells,28 and (b-2) presented the OD values analysis with several PAGE results (n = 5), indicating that the amount of PCR product was observed to increase and become saturated from C1st to C6th. P: positive control using phage library. N: negative control using ddH2O. L: 50-bp ladders. Lanes H1st to H6th and C1st to C6th represent the first to the sixth round of the selection, respectively. Reprinted with permission from Che et al., in 2014 9th IEEE International Conference on Proceedings of Nano/Micro Engineered and Molecular Systems (NEMS), Hawaii, USA, 13–16 April, 2014, pp. 440–443. Copyright 2014 IEEE.
The PCR product after 6 round of screening was used for TA cloning and DNA sequencing. The DNA sequences encoding peptide displayed from M13 phage screened using HCT-8 as target were screened into two peptide candidates, HOLC-1 and HOLC-2, as shown in Table I, and the ones using CRSC as target were screened into one peptide candidate, COLC-1, as shown in Table I. The amino acid sequences of HOLC-1 (N′-AELAKLPLF-C′), HOLC-2 (N′-GAISFDHVHP-C′), and COLC-1 (N′-MNAKWPIENLMN-C′) were translated from the screened DNA sequences.
TABLE I.
The DNA sequence of the phage displayed specific peptide and amino acid sequences of HCT-8 specific peptides: HOLC-1, HOLC-2, and CRSC specific peptide: COLC-1.
| Specific peptide | Sequence 5′→3′ | Amino acid sequence |
|---|---|---|
| HOLC-1 | GCGGAGCTTGCTAAGTTGCCGCTGTTTTAGATGCGC | AELAKLPLF |
| HOLC-2 | GGTGCTATTTCTTTTGATCATGTGCATCCTTAGAGG | GAISFDHVHP |
| COLC-1 | ATGAATGCTAAGTGGCCGATTGAGAATTTGATGAAT | MNAKWPIENLMN |
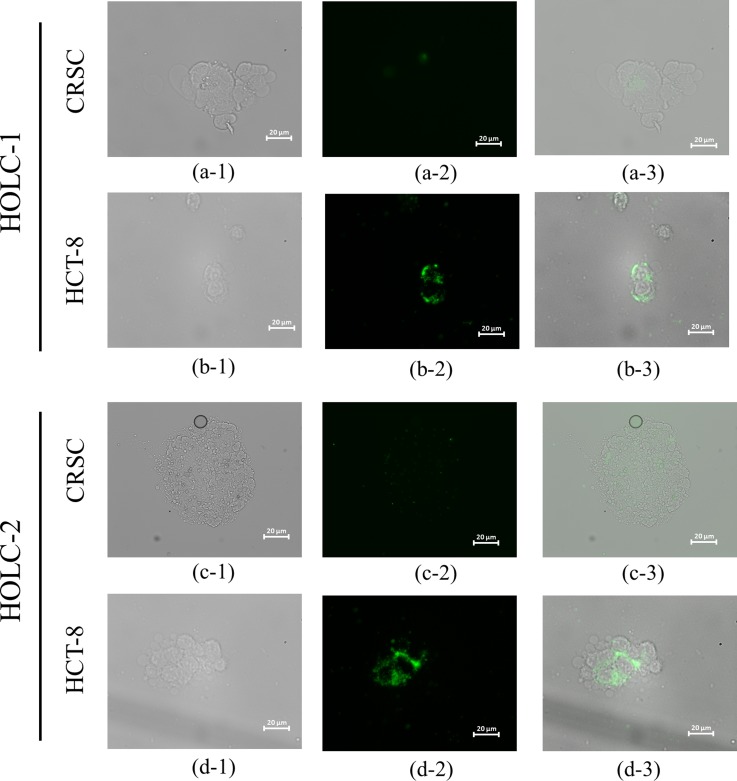
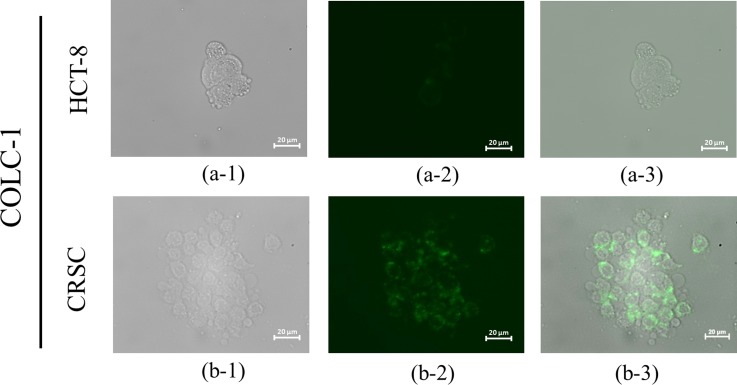
Furthermore, the screened peptides were synthesized and used for cell capturing tests. Figure 6 showed the microscopic images of fluorescent-labeled (FAM/5/6-carboxyfluorescein succinimidyl ester labeled) HOLC-1 and HOLC-2 peptides binding on the target cells, HCT-8, but showed weak to no green fluorescence signals toward the control cells, CRSC, indicating that the selected peptides could only recognize the target cells. The green florescence signal represents HOLC-1 and HOLC-2 peptides, which shows significant fluorescent signals due to the fact that these peptides could specifically bind to HCT-8. Experimental results clearly indicate that both HOLC-1 and HOLC-2 peptides can bind to the target cells, especially at the cell junction. Similarly, Figure 7 shows the microscopic images of fluorescence dye FAM-labeled CLOC-1 peptide binding on the target cells, CRSC. Experimental results also show that CLOC-1 peptide can bind to the target cells.
FIG. 6.
Microscopic images of FAM-labeled HOLC-1 and HOLC-2 peptides bound to CRSCs and HCT-8 cells. (a-1) and (c-1) Bright-field images of CRSCs. (a-2) and (c-2) Fluorescent microscope images of HOLC-1- and HOLC-2-bound CRSCs, respectively. (a-3) and (c-3) Merged images of HOLC-1- and HOLC-2-bound CRSCs, respectively. (b-1) and (d-1) Bright-field images of HCT-8 cells. (b-2) and (d-2) Fluorescent microscope images of HOLC-1- and HOLC-2-bound HCT cells, respectively. (b-3) and (d-3) Merged images of HOLC-1- and HOLC-2-bound HCT-8 cells.
FIG. 7.
Microscopic images of FAM-labeled CLOC-1 peptide bound to CRSCs and HCT-8 cells. (a-1) Bright-field images of HCT-8 cells. (a-2) Fluorescent images of CLOC-1-bound HCT-8 cells. (a-3) Merged images of CLOC-1-bound HCT-8 cells. (b-1) Bright-field images of CRSC cells. (b-2) Fluorescent images of CLOC-1-bound CRSC cells. (b-3) Merged images of CLOC-1-bound CRSC cells.
D. Specificity test
The specificity of the screened peptides was further examined by measuring the cell capture rate of magnetic beads surface-coated with the peptide for their target cells and other cell lines. Table II shows the specificity test results of the two HCT-8 specific peptides (HOLC-1 and HOLC-2) and CRSC specific peptide (COLC-1), respectively. The cell capture rate using a traditionally used antibody (EpCAM) on Epi-Enrich magnetic beads was also shown for comparison. The experimental results show that the capture rates of HOLC-1 and HOLC-2 have a comparable value to EpCAM antibody in terms of capture rates for their target cells, while they exhibit a better specificity towards other cell lines. Similarly, CLOC-1 shows a high capture rate on its target cell (CRSC) and a low capture rate toward the control cells (HCT-8) with capture rates of CRSCs and HCT-8 as 49.79% and 9.42%, respectively.
TABLE II.
Comparison of the capture rates of cells of different origins by three peptides and the Epithelial Enrich magnetic beads.
| Species | Organ source | Cell type | HOLC-1 | HOLC-2 | COLC-1 | Epi-Enrich |
|---|---|---|---|---|---|---|
| Capture rate (%) | ||||||
| Human | Colon | HCT-8 | 43.40 ± 7.23 | 45.16 ± 7.12 | 9.42 ± 2.67 | 34.51 ± 6.60 |
| Colon | CRSC | 22.97 ± 4.29 | 20.85 ± 0.61 | 49.79 ± 5.34 | 68.14 ± 5.11 | |
| Liver | HepG2 | 23.60 ± 3.50 | 23.02 ± 2.77 | 16.77 ± 6.49 | 6.13 ± 1.51 | |
| Pancreas | BxPC3 | 3.41 ± 1.14 | 3.13 ± 0.62 | 10.68 ± 5.08 | 4.44 ± 1.02 | |
| Lung | A549 | 28.51 ± 7.09 | 34.29 ± 5.22 | 17.87 ± 6.18 | 74.04 ± 4.43 | |
| Breast | MCF7 | 12.84 ± 7.67 | 20.45 ± 4.04 | 3.58 ± 1.18 | 66.82 ± 6.81 | |
| Mouse | Fibroblast | NIH3T3 | 6.46 ± 1.90 | 6.87 ± 2.83 | 19.53 ± 7.27 | 9.43 ± 1.22 |
As shown in Figures 6, 7, and Table II, this approach of increasing the shear force to reduce non-specific binding and weak-binding peptides might greatly improve the affinity and specificity of the screened peptides, since only highly specific and high-affinity peptides could remain after the selection experiment. Experimental data showed that only three and four candidates were screened after 6 rounds of panning for HCT-8 and CRSC, respectively. The specificity of HOLC-1 and HOLC-2 shows that relatively high affinity to colon cancer cells with capture rates of 43.40% and 45.16%, respectively. Note that the EpCAM antibody only has a capture rate of 34.51% after five washing. They also show high capture rates on human cells while exhibiting a low capture rate on mouse fibroblast cells. Moreover, these two screened peptides exhibited relatively low capture rates toward other cells lines, including the control cell (CRSC). Furthermore, the EpCAM antibody has a relatively high capture rate of 68.14% for CRSCs after five washing. However, EpCAM still have relatively high capture rates toward A549 and MCF7, indicating that this antibody does not exhibit high specificity for other cancer cells. Conversely, COLC-1 has relatively low capture rates on all cells lines (3.58%–19.53%), indicating that this screened peptide may be a better candidate for screening of CRSC.
E. Dissociation constants of screened peptides
The dissociation constant (Kd) values of the screened peptides were further measured. Before Kd measurement, we first tested these specific peptide candidates with various concentrations (10 nM, 100 nM, 1 μM, 10 μM, 100 μM, 1 mM, and 10 mM) by using their target cells and control cells to find the proper concentration range of peptides for Kd measurement, and the detailed data could be found in supplementary material.29 The Kd values of HOLC-1 and HOLC-2 were measured to be 60.6 ± 23.1 nM and 219.8 ± 11.0 nM, respectively. Comparing to the antibody used presently (EpCAM) and other screened aptamers with a Kd value at a range of 10−1–103 nM,27 the affinities of the HOLC-1 and HOLC-2 were comparable to these values. The Kd value of CLOC-1 was measured to be 507.9 ± 78.7 nM.
IV. CONCLUSIONS
In this study, we have successfully demonstrated an integrated microfluidic system for screening of peptides specific to colon cancer cells and colon cancer stem cells. The entire process including incubation, partition, and E. coli amplification could be automated with less human intervention. Furthermore, precise control of the shear force during each washing process was achieved to wash away non-specific or weak binding phages such that high-affinity and highly specific peptides could be screened. A negative selection process was further adopted to improve the cell-specificity of the screened peptide. Compared to the traditional method, the time spent on screening has been greatly shortened from 5–6 days (for each round) to 6 h. The developed system also greatly reduced the number of potential peptide candidates, thus alleviating the tedious work for subsequent peptide verification. Only three and four peptide candidates were screened after 6 round of panning for HCT-8 and CRSC, respectively. This integrated microfluidic system successfully screened cell-specific peptides, HOLC-1 and HOLC-2, to recognize HCT-8 and COLC-1 to recognize CRSC. HOLC-1 and HOLC-2 showed relatively high affinity and specificity on HCT-8 (43.40% and 45.16%, respectively), while COLC-1 showed a capture rate of 49.79% on CRSC. The Kd values of HOLC-1 and HOLC-2 were measured to be 60.6 ± 23.1 nM and 219.8 ± 11.0 nM, respectively, which is comparable to the existing antibody. The Kd value of CLOC-1 was measured to be 507.9 ± 78.7 nM. It may provide a useful platform for rapid screening of specific ligands for clinical diagnosis. Furthermore, the identified peptides may be applied in target therapy for colon cancer treatment. Note that the working process about the optimization of phage amplification time could be found in supplementary material.29
ACKNOWLEDGMENTS
The authors would like to thank the National Science Council of Taiwan for financial support (NSC 102-2221-E-007-054-MY3). Partial financial support from National Health Research Institutes, the “Innovative Research Grant” (IRG, NHRI-EX104-10428EI), the “Towards A World-class University Project,” and Nanomedicine Program, Academia Sinica is also greatly appreciated.
References
- 1. Vaiopoulos A. G., Kostakis I. D., Koutsilieris M., and Papavassiliou A. G., Stem Cells 30, 363 (2012). 10.1002/stem.1031 [DOI] [PubMed] [Google Scholar]
- 2. Fleming S. T., Mackley H. B., Camacho F., Seiber E. E., Gusani N. J., Matthews S. A., Liao J., Yang T. C., Hwang W., and Yao N., J. Rural Health 30, 27 (2014). 10.1111/jrh.12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Q., Cai G., Li D., Wang Y., Zhuo C., and Cai S., PLoS One 9, e93756 (2014). 10.1371/journal.pone.0093756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ven der Schouwm Y. T., Verbeek A. L. M., Wobbes Th., Segers M. F. G., and Thomas C. M. C., Br. J. Cancer 66, 148 (1992). 10.1038/bjc.1992.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu J., Ye X., Fan F., Xia L., Bhattacharya R., Bellister S., Tozzi F., Sceusi E., Zhou Y., Tachibana I., Maru D. M., Hawke D. H., Rak J., Mani S. A., Zweidler-McKay P., and Ellis L. M., Cancer Cell 23, 171 (2013). 10.1016/j.ccr.2012.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Puglisi M. A., Tesori V., Lattanzi W., Gasbarrini G. B., and Gasbarrini A., WJG 19, 2997 (2013). 10.3748/wjg.v19.i20.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cammareri P., Lombardo Y., Francipane M. G., Bonventre S., Todaro M., and Stassi G., Methods Cell Biol. 86, 311 (2008). 10.1016/S0091-679X(08)00014-9 [DOI] [PubMed] [Google Scholar]
- 8. Mukhopadhyay N. K., Gilchrist D., Gordon G. J., Chen C. J., Bueno R., Lu M. L., Salgia R., Sugarbaker D. J., and Jaklitsch M. T., Ann. Thorac. Surg. 78, 450 (2004). 10.1016/j.athoracsur.2004.01.042 [DOI] [PubMed] [Google Scholar]
- 9. Yang L. and Nolan J. P., Cytometry, Part A 71A, 625 (2007). 10.1002/cyto.a.20417 [DOI] [PubMed] [Google Scholar]
- 10. Smith G. P., Science 228, 1315 (1985). 10.1126/science.4001944 [DOI] [PubMed] [Google Scholar]
- 11. Haas H. J. and Smith G. P., Biotechnique 15, 422 (1993). [PubMed] [Google Scholar]
- 12. Hoen P. A. T., Jirka S. M., Ten Broeke B. R., Schultes E. A., Aguilera B., Pang K. H., Heemskerk H., Aartsma-Rus A., van Ommen G. J., and den Dunnen J. T., Anal. Biochem. 421, 622 (2012). 10.1016/j.ab.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y., Adams J. D., Turner K., Cochran F. V., Gambhir S. S., and Soh H. T., Lab Chip 9, 1033 (2009). 10.1039/b820985e [DOI] [PubMed] [Google Scholar]
- 14. Wang J., Liu Y., Teesalu T., Sugahara K. N., Kotamrajua V. R., Adams J. D., Ferguson B. S., Gong Q., Oh S. S., Csordas A. T., Cho M., Ruoslahti E., Xiao Y., and Soh H. T., PNAS 108, 6909 (2011). 10.1073/pnas.1014753108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cung K., Slater R. L., Cui Y., Jones S. E., Ahmad H., Naik R. R., and McAlpine M. C., Lab Chip 12, 562 (2012). 10.1039/C2LC21129G [DOI] [PubMed] [Google Scholar]
- 16. Wang C. H., Weng C. H., Che Y. J., Wang K., and Lee G. B., Theranostics 5, 431 (2015). 10.7150/thno.10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bitarte N., Bandres E., Boni V., Zarate R., Rodriguez J., Gonzalez-Huarriz M., Lopez I., Javier Sola J., Alonso M. M., Fortes P., and Garcia-Foncillas J., Stem Cells 29, 1661 (2011). 10.1002/stem.741 [DOI] [PubMed] [Google Scholar]
- 18. Breed R. S. and Dotterrer W. D., J. Bacteriol. 1, 321 (1916). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weng C. H., Huang T. B., Huang C. C., Yeh C. S., Lei H. Y., and Lee G. B., Biomed. Microdevices 13, 585 (2011). 10.1007/s10544-011-9529-3 [DOI] [PubMed] [Google Scholar]
- 20. Yang S. Y., Lien K. Y., Huang K. J., Lei H. Y., and Lee G. B., Biosens. Bioelectron. 24, 855 (2008). 10.1016/j.bios.2008.07.019 [DOI] [PubMed] [Google Scholar]
- 21. Mcdonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H. K., Schueller O. J. A., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- 22. Bhattacharya S., Datta A., Berg J. M., and Gangopadhyay S., J. Microelectromech. Syst. 14, 590 (2005). 10.1109/JMEMS.2005.844746 [DOI] [Google Scholar]
- 23. Sefah K., Shangguan D., Xiong X. L., O'Donoghue M. B., and Tan W. H., Nat. Protoc. 5, 1169 (2010). 10.1038/nprot.2010.66 [DOI] [PubMed] [Google Scholar]
- 24. Weng C. H., Hsieh I. S., Hung L. Y., Lin H. I., Shiesh S. C., Chen Y. L., and Lee G. B., Microfluid. Nanofluid. 14, 753 (2013). 10.1007/s10404-012-1095-3 [DOI] [Google Scholar]
- 25. Weng C. H., Lien K. Y., Yang S. Y., and Lee G. B., Microfluid. Nanofluid. 10, 301 (2011). 10.1007/s10404-010-0669-1 [DOI] [Google Scholar]
- 26. Herman P., El-Kirat-Chatel S., Beaussart A., Geoghegan J. A., Foster T. J., and Dufrene Y. F., Mol. Microbiol. 93, 356 (2014). 10.1111/mmi.12663 [DOI] [PubMed] [Google Scholar]
- 27. Shigdar S., Qiao L., Zhou S., Xiang D., Wang T., Li Y., Lim L. Y., Kong L., Li L., and Duan W., Cancer Lett. 330, 84 (2011). 10.1016/j.canlet.2012.11.032 [DOI] [PubMed] [Google Scholar]
- 28. Che Y. J., Wu H. W., Hung L. Y., Chang H. Y., Wang K., and Lee G. B., in 2014 9th IEEE International Conference on Proceedings of Nano/Micro Engineered and Molecular Systems (NEMS), Hawaii, USA, 13–16 April, 2014, pp. 440–443. [Google Scholar]
- 29.See supplementary material at http://dx.doi.org/10.1063/1.4933067E-BIOMGB-9-021505 for the optimization of conditions, on-chip panning process, and details of chip characterization.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4933067E-BIOMGB-9-021505 for the optimization of conditions, on-chip panning process, and details of chip characterization.