SUMMARY
The development of multicellular organisms requires the precisely coordinated regulation of an evolutionarily-conserved group of signaling pathways. Temporal and spatial control of these signaling cascades is achieved through networks of regulatory proteins and/or segregation of pathway components in specific subcellular compartments. In vertebrates, dysregulation of primary cilia function has been strongly linked to developmental signaling defects, yet it remains unclear whether cilia sequester pathway components to regulate their activation or whether cilia-associated proteins directly modulate developmental signaling events. To elucidate this question, we conducted an RNAi-based screen in Drosophila non-ciliated cells to test for cilium-independent loss-of-function phenotypes of ciliary proteins in developmental signaling pathways. Our results show no effect on Hedgehog signaling. In contrast, our screen identified several cilia-associated proteins as functioning in canonical Wnt signaling. Further characterization of specific components of Intraflagellar Transport complex A (IFT-A) uncovered a cilia-independent function in potentiating Wnt signals by promoting β-catenin/Armadillo activity.
Graphical abstract
INTRODUCTION
Signaling pathways that regulate embryonic development and adult homeostasis are highly conserved from Drosophila to humans. Many of these pathways were elucidated in Drosophila, in particular the Hedgehog (Hh), Wnt/Wingless (Wg), Notch (N), and RTK pathways, making the fly an ideal organism to identify new pathway regulators.
The primary cilium is an organelle involved in sensing the extracellular environment, and is required throughout development and for homeostasis in most eukaryotes. Cilia are microtubule-based protrusions arising from a modified centriole: the basal body (Pazour and Witman, 2003). Cilium biogenesis and function requires a complex network of cilia associated proteins, serving for example to anchor the basal body to the cell membrane or to selectively allow protein entry/exit, thus creating a distinct subcellular compartment (Gherman et al., 2006; Ishikawa and Marshall, 2011; Nachury et al., 2010; Pazour and Witman, 2003; Pedersen et al., 2008; Reiter et al., 2012; Taschner et al., 2012). The ability to function as a separate compartment appears key to the primary cilium acting as an environmental sensor and signaling hub (Berbari et al., 2009).
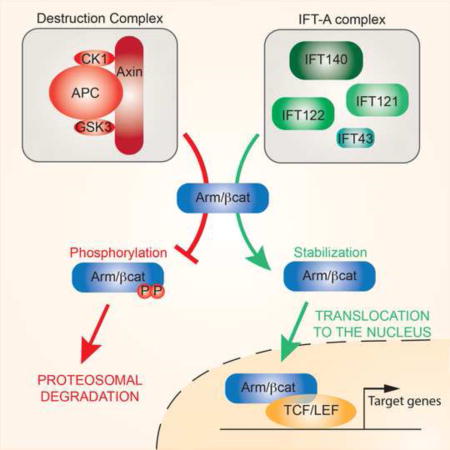
Cilia are critical for Shh/Hh signal transduction and potential roles in other developmental pathways are emerging (Berbari et al., 2009; Goetz and Anderson, 2010). Hh ligand binds to its receptor Patched (Ptc), to relieve Ptc inhibition of Smoothened (Smo), which then activates the downstream transcription factor Ci (Gli in vertebrates). Ptc, Smo and Gli all localize to the cilium in vertebrates in a ligand-regulated manner, and ciliary mutants disrupt Hh signaling (Briscoe and Therond, 2013; Goetz et al., 2009). It is unclear whether cilia facilitate Hh signaling by providing a specialized subcellular compartment, or whether (specific) ciliary components are directly required to transduce the Hh signal. In contrast, the cilium has been suggested to limit the response to Wnt signaling by affecting the stability and localization of β-catenin/Armadillo (β-cat/Arm) (Oh and Katsanis, 2013). When Wnt/Wg ligand binds to its receptor complex of Frizzled (Fz)/LRP5-6 (Arrow/Arr in Drosophila) it inhibits the function of the β-cat destruction complex, allowing β-cat accumulation and its nuclear translocation, and transcriptional activation via TCF/LEF transcription factors (Clevers and Nusse, 2012; MacDonald et al., 2009; Niehrs, 2012). Although roles for ciliary proteins in degrading β-cat and of limiting its nuclear entry have been reported (Corbit et al., 2008; Gerdes and Katsanis, 2008), there are many contradicting conclusions from analyses of Wnt signaling in the context of ciliary mutants, ranging from inactivation to over-activation of the pathway (Oh and Katsanis, 2013). Given the interest sparked by the primary cilium functioning in Hh and Wnt signaling, other signaling pathways, including Notch, RTK and TGFβ, have been examined. However, a specific requirement for cilia during signaling by each of these pathways remains to be confirmed (Christensen et al., 2012; Clement et al., 2013; Ezratty et al., 2011; Leitch et al., 2014; Ten Dijke et al., 2012).
The discovery that mutations in ciliary proteins can lead to several genetic disorders, termed ciliopathies, has led to great interest in this cellular compartment. It remains unclear, however, whether the affected ciliary proteins are directly required for signal transduction, or rather only to maintain the cilium as a functioning compartment. Interestingly, different roles for ciliary proteins in mitotic spindle orientation and immune synapse recycling that are independent of their function within the cilium have recently been described (Delaval et al., 2011; Finetti et al., 2009; Sedmak and Wolfrum, 2010). The main barrier to understanding how cilia-associated proteins could function independently of the cilium stems from the crucial role that most of these proteins play in biogenesis and maintenance of the cilium, and thus it is very difficult to distinguish cilia-dependent and cilia-independent effects. To overcome this problem we have used Drosophila as a model, as here all cell types are non-ciliated, with the exception of sensory neurons and sperm (Gogendeau and Basto, 2010). Consequently, any phenotypes associated with the loss of cilia-associated protein function should be due to a cilia-independent function(s).
We thus performed a two-step candidate RNAi-based screen in Drosophila, in vivo, to test for roles of cilia-associated proteins in developmental signaling pathways, independently of their ciliary function. We first assayed adult phenotypes in the wing and thorax for developmental defects. The respective hits were then analyzed using molecular markers in developing wing discs to determine which specific signaling pathways were altered. Wingless signaling was predominantly affected (17/63 tested genes) by knockdown of cilia-associated proteins, with small numbers modulating the Notch and EGFR/RTK pathways (4/63 and 2/63, respectively). Importantly, none of the candidates affected Hh signaling. We selected components of the Intraflagellar Transport complex A (IFT-A) for further study, as they can form a defined complex, and the majority of IFT-A components (4/5) appeared to have a comparable effects on Wg signaling. Strikingly, the IFT-A proteins act as positive regulators of Wg signaling. Epistasis experiments in vivo suggest that they function at the level of β-cat/Arm stabilization. Together, our data argue for a role of a specific subset of IFT-A proteins in the Wg pathway, distinct from their role in ciliary biogenesis and function.
RESULTS
An in vivo RNAi-based screen identifies a role for ciliary proteins in Wg/Wnt signaling in non-ciliated cells
To compile a list of candidate genes for the cilia-associated protein screen, we started with a total of 103 genes that were previously characterized in cilium and centriole biogenesis and function in vertebrates, or had been linked to ciliopathies in humans. Of these genes, 63 homologs are highly conserved in Drosophila and expressed during the imaginal disc patterning stages and were thus selected (Table S1 shows full listing). We then conducted an RNAi-based knockdown (KD) screen in two steps: a primary screen examining adult phenotypes for significant developmental defects, and a secondary screen examining specific molecular target genes to identify the respective signaling pathway(s) affected by each KD (Fig. 1, Suppl. Fig. S1, Table 1, and Suppl. Table S1). The Gal4-UAS system (Brand and Perrimon, 1993) was used to drive RNAi expression (KD), and wherever possible, we used at least two independent non-overlapping RNAi lines to eliminate false positive off-target effects. In the wing we used engrailed (en)-Gal4 in the posterior compartment and nubbin (nub)-Gal4 in the entire wing pouch. We additionally used apterous (ap)-Gal4 in the thorax (Fig. 1A–G, Suppl. Fig. S1A–B, and Suppl. Table S1). KDs of 38 genes of the 63 tested induced reproducible and specific phenotypes in the wing and/or thorax, which were categorized according to their appearance (Fig. 1B–G, Suppl. Table S1 and Fig. S1A–B). These effects suggested cilia-independent function(s) for several cilia-associated proteins in the development of the wing and/or thorax.
Figure 1. Primary and secondary screening outline and examples.
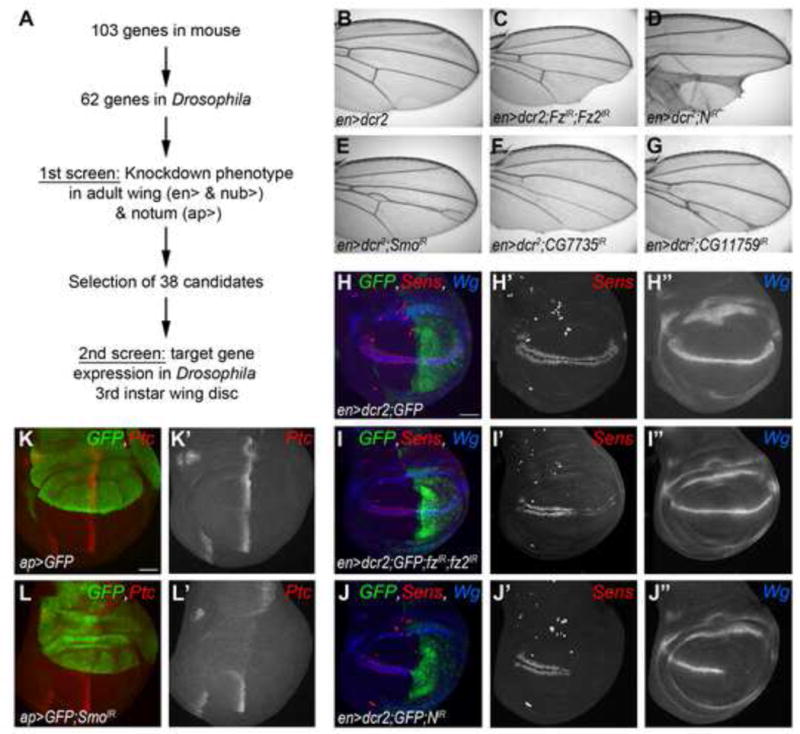
(A) Schematic representation of the screening strategy. (B) Wild-type (wt) control adult wing expressing enGal4, adult wings are oriented proximal on the left and anterior up. (C–E) Control phenotypes induced by knockdown of components of the different signaling pathways using en> (C) en-driven Fz/Fz2 RNAi phenotype displaying wing margin notches in the posterior compartment, a phenotype characteristic of canonical Wg/Wnt signaling. (D) Notch KD shows notches at the margin and vein thickening. (E) Smo RNAi (Hedgehog pathway) displays characteristic vein defects. (F–G) Examples of phenotypes of the primary screen.
(H–L) Wt and control examples of phenotypes observed in the secondary screen, wing discs are oriented anterior left and dorsal up. (H) Wt patterns of Sens (red, monochrome in H′) and Wg (blue, monochrome in H″), used as markers for activation of canonical Wg and Notch pathways, respectively, in flies expressing GFP under the control of en>. (I–J) fz/fz2-IR driven in the posterior compartment leads to loss of Sens but not Wg (I), whereas N-IR leads to loss of both Wg and Sens (J), compared to wt patterns (H). (K) Wt pattern of Ptc staining (red, monochrome in K′), used as a marker for Hh pathway activation, in flies expressing GFP under the control of ap>. (L) Loss of Hh signaling using ap>smo-IR induces loss of Ptc staining in GFP-marked compartment.
See also Figure S1.
Table 1.
Secondary screen results: Analysis of developmental signaling target genes in the 3rd instar wing discs.
| Gene name | Vertebrate ortholog | RNAi line # | Hedgehog pathway | Canonical Wnt pathway | Notch pathway | EGF pathway |
|---|---|---|---|---|---|---|
| Ptc | Sens | Wg | Aos–lacZ | |||
| Frizzled/Frizzled2 | v43075;tJF01259 | loss | wt | |||
| Notch | v27228 | loss | loss | |||
| Smoothened | tJF02363 | loss | ||||
| EGFR | tJF01368 | loss | ||||
| CG7735 | BBS3, ARL6 | v104462 | wt | loss | loss | wt |
| CG13232, BBS4 | BBS4 | v100571 | wt | loss | wt | wt |
| CG1126 | BBS5 | v18200 | wt | loss | wt | ND |
| CG15666 | BBS9 | v40013 | wt | loss | wt | wt |
| CG15923 | MKS3 | v37373;MS54 | wt | loss | wt | wt |
| CG14870 | B9D1 | v107330 | wt | loss | wt | wt |
| CG18631 | CC2D2A, CEP76 | v100302 | wt | wt | wt | wt |
| CG15161 | IFT46 | d15161R-1 | ND | wt | wt | ND |
| CG3259 | IFT54 | v46163 | wt | wt | wt | wt |
| CG5142 | IFT70, FLEER | v22015 | wt | loss | wt | ND |
| CG9333, Oseg5 | IFT80 | v100020 | wt | loss | wt | wt |
| CG13809, osm-1 | IFT172 | v107157 | wt | loss | wt | wt |
| CG7293, Klp68D | KIF3B | tJF03346 | wt | wt | wt | wt |
| CG11759, Kap3 | KIFAP3 | v103548 | wt | loss | wt | wt |
| CG2069, oseg4 | IFT121 | v109805 | wt | loss | wt | wt |
| CG7161, oseg1 | IFT122 | v103598 | wt | loss | wt | wt |
| CG11838, rempA | IFT140 | v31575 | wt | loss | wt | wt |
| CG5780 | IFT43 | v106366 | wt | loss | wt | wt |
| CG15148, btv | DYNCH2H1 | tJF03010 | wt | loss | wt | wt |
| CG3769 | DYNC2LI1 | v40469 | ND | wt | wt | ND |
| CG6998, ctp | DYNLL1 | v109084 | wt | loss | increase | wt |
| CG11356 | Arl13b | v33812 | wt | wt | wt | ND |
| CG1501, unc | OFD1 | tJF03403 | wt | wt | wt | ND |
| CG9398, king-tubby | TULP3 | v29110 | ND | wt | wt | ND |
| CG14367 | CCDC104 | v100799 | ND | wt | wt | ND |
| CG17599 | CLUAP1 | v14682 | wt | wt | wt | wt |
| CG4525 | TTC26 | v107708 | ND | wt | wt | ND |
| CG3265, Eb1 | EB1 | v24451 | wt | loss | wt | wt |
| CG6312, rfx | RFX | v10416 | wt | loss | wt | wt |
| CG33957, cp309 | Pericentrin | v100969 | wt | wt | wt | ND |
| CG15524, sas-6 | SAS-6 | v25073 | wt | wt | wt | increase |
| CG6129, rootletin | ROOTLETIN | d6129R-1 | wt | wt | wt | wt |
| CG6560, dnd | ARL3 | v104311 | wt | loss | loss | increase |
| CG17286, Spd-2 | CEP192 | v36623 | wt | wt | wt | ND |
| CG7186, Sak | SAK/PLK4 | v105102 | wt | loss | wt | ND |
| CG10061, sas-4 | SAS-4 | v106051 | wt | wt | wt | wt |
| CG14617, cp110 | CP110 | v101161 | wt | loss | loss | ND |
| CG17081, cep135 | CEP135 | v14194 | ND | wt | wt | ND |
Transgenic RNAi strains were from the following sources:
v – VDRC, Vienna, Austria; t – TRIP Collection, Bloomington; d – DGRC, Kyoto, Japan; MS – our lab at Mount Sinai.
To assess which signaling pathways were affected by candidate gene KDs, we first established control RNAi lines directed to receptors of the respective developmental signaling pathways: Frizzled/Frizzled2 (Fz/Fz2 - Wnt pathway[s]), Notch (Notch pathway), Smo (Hh pathway), Epidermal Growth Factor Receptor (EGFR - RTK pathway) and Thickveins (Tkv - Dpp/TGFβ pathway) and compared these effects to the candidate KD phenotypes in adults (Fig. 1B–G; and Suppl. Table S1 and Figure S1A–C). Based on the adult wing and thorax phenotypes from the primary screen, 38 candidates were selected as potential regulators of developmental signaling pathways. We tested these in a secondary screening strategy to monitor, at the molecular level, the expression of target genes of the respective signaling pathway. In developing wing imaginal discs: Senseless (Sens) was used as marker for canonical Wg activity; Wg ligand expression was used to monitor N activity; Ptc expression reflected Hh signaling activity; and argos-lacZ (aos-lacZ) was used as an EGFR-signaling reporter. Knockdowns of Fz/Fz2, Notch, Smo and EGFR were used as pathway specific controls (Table 1 and Fig 1H–L; also Suppl. Fig S1D–I). As N-signaling controls the transcription of Wg (which activates the canonical Wnt/Wg pathway), Notch and Wg signaling were always assayed simultaneously. Depending on the target genes’ expression pattern, we used either enGal4 or apGal4 to knockdown the 38 candidate genes (and control genes) identified in the primary screen (Table 1, and Suppl. Fig S1D–I).
The secondary screen revealed that none of the candidates had an impact on Ptc expression, implying Hh signaling was unaffected by impairing cilia-associated protein expression (see Discussion). Only a small number of candidate genes showed either loss or gain-of-function effects for Notch or EGFR signaling (4 and 2, respectively). However, a significant fraction displayed a Wg signaling loss-of-function-like phenotype, manifested by decreased Sens expression, without affecting Wg expression (17 genes, Table 1). Our results thus strongly suggest that several ciliary proteins are involved in regulating aspects of Wg signal transduction, independently of their function within the cilium.
Of particular note were the intraflagellar transport complex A (IFT-A) proteins, as their knockdown (with the exception of IFT144, see also Discussion) led to highly reproducible phenotypes with loss or reduction of Sens (Fig. 2A–F). Therefore, the components of the IFT-A protein complex were selected for further functional analyses.
Figure 2. Wg-specific markers are disrupted by knockdown of a subset of IFT-A components.
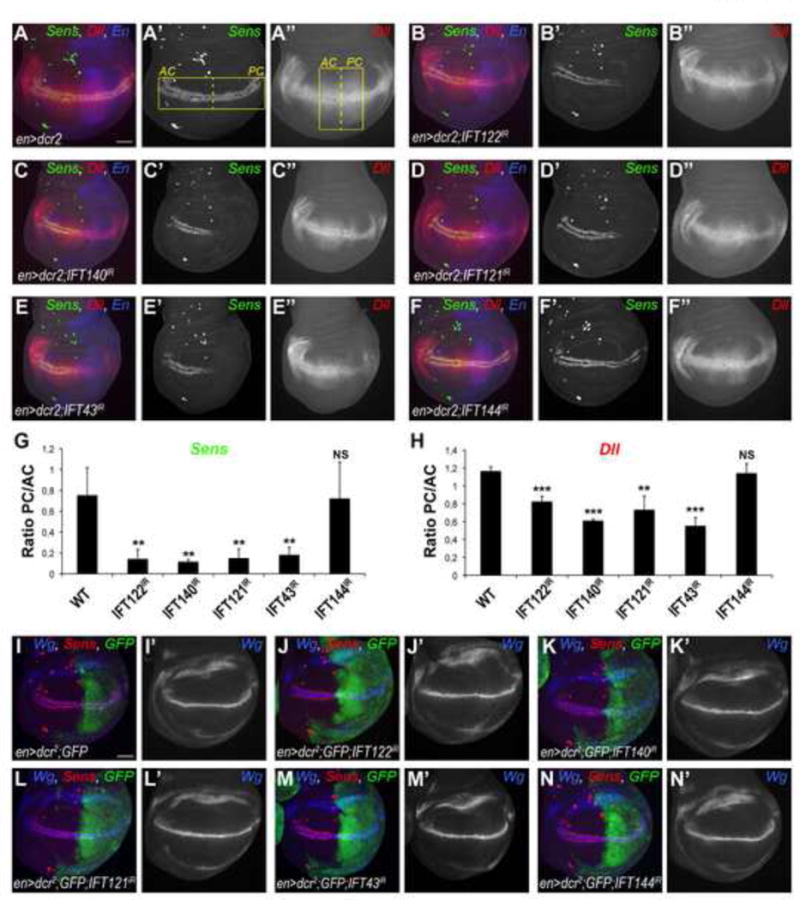
(A–A″) Wt expression pattern of Sens (green, monochrome in A′) and Dll (red, monochrome in A″) in 3rd instar wing imaginal discs of en> flies. en compartment (posterior) is marked in blue (A). Yellow rectangles in (A′) and (A″) indicate the areas used for quantifications of fluorescence intensity of Sens and Dll stainings (see G and H for graphs). (B–F) Sens staining is largely lost and Dll staining is reduced in en-driven RNAi based knockdowns of IFT122IR (B–B″), IFT140IR (C–C″), IFT121IR (D–D″), and IFT43IR (E–E″) at 29°C, whereas both remain WT in IFT144IR (F–F″) (n≥20 per genotype). Scale bar represents 25μm. (G–H) Quantification of fluorescence intensity ratio in the posterior compartment (PC) to the anterior compartment (AC) of Sens (G) and Dll (H) stained wing discs. The bars represent mean standard deviation (SD), n=4 representative stainings per genotype, **= p<0.01 and ***= p<0.001, NS = non significant; student t-test. (I–I′) Wt pattern of Sens (marker for canonical Wg signaling, in red) and Wg (marker for Notch signaling, in blue, monochrome in I′) in control wing disc expressing GFP under the control of enGal4. (J–N) Wg staining remains unaffected in all IFT-A KD (J′–N′) whereas Sens is lost in the posterior compartment of IFT122IR (J), IFT140IR (K), IFT121IR (L), IFT43IR (M) wing discs.
See also Figure S2.
IFT-A proteins are necessary for Wg signal transduction
Cilia assembly and function is dependent on the conserved bidirectional intraflagellar transport protein (IFT) complexes. IFT complexes carry axonemal and membrane proteins in and out of the cilium through anterograde transport driven by heterotrimeric Kinesin II (Cole et al., 1998; Kozminski et al., 1995) or retrograde transport driven by specific isoforms of cytoplasmic Dynein (Pazour et al., 1999; Signor et al., 1999). More specifically, a subgroup of IFT, the IFT-A protein complex, controls retrograde protein transport from the tip to the base of the cilium (Behal et al., 2012; Iomini et al., 2001; Iomini et al., 2009; Piperno et al., 1998). There are 6 IFT-A proteins in vertebrates, of which 5 are conserved in Drosophila: Oseg1/IFT122, Oseg3 or RempA/IFT140, Oseg4/IFT121, Oseg6/IFT144 and CG5780/IFT43. Adult wings of silenced IFT122, IFT140, IFT121 and IFT43 proteins all displayed growth defects, and wing notches or missing wing margin bristles (Suppl. Fig. S2A–J; IFT144 KD did not show a phenotype). No KD of any of the IFT-A proteins induced defects in the adult thorax (Suppl. Fig. S2K–O). The phenotypes observed were comparable to Fz/Fz2 KDs (Suppl. Fig. S1C) and thus suggested a loss/reduction of canonical Wg signaling (note that multiple cellular hairs and misoriented wing hair/thoracic bristles observed in the Fz/Fz2 double KD are due to disruption of the Wnt/Planar cell polarity [PCP] pathway; Suppl. Table S1, and Suppl. Fig. S1C). For function within the cilium, it has been hypothesized that IFT121 and IFT43 are peripheral components of the IFT-A complex, whereas IFT122, 140, and 144 form the core of the complex (Behal et al., 2012). Consistently, IFT122 and IFT140 KD showed strong phenotypes and IFT121 KD had a generally weaker effect. Strikingly, KD of the peripheral protein IFT43 displayed a strong phenotype and that of IFT144 did not seem to disrupt any signaling pathway tested (Suppl. Fig. S2). To measure the extent of IFT144 KD, and to confirm that the lack of signaling phenotypes is not due to inefficient KD, we analyzed two GFP-tagged expression constructs: IFT144-GFP, and for comparison IFT121-GFP (Avidor-Reiss et al., 2004) [ IFT121 KD served as control, as it displayed a Wg-signaling defect-like phenotype]. RNAi targeting of both genes reduced their protein expression to barely detectable levels, indicating that IFT144 was efficiently silenced, similarly to IFT121 (Suppl. Fig. S2S). Thus, these data suggest that the IFT-A complex has an alternative configuration and/or function during Wg signal transduction, as compared to its ciliary role.
Consistent with the adult phenotypes, silencing of IFT122, IFT140, IFT121 and IFT43 (but again not IFT144) markedly reduced expression of Sens and Distalless (Dll, a lower threshold target of the Wg-pathway) (Fig. 2A–H), two molecularly well-defined Wg-signaling target genes. The reduction in expression was quantified by comparing ratios of posterior [experimental] to anterior [control] Sens and Dll expression levels; Fig 2G–H. Whereas the high threshold level target Sens was strongly reduced (Fig. 2G), the expression of Dll, showed a significant but milder reduction in the IFT-A KD compartment (Fig. 2H). These data are consistent with a role of these IFT-A proteins in Wg signal transduction. In contrast, Hh, Notch, and EGFR signaling were not affected by KD of any IFT-A protein, as reflected by wild-type (wt) expression patterns and levels of Ptc/Ci, Wg/Notch-Reporter-Element (NRE)-GFP and aos-lacZ stainings (Table 1, Suppl. Fig. S2P–R). Importantly, none of the KDs of the IFT-A components affected Wg ligand expression (Fig. 2I–N, Suppl. Fig. S2P), a target of Notch signaling, indicating that their function is linked to downstream signaling events during Wg-signal transduction, but not to Wg expression itself.
Taken together, these data suggest that four out of five IFT-A proteins are specifically involved in Wg signal transduction in non-ciliated epithelial cell in Drosophila.
IFT-A mutants recapitulate RNAi-mediated phenotypes
To confirm the RNAi-mediated phenotypes, we generated loss-of-function clones for existing mutant alleles of two IFT-A components, IFT122/oseg1179 (Avidor-Reiss et al., 2004) and IFT140/rempA21Ci (Lee et al., 2008) and analyzed these in third instar wing discs. Consistent with the RNAi KD results, Sens and Dll expression was reduced in IFT122 null cells (Fig. 3A–B′″ and Suppl. Fig. S3A–A′″). Ptc and Wg expression was again not affected in IFT122/oseg1179 clones (Fig 3A″; and Suppl. Fig. S3B–C″), indicating that IFT122 specifically modulates canonical Wg signaling downstream of Wg with no effect on other developmental pathways. Similarly, mutants in the IFT-A component IFT140/rempA21Ci caused a reduction or loss of Sens expression (Fig. 3C–C′″; compare Sens levels in mutant cells to neighboring wt tissue), and consistently such clones caused partial loss of margin bristles in adult wings, as these are specified by Sens expression (Suppl. Fig. S3E–F′). Again, Wg expression was not affected in IFT140/rempA21Ci clones (Fig. 3D–D′), in accordance with a role for IFT140 in canonical Wg signaling downstream of Wg. In addition, IFT140 mutant clones sometimes induced apoptosis in mutant cells at the border of clones (detected via cleaved-Caspase 3 staining; Suppl. Fig. S3D–D′), a feature that has been reported to occur at sharp Wg signaling borders (Vincent et al., 2011) and thus consistent with a reduction in Wg signaling in IFT140/rempA21Ci mutant cells.
Figure 3. IFT122 and IFT140 mutant clones disrupt Wg signaling.
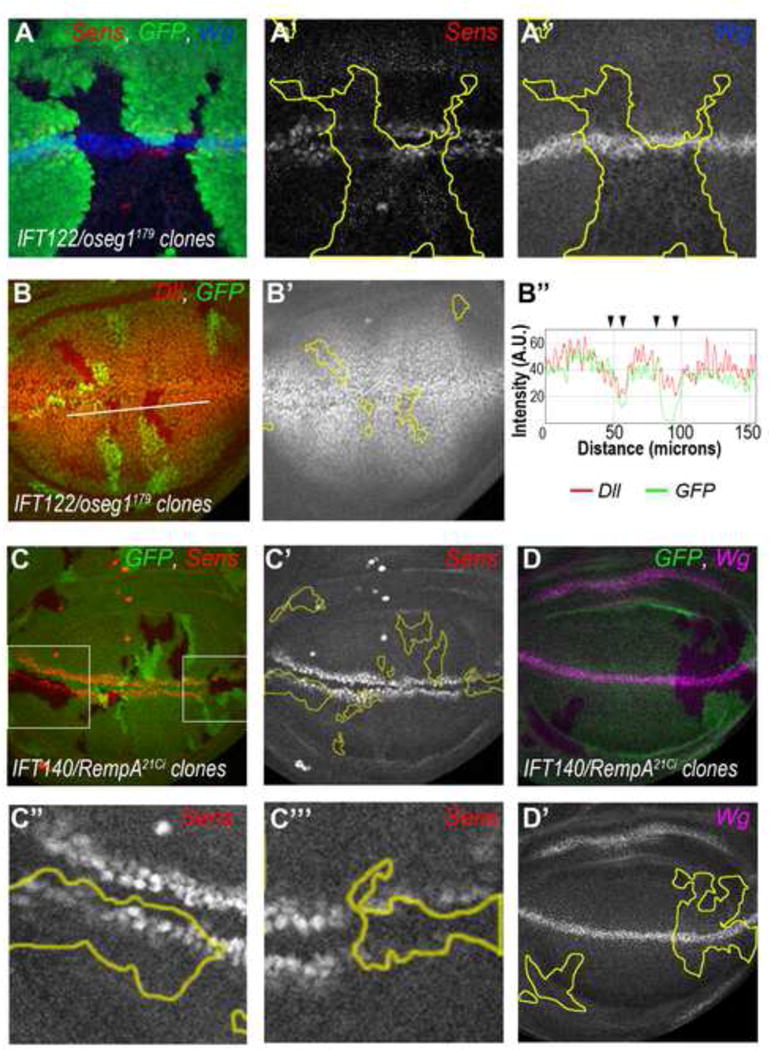
(A–B) Expression of Wg signaling targets Sens and Dll is reduced in IFT122/oseg1179 mutant cells whereas the N pathway target Wg remains unaffected. (A) IFT122/oseg1179 cells (marked by absence of GFP, in a minute background) display reduced levels of nuclear Sens (red, monochrome in A′) whereas Wg staining is wt (blue, monochrome in A″). (B) IFT122/oseg1179 cells (marked by the absence of GFP) show reduced levels of Dll (red, monochrome in B′); quantified in B″ (along white line in B, Intensity/A.U. = arbitrary units).
(C–C″) IFT140/rempA21Ci mutant cells (marked by absence of GFP) display reduction or loss of Sens expression (red, monochrome in C′–C′″). High magnification of two areas (boxed in C) are shown as monochrome in C″ (with reduced Sens expression) and C″′ (with loss of Sens expression). (D–D′) Wg expression is not affected is IFT140/rempA21Ci mutant cells (magenta, monochrome in D′).
See also Figure S3.
In parallel to analyzing existing mutant alleles of IFT-A components, we generated IFT121/oseg4 mutants using the CRISPR technique (Suppl. Fig. S3G–J). Wings of adults homozygous mutant for IFT121/oseg4 displayed wing margin defects with missing bristles (Suppl. Fig. S3H–J), reminiscent of hypomorphic Wg signaling mutant effects and consistent with the weaker effect of IFT121 in RNAi KD experiments (for example Suppl. Fig. S2C,H as compared to B, G).
Taken together, the analyses of mutant alleles of IFT-A components confirmed the phenotypes observed in IFT-A KD backgrounds, indicating that specific IFT-A components act in a cilia-independent context and are required for high-levels of canonical Wg/Wnt signaling.
IFT-A proteins act downstream of Frizzled2/Arrow and upstream of Armadillo
To begin to characterize the mechanistic involvement of IFT-A proteins in Wg signaling, we performed epistasis experiments in vivo. A Fz2-Arr chimeric protein, in which full length Fz2 is fused to the cytoplasmic tail of Arr, constitutes an activated form of the two receptors (Tolwinski et al., 2003). Expression of Fz2-Arr under enGal4 (en>Fz2-myc-Arr) leads to induction of extra Sens positive cells in wing discs and margin bristles in adult wings in the posterior compartment (Fig. 4A, G and Suppl. Fig. S4A–A″, G). Co-expression of RNAi against IFT122, IFT140, IFT121 and IFT43 suppressed this phenotype (for wing discs see: Suppl. Fig S4B–E; and adult wings see: Fig. 4A–E, quantified in Fig. 4G, and Suppl. Fig. S4H–K). IFT144 did not interact, as expected from earlier observations, and served as a control (Fig. 4F, and Suppl. Fig S4F, L). These data indicate that the specific IFT-A proteins act downstream of the receptor complex.
Figure 4. In vivo epistasis analyses reveal IFT-A proteins function downstream of the Fz2-Arrow receptor complex and upstream of Arm/β-cat.
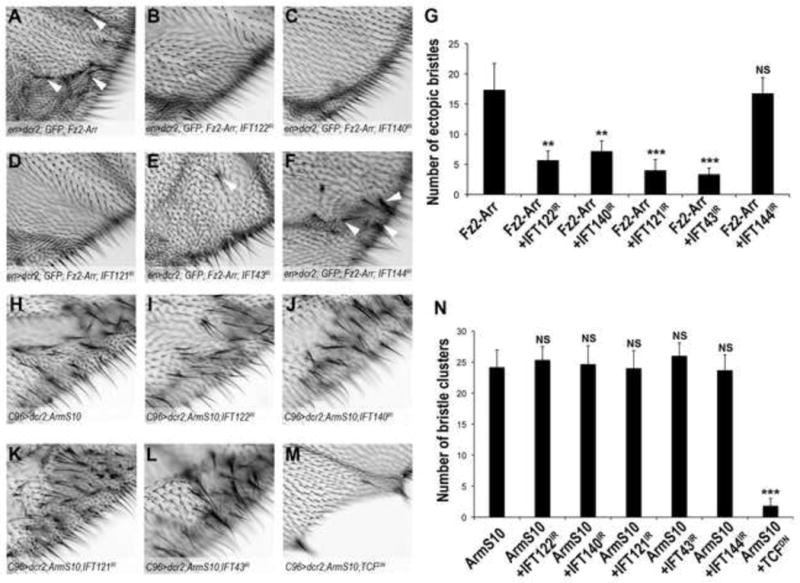
(A–G) High magnification view of extra margin bristle phenotype (examples indicated by white arrowheads) observed near margins of adult wings between L3 and L4 from flies expressing activated Fz2-Arr chimeric receptors (A) and Fz2-Arr combinations with IFT-A protein RNAi KD (B–F; genotypes are indicated in panels) under the control of enGal4 (image field as indicated in overview in Suppl. Fig. S4G). (G) Quantification of number of extra bristles/wing area shown, as induced by expression of Fz2-Arr (A) or Fz2-Arr and IFT-A KD factors (B–F) as indicated. All IFT-A KDs, except IFT144, suppress the extra- bristle phenotype. The bars represent mean SD, n≥10 fields per genotype, **= p<0.01 and ***= p<0.001, NS = non significant (student t-test).
(H–N) High magnification view of extra-margin bristles phenotype near margin of adult wings in C96>ArmS10 flies (image field as indicated in Suppl. Fig. S4M). (I–L) Co-expression of ArmS10 and RNAi against IFT-A components does not affect C96>ArmS10 phenotype (see also Suppl. Fig. S4N–S for whole adult wing images). (M) Co-expression of TCFDN suppresses C96>ArmS10 phenotype. (N) Quantification of number of extra bristles/wing area in the respective IFT-A protein KD backgrounds. The bars represent mean SD, n≥10 fields per genotype, ***= p<0.001, NS = non significant (student t-test).
See also Figure S4.
We next sought to establish whether IFT-A proteins act upstream of β-cat/Arm. A constitutively active form of Arm, ArmS10, which bears an N-terminal truncation inhibiting its phosphorylation by the destruction complex was selected (Pai et al., 1997). Expression of ArmS10 driven by C96Gal4 along the developing wing margin induced extra margin bristles in adult wings (Fig. 4H, Suppl. Fig. S4M; other drivers could not be used for ArmS10 expression as they caused early lethality). None of the IFT-A component KDs had a detectable effect on the ArmS10 phenotype (Fig. 4H–L; quantified in Fig. 4N, see also Suppl. Fig. S4N–R). Nonetheless, as a control, known downstream components can suppress the ArmS10 phenotype, for example a dominant negative isoform of the transcription factor TCF (Fig. 4M–N, and Suppl. Fig. S4S). These data are consistent with a role for the IFT-A proteins upstream of nuclear Arm/β-catenin transcriptional activation.
IFT-A proteins modify phosphorylation levels of Armadillo
Next, we addressed whether IFT-A proteins acted upstream or downstream of the destruction complex. en-driven knockdown of Axin, a key component of the destruction complex, leads to constitutive activation of Wg signaling in almost every cell in the posterior compartment of the wing pouch (Fig. 5A), causing pupal lethality. Co-knockdown of IFT122, IFT140, IFT121 and IFT43 together with Axin suppressed the over-activation of the Wg pathway and associated Sens expression (Fig. 5B–F, quantified in Fig. 5G; IFT144 served as control). Viability was also restored and adult flies were recovered (Suppl. Fig. S5A–D). Importantly, double mutant clones for IFT140/rempA21Ci; axnE77 also suppressed ectopic activation of Sens and Dll normally observed in axnE77 single mutant clones (Fig. 5H–H″ and Suppl. Fig. S5E–E′ and G–G′″, note that only the double mutant clones lack GFP as both FRT chromosomes are marked by GFP; single mutant axnE77 patches can be identified by high Sens and Dll levels; see Suppl. Fig S5F–F′ for axn clones positively labeled with GFP). Together, these analyses suggest that IFT-A proteins function downstream of or in parallel to the destruction complex, and upstream of Arm nuclear function, suggesting a role in Arm stabilization in the cytoplasm prior to its nuclear transcriptional activity.
Figure 5. IFT-A proteins act downstream or in parallel to the destruction complex.
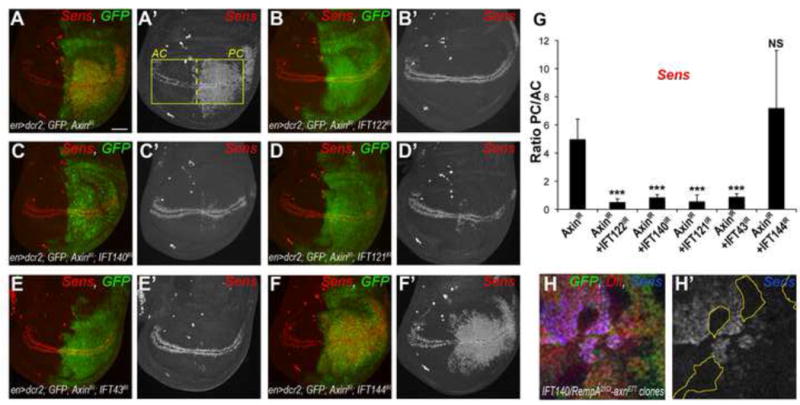
(A–A′) Axin RNAi knockdown in the posterior compartment of the wing disc induces a strong increase in Sens positive cells at 29°C (red, monochrome in A′). (B–E) Co-expression of RNAi against Axin and IFT122 (B), IFT140 (C), IFT121 (D), or IFT43 (E) suppresses extra Sens positive cells in the posterior compartment (PC). (F) Knockdown of Axin and IFT144 resembles Sens pattern in Axin KD alone. (G) Quantification of the ratio of PC/AC of fluorescence intensity in A′–F′ in 3rd instar wing discs of Axin KD alone and in combinations with IFT-A RNAi. The bars represent mean SD, n≥20 wings per genotype, *** = p<0.001, NS = non significant (student t-test). Scale bar in A = 25μm.
(H) Dll and Sens expression in axnE77 and IFT140/rempA21Ci double mutant clones. Within the GFP negative cells (double mutant), Sens (blue, monochrome in H′) and Dll (red, monochrome in Suppl. Fig. S5E′) are not highly induced compared to axnE77 clones (visible as high levels of Sens in margin proximal regions, see also Suppl. Fig S5F), which is consistent with the RNAi data above (A–E′).
See also Figure S5.
In Drosophila imaginal discs, like all epithelial tissues, Arm/β-catenin localizes both to apical adherens junctions (via interaction with E-cadherin) and in the cytoplasm, with the cytoplasmic Arm/β-catenin pool being used in Wg signaling. Within the cytoplasm, Arm/β-catenin is present in two forms: the unphosphorylated “active” form that can translocate to the nucleus to promote target gene activation and the phosphorylated (“inactive”) form, which is targeted for degradation by the proteasome. To remove junctional Arm and allow specific analysis of the cytoplasmic fraction, we used concanavalin-A (Con-A)-coupled beads, which bind to cadherins and thus sequester molecules associated with E-cad, i.e. Arm (Peifer, 1993). In wt 3rd instar wing discs, equal amounts of the “active” and “inactive” form of cytoplasmic Arm were present, in the fraction of Arm not bound to E-cad. When the destruction complex was inhibited in nubGal4-driven Axin KD wing discs, this balance changed to 80% of “active” Arm (Fig. 6A–B), leading to activated Wg signaling and causing the induction of Sens expression throughout the wing pouch (Suppl. Fig. S6A–B; nub-Gal4 driven Axin KD does not completely disrupt the destruction complex with basal phosphorylation of Arm still observed). In this background we tested whether IFT-A protein KD could affect Arm phosphorylation levels. In wing discs with combined nub>IFT-A and Axin KDs the ratio of “active/inactive” Arm is significantly decreased (Fig. 6A–B; i.e. “inactive”/phosphorylated Arm levels are increased relative to Axin KD alone), which is consistent with Sens expression patterns reverting to similar to the wt (Suppl. Fig. S6A–D). To address this further, we tested whether endogenous (cytoplasmic) Arm levels are affected in IFT-A component knock-downs or mutants. Generally, the levels of Arm were reduced in such mutant scenarios. For example, in enGal4 driven KD of IFT43 or IFT140 in the posterior compartment (Fig. 6D–E′; compare to control in Fig. 6C–C′, quantified in Suppl. Figure S6E–G′) or in mutant clones of IFT122 (Oseg1179; Fig. 6F–F′, compare Arm levels to surrounding wt cells) the intracellular, cytoplasmic Arm/β-catenin levels were reduced. These results are in agreement with the increased proportion of phosphorylated Arm in axin, IFT-A double knockdowns, which is targeted for degradation (Fig. 6A–B). As double mutant clones for IFT140/rempA21Ci; axnE77 suppressed the elevated expression of Sens and Dll seen in their neighboring axnE77 single mutant clones (Fig 5H–H′ and Suppl. Fig. S5E–E′), we examined the level of cytoplasmic Arm staining in these clones compared to either axnE77 single mutant clones or the surrounding tissue. Cytoplasmic arm staining was elevated in double mutant clones, similarly to neighbouring axnE77 clones, when compared to the surrounding tissue (Fig. 6G; surrounding tissue is heterozygous and so phenotypically wt). The elevated levels of Arm detected by immunofluorescence comprise both phosphorylated “inactive” and unphosphorylated “active” forms. Based on the Western blot data and ratiometric analysis, we infer that the elevated Arm staining in the double mutant clones represents more of the phosphorylated “inactive” form and less “active” Arm, thus explaining the reduction in Arm transcriptional activity (as shown previously in Fig 5H–H′ and Suppl. Fig. S5E–E′). Taken together with the epistasis analyses, these data suggest that IFT-A components are necessary for proper stabilization of unphosphorylated Arm/β-catenin in the cytoplasm to allow its translocation to the nucleus, downstream or in parallel to the destruction complex.
Figure 6. IFT-A proteins regulate Wg signaling by affecting the stability of excess Arm resulting from the destruction complex inhibition.
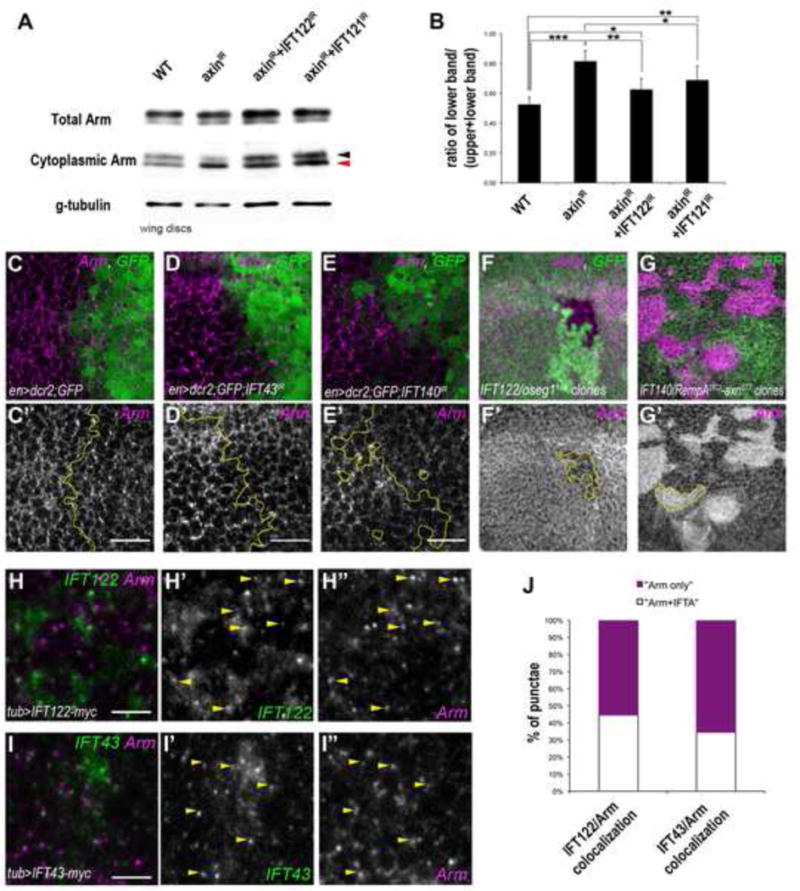
(A–B) Western blot analysis of cytoplasmic levels of Arm in wt, Axin-IR and Axin-IR combined with IFT122-IR or IFT121-IR wing disc extracts (black and red arrowheads indicate upper “inactive”/phosphorylated Arm band and lower “active”/stable Arm band, respectively (n=300 wing discs/genotype). (B) Quantification of 3 independent experiments of the ratio of lower band (“stable” Arm, red arrowhead) compared to the total cytoplasmic Arm levels. The bars represent mean SD, *= p<0.05, **= p<0.01 and ***= p<0.001; Student t-test.
(C–E) Cytoplasmic levels of Arm are reduced in cells of IFT43IR (magenta in D, monochrome in D′) and IFT140IR (magenta in E, monochrome in E′) compared to the wt control levels (magenta in C, monochrome in C′). See also the line scan quantification of protein staining in Suppl. Figure S6E–G. (F–F′) Cytoplasmic Arm levels are reduced in IFT122/oseg1179 clones (marked by GFP absence, below the junctional level; red F, monochrome in F′).
(G–G′) Cytoplasmic Arm levels are elevated in axnE77; IFT140/rempA21Ci double mutant clones (GFP negative cells; magenta in G, monochrome in G′) and indistinguishable from neighboring axnE77 single mutant clones (GFP positive cells with high levels of cytoplasmic Arm).
(H–J) IFT122 and IFT43 partially colocalize with Arm. tub-promoter driven IFT122-myc (green in H, monochrome in H′) and IFT43-myc (green in I, monochrome in I′) show partial co-localization (in punctate structures) with Arm (scan is just below the apical junctional region; see also apical staining of Arm and IFT in Suppl. Fig. S6H–I). (J) Quantification of the number of punctae containing Arm alone (magenta) or Arm with IFT122 or IFT43 (white; n≥200 punctae per genotype, from 4 different wing discs). See also line scan quantification of respective co-localization in Suppl. Fig. S6H″–I″. Scale bar represents 15μm in C–E and 3μm in H–I.
See also Figure S6.
To analyze the localization of IFT-A proteins in Drosophila wing cells and test for potential co-localization with Arm, we expressed myc-tagged IFT122 and IFT43 using tubGal4, which is within the physiological range of their endogenous expression levels. Both, IFT122-myc and IFT43-myc localize in punctae below the apical junctions (Fig. 6HI″). Importantly, these punctae are often co-labeled with Arm: approximately 40–50% of the pool of cytoplasmic Arm punctae co-stain with the IFT-A components (Fig. 6H–J, and Suppl. Fig. S6H–I″; note that in particular the brightest Arm spots overlap with IFT-A staining).
Taken together, our data argue for a role of IFT-A proteins in Wg/Wnt signaling in Drosophila via an effect on the stabilization or localization of “active”, non-phosphorylated Arm upstream of its nuclear translocation and downstream or in parallel to the destruction complex.
DISCUSSION
We have performed a systematic screen to test for potential roles of ciliary proteins in non-ciliated epithelial cells and developmental signaling pathway contexts. This approach identified roles for several cilia-associated proteins in regulating Notch, Wg, and EGFR signaling (Table 1), demonstrating cilia-independent functions in vivo in Drosophila. We show that ciliary proteins have no effect on Hh signaling in non-ciliated cells.
Components of the Hh/Shh pathways are highly conserved between Drosophila and vertebrates, with the main difference being that Shh/Hh signaling takes place in the cilium in vertebrate cells (Goetz and Anderson, 2010). The lack of a requirement for conserved ciliary proteins in Hh signaling in Drosophila epithelial cells thus suggests that the cilium provides a structural compartment for transducing Hh signals, rather than that specific cilia-associated proteins are required within the pathway. Consistent with this, it was recently demonstrated that Hh signaling components are localized within the cilium in ciliated Drosophila neuronal cells (Kuzhandaivel et al., 2014). The IFT-B protein IFT25, which has no effect on cilia assembly, but is required for Hh signaling in vertebrates, is the exception. However, IFT25 is not conserved in C. Elegans or Drosophila (Keady et al., 2012). The importance of IFT25 suggests that moving Shh-signaling components along the axoneme (in and out of the cilium) is also critical for Shh signaling in vertebrates, which is again consistent with the notion that Shh signaling needs to take place inside the cilium when cilia are present. Evolutionary aspects of the Hh/Shh pathway components, studied in planarians, revealed that Hh signaling might have originally been organized by the cilium, with cilia serving as a signaling compartment (Rink et al., 2009). The evolutionary loss of IFTs in planaria does not affect Hh signaling (Rink et al., 2009), consistent with the notion that the cilium serves as a Hh/Shh signaling hub, but that specific components of ciliogenesis are not required for Hh signaling per se.
Our screen for developmental signaling requirements of ciliary proteins in non-ciliated cells identified several such factors as being required for canonical Wg/Wnt signaling. Previous studies have proposed conflicting effects of the loss of cilia on canonical Wnt signaling in vertebrates, ranging from reduction or loss of signaling to over-activation of the Wnt pathway (rev. in (Oh and Katsanis, 2013). These different, or even opposing, outcomes were possibly caused by pathway crosstalk(s) and/or context specific interactions. For example, Shh and Wnt signaling often mutually affect each other (Borday et al., 2012; Rink et al., 2009; van den Brink et al., 2004; Yanai et al., 2008), and when cilium biogenesis is impaired, Shh signaling is reduced or abolished. Thus, the potential positive effect of impaired ciliary function on Wnt signaling could be secondary to the loss of Shh signaling, leading to an over-activation of Wnt signaling in ciliary mutants. In particular, Lancaster et al. (2011) showed in vitro and in vivo that mutations in a dynein subunit of the retrograde transport complex (Dync2h1) display cell-type specific effects on cilium integrity and canonical Wnt signaling. dync2h1 MEFs and regions of dync2h1 mutant embryos that exhibit loss of cilia, and thus loss of Shh signaling, induce over-activation of Wnt signaling. However, in dync2h1 mutant embryos or siRNA in MEFs that disrupt retrograde transport but leave the cilium intact (and thus do not disrupt Shh signaling) display reduced canonical signaling levels (Lancaster et al., 2011).
Here we demonstrate that a subset of IFT-A components and also other ciliary proteins (Table 1), act positively in canonical Wnt/Wg signaling in a cilium-independent manner. This is consistent with the above data, in particular the effects of the dync2h1 mutants that leave cilia intact. Importantly, our data provides insight into the role of cilia-associated proteins outside of the ciliary structure. We have focused our mechanistic studies in Drosophila, on the IFT-A proteins, and demonstrate that a subset of these is required for stabilization and/or localization of β-cat/Arm prior it its activity in the nucleus. Taken together with the Lancaster et al. (2011) observations, this non-ciliary function is likely conserved in vertebrates. Due to the omnipresence of primary cilia in vertebrate cells and the associated difficulty of uncoupling ciliary and non-ciliary functions of IFT-A proteins, Drosophila provides a unique and ideal model system to dissect their non-ciliary function. Our data also suggest that the IFT-A complex functions in a different configuration(s) in ciliary vs. non-ciliary contexts, as IFT144 (although efficiently knocked down; Fig. S2S) did not affect any detectable aspects of developmental signaling pathways tested. IFT144 is a core structural component of the IFT-A complex in cilia, thus suggesting a different composition of the IFT-A complex outside of the ciliary compartment. However, IFT43 and IFT121, which are described as peripheral within the IFT-A complex in the cilium, show effects (albeit IFT121 effects being generally weaker, both in knock downs and in mutants, as compared to other IFT-A components), suggesting that some aspects of the IFT-A complex are nonetheless preserved between the cilia and cytoplasmic locations.
Can these observations be related to disease aspects of the broad spectrum of “ciliopathies”? Interestingly, mutations in human IFT-A components cause a specific subcategory of ciliopathies, called “skeletal ciliopathy”, which are characterized by limb morphogenesis defects as well as extra-skeletal abnormalities, including retinal or renal defects (Huber and Cormier-Daire, 2012). IFT-A mutations often lead to Shh/Hh pathway disruption (Liem et al., 2012; Qin et al., 2011), which in turn induces skeletal morphogenesis problems similar to the ones observed in skeletal ciliopathies (Ashe et al., 2012; Cortellino et al., 2009; Mill et al., 2011). Importantly, the canonical Wnt-signaling pathway has been proposed to act downstream of Hh and BMP signaling in bone morphogenesis (Baron et al., 2006), and it is therefore possible that defects observed in these syndromes are due to both impaired Hh and Wnt signaling. Retinal and renal defects can also be induced by defective Wnt signaling levels (Kawakami et al., 2013; Lad et al., 2009). Our work supports this concept and adds further insight into the function of ciliary proteins with regards to Wnt signaling.
It remains unclear whether the cytoplasmic IFT-A proteins (in their possibly altered configuration) associate with microtubular structures and whether such association is required for function in Wnt/Wg signaling. Interestingly, a role for Kinesin-II in Wg signaling and the transport of β-cat/Arm was recently reported (Vuong et al., 2014), consistent with a link to microtubules. One hypothesis is that the IFT-A proteins associate with microtubules and play a similar role to Costal-2 (Cos-2)/Kif7 in Hh signaling. In the absence of Hh signals, Cos-2 binds to the Hh/Shh effector Ci/Gli and tethers it to microtubules together with Fused, Suppressor of Fused and several kinases (He et al., 2014; Robbins et al., 1997). Hh activation reverses this binding and frees Ci from the complex (Goetz and Anderson, 2010). Cos-2 is a negative regulator of Hh signaling, and our data suggest that IFT-A proteins act antagonistically to the destruction complex by promoting β-cat/Arm stabilization. Sequestering Arm away from the destruction complex to prevent its phosphorylation and allow activation of the pathway makes this an attractive hypothesis for future study.
EXPERIMENTAL PROCEDURES
Fly stocks
Drosophila strains were raised at 25° on standard medium unless otherwise indicated. The following lines used were obtained from: axin-IR (Bloomington stock center, 31705), UAS-IFT121-GFP and UAS-IFT144-GFP, oseg1179/TM6b (gifts from T. Avidor-Reiss), rempAl(2)21Ci-1 (gift from M. Kernan), UAS-Fz2-myc-Arr (gift from M. Wehrli), UAS-ArmS10 (Pai et al., 1996), UAS-TCFDN (Bloomington stock center, 4784), FRT82B, axnE77/TM6b and aoslacZ/TM6b (gifts from J. Treisman).
The following lines were used to induce and analyze mutant clones:
y, w, hsFlp; armlacZ, FRT40
y, w, hsFlp; ; P(w+), min55, UbiGFP, FRT80B
y, w, hsFlp; ; UbiGFP(nls), FRT80B
y, w. UbxFlp; UbiGFP(nls), FRT40; UbiGFP(nls), FRT82B/SM5:TM6b
y, w, hsFlp, tubGal4, UAS-GFP; ; FRT82B, tubGal80, CD21/TM6b. See Supplemental Experimental Procedures for generation of UAS-IFT122-myc, UAS-IFT43-myc and oseg1 CRISPR alleles.
The Gal4/UAS system (Brand and Perrimon, 1993) was used for gene expression studies, with the following Gal4 drivers used: UAS-dcr2; enGal4/CyoGFP,
UAS-dcr2;enGal4,
UAS-GFP,
UAS-dcr2; nubGal4,
apGal4; UAS-GFP/S-T,
UAS-dcr2; neurGal4/S-T,
tubGal4/TM3,
UAS-dcr2; C96Gal4,
enGal4, UAS-myrRFP, NRE-GFP.
Our 62 screen candidates were identified using the NCBI protein-blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and expression levels were verified on Flybase (Flybase.org). RNAi lines used for the screen were ordered from VDRC, DGRC Kyoto or Bloomington stock centers (Table S1).
Immunohistochemistry and histology
Imaginal wing discs were dissected at the 3rd instar stage in PBS and fixed in PBS-4%PFA unless otherwise indicated. Discs were washed 3 times in PBS 0.1% Triton X-100 (PBT), incubated in primary antibodies at RT for 3h or overnight at 4°. After 3 washes in PBT, incubation with secondary antibodies was performed at RT for 2h. Antibody dilutions can be found in Supplemental Experimental Procedures. Samples were mounted in Vectashield (Vector Laboratories). Wing discs images were acquired at room temperature using a confocal microscope (40X~ oil immersion, 1.4 NA; SP5 DM; Leica) with LAS AF (Leica) software. Images were processed with ImageJ (National Institutes of Health) and Photoshop (CS4; Adobe). Fluorescent intensity quantification was performed using ImageJ software by drawing a rectangular area as shown in figure 1 and 2 and measuring the profile intensity using the Plot Profile tool.
For analysis of adult wing, wings were removed, incubated in PBT, and mounted on a slide in 80% glycerol in PBS and imaged at room temperature (RT) using a Zeiss Axioplan microscope (Carl Zeiss). For analysis of thorax, whole flies were washed in 70% ethanol, mounted on gelatine plates and imaged at room temperature using a stereomicroscope. All adult images were acquired using Zeiss Axiocam color-type 412-312 (Carl Zeiss) camera and the axiocam software.
Armadillo assay (ConA-sepharose fractionation)
Assay was performed according to the protocol from (Peifer, 1993). Briefly, Drosophila 3rd instar wing discs (300/genotype) were isolated and lysed for 45min at 4° under constant rotation with lysis buffer containing 20mM HEPES, 100mM NaCl, 1mM EDTA, 1% NP40, 10% glycerol, 1mM DTT, 50mM NaF, 10mM NaVO4 and protease inhibitor cocktail (Roche). Supernatants were cleared by 20min centrifugation at 4° and soluble fraction was incubated for 2h with 50μL ConA-Sepharose (Sigma) (an aliquot was kept before incubation as total lysate). Supernatant was saved as the cytoplasmic fraction, and bead samples (containing the junctional Arm) were washed 3 times with lysis buffer. Protein extracts were boiled for 10min at 95° in SDS-sample buffer, separated by 8% SDS-page gel and transferred to nitrocellulose membrane. Protein levels were analyzed by immunoblotting with the corresponding antibodies.
Supplementary Material
Acknowledgments
We thank the Bloomington Stock Center, Kyoto DGRC stock center, Vienna Drosophila RNAi center, Drosophila Genomics Resource Center (DGRC, supported by NIH grant 2P40OD010949-10A1) and Developmental Studies Hybridoma Bank (DSHB, created by the NICHD of the NIH and maintained at the University of Iowa) for fly strains and antibodies. We are grateful to Ursula Weber for help with the generation of CRISPR allele. We thank H. Bellen for the Sens antibody, J. Treisman and K. Legent for fly strains and discussions, T. Avidor-Reiss for IFT-A-GFP and oseg1179 fly strains, M. Wehrli for Fz2-myc-Arr fly strains, M. Kernan for rempA21Ci fly strain and J.R. Huynh, B. Mollereau, A. Benmerah, A. Guichet and A. Audibert for discussions. We thank all Mlodzik and Iomini lab members for help and discussions, and A. Humphries and N. Founounou for helpful comments on the manuscript. Confocal microscopy was performed at the Icahn School of Medicine at Mount Sinai Microscopy Shared Resource Facility. This work was supported by NIH/NIGMS grant GM102811 to M.M. and grant EY022639 to C.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, S.B., E.B., C.I., M.M.; Methodology S.B., G.C., E.B., M.M.; Investigation S.B., A.D., G.C., E.B.; Visualization S.B.; Writing – original draft S.B. and M.M; Writing – review and editing S.B., G.C., C.I. and M.M.; Supervision C.I. and M.M.
References
- Ashe A, Butterfield NC, Town L, Courtney AD, Cooper AN, Ferguson C, Barry R, Olsson F, Liem KF, Jr, Parton RG, et al. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Human molecular genetics. 2012;21:1808–1823. doi: 10.1093/hmg/ddr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Baron R, Rawadi G, Roman-Roman S. Wnt signaling: a key regulator of bone mass. Current topics in developmental biology. 2006;76:103–127. doi: 10.1016/S0070-2153(06)76004-5. [DOI] [PubMed] [Google Scholar]
- Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. The Journal of biological chemistry. 2012;287:11689–11703. doi: 10.1074/jbc.M111.287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Current biology: CB. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borday C, Cabochette P, Parain K, Mazurier N, Janssens S, Tran HT, Sekkali B, Bronchain O, Vleminckx K, Locker M, et al. Antagonistic cross-regulation between Wnt and Hedgehog signalling pathways controls post-embryonic retinal proliferation. Development. 2012;139:3499–3509. doi: 10.1242/dev.079582. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Clement CA, Satir P, Pedersen LB. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. The Journal of pathology. 2012;226:172–184. doi: 10.1002/path.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MP, Pedersen LB, Benmerah A, Andersen CY, Larsen LA, et al. TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell reports. 2013;3:1806–1814. doi: 10.1016/j.celrep.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. The Journal of cell biology. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nature cell biology. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Wang C, Wang B, Bassi MR, Caretti E, Champeval D, Calmont A, Jarnik M, Burch J, Zaret KS, et al. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Developmental biology. 2009;325:225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Bright A, Lawson ND, Doxsey S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nature cell biology. 2011;13:461–468. doi: 10.1038/ncb2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE, Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, Rosenbaum JL, Baldari CT. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nature cell biology. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Katsanis N. Ciliary function and Wnt signal modulation. Current topics in developmental biology. 2008;85:175–195. doi: 10.1016/S0070-2153(08)00807-7. [DOI] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nature genetics. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nature reviews Genetics. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Ocbina PJ, Anderson KV. The primary cilium as a Hedgehog signal transduction machine. Methods in cell biology. 2009;94:199–222. doi: 10.1016/S0091-679X(08)94010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogendeau D, Basto R. Centrioles in flies: the exception to the rule? Seminars in cell & developmental biology. 2010;21:163–173. doi: 10.1016/j.semcdb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Jr, Kapoor TM, Anderson KV. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nature cell biology. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C, Cormier-Daire V. Ciliary disorder of the skeleton. American journal of medical genetics Part C, Seminars in medical genetics. 2012;160C:165–174. doi: 10.1002/ajmg.c.31336. [DOI] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. The Journal of cell biology. 2001;153:13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Till JE, Dutcher SK. Genetic and phenotypic analysis of flagellar assembly mutants in Chlamydomonas reinhardtii. Methods in cell biology. 2009;93:121–143. doi: 10.1016/S0091-679X(08)93007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nature reviews Molecular cell biology. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Ren S, Duffield JS. Wnt signalling in kidney diseases: dual roles in renal injury and repair. The Journal of pathology. 2013;229:221–231. doi: 10.1002/path.4121. [DOI] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, Pazour GJ. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Developmental cell. 2012;22:940–951. doi: 10.1016/j.devcel.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. The Journal of cell biology. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhandaivel A, Schultz SW, Alkhori L, Alenius M. Cilia-mediated hedgehog signaling in Drosophila. Cell reports. 2014;7:672–680. doi: 10.1016/j.celrep.2014.03.052. [DOI] [PubMed] [Google Scholar]
- Lad EM, Cheshier SH, Kalani MY. Wnt-signaling in retinal development and disease. Stem cells and development. 2009;18:7–16. doi: 10.1089/scd.2008.0169. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Schroth J, Gleeson JG. Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nature cell biology. 2011;13:700–707. doi: 10.1038/ncb2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Sivan-Loukianova E, Eberl DF, Kernan MJ. An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Current biology: CB. 2008;18:1899–1906. doi: 10.1016/j.cub.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch CC, Lodh S, Prieto-Echague V, Badano JL, Zaghloul NA. Basal body proteins regulate Notch signaling through endosomal trafficking. Journal of cell science. 2014;127:2407–2419. doi: 10.1242/jcs.130344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, Anderson KV. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. The Journal of cell biology. 2012;197:789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill P, Lockhart PJ, Fitzpatrick E, Mountford HS, Hall EA, Reijns MA, Keighren M, Bahlo M, Bromhead CJ, Budd P, et al. Human and mouse mutations in WDR35 cause short-rib polydactyly syndromes due to abnormal ciliogenesis. American journal of human genetics. 2011;88:508–515. doi: 10.1016/j.ajhg.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annual review of cell and developmental biology. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nature reviews Molecular cell biology. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Context-dependent regulation of Wnt signaling through the primary cilium. Journal of the American Society of Nephrology: JASN. 2013;24:10–18. doi: 10.1681/ASN.2012050526. [DOI] [PubMed] [Google Scholar]
- Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M. Drosophila alpha-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. The Journal of biological chemistry. 1996;271:32411–32420. doi: 10.1074/jbc.271.50.32411. [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. The Journal of cell biology. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Witman GB. The vertebrate primary cilium is a sensory organelle. Current opinion in cell biology. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Veland IR, Schroder JM, Christensen ST. Assembly of primary cilia. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- Peifer M. The product of the Drosophila segment polarity gene armadillo is part of a multi-protein complex resembling the vertebrate adherens junction. Journal of cell science. 1993;105(Pt 4):993–1000. doi: 10.1242/jcs.105.4.993. [DOI] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. The Journal of cell biology. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO reports. 2012;13:608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink JC, Gurley KA, Elliott SA, Sanchez Alvarado A. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326:1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. The Journal of cell biology. 2010;189:171–186. doi: 10.1083/jcb.200911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. The Journal of cell biology. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation; research in biological diversity. 2012;83:S12–22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Dijke P, Egorova AD, Goumans MJ, Poelmann RE, Hierck BP. TGF-beta signaling in endothelial-to-mesenchymal transition: the role of shear stress and primary cilia. Science signaling. 2012;5(pt2) doi: 10.1126/scisignal.2002722. [DOI] [PubMed] [Google Scholar]
- Tolwinski NS, Wehrli M, Rives A, Erdeniz N, DiNardo S, Wieschaus E. Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta activity. Developmental cell. 2003;4:407–418. doi: 10.1016/s1534-5807(03)00063-7. [DOI] [PubMed] [Google Scholar]
- van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nature genetics. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- Vincent JP, Kolahgar G, Gagliardi M, Piddini E. Steep differences in wingless signaling trigger Myc-independent competitive cell interactions. Developmental cell. 2011;21:366–374. doi: 10.1016/j.devcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong LT, Mukhopadhyay B, Choi KW. Kinesin-II recruits Armadillo and Dishevelled for Wingless signaling in Drosophila. Development. 2014;141:3222–3232. doi: 10.1242/dev.106229. [DOI] [PubMed] [Google Scholar]
- Yanai K, Nakamura M, Akiyoshi T, Nagai S, Wada J, Koga K, Noshiro H, Nagai E, Tsuneyoshi M, Tanaka M, et al. Crosstalk of hedgehog and Wnt pathways in gastric cancer. Cancer letters. 2008;263:145–156. doi: 10.1016/j.canlet.2007.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


