Abstract
Although new serotypes of enterohemorrhagic Escherichia coli (EHEC) emerge constantly, the mechanisms by which these new pathogens arise and the reasons emerging serotypes tend to carry more virulence genes than other E. coli are not understood. An insertion sequence (IS) excision enhancer (IEE) was discovered in EHEC O157:H7 that promoted the excision of IS3 family members and generating various genomic deletions. One IS3 family member, IS629, actively transposes and proliferates in EHEC O157:H7 and enterotoxigenic E. coli (ETEC) O139 and O149. The simultaneous presence of the IEE and IS629 (and other IS3 family members) may be part of a system promoting not only adaptation and genome diversification in E. coli O157:H7 but also contributing to the development of pathogenicity among predominant serotypes. Prevalence comparisons of these elements in 461 strains, representing 72 different serotypes and 5 preassigned seropathotypes (SPT) A to E, showed that the presence of these two elements simultaneously was serotype specific and associated with highly pathogenic serotypes (O157 and top non-O157 Shiga toxin-producing Escherichia coli [STEC]) implicated in outbreaks and sporadic cases of human illness (SPT A and B). Serotypes lacking one or both elements were less likely to have been isolated from clinical cases. Our comparisons of IEE sequences showed sequence variations that could be divided into at least three clusters. Interestingly, the IEE sequences from O157 and the top 10 non-O157 STEC serotypes fell into clusters I and II, while less commonly isolated serotypes O5 and O174 fell into cluster III. These results suggest that IS629 and IEE elements may be acting synergistically to promote genome plasticity and genetic diversity among STEC strains, enhancing their abilities to adapt to hostile environments and rapidly take up virulence factors.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) strains are important food pathogens responsible for serious human disease worldwide (1–3). Although new STEC serotypes constantly emerge (4), the mechanisms by which these strains acquire virulence are not entirely understood. Elements such as phages, pathogenicity islands (PAIs), and plasmids can introduce new virulence genes (such as those for producing Shiga toxin) that contribute to the pathogenic potential of strains, but the existence of horizontal transfer is not sufficient to explain the subsequent evolution of these strains or why some strains carry more virulence genes than others.
Karmali et al. (5) classified STEC into seropathotypes (SPT) from A to E in descending order of virulence. Serotypes that are frequently linked to outbreaks and severe disease, for example, hemolytic-uremic syndrome (HUS), are classified as SPT A. Serotypes in SPT B are also associated with outbreaks and severe disease, but these associations occur less frequently in SPT B serotypes than those in SPT A serotypes. Serotypes associated with sporadic cases of HUS, but not known to be responsible for outbreaks, are classified as SPT C, and serotypes classified as SPT D are known to have caused diarrhea but not HUS or outbreaks. The last category, SPT E is reserved for serotypes not yet implicated in human disease. These SPT classifications are dynamic since serotypes may shift from one category to another on the basis of new epidemiological data (6). Since 2009, the CDC has recognized the importance of STEC serotypes other than O157:H7 (SPT A) and has recommended the detection of the presence of the stx toxin in every stool sample to diagnose Shiga toxin-producing E. coli infections for clinical laboratories. This is because it has become evident that STEC serotypes other than O157:H7 can cause severe disease, such as hemolytic uremic syndrome (7).
The evolution of pathogens is also influenced by insertion sequence (IS) elements that have contributed to the diversification of enterohemorrhagic Escherichia coli (EHEC) O157 (8, 9) (10, 11). These IS elements move by inserting several copies of themselves randomly in the same genome, generally by a replicative mode (12). It is possible for the genomes of E. coli O157:H7 strains to contain many different IS elements; strain Sakai carries 25 different types of IS elements and a total of 116 copies of IS elements, of which 23 copies are IS629 (13, 14). Some authors have speculated that IS elements are kept in the genome because they are not simply parasitic DNA but also have a cooperative function, supporting the evolutionary fitness of the cells that carry them (15). IS elements also have been linked to a series of genomic rearrangements; simple insertions, excisions and deletions, and inversions and duplications have been described (16, 17).
An IS excision enhancer (IEE) was recently discovered in EHEC O157 that promotes the excision and deletion of IS3 family members (such as IS629) and also causes genomic rearrangements and strain diversification (16, 18). Kusumoto et al. observed four different kinds of IEE-promoted genomic deletions: (i) IS629 can be completely deleted with a short (1 to 7 bp) or (ii) long contiguous sequence. Alternatively, (iii) IS629 can be only partially deleted, or (iv) a long adjacent sequence to IS629 can be deleted (9). Similar IEE genes have been found in a broad range of bacteria, frequently forming parts of larger integrative elements, such as SpLE1 (as seen in O157:H7 strains Sakai and EDL933), or other, similar elements (9). Long-term experimental studies of E. coli have shown that IS-mediated mutations can contribute to E. coli adaptability or otherwise improve cell fitness (19, 20). The simultaneous presence of IEE and IS3 family members in the same genome may, therefore, be part of a mechanism that synergistically promotes adaptation and genome diversification, with IEE accelerating this process.
In a previous study, we analyzed strains of the E. coli O157:H7 stepwise evolutionary model (21, 22) and determined different IS629 excision frequencies for each clonal complex (CC) A1 to A6 (11). Strains belonging to the most common, non-sorbitol-fermenting (NSF), glucuronidase (GUD)-negative O157:H7 lineage exhibited a high IS629 excision frequency, which was linked to the presence of IEE in their genomes (EDL933 [A6] and G5101 [A5]) (11). The O157:H7 genomes we studied showed a highly diverse IS629 distribution, which might be related to the elevated IS629 transposition frequency (10). In contrast, another study showed that two O55:H7 strains (from A1 and A2) and a closely related sorbitol-fermenting (SF) strain (A4), which did not carry IEE, had lower IS629 excision frequencies (11).
To establish whether IEE and IS629 play significant roles in the genomic plasticity and genetic diversity of STEC, as they do in O157:H7, we screened 461 E. coli strains for the prevalence of these two elements. Our strain collection included 66 different serotypes from all 5 SPT to test whether they are present in the most pathogenic and diverse E. coli SPT, indicating a possible role in enhancing the pathogenicity potential of those strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli isolates (n = 461) from 72 different serotypes (see Tables S1 and S2 in the supplemental material) included O157:H7 and non-O157 STEC. We assigned serotypes into seropathotypes (SPT) based on current data by combining the original classification from Karmali et al. with other data found in the works of Bosilevac et al. and the European Food Safety Authority (EFSA) panel on Biological Hazards (BIOHAZ/EFSA) (current for 2013) (5, 6, 23). Most strains (n = 415 [90%]) were assigned to SPT A to E (see Table S1 in the supplemental material). Serotypes with no data for human disease were classified as SPT E according to Karmali's scheme (5). Strains for which only partial information was available were classified as unknown. All strains were plated on sorbitol MacConkey agar overnight at 37°C. DNA from overnight cultures grown on tryptic soy agar (TSA) at 37°C was extracted with InstaGene matrix (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions.
DNA preparation.
Genomic DNA from each strain was isolated from overnight cultures using the DNeasy blood and tissue kit (Qiagen, Valencia, CA), following the manufacturer's instructions. The resultant DNA solution was stored at −20°C until it was used as a PCR template.
Detection of IS629 and IEE.
The presence of IS629 and IEE was determined by PCR using internal IS629 primers (IS629-TF1 5′-ACT AAA AAT ACT CGT TTT TCC CCC GAA G-3′ and IS629-TR1 5′-GGCTGCCAGATCATCGTTTCCGATG-3′; 1,208 bp) and IEE-specific primers (IEEF4 5′-GAT AAA CGA TTT GCC GGG ACG GAA TG-3′ and IEER4 5′-GTT TAC CTC CCC CGA TAA TAC CAA TAC-3′, 280 bp) as described previously (8). The 25-μl PCR mix contained 1 U of GoTaq DNA polymerase (Promega, Madison, WI), 1× polymerase buffer, 3.5 mM MgCl2, 200 μM deoxynucleoside triphosphate (dNTP), 200 nM each primer, and ∼1 ng of template DNA. PCR conditions were as follows: 3 min at 90°C to activate the Taq polymerase, followed by 30 cycles of denaturation at 95°C for 15 s, then primer annealing and extension at 62°C for 30 s and 72°C for 1.5 min, and a final extension at 72°C for 1 min. Products were examined on a 1% agarose gel in 1× Tris-borate-EDTA (TBE) buffer.
Sequencing of IEE elements.
The IEE core sequences were determined using IEE primers designed for this study (IEE LR SFW 5′-CCC GTC TGG TCT CAT TAC TTG-3′ and IEE LR SREV 5′-TAT CCC ACA TTC TGA GCA GCA-3′) to retrieve a 2,241-bp product (conditions as described above). The partial IEE element was sequenced using internal sequencing primers: IEE LR SF1 (5′-GCATGTCTGGATATTTTTTGCCTCA-3′), IEE LR SR1 (5′-TGAGGCAAAAAATATCCAGACATGC-3′), IEE LR SF2 (5′-TGAAAATCACGCTGGCAAACCAGA-3′), IEE LR SR2 (5′-TCTGGTTTGCCAGCGTGATTTTCA-3′), IEE LR SF3 (5′-AGTGATTGCCAAGCGGAAAGTGAATA-3′), IEE LR SR3 (5′-TATTCACTTTCCGCTTGGCAATCACT-3′), IEE LR SF4 (5′-GCGAAAAGTTCTGGTACTGACTGAAC-3′), and IEE LR SR4 (5′-GTTCAGTCAGTACCAGAACTTTTCGC-3′). The IEE PCR products were sequenced by Genewiz Inc. using the same primers (Germantown, MD). DNA sequences were individually inspected and manually assembled. Alignments of these sequences were performed with BioEdit software version 7.2.5 (24).
IEE phylogenetic tree.
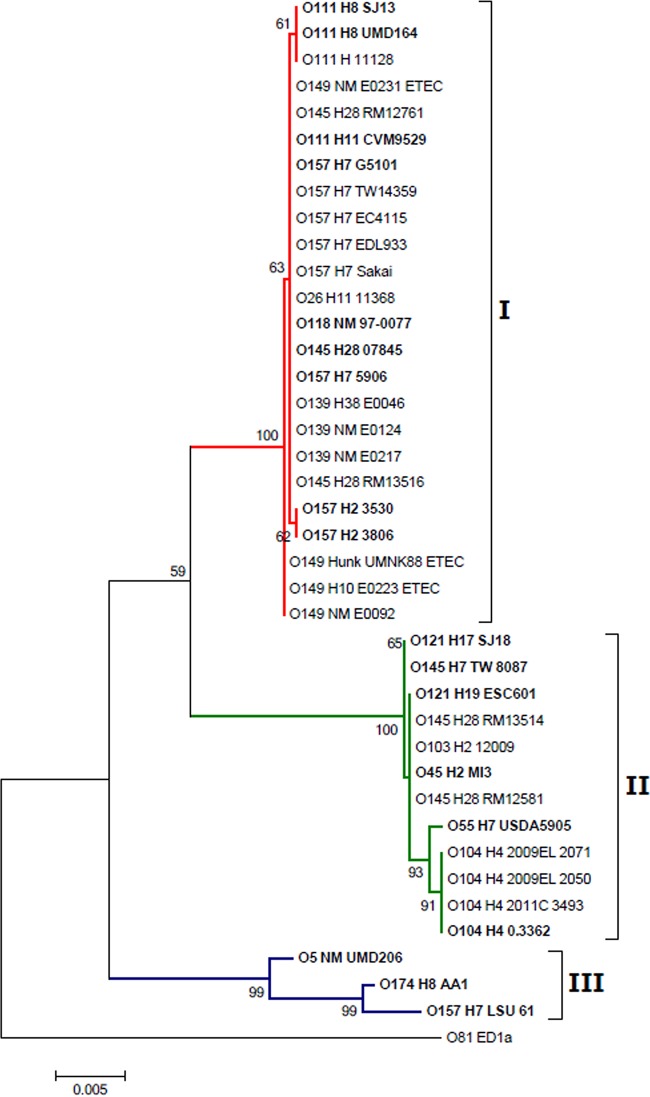
A maximum likelihood phylogenetic tree was constructed using MEGA software version 6.06 (25) based on the IEE sequences from different STEC strains. The neighbor-joining algorithm (26) was used to generate the initial tree. Then, the statistical support of the nodes in the ML tree was assessed by 1,000 bootstrap resampling. The general time reversible (GTR) model and gamma distribution with invariant sites (G+I) were selected. Nearest-neighbor-interchange (NNI) was used as a heuristic method. IEE sequences available publicly at NCBI in the GenBank databse were used to build the phylogeny: E. coli O111:H-str. 11128 (AP010960.1), E. coli O26:H11 str. 11368 (AP010953.1), E. coli O157:H7 str. Sakai (BA000007.2), E. coli O157:H7 str. EDL933 (AE005174.2), E. coli O157:H7 str. EC4115 (CP001164.1), E. coli O157:H7 str. TW14359 (CP001368.1), E. coli O145:H28 str. RM12761 (CP007133.1), E. coli O145:H28 str. RM13516 (CP006262.1), E. coli O145:H28 str. RM12581 (CP007136.1), E. coli O145:H28 str. RM13514 (CP006027.1), E. coli O139:H38 str. E0046 (AB786874.1), E. coli O139:NM str. E0124 (AB786876.1), E. coli O139:NM str. E0217 (AB786877.1), E. coli O149:NM str. E0231 (AB786879.1), E. coli O149:H10 str. E0223 (AB786878.1), E. coli O149:H? str. E0092 (AB786875.1), E. coli O149:H? str. UMN K88 (CP002729.1), E. coli O104:H4 str. 2011C-3493 (CP003289.1), E. coli O104:H4 str. 2009EL-2050 (CP003297.1), E. coli O104:H4 str. 2009EL-2071 (CP003301.1), and E. coli O103:H2 str. 12009 (AP010958.1). Since a BLAST analysis of whole genomes available in GenBank showed that more than 100 other O157:H7 strains also contained IEE sequences that were 100% identical to the IEE found in Sakai, we included only a few O157:H7 IEE sequences for illustrative purposes (Fig. 1). The IEE sequence of ED1a-O81 (GenBank accession number NC_007519.1) was used as an out group (see Table S3 in the supplemental material).
FIG 1.
Maximum likelihood phylogenetic tree with 3 IEE types observed in O157 and non-O157 STEC and their phylogenetic relationships. The IEE sequence of ED1a-O81 (NC_007519.1) was used as an out group. IEEs sequenced in this study are indicated in bold.
Whole-genome phylogenetic SNP analysis of E. coli genomes.
In order to study the lateral transfer acquisition of the IEE elements, we performed a whole-genome single nucleotide polymorphism (SNP) analysis of eight complete E. coli genomes available at NCBI (O157:H7 strains Sakai, EDL933, TW14359, and EC4115; O145:H28 strain RM13514; O111:H strain 11128; O26:H11 strain 11368; O104:H4 strain 2011C-3493; and O139:H28 strain E24377A) using the free snpTree server (https://cge.cbs.dtu.dk/services/snpTree) (27). This broad SNP study provided an evolutionary context for some E. coli isolates used for the analysis. All E. coli genomes analyzed carried IEE type I (Fig. 1). The snpTree server generated a matrix of core SNPs (i.e., SNPs that were present in all eight genomes) for each lineage using the reference-based SNP-finding program (27). The genome of E. coli Sakai was used as a reference, with the following filtering parameters: minimum distance between SNPs (prune), 10 bp, and minimum distance to end of sequence, 20 bp. The final matrix consisted of 70,282 SNPs.
Nucleotide sequence accession numbers.
All newly identified IEE sequences determined in this study were deposited in GenBank under accession numbers JF330836 to JF330838 and KP235311 to KP235328.
RESULTS
In the present study, we screened 461 STEC strains representing 72 different serotypes and all five seropathotypes (SPT) for the presence of IS629 and IEE. We also studied the lateral acquisition of IEE elements by analyzing the sequence of several IEE elements and comparing them to those of the whole-genome phylogeny of the strains.
Distribution of IS629 and IEE elements among the SPTs.
In our study, most strains were positive for IS629 (405 [88%]), and more than half carried IEE (277 [60%]). The two elements were simultaneously present in 271 strains (59%); most strains carrying the two elements belonged to SPT A or B. The presence of either element did not appear to be serogroup-specific (Table 1); however, in most of the cases (62%), it was serotype (O type and H types combined) specific (see Table S2 in the supplemental material). Examples include serogroup O103, with O103:H2 (IEE-positive [IEE+] and IS629-positive [IS629+]) and O103:H6 (IEE-negative [IEE−] and IS629+), and serogroup O104, with O104:H4 (IEE+ and IS629+) and O104:H21 (IEE− and IS629+). Strains from seropathotypes C, D, E, and unknown (UNK) contained either IEE or IS629 but not the two elements together. This either/or pattern was seen in only one SPT A strain (LSU-61) and two SPT B strains. Most of the strains classified as SPT E carried only IS629, if they carried any element at all (121/138).
TABLE 1.
IEE and IS629 prevalence by seropathotype
| Presence IEE and/or IS629 | Seropathotypea |
|||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | UNKb | |
| IEE and IS629 | O157:H7 | O26:H11 | O55:H7 | O50:H7 | O104:?c | |
| O45:H2 | O111:H11O | O121:? | ||||
| O103:H2 | O118:NMd | O145:? | ||||
| O104:H4 | O121:H17 | |||||
| O111:H8 | O125:NM | |||||
| O121:H19 | 157:H2 | |||||
| O145:H28 | O157:H16 | |||||
| O157:H19 | ||||||
| O157:H29 | ||||||
| O157:H43 | ||||||
| O139:NMe | ||||||
| O149:NMe | ||||||
| IEE | O157:H7f | g | O5:NM | O174:H8 | g | |
| O113:H21 | O55:H7 | |||||
| IS629 | g | O103:H2 | O104:H21 | O55:H7 | O2:H7 | O91:? |
| O145:H28 | O5:NM | O73:H18 | O2:H25 | O106:? | ||
| O157:H11 | O2:H27 | O28ac | ||||
| O8:H16 | ||||||
| O8:H30 | ||||||
| O15:H16 | ||||||
| O22:H5 | ||||||
| O28ac:H7 | ||||||
| O103:H6 | ||||||
| O104:H6 | ||||||
| O104:H7 | ||||||
| O104:H11 | ||||||
| O104:H12 | ||||||
| O104:H27 | ||||||
| O126:H8 | ||||||
| O127:NM | ||||||
| O157:H1 | ||||||
| O157:H16 | ||||||
| O157:H42 | ||||||
| O157:H43 | ||||||
| None | g | O45:H2 | O91:H21 | O4:NM | O8:H2 | O9:? |
| O128:H2 | ||||||
| O113:H21 | ||||||
| O15:H27 | O8:H28 | O92:? | ||||
| O118:H16 | ||||||
| O146:H21 | O22:H8 | O105:? | ||||
| O168:H8 | O28ac:H7 | O122:? | ||||
| O36:H14 | ||||||
| O46:H38 | ||||||
| O83:H8 | ||||||
| O88:H38 | ||||||
| O104:H49 | ||||||
| O113:H36 | ||||||
| O128:H45 | ||||||
| O157:H1 | ||||||
| O157:H32 | ||||||
| O157:H46 | ||||||
| O157:Ha18d | ||||||
| O174:H36 | ||||||
| O174:H36 | ||||||
| O28ac:H7 | ||||||
Genetic variation among IEE sequences found among E. coli strains in this study.
In order to define the genetic differences among IEE sequences, we sequenced the IEE from a representative strain from each individual non-O157 serotype that was positive for IEE. Then, by comparing 18 IEE sequences from our current project with 22 additional IEE sequences available in GenBank, we were able to group IEEs into three clusters (Fig. 1); clusters I (9 IEE serotypes) and II (7 IEE serotypes) appear to be the most prevalent in this study.
Cluster I includes IEE sequences from STEC serotypes O157:H7, O26:H11, O111:H11, O118:NM, O145:H28, and O139:NM/H38, which were identical. Serotypes O111:H8, O157:H2, and O149:NM also group within IEE cluster I but with minor differences (1 to 2 SNPs). The IEE cluster II includes IEE sequences from serotypes O104:H4, O55:H7, O121:H17, O145:H28, O45:H2, O103:H2, and O121:H19. In this group, IEE sequences cluster by serotype but differ among each other more than cluster I sequences (Fig. 1). The IEE found in the O55:H7 group clustered closely together with O104:H4 more than sequences of any other IEE in this group, and they cluster far from the IEE from O157:H7. Serotypes O145:H28, O45:H2, O103:H2, and O121:H19 display identical IEE sequences and cluster separately from that of serotype O121:H17. The IEE cluster III includes IEE sequences from serotypes O5:NM and O174:H8 (both lacking IS629) and E. coli O157:H7 strain LSU-61 (which also carries IS629). However, this last E. coli O157:H7 strain also lacks stx genes and clusters apart from the other O157:H7 strains in our study.
In this study, serotype O145:H28 appears to carry two different IEE sequences: strains RM12761, 07845, and RM13516 carried IEE cluster I and two others (RM13514 and RM12581) carried IEE cluster II (Fig. 1).
DISCUSSION
E. coli O157:H7 has been frequently associated with severe diseases and outbreaks (28), and its genome is highly diverse mainly due to the acquisition of mobile genetic elements, such as phages and other mobile elements (like ISs) (22). While phages contribute to genome plasticity, occasionally carrying virulence genes such as Shiga-toxins, gene disruptions and rearrangements caused by ISs result in those virulence genes becoming permanently fixed in the genome (15, 23). In a previous study, we observed that highly pathogenic O157:H7 carries IS629 and IEE, suggesting that a synergistic mechanism between these two elements may contribute to genome diversification (9, 10).
In this study, we hypothesized that there is an association between the simultaneous presence of IS629 and IEE elements in STEC strains and their pathogenic potential (represented by their seropathotype). Our main finding was that almost all SPT A and SPT B strains contained IS629 and IEE (249 [98%]). These two SPTs include the most pathogenic and clinically relevant non-O157:H7 STEC serotypes (O26:H11, O103:H2, O111:H8, O121:H19, and O145:H28). These IS629 and IEE elements were not found together in the majority of those serotypes, which have rarely, or never, been implicated in human illness (6, 29). Notably, strains of serotype O104:H4, the causative agent of the devastating E. coli outbreak in Germany in 2011, also carried the two elements (3, 4). Analyses showed that the outbreak strain had evolved from an enteroaggregative E. coli (EAEC) that had acquired the stx gene through horizontal gene transfer (4).
We found that serogroups O139 and O149 and serotypes O50:H7, O111:H11, O118:NM, O121:H17, O125:NM, O157:H2, and O157:H16 also carried the IEE and IS629 combination, which suggests that these strains may have an enhanced pathogenic potential, despite the fact that they have not yet been associated with human illness. The established behaviors of two SPT E strains, enterotoxigenic E. coli (ETEC) serotypes O139 and O149, support this hypothesis. They have already been recognized as major causes of diarrhea and edema disease in swine (30). It may be that these ETEC serotypes are more able than others to acquire virulence elements thorough horizontal gene transfer.
Conversely, strains from seropathotypes C, D, E, and unknown (UNK) contained either IEE or IS629 but not the two elements together. This either/or pattern was seen in only one SPT A strain and two SPT B strains. Strain LSU-61 (SPT A) is considered an atypical O157:H7 strain because it displays a mix of different traits from various clonal complexes, including SF-positive (SOR+) and β-glucuronidase-positive (GUD+) traits (10, 11, 21, 31). However, it is possible that the absence of IEE in the 2 SPT B strains might be explained as lineage differences; there are two different lineages within this serotype (e.g., O103:H2).
A previous study in swine isolates reported that the presence of either element (IS629 or IEE) is serotype specific (8); results from Kusumoto et al. suggest that IEEs spread to different lineages of ETEC and EHEC by horizontal gene transfer with a highly lineage-specific occurrence (8). Our results confirm those findings, as isolates from the same serogroup but different serotypes carry differences in the presence of the two elements. For example, strains of serogroups O103 and O104 displayed different IEE and/or IS629 combinations on the serotype (Table 1). These results show that the presence of IEE and IS629 appears to be related to a strain H-type (lineage specific) and that this IEE and IS629 combination appears to be associated mostly with serotypes related to high pathogenicity and human disease (SPT A and B). This supports other findings by Stanton et al., who found an association between the presence of IS629 and elements with an effect on virulence and phage production (32).
Some serotypes—O5:NM and O174:H8—carried IEE alone (these strains represented SPT C and D, respectively). Although IS629 was absent in those strains, they did appear to carry IS2, another IS3 family member (data not shown) prevalent among pathogenic E. coli and the excision of which is also enhanced by IEE (8, 9).
As suggested by Kusumoto et al., our observations confirm that IEEs within each cluster evolved independently of their current host genomes and were most likely acquired via horizontal gene transfer (9). Finding identical sequences in disparate E. coli lineages that have evolved separately confirms that the same IEE element has been acquired multiple times by different E. coli lineages, most likely by horizontal gene transfer (see Fig. S1 in the supplemental material).
Strains of different serotypes clustered together to form cluster I, and many of them displayed identical sequences (O157:H7, O26:H11, O111:H11, O118:NM, O145:H28, and O139:NM/H38). Those serotypes have frequently been involved in serious outbreaks, whereby those with minor differences rarely cause illness (serotype O111:H8/NM) or have not yet been implicated in human illness (O157:H2, O149:NM) (33, 34, 35). Although less frequent in human outbreaks than E. coli O111:H11, serotype O111:H8 has been implicated in severe disease, such as hemolytic uremic syndrome (HUS) (36). Cluster II also includes serotypes frequently associated to human disease; however, cluster III groups strains with a lower pathogenic potential.
Although most of O157:H7 strains belong to cluster I, the IEE of LSU-61 is located in cluster III. Previous researchers have debated whether LSU-61 may be a potential intermediate between serogroups O55:H7 and O157:H7 (21). However, its IEE sequence neither clusters close to other O157:H7 nor to ancestral O55:H7, reducing the likelihood that it is a missing link. A similar finding was shown by analyzing these two serotypes using clustered regularly interspaced short palindromic repeats (CRISPR); LSU-61 carries repeats not present in either serotype (37). These findings suggest that LSU-61 evolved on a separate path from typical O157:H7 and might be better considered as part of a different O157:H7 lineage rather than an ancestor of highly pathogenic O157:H7. Our data suggest that IEEs were acquired by O55:H7, typical O157:H7, atypical O157:H7, and LSU-61 independently and that these strains diverged earlier. Intriguingly, an O157:H7 strain similar to LSU-61 was isolated from a red deer in 2001 in Europe, but it has not yet been reported to cause human disease (38). This suggests that IEE was acquired separately by each serotype.
Interestingly, serotype O145:H28 appears to carry two different IEE sequence types (Fig. 1) even when all four O145:H28 strains are closely related; however, these strains differed by food matrix: RM13516 was isolated in Belgium in 2007 from an ice-cream outbreak (39, 40), whereby RM12581 was a lettuce-associated outbreak strain identified in the United States in 2010 (41). Little is known about serotype O145:H28 genetic diversity, but this finding suggests that it might have two different lineages that acquired IEE separately. Since the two IEEs belong to the pathogenic IEE clusters (I and II), those differences might not affect the likelihood that these lineages may evolve to become more virulent.
Since IS elements (particularly IS629) are important driving forces behind genomic diversity in O157:H7 (16, 42), if IEE accelerates IS629-mediated mutations, the presence of the two elements in a single genome may improve the plasticity and fitness of major STEC serotypes over strains lacking them (19, 20). This might occur because the preferred substrates for IEE are IS3 family members (specifically IS629) resulting in a negative feedback loop. When IS629's OrfB overexpresses, its transposase (OrfAB), which is obtained by a −1 frameshift signal, is repressed. As a consequence, the expected transposition is inhibited (43). Genomes having a high copy number of IS629 will also overexpress OrfB, and IS629 transposition will be reduced or inhibited. When IEE removes (using any of the 4 types of genomic deletions) some of those IS629 copies (9), the levels of OrfB will be reduced and the transposition of IS629 will be indirectly promoted. This, in turn, increases the probability that IS629 will be inserted into a new region of the genome, subsequently silencing or fixating new genes, and thereby increasing genome plasticity. Only those strains that gain survival benefits will thrive, but past observations have shown that enhancements to virulence or persistence in the environment have often been associated with increased survival.
Some researchers have suggested that new clones may emerge due to the intervention of IS629 (44) and, further, that IS629 can inactivate inserted genes, thereby promoting the fixation of mobile elements, such as phages (45). For example, in E. coli O157:H7 (KE531) the stx2 gene has been silenced by an IS629 (45). Further activation of transposition/excision mechanisms produced a perfect excision of the IS629, resulting in a strain with a functional stx2 gene (45). Many other non-O157 serotypes appear to be emerging to highly pathogenic serotypes and are being isolated from unusual sources (39–41). Genomic plasticity allows rapid adaptation to changing environments, which provides an evolutionary advantage to virulent E. coli serotypes.
The genomes of STEC strains are much larger (5.5 to 5.9 Mb) than those of other E. coli strains because they contain virulence factors located in prophages, pathogenicity islands (PAIs) (46), and other integrative elements (IEs) (47) typically acquired through horizontal gene transfer (4). After up-take of an Stx2 bacteriophage, enteroaggregative E. coli (EAEC) O104:H4 evolved to a highly virulent clone (4). Therefore, another way that the IEE and IS629 system may increase pathogenicity is by inserting previously IEE-excised IS elements into genes required for the mobility of mobile elements, thereby integrating phages carrying virulence markers and PAIs permanently into the hosts' genome (16). The process of evolution is accelerated by rapid uptake of phages and other mobile elements carrying virulence factors, increasing the likelihood of pathogenic clones.
In conclusion, the combined presence of IEE and IS629 in highly pathogenic serotypes may have the potential to leverage the evolution to strains with higher virulence due to beneficial mutations to enhance adaptation. Therefore, it is possible that serotypes not currently associated with human disease but carrying IS629 and IEE may evolve to become new, highly virulent serotypes. Similarly, less predominant serotypes may likely, due to their more rapidly changing genomes, evolve to more pathogenic serotypes.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by the FDA Foods Program Intramural Funds and the ORISE fellowship program.
We thank Masahiro Kusumoto and his colleagues from the Bacterial and Parasitic Disease Research Division, National Institute of Animal Health Kannondai, Japan, for providing further E. coli O149 and O139 strain information used in their IEE study. We thank Lili Fox Vélez for her editorial assistance on the manuscript.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Food and Drug Administration.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01349-15.
REFERENCES
- 1.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Muller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 2.Frank C, Faber MS, Askar M, Bernard H, Fruth A, Gilsdorf A, Hohle M, Karch H, Krause G, Prager R, Spode A, Stark K, Werber D. 2011. Large and ongoing outbreak of haemolytic uraemic syndrome, Germany, May 2011. Euro Surveill 16(21):pii=19878 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19878. [PubMed] [Google Scholar]
- 3.Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 4.Beutin L, Martin A. 2012. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J Food Prot 75:408–418. doi: 10.4315/0362-028X.JFP-11-452. [DOI] [PubMed] [Google Scholar]
- 5.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol 41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EFSA Panel on Biological Hazards (BIOHAZ). 2013. Scientific opinion on VTEC-seropathotype and scientific criteria regarding pathogenicity assessment. EFSA J 11:3138. [Google Scholar]
- 7.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D'Angelo M, Griffin PM, Gerner-Smidt P. 2009. Recommendations for diagnosis of Shiga toxin–producing Escherichia coli infections by clinical laboratories. MMWR Recomm Rep 58:1–14. [PubMed] [Google Scholar]
- 8.Kusumoto M, Fukamizu D, Ogura Y, Yoshida E, Yamamoto F, Iwata T, Ooka T, Akiba M, Hayashi T. 2014. Lineage-specific distribution of insertion sequence excision enhancer in enterotoxigenic Escherichia coli isolated from swine. Appl Environ Microbiol 80:1394–1402. doi: 10.1128/AEM.03696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusumoto M, Ooka T, Nishiya Y, Ogura Y, Saito T, Sekine Y, Iwata T, Akiba M, Hayashi T. 2011. Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. Nat Commun 2:152. doi: 10.1038/ncomms1152. [DOI] [PubMed] [Google Scholar]
- 10.Rump LV, Fischer M, Gonzalez-Escalona N. 2011. Prevalence, distribution and evolutionary significance of the IS629 insertion element in the stepwise emergence of Escherichia coli O157:H7. BMC Microbiol 11:133. doi: 10.1186/1471-2180-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rump LV, Fischer M, Gonzalez-Escalona N. 2011. Different IS629 transposition frequencies exhibited by Escherichia coli O157:H7 strains in the stepwise evolutionary model. Appl Environ Microbiol 77:5030–5033. doi: 10.1128/AEM.00249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duval-Valentin G, Marty-Cointin B, Chandler M. 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J 23:3897–3906. doi: 10.1038/sj.emboj.7600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han CG, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res 5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Sinzelle L, Izsvak Z, Ivics Z. 2009. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci 66:1073–1093. doi: 10.1007/s00018-009-8376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooka T, Ogura Y, Asadulghani M, Ohnishi M, Nakayama K, Terajima J, Watanabe H, Hayashi T. 2009. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res 19:1809–1816. doi: 10.1101/gr.089615.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G III, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun 71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci U S A 96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng PC, Monday SR, Lacher DW, Allison L, Siitonen A, Keys C, Eklund M, Nagano H, Karch H, Keen J, Whittam TS. 2007. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis 13:1701–1706. doi: 10.3201/eid1311.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol 187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosilevac JM, Koohmaraie M. 2011. Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl Environ Microbiol 77:2103–2112. doi: 10.1128/AEM.02833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp 41:95–98. [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 27.Leekitcharoenphon P, Kaas RS, Thomsen MC, Friis C, Rasmussen S, Aarestrup FM. 2012. snpTree–a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics 13(Suppl):S6. doi: 10.1186/1471-2164-13-S7-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol 295:405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention. 2013. National Shiga toxin-producing Escherichia coli (STEC) surveillance. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 30.Frydendahl K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet Microbiol 85:169–182. doi: 10.1016/S0378-1135(01)00504-1. [DOI] [PubMed] [Google Scholar]
- 31.Rump LV, Strain EA, Cao G, Allard MW, Fischer M, Brown EW, Gonzalez-Escalona N. 2011. Draft genome sequences of six Escherichia coli isolates from the stepwise model of emergence of Escherichia coli O157:H7. J Bacteriol 193:2058–2059. doi: 10.1128/JB.00118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanton E, Park D, Dopfer D, Ivanek R, Kaspar CW. 2014. Phylogenetic characterization of Escherichia coli O157: H7 based on IS629 distribution and Shiga toxin genotype. Microbiology 160:502–513. doi: 10.1099/mic.0.073437-0. [DOI] [PubMed] [Google Scholar]
- 33.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 34.Malloy CD, Marr JS. 2001. Evolution of the Control of Communicable Diseases Manual: 1917 to 2000. J Public Health Manag Pract 7:97–104. doi: 10.1097/00124784-200107050-00013. [DOI] [PubMed] [Google Scholar]
- 35.Centers for Disease Control and Prevention. 2008. Shiga toxin-producing Escherichia coli: burden and trends. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/FoodNet/news/2008/January_FoodNet_News.pdf. [Google Scholar]
- 36.Brooks JT, Bergmire-Sweat D, Kennedy M, Hendricks K, Garcia M, Marengo L, Wells J, Ying M, Bibb W, Griffin PM, Hoekstra RM, Friedman CR. 2004. Outbreak of Shiga toxin-producing Escherichia coli O111:H8 infections among attendees of a high school cheerleading camp. Clin Infect Dis 38:190–198. doi: 10.1086/380634. [DOI] [PubMed] [Google Scholar]
- 37.Yin S, Jensen MA, Bai J, Debroy C, Barrangou R, Dudley EG. 2013. The evolutionary divergence of Shiga toxin-producing Escherichia coli is reflected in clustered regularly interspaced short palindromic repeat (CRISPR) spacer composition. Appl Environ Microbiol 79:5710–5720. doi: 10.1128/AEM.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn JR, Keen JE, Moreland D, Alex T. 2004. Prevalence of Escherichia coli O157:H7 in white-tailed deer from Louisiana. J Wildl Dis 40:361–365. doi: 10.7589/0090-3558-40.2.361. [DOI] [PubMed] [Google Scholar]
- 39.Buvens G, Posse B, De Schrijver K, De Zutter L, Lauwers S, Pierard D. 2011. Virulence profiling and quantification of verocytotoxin-producing Escherichia coli O145:H28 and O26:H11 isolated during an ice cream-related hemolytic uremic syndrome outbreak. Foodborne Pathog Dis 8:421–426. doi: 10.1089/fpd.2010.0693. [DOI] [PubMed] [Google Scholar]
- 40.De Schrijver K, Buvens G, Posse B, Van den Branden D, Oosterlynck O, De Zutter L, Eilers K, Pierard D, Dierick K, Van Damme-Lombaerts R, Lauwers C, Jacobs R. 2008. Outbreak of verocytotoxin-producing E. coli O145 and O26 infections associated with the consumption of ice cream produced at a farm, Belgium, 2007. Euro Surveill 13(7):pii=8041 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8041. [DOI] [PubMed] [Google Scholar]
- 41.Taylor EV, Nguyen TA, Machesky KD, Koch E, Sotir MJ, Bohm SR, Folster JP, Bokanyi R, Kupper A, Bidol SA, Emanuel A, Arends KD, Johnson SA, Dunn J, Stroika S, Patel MK, Williams I. 2013. Multistate outbreak of Escherichia coli O145 infections associated with romaine lettuce consumption, 2010. J Food Prot 76:939–944. doi: 10.4315/0362-028X.JFP-12-503. [DOI] [PubMed] [Google Scholar]
- 42.Ooka T, Terajima J, Kusumoto M, Iguchi A, Kurokawa K, Ogura Y, Asadulghani M, Nakayama K, Murase K, Ohnishi M, Iyoda S, Watanabe H, Hayashi T. 2009. Development of a multiplex PCR-based rapid typing method for enterohemorrhagic Escherichia coli O157 strains. J Clin Microbiol 47:2888–2894. doi: 10.1128/JCM.00792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CC, Hu ST. 2006. Two frameshift products involved in the transposition of bacterial insertion sequence IS629. J Biol Chem 281:21617–21628. doi: 10.1074/jbc.M602437200. [DOI] [PubMed] [Google Scholar]
- 44.Hirai S, Yokoyama E, Yamamoto T. 2013. Linkage disequilibrium of the IS629 insertion among different clades of enterohemorrhagic Escherichia coli O157:H7/H-strains. Infect Genet Evol 18:94–99. doi: 10.1016/j.meegid.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Kusumoto M, Okitsu T, Nishiya Y, Suzuki R, Yamai S, Kawamura Y. 2001. Spontaneous reactivation of Shiga toxins in Escherichia coli O157:H7 cells caused by transposon excision. J Biosci Bioeng 92:114–120. doi: 10.1016/S1389-1723(01)80210-3. [DOI] [PubMed] [Google Scholar]
- 46.Herold S, Karch H, Schmidt H. 2004. Shiga toxin-encoding bacteriophages–genomes in motion. Int J Med Microbiol 294:115–121. doi: 10.1016/j.ijmm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 47.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.