Abstract
Metastasis is the defining feature of advanced malignancy, yet remains challenging to study in laboratory environments. Here we describe a high-throughput zebrafish system for comprehensive, in vivo assessment of metastatic biology. First, we generated several stable cell lines from melanomas of transgenic mitfa-BRAFV600E;p53−/− fish. We then transplanted the melanoma cells into the transparent casper strain to enable highly quantitative measurement of the metastatic process at single cell resolution. Using computational image analysis of the resulting metastases, we generated a metastasis score, μ, that can be applied to quantitative comparison of metastatic capacity between experimental conditions. Furthermore, image analysis also provided estimates of the frequency of metastasis-initiating cells (~1/120,000 cells). Finally, we determined that the degree of pigmentation is a key feature defining cells with metastatic capability. The small size and rapid generation of progeny combined with superior imaging tools make zebrafish ideal for unbiased high-throughput investigations of cell-intrinsic or microenvironmental modifiers of metastasis. The approaches described here are readily applicable to other tumor types and thus serve to complement studies also employing murine and human cell culture systems.
Quick Guide to Equations and Assumptions
Estimating metastasis initiating cell (MIC) frequency:
We assume that only a rare subpopulation of cells possesses the combination of traits, which enables them to leave the primary site, survive in circulation and establish a metastasis large enough to be detected within the 14 days of the experiment – we dub these ‘Metastasis initiating cells’ (MIC's). We may then assume that when picking ZMEL1 cells at random there is a fixed probability p, of getting a cell with a stable metastasis initiating phenotype. The probability of getting k MIC′s out of N randomly picked ZMEL1 cells must thus be given by the binomial distribution:
The probability of getting no MIC's out of N cells (k = 0) is thus:
Poisson approximation to the binomial distribution:
When N is large and p is small the binomial distribution is well approximated by the Poisson distribution, with mean Np. This approximation is typically used if N>20 and p<0.05, or if N>100 and Np<10. Since we have N>104 and most likely p<<0.05, we are able to use this approximation, so the probability of picking k MIC's when randomly picking N cells is:
For k=0 this becomes:
Symbol explanation:
N: number of randomly picked cells from initial ZMEL1 population.
k: number of times a MIC (a cell with a metastasis initiating phenotype) was picked out of the N random picks.
p: probability of getting a cell with a MIC.
Metastasis score (μ score):
We wished to develop a score that captured metastatic burden across different sets of fish. We developed a weighted score, called the μ score, which incorporates multiple parameters captured from imaging. The measures mi, were chosen since they where the three top contributors to the first principal component (PC1) of the entire data set. Further the weights wi where chosen to be similar to the weights of the three measures in PC1. Each measure is normalized by a fish specific measure ni of the same unit as mi to make the terms of the sum dimensionless. These ni where picked to be approximately equal to the upper limit which the measures mi can reach. The score can easily be modified to include additional measures m4, m5, ... if later needed. (Note that although it is numerically possible to reach a score of 100 it is not actually practically possible. Our highest scoring fish gets a score of 50).
m1 : total summed area of metastases on day 14.
n1 : area of fish body on day 14
w1 : 1/2
m2 : AP distance on day 14
n2 : length of fish body on day 14
w2 : 1/4
m3: number of metastatic events on day 14
n3 : L/2l , (where L length of fish and l is the threshold for clustering metastases)
w3 : 1/4
INTRODUCTION
Despite remarkable advances in elucidating the mechanisms of tumor initiation and growth, improvements in survival from metastatic cancer have remained elusive. In part, this is due to the difficulty of studying metastasis in vivo at large scale. Studies in murine systems have helped establish key steps in metastasis (1): local invasion at the primary site, intravasation into blood vessels at the primary site, circulation in the bloodstream, extravasation from blood vessels at distant sites, and the transition from micro to macrometastatic growth at distant sites after a period of dormancy. In individual patients, each of these steps is highly variable, likely in part due to the extreme heterogeneity across tumors. Moreover, it is increasingly recognized that the metastatic phenotype is intrinsically dependent upon interacting signals from the tumor and microenvironment (2, 3). Because of these factors, the study of metastasis requires an experimental system that allows for high-throughput manipulation of both tumor-cell and microenvironmental compartments.
In recent years, the zebrafish has emerged as an important model in cancer research(4), particularly in melanoma where transgenic expression of the human BRAFV600E gene leads to a fully penetrant disease that is similar to the human disease(5-7). Building upon these transgenic models, we have developed a high-throughput system for studying metastasis that is composed of two separate toolsets: 1) zebrafish melanoma cell lines with defined genetic and phenotypic characteristics as a source of donor tumor cells, and 2) a highly quantitative metastasis transplantation assay using the transparent casper strain(8, 9) of zebrafish as a recipient host. The casper strain maintains relative transparency throughout life and is particularly suited to quantitative assessment of spatio-temporal dynamics of metastasis, allowing us to build statistical pictures of metastatic patterns with unprecedented detail. Dramatic advances in zebrafish genome manipulation using CRISPR (10, 11) technologies allow us to easily modify both our zebrafish melanoma cell lines as well as the casper recipient. Taken together, our system provides the first high-throughput method to probe metastatic biology in vivo, which will be broadly applicable to researchers across the cancer spectrum.
MATERIALS AND METHODS
MiniCoopR transgenic melanoma fish and isolation of the ZMEL1 cell line
Transgenic melanoma zebrafish using the MiniCoopR system were created as previously described (7). Briefly, a plasmid was created in which the zebrafish mitfa promoter drives a zebrafish MITF minigene devoid of introns. On the same plasmid was a second cassette in which the mitfa promoter drives EGFP. Flanking both of these genes are Tol2 transposon arms. This plasmid was injected into fish with the following genotype: mitfa-BRAFV600E;p53−/−;mitfa−/−. This strain of fish is devoid of all melanocytes (due to the mitfa−/− mutation), but upon mosaic rescue with the mitfa-MITF minigene will develop “patches” of rescued melanocytes, some of which will go on to develop melanoma during adulthood. Because the rescued melanocytes all contain the MiniCoopR plasmid, they will necessarily also express mitfa-EGFP, resulting in melanomas which are entirely EGFP positive. For the isolation of the cell lines, tumors were cleanly dissected with a scalpel from melanoma bearing MiniCoopR fish and transferred to a small petri dish containing 2 ml dissection medium (50% Ham's F12/50% DMEM, 10X Pen/Strep, 0.075 mg/ml Liberase). They were then manually disaggregated for 30 minutes at room temperature. An inactivating solution (50% Ham's F12/50% DMEM, 10X Pen/Strep, 15% heat inactivated FCS) was then added, and the suspension filtered 2-3X in a 40μM filter. This was then centrifuged for 5 minutes@500rcf, and resuspended in 500μl of complete zebrafish media (see Supplemental Methods for further details). This 500μl was then plated in a single well of a 48-well plate that be been previously coated with fibronectin.
Proliferation assays/drug treatments
Cells were plated at a density of 25,000-50,000 cells per well in a 96 well plate in 100μL of DMEM/10. The cells were allowed to adhere for 24 hours, and then media changed to fresh media containing either DMSO or drugs at the indicated doses. The final concentration of all wells contained equivalent amounts of DMSO solvent (1%). The media was refreshed every 2 days, and at day 5, Alamar blue was added and fluorescence read using a 96-well plate reader. All values were normalized to the DMSO control well, and done in at least triplicate for each day of experiments.
RNA-seq of ZMEL1
Reads from each RNA-Seq run were mapped to the zebrafish reference genome version danRer7 from the UCSC Genome Browser(12) using GSNAP and quantified on the gene level using HTSeq and Ensembl version 75. Differential expression analysis was performed using DESeq2. The 40 bp single-end and 100 bp paired-end runs of ZMEL1 were used as separate replicates. Runs ERR004009, ERR004010, ERR004011, ERR004012, ERR015568 from ENA study ERP000016(13) were used as normal samples.
Reagents
The plasmids used for the MiniCoopr transgenics were obtained as a gift from Yariv Houvras (Weill-Cornell Medical College). The Cas9 plasmid was obtained from Addgene (#42251). All cell culture media (as outlined in the Supplemental Methods) were obtained from Life Technologies. PLX4032 was a gift from Plexxikon, and CI1040 was obtained from Selleckchem (Catalog number S1020).
Animal husbandry
All zebrafish were housed in a temperature (28.5C) and light-controlled (14h on, 10h off) room. Fish were initially housed at a density of 5-10 fish per liter, and fed 3 times per day using brine shrimp and pelleted zebrafish food. After transplantation, the fish were housed in individual chambers for serial imaging. All anesthesia was done using Tricaine (Western Chemical Incorporated) with a stock of 4g/L (protected for light) and diluted until the fish was immobilized. All procedures adhered to IACUC protocol #12-05-008 through Memorial Sloan Kettering Cancer Center.
Imaging and image analysis
Equipment
All fish were anesthesized with Tricaine and placed onto an agar coated petri dish. The fish were imaged from above using a Zeiss Axio Zoom V16 Fluorescence Stereo Zoom Microscope with a 0.6x or 1.6X lens. Each fish was successively imaged using brightfield, GFP and Rhodamine filter sets on both sides. The exposure times for each group were determined at day 1 and kept fixed throughout the entire experiment. If the fish was larger than a single field, multiple images for each fish could be taken using a motorized stage and the stitched together using the Zeiss Zen software. Raw image files (CZI) for each fish were then exported using Zen into high resolution TIFFs which could then be used for downstream image analysis in MatLab. The MatLab code used for all analyses is available online(14).
Image registration / image transformations
Each adult fish in the study (n=106) was imaged at three different time points (day 1, 7 and 14 post implant). At each time point bright field, GFP and RFP channel images where taken of both the right and the left side of the fish. After the images had be transformed and registered using landmarks they could be superimposed allowing for comparison of (i) left and right side image of each fish, (ii) same fish imaged at a different time points and (iii) images of different fish with each other. The transformations where done using a custom fully automated image registration pipeline, see further details in the Supplemental Methods and Supplemental Figures 16-20. The Matlab code for all image analysis is available online(14).
Principle component analysis
We were interested in extracting features from the segmented images, which in different ways characterized/quantified the growth of the tumor and the metastasis formation over time. We initially extracted 15 features (Supplemental Table 3), and from this initial pool found the following 5 to be most informative:
Solidity (ratio of total area of GFP region to area of smallest convex polygon, which will encapsulate the GFP region. This measure will give a value close to one for e.g. a solid sphere or triangle and a low value for a very fragmented or fractal like region).
Area of primary tumor.
Total area of all metastasis.
Number of metastatic events (number of times a new metastasis occurs).
Anterior–posterior distance of tumor/metastasis (the length between the two pixels which are the furthest apart (regardless of whether they belong to primary or metastatic regions) measured along the anterior – posterior axis of the fish).
These 5 features were extracted from the images taken at the 3 different time points, meaning that each fish became represented by a point in a 15-dimensional feature space. In order to determine which of these features where primarily responsible for the variance across the group and also to determine which features where correlated/anti-correlated/uncorrelated we did principle component analysis (PCA) on the data. Since PCA is sensitive to the scaling of the variables we normalized all measures by the variance in the group of that measure on the day 14 time point, before performing the analysis. E.g.:
With this normalization the PCA will reveal which features contributed most to the overall variance in the dataset while still keeping different measures that are sharing the same basic unit on the same scale (like AreaPrimary_D7 and TotalMetsArea_D14 which both have unit length2).
Metastasis initiating cell (MIC) frequency
One hypothesis about metastasis formations is that only a rare subpopulation of cells possesses the combination of traits, which enables them to leave the primary site, survive in circulation and establish a metastasis large enough to be detected within the 14 days of the experiment. If this hypothesis holds we may assume that when picking ZMEL1 cells at random there is a fixed probability p, of getting a cell with a stable metastasis initiating phenotype. The probability of getting k MIC′s out of N randomly picked cell must thus be given by the binomial distribution:
The probability of getting no MIC's (k = 0) is thus:
| (equation 1) |
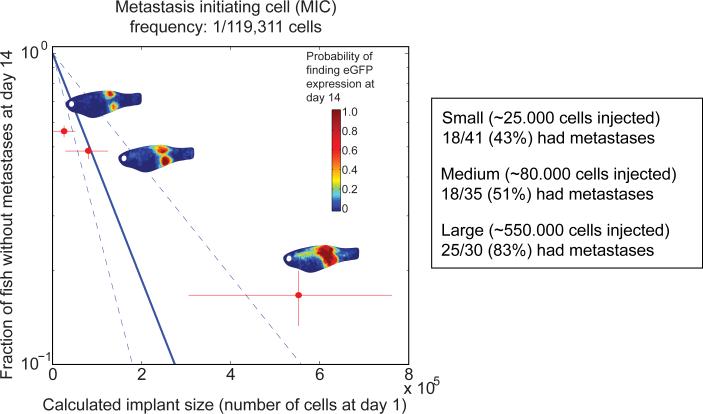
Based on the images taken on day 1 and the knowledge of which original implant size group a fish came from estimated the number of cells which were successfully implanted in fish nr i at day 0, Ni. (See Supplemental Methods for the details of how we did this estimate). By day 14 each fish either has at least one metastasis or none, i.e. Pri(k=0) of fish i is either 1 or 0. We fit the points (Ni , Pri(k=0)), using nonlinear regression (to equation 1) and was thus able to estimate the parameter p, which is the frequency of MIC's in the ZMEL1 population. We found this frequency to be p=8.4×10-6 (i.e. 1 out of ~120,000 cells are capable of forming a macro metastasis within the timespan on 14 days).
Poisson approximation
When N is large and p is small the binomial distribution is well approximated by the Poisson distribution, with mean Np. This approximation is typically used if N>20 and p<0.05, or if N>100 and Np<10. Since we have N>104 and most likely p<<0.05, we are able to use this approximation, so the probability of picking k MIC's when randomly picking N cells is:
We expect the probability of having no metastasis to depend on N in the following manner:
| (equation 2) |
We can estimate the probability of picking no MIC′s for a certain implant dose by counting the fish within a certain implant size group (a certain approximate N), which did not have any metastasis. Assuming existence of a rare subpopulation of MIC's, we see from equation 2 that we can expect that log (Pr(k=0)) for the three implant size groups small, medium and large, plotted versus N to follow a straight line going through (0,1), and p, the frequency of MIC′s, will be the slope of this straight line. As seen in Figure 5, the points for three size groups small, medium and large do not show a linear dependency, but rather suggest a convex dependency. Nonetheless, the precision of the estimate is not sufficient to refute the existence of a subpopulation of MIC's.
RESULTS
Generation of zebrafish melanoma cell lines
Previous work (5-7) has established a transgenic zebrafish model of human melanoma, in which expression of human BRAFV600E, under the melanocyte-specific mitfa promoter leads to rapid formation of pigmentation abnormalities and nevi. When crossed with p53−/− fish, 100% of the resultant animals (mitfa-BRAFV600E;p53−/−) develop melanomas in highly stereotyped locations including the head, dorsal skin and caudal fin. Although these animals have previously been used to identify genes and chemicals (6, 7), which affect melanoma initiation, the metastatic characteristics of these tumors have not been defined. To assess this, we performed a series of transplant studies of primary tumors into the transparent casper strain of zebrafish. The recipient fish developed highly variable degrees of metastatic dissemination (Supplemental Figure 1). This metastatic heterogeneity is likely due to the tremendous genetic heterogeneity that we have previously found to be present in the transgenic zebrafish melanomas(15). This observation prompted us to develop stable cell lines from the zebrafish tumors, which would lead to more reproducible metastatic behavior upon transplantation, as has been shown for human tumors (16, 17). Adopting methods commonly used for isolation of human melanoma lines, we developed fluorescently labelled stable zebrafish melanoma cell lines. We generated a large number of primary transgenic tumors using the MiniCoopR transposon system, which allows mosaic expression of the BRAFV600E in a p53−/− background, and generates transgenic animals with melanoma within 2-3 months (7). The MiniCoopR transposon also carries a mitfa-GFP cassette; because mitfa is only expressed by melanocyte derivatives, the presence of GFP confirms its lineage identity as a bona fide melanoma line. We isolated a series of these transgenic mitfa-BRAFV600E;p53−/−;mitfa-GFP tumors (an example is shown in Figure 1a), disaggregated them into single cell suspension, and plated them on fibronectin- coated plates in a media formulation similar to that used to isolate human melanoma cell lines from patients (18, 19). We then allowed these cells to propagate over time in order to select tumors that ultimately gave rise to stable cell lines.
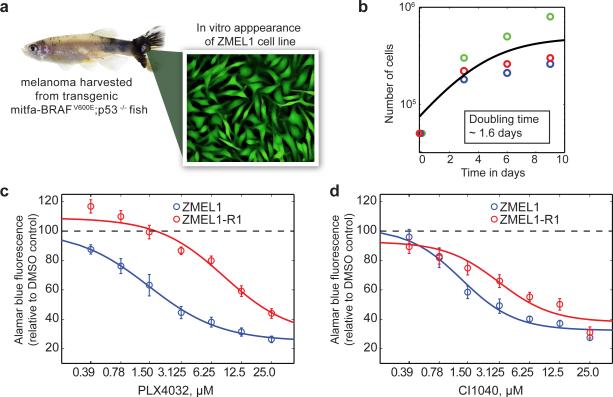
Figure 1.
Derivation and characterization of the ZMEL1 zebrafish melanoma cell line. a) Representative melanoma bearing fish (left) from the MiniCoopr background which mosaically expresses BRAFV600E in a mitfa-GFP;p53−/− background, and yielded the stable cell line ZMEL1, which is uniformly GFP positive. b) Growth curves of the ZMEL1 line demonstrate a population doubling time of 1.6 days. Individual colors representative replicate experiments. c,d) The response of the ZMEL1 and ZMEL-R1 lines to either the BRAFV600E inhibitor PLX4032 (c) or the MEK inhibitor CI1040 (d). The resistant line demonstrates an 8-fold increase in the EC50 to the BRAF inhibitor, but only a 2-fold increase in the EC50 to the MEK inhibitor.
Overall, we successfully established 31 stable cell lines, out of 43 attempts, for a success rate of 72% (Supplemental Table 1). All but one of these could be transitioned off of fibronectin onto plastic plates. For the purposes of demonstration, we focused on one particular line, which we refer to as ZMEL1 (Zebrafish Melanoma line 1, as shown in Figure 1a) because of the following key characteristics: After the 10th passage, we were able to transition the line off the more complex isolation medium to standard DMEM with 10% FCS, and could eliminate the need for fibronectin coating of the plates. By FACS sorting, ZMEL1 showed greater than 99.5% GFP-positive cells, indicating essentially no contaminating stromal cell elements. The population doubling time (Figure 1b) of ~1.6 days makes it amenable to generating large numbers of cells rapidly. We maintain the ZMEL1 line at 28.5C/5% CO2 in a standard tissue culture incubator. The ZMEL1 line can be transfected with expression vectors of interest using nucleofection technology. For example, nucleofection of an ubiquitin-EGFP-2A-tdTomato plasmid, followed by repeated FACS sorting, led to establishment of a stable cell ZMEL1 line expressing both eGFP and tdTomato (Supplemental Figure 2). We repeated this for additional genes including CFP and YFP, all using the 2A system, and were able to generate either transient or stable cell lines for all constructs (data not shown). We also asked whether we could knockout gene function using CRISPR technology. We nucleofected a CMV-Cas9 plasmid along with a plasmid containing a guide RNA against eGFP (driven by the zebrafish U6 promoter). After blasticidin selection, we were able to obtain a stable cell line in which ~99% of the cells no longer expressed eGFP (Supplemental Figure 3). We confirmed mutagenic efficiency of the eGFP gene using both FACS and the Surveyor nuclease assay (Supplemental Figure 4). Taken together, these data indicate that the ZMEL1 line can be genetically modified, and will readily allow both overexpression (cDNA) and knockout (CRISPR) screens.
Cross-species transcriptomic analysis
To determine the similarity between the ZMEL1 line and human melanoma, we compared the transcriptomic profile of ZMEL1 to human cancer cell lines using Gene Set Enrichment Analysis (GSEA) (20). We performed RNA-seq analysis of the ZMEL1 line (Supplemental Table 2), and identified the 250 most up- or down-regulated genes in ZMEL1 (compared to pooled normal reference RNA). We then compared that signature via GSEA to the human NCI60 gene signatures (21) (Supplemental Figures 5 and 6, and Supplemental Table 2). This revealed a striking enrichment of the ZMEL1 signature in human melanoma cell lines (i.e. MALME-3M and SK-Mel28) compared to all other tumors (NES=1.702, FDR=0.0012). Amongst the most up-regulated genes in both human and zebrafish melanoma are factors known to be expressed in neural crest-derived melanocytes, including sox10, ednrb and mitfa itself. These are genes also known to play pathogenic roles in human melanoma, either through overexpression or amplification (22-28). We also used GSEA to compare the ZMEL1 signature to previously established gene signatures from transgenic mitf-BRAFV600E;p53−/− primary tumors (6), and again found a strong similarity to the original tumors (Supplemental Figures 7 and 8 and Supplemental Table 2). Finally, we analyzed the ZMEL1 RNA-seq signature using Ingenuity Pathway Analysis (Supplemental Figures 9 and 10). This revealed a strong enrichment for pigment cell signaling, as well as a dependence upon MYC signaling, as expected for a tumor driven by BRAFV600E. Taken together, the transcriptomic data demonstrates that the ZMEL1 line strongly resembles well-characterized transgenic zebrafish melanomas as well as human melanoma cell lines.
ZMEL1 sensitivity to MAP kinase inhibition
We next confirmed functional dependency on BRAF-MAP kinase signaling using pharmacologic inhibition. We treated the ZMEL1 line with either the BRAFV600E inhibitor PLX4032 (29) or the MEK inhibitor CI1040 (30) and measured proliferation at 5 days using the Alamar blue assay (Figure 1c,d). Both of these drugs caused a dose-dependent inhibition of ZMEL1 proliferation, with an EC50 of 1.12μM and 1.30μM, respectively. To further confirm the functional similarity to human melanoma cell lines, we determined whether ZMEL1 could become BRAF inhibitor resistant in vitro, as has been widely reported for human melanoma lines (31, 32). We treated the ZMEL1 line with 1μM PLX4032 over a period of several months, which led to a rapid loss of most viable cells by 1 week, as expected, but a small population of persister cells remained. By 3-4 months, these cells had become resistant to this concentration of PLX4032 and reconstituted the culture. We then retested sensitivity to both PLX4032 and CI1040 (Figure 1c,d). The EC50 for PLX4032 increased 7.6-fold (from 1.13μM to 8.56μM). The cells remained moderately sensitive to MEK inhibition, with only a 2.3-fold increase in the EC50 for CI1040 (from 1.30μm to 3.06μM). This derivative line, which we refer to as ZMEL1-R1 (ZMEL1-Resistant Line 1), demonstrates that zebrafish melanoma cell lines react to MAP kinase inhibition in a manner analogous to their human counterparts. The genetic mechanism resulting in such resistance remains to be elucidated and will be the subject of future studies.
Metastatic behavior of the ZMEL1 line
We next determined the metastatic capacity of the ZMEL1 line. We took advantage of the previously described casper strain of zebrafish, which maintains relative transparency throughout its entire life cycle, and allows highly sensitive detection of fluorescently labeled cells anywhere in the animal at single-cell resolution (8, 9). We transplanted ZMEL1 cells into either adult or embryonic casper recipients using a quartz glass microcapillary needle attached to a microinjection apparatus (Figure 2). For the adult fish, we transplanted 500,000 ZMEL1 cells into the subcutaneous tissue of the ventral flank of a recipient that had been previously irradiated with a total of 30 Gy (15 Gy × 15 Gy split dose over 2 days). We found that irradiation of the adult recipient was required due to MHC mismatch between the ZMEL1 line and casper recipients. For the embryos, we transplanted 150 ZMEL1 cells directly into the vasculature (via the Duct of Cuvier). The embryos recipients did not require any preconditioning radiation since they do not yet have a mature adaptive immune system. MHC mismatch, the major cause of implant rejection, depends upon the adaptive response, which does not develop in the zebrafish until ~14 days of life. The adult assay allows for full assessment of metastasis from an orthotopic site but requires immunosuppression; the embryonic assay allows for direct intravascular injection of tumor cells and requires no immunosuppression.
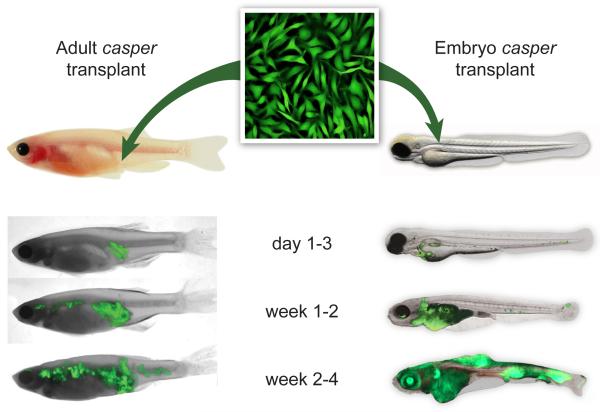
Figure 2.
Evaluating metastasis of the ZMEL1 line using transplantation into the transparent casper recipient line. ZMEL1-GFP cells can be transplanted either subcutaneously into the flank of an irradiated casper recipient (left) or directly into the vasculature of an unirradiated casper embryo at 2 days post fertilization (right). The fish are then imaged over a period of ~1 month using GFP and brightfield imaging. Representative fish for both assays are shown. For the adults, ~500,000 cells were transplanted, and for the first 1-3 days after transplant the cells remain localized, but by weeks 1 to 4, they widely disseminate anteriorly and posteriorly from the initial implant site. A similar pattern is seen in the embryo transplants, but because the cells are injected directly into the circulation, extravasation and formation of disseminated masses occurs more rapidly.
Representative fish are shown in Figure 2. In the adult recipients (Figure 2, left), at 1 day post-transplant a brightly GFP+ mass can be seen at the site of injection with little or no distant metastases. By 1 week, and then 2 weeks post-transplant, the size of the mass at the injection spot continued to grow, as expected, along with the appearance of multiple new anterior masses. These masses are clearly distinct from the site of implantation, and represent distant metastases. In the embryo recipients (Figure 2, right), at day 1 post-transplant the cells circulate primarily within the ventral vasculature (Supplemental Video 1) and have begun to extravasate into the caudal hematopoietic tissue, the first site of definitive hematopoiesis in the fish. By 1 week and then 2 weeks, all of the cells have extravasated, and the fish develops GFP+ tumors in the eye, kidney, muscle and head. This leads to the death of the animals by 30-60 days post-transplant. One major advantage of these embryo transplants is that single cell behavior is easily observed after transplantation. Taken together, these data indicate that the ZMEL1 line is capable of performing all of the canonical steps of metastasis.
Patterns of metastatic spread
In order for this assay to be useful as a screening tool, we needed to develop automated quantitative imaging of metastasis. We reasoned that the degree of metastatic dissemination would be dependent upon the number of cells transplanted into the casper recipient, and should vary with both site of implantation as well as time after transplant. We therefore performed a series of limiting dilution transplantation studies in which we varied the cell number (1×105, 5×105, 1×106) and the transplant site (dorsal vs. ventral skin). We then imaged each fish at 1 day post-transplant (dpt), 7dpt, and 14 dpt using both brightfield and GFP, on both the left and right side of the fish. We developed a custom-designed computer image analysis program to align each image such that it could be precisely overlaid with all of the others. Landmarks such as the eye, the anterior end and dorsal and ventral boundary points were used to guide image alignment transformations and the images from the GFP channel were subsequently analyzed using a custom image segmentation algorithm to detect location of primary tumor and metastasis. Because transplantation itself has some degree of mechanical variability, we imaged each fish on day 1 and used information from these images along with the information of the original cell dose to better estimate the number of cells that each recipient actually received. This is a major advantage of the fish compared to mouse studies, where accounting for such mechanical variability is not possible. After this determination, we appropriately regrouped the fish into small (25,700 ±24,560 cells), medium (80,544 ±52,358 cells) and large (552,720 ±272,030 cells) implant sizes (See Supplemental Methods for details on estimation procedure). We used data collected from 106 fish to create composite heatmap images of metastatic progression from cells transplanted either ventrally (Figure 3a) or dorsally (Supplemental Figure 11). This revealed a strong correlation between size of the initial implant and the likelihood of metastasis. Small implants on day 1 generally produced fish with few metastases at day 14, whereas those with large masses on day 1 led to a large increase in the metastatic burden, particularly in the anterior region just behind the gill structure, as well as scattered other metastases to the posterior tail musculature and eye. This dissemination pattern was not due to the implant site, as we saw similar patterns whether the cells were transplanted dorsally or ventrally. Overall, 83% (25/30) of fish in the large implant group had metastases at day 14, compared to 44% (18/41) in the small implant group. We noted on the heatmaps that some of the anterior metastases anatomically corresponded to the area of the kidney marrow, the region of hematopoiesis in the zebrafish (33). To confirm this, we performed histological analysis on a series of fish at 2 weeks post-transplant (Supplemental Figure 12). This showed that fish with higher cell doses had clear metastases in the kidney marrow (by both H&E and anti-GFP imaging), an example of which is shown in Figure 3b. To determine if this tropism was an artifact of the transplant technique, we performed RNA in situ staining in three of the original mitfa-BRAFV600E;p53−/− transgenic fish from which ZMEL1 was derived. We stained for expression of the neural crest marker crestin, a retroelement normally only expressed in embryonic neural crest cells (34), but which becomes aberrantly expressed exclusively in adult melanomas due to their neural crest origins (6). One of the three fish had evidence of metastases to the kidney (Supplemental Figure 13), indicating that the transplanted cells have a tropism for the kidney marrow similar to melanoma developed spontaneously in the stable transgenic mitfa-BRAF;p53−/− fish line. These data are consistent with other reports indicating a tropism of human melanoma cells for the bone marrow in mammalian systems (35-37).
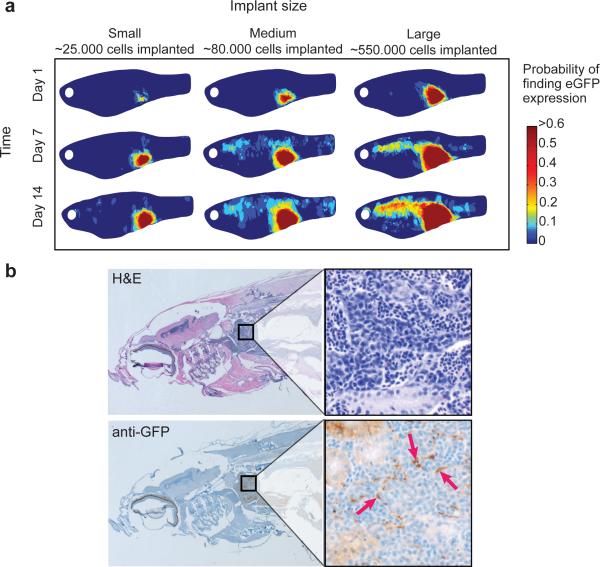
Figure 3.
Analyzing the pattern of metastatic spread after ZMEL transplantation. a) Composite heatmap images of a group of fish (n=53) transplanted with small (~25,000 cells, n=19), medium (~50,000 cells, n=20) or large cell numbers (~500,000 cells, n=14) at day 1, and then imaged at days 7 and 14. The heatmap corresponds to the probability of finding GFP+ ZMEL1 cells at the given location. This demonstrates that the likelihood of metastasis varies with the number of cells transplanted. The GFP positive masses seen anteriorly at day 14 are suggestive of localization within the kidney marrow, the site of hematopoiesis in the zebrafish. This localization was confirmed using histological sectioning of a representative fish (b), stained with either hematoxylin/eosin (b, top) or an anti-GFP antibody (b, bottom). Brown staining (denoted by red arrows) indicates the presence of ZMEL1-GFP cells in the kidney marrow compartment.
Development of a metastasis score (μ score)
Although the heatmap views are useful for determining overall patterns of spread, they do not allow for a simple quantification of overall metastatic burden. We wished to develop a straightforward “metastasis score”, μ, which would be of general use to the community and to determine how this score differed between the three implant size groups. Initially we extracted 15 parameters from the images taken at 3 separate time points (e.g. area of primary tumor, area and number of metastasis, length of fish body, see Supplemental Table 3 for the complete list of features) and then used Principal Components Analysis to determine which parameters best explained the variance in the data. The PCA analysis (Figure 4a) demonstrated that just 3 of these parameters at day 14 accounted for a substantial amount of the variance in the data which was related to formation of metastasis: 1) total number of metastases, 2) total area of metastases, and 3) antero-posterior distance spanned by both primary and metastases. We combined the three parameters into a simple dimensionless μ score, which can easily be applied in other studies (Supplemental Figure 14). We calculated the median μ score for each of the three implant size groups (Figure 4b): At day 14 for the group with small implants it was 5.35, whereas for the medium group it was 12.94, and for the large group 21.42. The difference between the μ score distributions was significant for small versus medium (p=0.026, two-sample Kolmogorov-Smirnov (KS) test), medium versus large (p=0.024, two-sample KS test) and small versus large (3.75×10-6, two-sample KS test). The μ score can be applied to future experimental interventions (i.e. overexpressing or knocking down specific genes) allowing for a rapid and quantifiable assessment of metastatic efficiency.
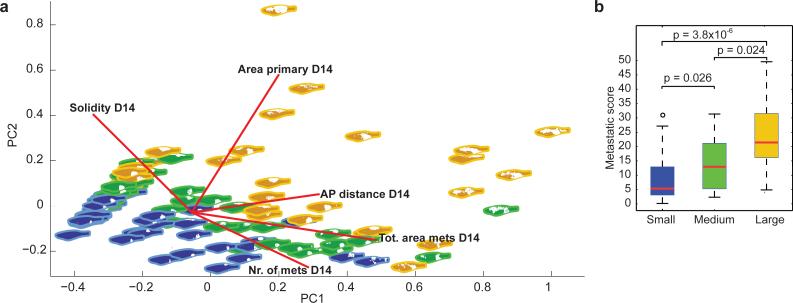
Figure 4.
Quantification of metastatic burden in the zebrafish. a) PCA plot of the major variables measured in each fish at days 1, 7 and 14 (area of the primary, total area of metastases, total number of metastases, solidity of primary tumor and antero-posterior (AP) distance from primary to metastases). Blue fish correspond to “small” implant size at day 1, green to “medium” implant, and yellow to “large” implant. b) The µ score as a measure of metastases. Using the key principal components described in Figure 4a and Supplementary Figure 10, we calculated a µ score (metastasis score) for fish with small, medium or large implant tumors at day 1. See Supplemental Figure 14 for the equation used to calculate the µ score. This demonstrates that the size of the implant at day 1 strongly predicts metastasis at day 14, with a significantly greater µ score in medium or large groups compared to the small group (p values as indicated from two-sample Kolmogorov-Smirnov test) .
Quantification of metastasis with MIC frequency
We next wished to estimate the frequency of metastasis-initiating cells (MICs), analogous to the tumor-initiating cell calculations frequently employed for quantifying cancer stem cells. Using standard limiting dilution analysis (38, 39), we determined the number of animals with or without metastases at day 14 using either the intended cell numbers (Supplemental Figure 15) or after regrouping into the estimated small/medium/large groups (Figure 5). Since we found that regrouping more accurately reflects the actual number of cells the fish received, we used this to calculate MIC. We estimate 1 MIC per 119,311 cells (lower bound=1/78,019; upper bound=1/228,233), meaning that this is the frequency of cells within the ZMEL1 population capable of giving rise to measurable distant metastases within 14 days. This is in agreement with other reports indicating that the frequency of cells capable of completing all steps in the metastatic cascade are quite rare (40, 41). These two measures, the μ score and the MIC frequency, are key measures of metastasis in the zebrafish. Having such quantitative measures is essential to detecting changes in metastatic efficiency during experimental perturbations to either the cells or the recipient host background.
Figure 5.
An estimation of metastasis initiating cell frequency. The estimated number of cells transplanted at day 1 (x-axis) is correlated with the proportion of fish that do or do not have metastasis at day 14 (y-axis). Limiting dilution analysis allows for an estimation of Metastasis Initiating Cell frequency of 1/119,311 cells for the ZMEL1 line.
Pigmentation and metastatic progression
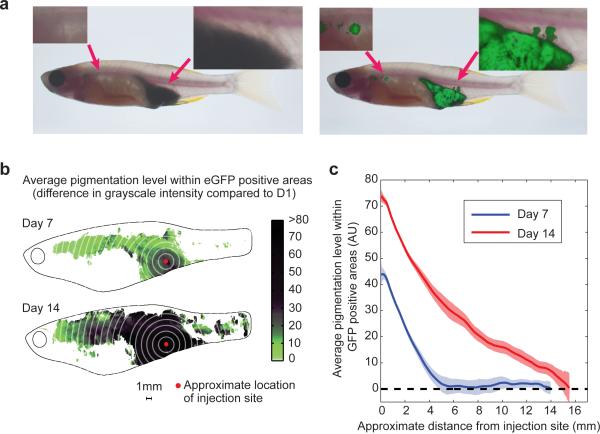
Finally, we wished to understand the characteristics of these rare cells capable of metastasis. We noted a significant discrepancy between the appearance of pigmentation and GFP in the recipient animals (Figure 6a), suggesting that pigment status and metastatic progression were related. In the representative fish shown in Figure 6a, at 2 weeks post-transplant, brightfield imaging (Figure 6a, left) showed a deeply pigmented, black mass at the implant site, with sharp borders and no obvious metastatic lesions. In contrast, examination of the same fish under GFP (Figure 6a, right) showed that the implanted mass has several protrusions in the dorsal direction, along with multiple anterior metastases. This suggested that the cells which leave the implant site are likely unpigmented and less differentiated melanocytes. To quantify this, we created composite heatmap images depicting the average level of black pigmentation within the GFP positive regions at day 7 and 14 (Figure 6b). This demonstrated that the primary implant site (the center of which is denoted by the red dot) was far more pigmented than the distant anterior metastases. We then determined the distance from the implant site versus the degree of pigmentation (Figure 6c). This demonstrates a clear inverse relationship: the cells furthest away from the implant site are nearly all unpigmented, whereas those at the implant site are strongly pigmented.
Figure 6.
Pigmentation status as related to metastatic capacity. a) Representative fish is shown under brightfield (left) and GFP (right) imaging. This reveals a significant discrepancy between pigmented, melanized cells and metastases. The tumor under brightfield appears smooth bordered and deeply pigmented, with no metastatic lesions. In contrast, GFP imaging of the same fish shows that the local tumor has irregular protrusions dorsally and several anterior metastases. b) Composite heatmap of a group of 53 fish showing the relationship between pigmentation and metastasis at day 7 and day 14 post transplant, suggesting that further the cells are from the implant site (red dot), the more likely they are to be unpigmented. c) Quantification of the heatmap image shown in (c), measuring both distance from injection site along with pigmentation level. This demonstrates a clear inverse relationship, such that the cells capable of furthest travel are largely unpigmented.
Although the tumor from which ZMEL1 was originally derived was highly pigmented, the ZMEL1 line in culture shows no evidence of pigmentation, suggesting the cells only become pigmented in vivo. To confirm this, we examined the behavior of the transplanted ZMEL1 cells using time lapse imaging, in which we took one picture per day over a range of 21-30 days. As shown in Supplemental Video 2, the implant site becomes large and pigmented prior to the metastases, which are initially only GFP positive before becoming pigmented as well. Over time, once the metastases are completely engrafted, usually even those cells too become pigmented. To determine if this effect was specific to the ZMEL1 line, we also performed transplantation of several BRAFV600E;p53−/− primary transgenic melanomas (Supplemental Figure 1) and found that both the primary engraftment site and metastases become pigmented as well, indicating that melanoma cells retain significant differentiation plasticity: they are unpigmented in vitro, become pigmented when implanted into the primary site in the fish, are unpigmented while metastasizing, and then again become pigmented in the new site of dissemination. These observations are consistent with data from mammalian systems showing that metastatic cells are initially less differentiated (42).
DISCUSSION
We have described a zebrafish system for studying melanoma metastasis which has all of the components necessary to study both cell intrinsic and microenvironmental regulators of this process at large scale. The isolation of fluorescent zebrafish cell lines which closely resemble the human disease, and which can be easily genetically modified using overexpression or CRISPR cassettes, opens up the possibility to perform genome-wide screens to find modifiers of metastasis. The capacity for in vivo imaging using the transparent casper recipient, which allows for visualization at the single cell level, enables a finer resolution view of micro to macrometastatic progression than is currently achievable in murine models. A particular strength of such high-resolution imaging may be the study of tumor cell extravasation at distant sites, one of the most difficult parts of the metastatic cascade to analyze using current murine models.
We find that the embryonic microenvironment augments metastatic growth compared to the adult, strongly suggesting that the microenvironment is a dominant force in establishing successful metastatic spread. Because it is now straightforward to make CRISPR zebrafish recipient fish carrying germline or somatic mutations of nearly any microenvironmental gene, we envision that a major use of our model will be in screens in which ZMEL1 cells are transplanted into these modified recipient fish. This will allow for a direct assessment of which genes in the microenvironment act as modifiers of disseminated tumor growth at a scale not achievable in other vertebrate systems.
Each of the assays described here has distinct strengths and caveats. The adult casper assay is robust and recapitulates the microenvironment present when most melanomas in humans form, i.e. during the postembryonic period. However, because the animals must be immunocompromised to prevent rejection of the ZMEL1 cells, its capacity for studying immune regulation of metastasis is limited. The embryo assay has the distinct advantage of a largely intact immune system, and because the 2 day old animal is much smaller than the adults at 4-6 months described above (where we used 100,000 – 1,000,000 cells) we need only transplant between 50-150 ZMEL1 cells into each 2 day old recipient. These advantages are partially offset by the obvious growth-promoting effects of the embryonic microenvironment, which likely accounts for the rapidity of disease progression. It is likely that different investigators will find advantages to either of these assays, depending upon the particular question being addressed.
One of the major strengths of studying metastasis in the zebrafish is related to the achievable scale of the experiments. Each individual fish at 6 weeks of age measures ~0.021cm3, in contrast to an average mouse at that age which measures ~73cm3. On a size basis, for every 1 mouse, it is possible to study ~3000 young zebrafish. Even in a modest sized zebrafish facility, this opens up a wide range of studies probing metastatic biology that would not be possible in traditional murine systems. Because of the intrinsic heterogeneity of the metastatic state, the zebrafish offers considerable statistical power to discern even moderate modifiers of the metastatic phenotype.
It is likely that some aspects of metastatic biology in a zebrafish will be mechanistically distinct from that in human patients, a caveat for all model systems. In some cases, findings in fish have not translated well to humans (43), yet in others we have seen remarkable conservation of core pathways and genes. For example, findings in zebrafish have resulted in at least two clinical trials in human cancer patients (6, 44, 45), including one specifically focused on melanoma. Because the number of labs currently using zebrafish to study metastasis is limited, it will require a great deal of study before we can begin to understand the similarities, and differences, between fish and human metastasis. The tools described here are an integral component of opening up this line of studies. We anticipate that these methods will be readily extended outside of melanoma into other tumor types, making it broadly applicable to cancer investigators with diverse interests.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yariv Houvras for supplying the photo of the MiniCoopR fish in Figure 2, Sara Fischer for editorial contributions, and Wenjing Wu for assistance with illustrations . This work was supported by the NIH Directors New Innovator Award (DP2CA186572), K08AR055368, the Melanoma Research Alliance Young Investigator Award, an AACR/ASCO Young Investigator Award, the Alan and Sandra Gerry Metastasis Research Initiative, and the Howard Hughes Medical Institute of which L.I.Z. and R.O.H. are Investigators. S.H. is a James S. McDonnell postdoctoral fellow, L.P. is a TROT Fellow (5T32CA160001-Translational Research Oncology Training Program) and A.M. was a Helen Hay Whitney Fellow.
Footnotes
CONFLICT OF INTEREST DISCLOSURE:
Leonard Zon is a founder and Scientific Advisory Board member of FATE Therapeutics and Scholar Rock. He has stock in both companies.
REFERENCES
- 1.Bos PD, Nguyen DX, Massague J. Modeling metastasis in the mouse. Current opinion in pharmacology. 2010;10:571–7. doi: 10.1016/j.coph.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine. 2013;19:1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer cell. 2011;20:576–90. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nature reviews Cancer. 2013;13:624–36. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Current biology : CB. 2005;15:249–54. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 6.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–22. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–7. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell stem cell. 2008;2:183–9. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Alt C, Li P, White RM, Zon LI, Wei X, et al. An optical platform for cell tracking in adult zebrafish. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2012;81:176–82. doi: 10.1002/cyto.a.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–7. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- 11.Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome research. 2014;24:125–31. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UCSC Genome Browser Available from: http://genome.ucsc.edu/
- 13.EMBL EBI Available from: http://www.ebi.ac.uk/ena.
- 14.Heilmann S. Matlab-fish-image-analysis. [Online repository] 2015 2015 [cited 2015 2015]; Available from: https://github.com/SnowballTheThird/Matlab-fish-image-analysis.
- 16.Yen J, White RM, Wedge DC, Van Loo P, de Ridder J, Capper A, et al. The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome biology. 2013;14:R113. doi: 10.1186/gb-2013-14-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soo JK, Ross AD, Bennett DC. Isolation and culture of melanoma and naevus cells and cell lines. Methods in molecular biology. 2011;731:141–50. doi: 10.1007/978-1-61779-080-5_13. [DOI] [PubMed] [Google Scholar]
- 20.Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. Journal of the National Cancer Institute. 1977;59:221–6. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13086–91. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graf SA, Busch C, Bosserhoff AK, Besch R, Berking C. SOX10 Promotes Melanoma Cell Invasion by Regulating Melanoma Inhibitory Activity. The Journal of investigative dermatology. 2014 doi: 10.1038/jid.2014.128. [DOI] [PubMed] [Google Scholar]
- 24.Cronin JC, Watkins-Chow DE, Incao A, Hasskamp JH, Schonewolf N, Aoude LG, et al. SOX10 ablation arrests cell cycle, induces senescence, and suppresses melanomagenesis. Cancer research. 2013;73:5709–18. doi: 10.1158/0008-5472.CAN-12-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronnstrand L, Phung B. Enhanced SOX10 and KIT expression in cutaneous melanoma. Medical oncology. 2013;30:648. doi: 10.1007/s12032-013-0648-y. [DOI] [PubMed] [Google Scholar]
- 26.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Munoz W, Jaramillo ML, Man S, Xu P, Banville M, Collins C, et al. Roles for endothelin receptor B and BCL2A1 in spontaneous CNS metastasis of melanoma. Cancer research. 2012;72:4909–19. doi: 10.1158/0008-5472.CAN-12-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahav R, Suva ML, Rimoldi D, Patterson PH, Stamenkovic I. Endothelin receptor B inhibition triggers apoptosis and enhances angiogenesis in melanomas. Cancer research. 2004;64:8945–53. doi: 10.1158/0008-5472.CAN-04-1510. [DOI] [PubMed] [Google Scholar]
- 30.Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews Drug discovery. 2012;11:873–86. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 31.Huang MH, Lee JH, Chang YJ, Tsai HH, Lin YL, Lin AM, et al. MEK inhibitors reverse resistance in epidermal growth factor receptor mutation lung cancer cells with acquired resistance to gefitinib. Molecular oncology. 2013;7:112–20. doi: 10.1016/j.molonc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma D, Zhang J, Lin HF, Italiano J, Handin RI. The identification and characterization of zebrafish hematopoietic stem cells. Blood. 2011;118:289–97. doi: 10.1182/blood-2010-12-327403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Developmental dynamics : an official publication of the American Association of Anatomists. 2001;220:169–74. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1097>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 36.Leong SP, Tseng WW. Micrometastatic cancer cells in lymph nodes, bone marrow, and blood: Clinical significance and biologic implications. CA: a cancer journal for clinicians. 2014;64:195–206. doi: 10.3322/caac.21217. [DOI] [PubMed] [Google Scholar]
- 37.Kaliks RA, Silveira PA, Osawa A, Campregher PV, Bacal NS, Velloso ED. Metastatic melanoma mimicking acute leukaemia. British journal of haematology. 2014;165:1. doi: 10.1111/bjh.12669. [DOI] [PubMed] [Google Scholar]
- 38.Serrier C, Lesesve JF. Metastatic malignant melanoma in the bone marrow. Blood. 2013;121:721. doi: 10.1182/blood-2012-09-454546. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of immunological methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Blackburn JS, Liu S, Langenau DM. Quantifying the frequency of tumor-propagating cells using limiting dilution cell transplantation in syngeneic zebrafish. Journal of visualized experiments : JoVE. 2011:e2790. doi: 10.3791/2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh M, Manoranjan B, Mahendram S, McFarlane N, Venugopal C, Singh SK. Brain metastasis-initiating cells: survival of the fittest. International journal of molecular sciences. 2014;15:9117–33. doi: 10.3390/ijms15059117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJ, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–16. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, et al. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer research. 2009;69:7969–77. doi: 10.1158/0008-5472.CAN-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nature chemical biology. 2005;1:366–70. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- 45.Goessling W, Allen RS, Guan X, Jin P, Uchida N, Dovey M, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell stem cell. 2011;8:445–58. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–11. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.