Abstract
In the course of investigating anti-DNA autoantibodies, we examined IgM and IgG antibodies to poly-G and other oligonucleotides in the sera of healthy persons and those diagnosed with systemic lupus erythematosus (SLE), scleroderma (SSc), or pemphigus vulgaris (PV); we used an antigen microarray and informatic analysis. We now report that all of the 135 humans studied, irrespective of health or autoimmune disease, manifested relatively high amounts of IgG antibodies binding to the 20-mer G oligonucleotide (G20); no participants entirely lacked this reactivity. IgG antibodies to homo-nucleotides A20, C20 or T20 were present only in the sera of SLE patients who were positive for antibodies to dsDNA. The prevalence of anti-G20 antibodies led us to survey human, mouse and Drosophila melanogaster (fruit fly) genomes for runs of T20 and G20 or more: runs of T20 appear > 170 000 times compared with only 93 runs of G20 or more in the human genome; of these runs, 40 were close to brain-associated genes. Mouse and fruit fly genomes showed significantly lower T20/G20 ratios than did human genomes. Moreover, sera from both healthy and SLE mice contained relatively little or no anti-G20 antibodies; so natural anti-G20 antibodies appear to be characteristic of humans. These unexpected observations invite investigation of the immune functions of anti-G20 antibodies in human health and disease and of runs of G20 in the human genome.
Keywords: antigens/peptides/epitopes, autoantibodies, human
Introduction
We have developed an antigen microarray device and informatics analyses that make it possible to profile microlitre amounts of serum for antibodies quantitatively binding to hundreds of different molecules.1–3 Our studies of autoantibodies binding to DNA in humans with systemic lupus erythematosus (SLE), scleroderma (SSc) or pemphigus vulgaris (PV) led us to search for antibodies binding to synthetic oligonucleotides. In a previous study, we found that a monoclonal anti-DNA antibody raised as an anti-idiotype to an antibody to a DNA-binding domain of p53 could, like p53, bind to a 20-mer poly-G homo-oligonucleotide (G20) but not to a poly-T 20-mer (T20).4 We therefore tested whether healthy participants or those with autoimmune diseases might harbour antibodies to G20, to T20 or to other synthetic oligonucleotides. We now report that all human sera show relatively large amounts of IgG antibodies binding to G20; some humans are negative for IgM anti-G20; in contrast, antibodies to T20 were low or undetectable; in contrast to humans, mice, healthy or afflicted with SLE, did not show significant reactivities to G20.
Here, we characterized the importance of length and nucleotide composition on the binding of IgG and IgM antibodies to oligonucleotides in healthy human participants and in patients with SLE, SSc or PV. We also surveyed the numbers of runs of G20 or more compared to runs of T20 or more in the genomes of humans, mice and the fruit fly Drosophila melanogaster. We found that the high frequency in humans of anti-G20 antibodies was associated with a relatively low frequency of runs of G20 compared with T20 in the human genome; mice, in contrast to humans, manifested a higher frequency of runs of G20 in the genome and little or no anti-G20 antibodies. Hence, there appeared to be a negative association between runs of genomic G20 and natural anti-G20 antibodies. This paper calls attention to these unexpected findings.
Materials and methods
Human participants
The study was approved by the Institutional Review Boards of each participating clinical unit; informed consent was obtained from all participants. In our initial study, we tested sera from 22 healthy participants, 18 PV patients, 15 SSc patients and 34 SLE patients using an antigen microarray that included A20, C20, G20 and T20. In the follow-up study, we expanded the oligonucleotides tested to 58 and examined sera from 49 SLE patients, 24 SSc patients and 23 healthy participants. Overall, 60 SLE patients, 26 SSc patients, 18 PV patients, and 31 healthy participants were tested. The patients with SLE or SSc were diagnosed according to clinically accepted criteria.5,6 The diagnosis of PV was based upon clinical features and laboratory tests: suprabasal separation on histology of skin lesions, positive direct and indirect immunofluorescence microscopy, and/or ELISA detection of anti-desmoglein antibodies.7 Blood samples and clinical data were collected from patients arriving at the Rheumatology and Nephrology Units at Rabin Medical Centre, Petach Tikva, Israel; the Rheumatology Unit and the Haematology Department of the Sheba Medical Centre, Israel; the Department of Dermatology, Tel Aviv Sourasky Medical Centre; and the Dipartimento di Scienze Mediche e Chirurgiche, Sezione di Clinica Medica, Polo Didattico, Ancona, Italy.
Mouse antibody testing
We tested sera, by tail bleeding, from three different groups of mice. A healthy control group that consisted of 10 female BALB/c mice, 2 months old; a ‘young SLE’ group that consisted of 17 female NZB/WF1 mice, 2 months old, that had not yet developed proteinuria; and an ‘old SLE’ group that consisted of 16 female NZB/WF1 mice, 5–6 months of age, that had proteinuria of at least +3 by Albustix (Bayer, Leverkusen, Germany). The NZB/WF1 and the BALB/c mice were purchased from Harlan Laboratories (Indianapolis, IN), and were housed at the animal facilities of the Weizmann Institute of Science under veterinary supervision. The experiments were approved by the Institutional Animal Care and Use Committee. The mouse sera were tested as described for human serum testing.
Antigens and human serum testing
We spotted 58 different oligonucleotides, and double- and single stranded DNA, some in various chain lengths (104 different preparations overall), on epoxy-activated glass substrates (ArrayIt SuperEpoxi microarray substrate slides, Sunnyvale, CA) using a 48-pin robot (Microgrid 600; Genomics Solutions, Ann Arbor, MI). The oligonucleotides were purchased from SBS Genetech Co., Ltd. (Shanghai, China) and are shown in the Supplementary material (Appendix S1). The microarrays were then blocked for 1 hr at 37° with 1% BSA. Test serum in 1% BSA blocking buffer (1 : 10 dilution) was incubated under a coverslip for 1 hr at 37°. The arrays were then washed and incubated for 1 hr at 37° with a 1 : 500 dilution of two detection antibodies, mixed together: a goat anti-human IgG Cy3-conjugated antibody, and a goat anti-human IgM Cy5-conjugated antibody (both purchased from Jackson ImmunoResearch Laboratories Inc., West Grove, PA). Image acquisition was performed by laser (Agilent Technologies, Santa Clara, CA) and the results were analysed using quantarray software (Packard BioChip Technologies, Billerica, MA). The quantitative range of signal intensity of binding to each antigen spot was 0–65 000; this range of detection made it possible to obtain reliable data at a 1 : 10 dilution of test samples.
Image analysis and data processing
The foreground and background intensities of multiple spots of each antigen were averaged, and the difference between the foreground and the background was calculated. The resulting value was taken as the antigen reactivity of the antibodies binding to that spotted antigen. All antigens showed meaningful reactivity in a significant number of slides; so no antigen was excluded.
Statistical analysis of antibody results
We sought to identify antigens whose reactivity was higher or lower in a specific study subgroup compared with other subgroups. Antigens that passed a threshold of positive predictive value ≥ 90% and had a sensitivity of at least 20% were determined to significantly characterize a specific subgroup. Patients with SLE were marked positive for dsDNA if their reactivity to dsDNA passed this threshold.8 To correct for multiple hypothesis testing, we only selected antigens that also passed a false discovery rate of 5%.9
Characterization of runs of poly-G and poly-T in the genomes of the human, mouse and fruit fly
Nucleotide patterns were identified in the genome sequences using the program fuzznuc from the emboss package,10 with the complement parameter. For human, mouse and fruit fly, the G10 and T10 sequences were taken for ‘collapse’ to provide a non-redundant set of pure G or T stretches in the genome. This was performed in Galaxy11 as follows. The file was converted to interval format, filtered to separate + and − strands; each strand was clustered separately, a column was added to return the strand to the results, and the results of both strands were concatenated into one file. The resulting sequences were filtered by length, and only those that were 20 bp or more were taken for intersecting with genes. Gene coordinates (from transcription start to transcription end) were taken from the RefSeq track of the UCSC genome browser via the Galaxy server (human 43 801 transcripts, mouse 30 997 transcripts, fruit fly 30 551 transcripts). The gene and repeat data were intersected with Galaxy, and non-redundancy on the level of the gene symbol was performed using Venny.12
Genomic GC percentages were calculated on the sequences used for the pattern search, after capitalizing the soft-masked sequence, and using the following unix command: cat sequence.fa ¦ grep -v “>”¦ awk ‘BEGIN{a = 0; c = 0; g = 0; t = 0;} {a+=gsub(“A”,”“); c+=gsub(“C”,”“); g+=gsub(“G”,”“); t+=gsub(“T”,”“);} END{print a,c,g,t}'. Tissue distribution was checked using the UP_TISSUE analysis in the DAVID server.13 The T20+ gene lists were divided into groups of 3000, and the results were compiled to check for enrichment. The DAVID server has 18 201 and 19 868 total genes defined in human and mouse, respectively, and 7789 and 7313 brain-expressed genes, in human and mouse, respectively.
Genomes were taken as follows from the UCSC Genome browser (http://genome.ucsc.edu/): human – hg19; mouse – mm10; fruit fly – dm3.
Results
Human antibodies binding to homo-nucleotide 20-mers
Figure1 shows the binding of human serum IgG and IgM antibodies to the four 20-mer homo-nucleotides – G20, A20, C20 and T20; sera were tested from healthy persons, and from PV, SSc and SLE patients. The reactivities were ordered by each participant's reactivity to dsDNA, from left to right. It can be seen that IgG reactivities to G20 were very high in all participants, significantly higher than the very low reactivities to the other oligonucleotides. However, patients with PV were found to have significantly lower IgG and IgM reactivities to G20 than did patients with SSc. Apart from that, no difference was found between the study groups. Healthy participants, and SSc and PV patients had very low or no reactivities to A20, C20 and T20. Reactivities to A20, C20 and T20 in patients with SLE correlated with their reactivities to dsDNA; patients with low reactivities to dsDNA did not manifest reactivities to A20, C20 and T20, but patients with higher reactivities to dsDNA showed some reactivities to A20, C20 and T20 (Fig.1).
Figure 1.
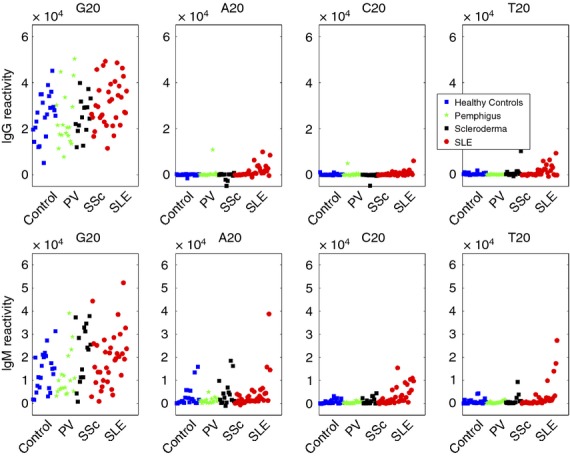
Individual IgM and IgG reactivities to A20, C20, G20 and T20 in sera from healthy participants (blue boxes), pemphigus vulgaris (PV) patients (green stars), scleroderma (SSc) patients (black boxes), and systemic lupus erythematosus (SLE) patients (red circles). Participants were ordered from left to right according to their reactivity to dsDNA.
IgM reactivities to the four homo-nucleotide sequences were more diffuse: some participants in each group showed high reactivities to G20, but, in contrast to the IgG reactivities to G20, some of the sera showed little or no IgM binding to G20.
To further characterize the antibodies against poly-G oligonucleotides, we performed an additional study on 23 healthy participants, 24 SSc patients and 49 SLE patients. We used an extended microarray antigen chip that contained 58 oligonucleotides, including poly-G and poly-T sequences with and without modifications (see list of oligonucleotides in the Supplementary material, Appendix S1). We divided the SLE patients according to their reactivity to dsDNA to see if dsDNA positivity or negativity was associated with antibodies to the synthetic oligonucleotides. The results of the IgM and IgG reactivities to G20, A20, C20 and T20 of the first study were confirmed. Combining both studies yielded a total of 60 SLE patients, 26 SSc patients, 18 PV patients and 31 healthy participants who all displayed high IgG reactivities to G20 and relatively low reactivities to A20, C20 and T20. Mean IgG reactivities where highest by far to G20 in all the study groups (data not shown). In healthy participants, IgG reactivities to G20 were almost always the highest reactivities in each individual compared with their reactivities to the other oligonucleotides (see Supplementary material, Data S1 and Fig. S1).
Mouse sera manifest little or no antibody binding to G20
To learn whether antibodies binding to G20 might also be detected in mouse sera, we obtained serum from 10 healthy mice of the BALB/c strain and from 33 NZB/WF1 female mice, which spontaneously develop SLE.14 Sixteen of the NZB/WF1 mice, aged 5–6 months had advanced SLE with proteinuria (at least +3 by Albustix) and 17 NZB/WF1 mice, aged 2 months had not yet manifested proteinuria. Figure2 shows that, in contrast to the human sera, none of the healthy BALB/c mice and very few of the NZB/WF1 mice manifested serum antibodies, IgG or IgM, to G20. Some of the SLE mice manifested IgG or IgM serum antibodies binding to A20, C20, or T20 – these antibodies tended to be associated with the presence of higher amounts of anti-DNA antibodies. Mouse sera that were negative for antibodies to G20 were strongly positive for antibodies to various self-antigens (not shown). Hence, anti-G20 antibodies characterized humans but not mice. We studied the human antibodies in greater detail below.
Figure 2.
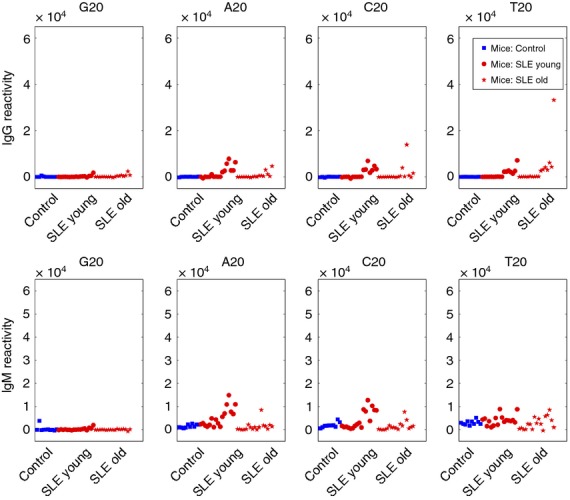
Individual IgM and IgG reactivities to A20, C20, G20 and T20 in mouse sera. Reactivities of healthy BALB/c mice (blue boxes) and either young or old NZB/W mice (red dots and red stars, respectively) were ordered from left to right according to their reactivity to dsDNA.
Human antibody binding to poly-G or poly-T is related to the length of the homo-nucleotide
Figure3 shows the effects of reducing the lengths of the nucleotide oligomers on the mean IgG and IgM binding of each group to the T or G homo-nucleotides. It can be seen that, except for SLE patients positive for anti-dsDNA who showed reactivities to T20, none of the other groups showed appreciable IgG or IgM mean reactivities to any of the poly-T homo-nucleotides. In contrast, mean IgG reactivities to poly-G in all of the sera were high to G20 and fell significantly as the lengths of the nucleotide chains were reduced to G17 and below. The SLE patients positive for anti-dsDNA manifested higher mean IgG reactivities to the shorter G polymers than did the other groups.
Figure 3.
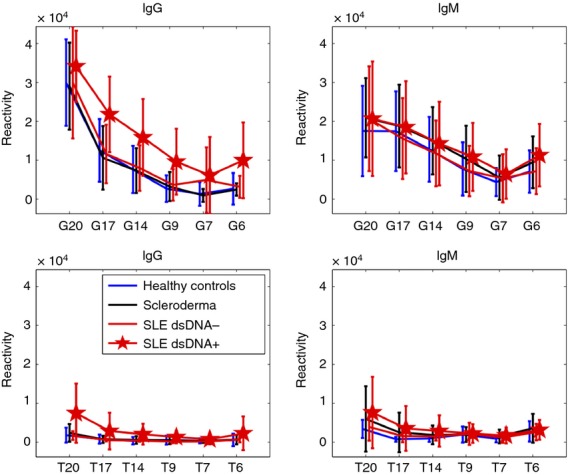
Mean IgM and IgG binding to poly-G and poly-T oligonucleotides as a function of chain length.
Mean IgM binding to G20 was lower than the IgG binding, but IgM binding too was affected by shortening the length of the oligomer. The mean IgM binding of the SLE patients positive to dsDNA did not differ from that of the other groups.
The effects of adding a single T to either the 5′ or 3′ termini of G16
To test the specificity of the binding to poly-G, we tested the degree of binding of IgG or IgM to G17 compared with G16 to which a single T had been added either at the 5′ or 3′ end of the G-oligonucleotide chain. Figure4 shows the results for individual participants. It can be seen that both the IgG and IgM binding to T1–G16 was essentially equal to the binding to the G17 chain; note the diagonal between G17 and T1–G16. However, the binding for each participant to G16–T1 was considerably less than the binding to G17; a diagonal relationship was no longer present. Hence, it would appear that the reactivities to poly-G in each of the groups was highly influenced by the addition of a single T moiety to the 3′ end of the poly-G chain but not by the addition of a T to the 5′ end of the G chain; the spatial order of the nucleotides would appear to form an antigen structure critical to antibody binding.
Figure 4.
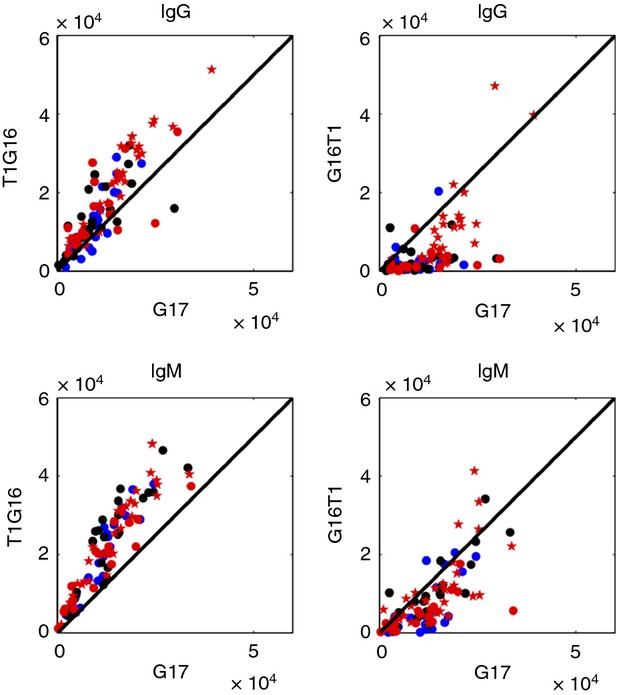
IgM and IgG reactivities to the G17 oligonucleotide compared with T1G16 and G16T1. The designations of the participants are as described in the legend to Figure3.
Single G additions to the ends of poly-T sequences enhance IgG and IgM antibody binding
In view of the marked effects of adding a single T to the 3′ end of a poly-G chain (Fig.4), we tested the effects on antibody binding by adding a single G to either the 5′ or 3′ end of a poly-T chain. Figure5 shows that although both IgM and IgG reactivities to G1T16 and to T16G1 increased in almost all the participants (most points were above the diagonal), the increases were much more pronounced when the guanine was added to the 5′ end. Similarly IgM and IgG reactivities to G2T16 and T16G2 were also increased compared with T17 (see Supplementary material, Fig. S2).
Figure 5.
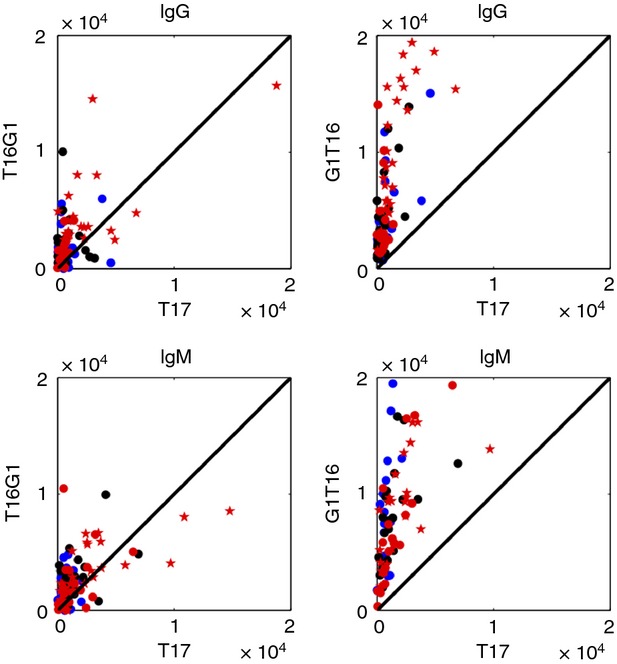
IgM and IgG reactivities to the modified T17 oligonucleotides G1T16 and T16G1 compared with T17 reactivities. The designations of the participants are as described in the legend to Figure3.
In summary, human reactivities to poly-T oligonucleotides were found to be very low or undetectable, but could be increased significantly by the addition of even a single G to either end of the chain; this was in marked contrast to the inhibition of antibody binding to poly-G by the addition of a single T to the 3′ end of the chain.
IgM reactivities to CpG repeats
We measured IgM reactivities in all the four human subgroups to a 20-mer formed by a chain of 10 alternating C-G dinucleotides (CG)10. IgM reactivities to (CG)10 were high in all but one of the healthy participants, in all of the SSc patients and in most of the SLE patients. Indeed, a subgroup of SLE patients manifested low IgM reactivities to (CG)10, a significant difference from the SSc patients (Fig.6). We did not find any significant clinical characteristics or overlap with other reactivities in this subgroup of SLE patients. A group of human SLE patients, mainly those positive for anti-dsDNA, manifested high IgG reactivities to (CG)10.
Figure 6.
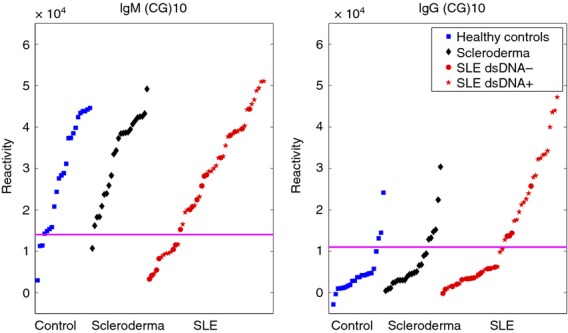
IgG and IgM binding to (CG)10 in healthy human participants, and patients with scleroderma (SSc) or systemic lupus erythematosus (SLE) patients.
IgG and IgM reactivities to dsDNA, ssDNA and synthetic oligonucleotides in SLE patients compared with those of healthy participants and SSc patients
Table S1 (see Supplementary material) lists the oligonucleotides with increases or decreases of antibody binding in the three subgroups of participants. A broad spectrum of oligonucleotides were found to bind both IgM and IgG antibodies in SLE patients compared with healthy participants. IgM and IgG reactivities to dsDNA overlapped; 18 of 23 (78%) SLE patients positive for IgM anti-dsDNA were also positive for IgG anti-dsDNA. Furthermore, the increased IgM and IgG reactivity to the oligonucleotides in the SLE participants overlapped with the IgM and IgG reactivity to dsDNA, respectively. Hence, the subgroup of dsDNA-positive SLE patients showed increased reactivities to oligonucleotides generally, perhaps to a backbone structure, compared with the dsDNA-negative patients. Indeed, we have found that the addition of oligonucleotide serology did not significantly increase the detection rate of SLE patients above that detected by dsDNA reactivity alone (data not shown).
IgG reactivities, but not IgM reactivities, to oligonucleotides were found to be significantly increased in patients with SLE compared with SSc patients, but we did not find any particular oligonucleotide to consistently and significantly distinguish SSc patients from healthy participants.
Repeats of G20 and T20 differ greatly in the human genome
The marked difference in the prevalence and amounts of antibodies binding to G20 compared with T20 in human sera, irrespective of health or autoimmune disease, led us to examine whether there might be a difference in the frequencies of G20 and T20 represented in the genome. Poly-G runs present a particular problem for sequencing: they can cause sequencing reactions to fail. Heterochromatin, which has a high repeat content, presumably containing poly-G among others, has also not been sequenced reliably. Nevertheless, we chose to look at the available sequences of the most completely and reliably sequenced genomes, the human, mouse and fruit fly; less completely sequenced genomes are more likely to show errors in runs of poly-G, or to miss them entirely. Table1 shows the results of the genomic search for runs of 20 nucleotides or more – G20+ and T20+.
Table 1.
Numbers of runs of G20+ and T20+ repeats in the designated genomes
| Species | Human | Mouse | Fruit fly |
|---|---|---|---|
| G20+ | 93 | 1392 | 66 |
| T20+ | 170 561 | 71 219 | 1889 |
| T/G ratio | 1835 | 51 | 29 |
The T20+ repeats were significantly more prevalent than runs of G20+ in the three genomes. In particular, the ratio of T to G runs in the human (outlined in bold) was more than 35 times higher than the mouse and 63 times greater than that of the fruit fly. Note that the increased ratio of T20+ to G20+ is due mostly to a reduction in runs of G20+. Figure S3 (see Supplementary material) shows the uncollapsed runs of G20 and T20 in other species – these numbers need to be taken with caution because sequencing of these other genomes is much less reliable than those of the human, mouse or fruit fly. Nevertheless, it is remarkable that no other species shows such a relative reduction of G20 runs as does the human, not even the chimpanzee, even (or especially) when taking the genomic GC% into account.
To learn more about G20+ repeats in the human genome, we searched for genes that overlap with T20+ or G20+ runs. Genes were defined according to RefSeq for each species, and a gene was considered overlapping if there was any overlap with the entire gene locus, from transcription start to transcription end. We found that 40 genes overlap the 93 G20+ repeats in humans, whereas 13 643 genes overlap the T20+ runs. In the mouse, 9841 genes overlapped with T20+ while 694 genes overlapped with G20+, and in the fruit fly 731 genes overlapped with T20+ and 15 with G20+. We analysed the gene lists with DAVID13 in order to gain a deeper understanding of these genes, despite the large discrepancy in the number of genes between the two groups and the difficulty in finding relevant annotations for such large (T20+) and small (G20+) numbers of genes. Only one feature was notable: 24 of the 35 G20+ containing genes recognized by DAVID (69%) are expressed in the brain (Table2). Although genes containing T20+ were also significantly associated with genes expressed in the brain (1·1 times higher than expected, P ≤ 0·001), the genes associated with G20+ in the human brain were 1·6 times more than expected by chance alone (P = 0·0021). Similar findings could be seen in the mouse, although to a lesser extent. We could not compare these findings with the fruit fly, as fruit fly brain expression is not defined in DAVID. As a general rule, one should not compare gene lists of such different sizes; nevertheless, these differences are statistically significant despite their size differences.
Table 2.
Numbers of genes associated with G20+ or T20+
| Category | Human T20+ | Human G20+ | Mouse T20+ | Mouse G20+ |
|---|---|---|---|---|
| Genes with overlaps | 13 643 | 40 | 9841 | 694 |
| Genes recognized by DAVID | 11 206 | 35 | 8366 | 596 |
| Genes expressed in the brain (UP_TISSUE) | 5440 | 24 | 3661 | 298 |
| % Observed | 49% | 69% | 44% | 50% |
| % Expected | 43% | 43% | 37% | 37% |
| % Fold enrichment T/G20+ | 1·1 | 1·6 | 1·2 | 1·4 |
| P value for fold enrichment | < 0·001 | 0·0021 | < 0·001 | < 0·001 |
We looked at the location of the G20+ repeats in the genes and found that all of the overlapping repeats in the human and mouse were located in non-coding regions – introns, untranslated regions or long non-coding RNA. These findings suggest that poly-G repeats might have a regulatory role.
Discussion
The experiments undertaken in this paper were originally designed to characterize antibodies to DNA in human SLE patients, but the results led us to discover in addition the apparently universal prevalence of antibodies to the homo-nucleotide G20 in healthy human participants as well as in patients with the autoimmune diseases SLE and SSc; some patients with PV manifested lower amounts of these antibodies, and we shall return to this finding below. Surprisingly, SLE mice and healthy mice manifested little or no serum antibodies to G20; we do not know whether other non-human mammals also lack these antibodies, but anti-G20 antibodies might just be a feature of the human.
We must note that the synthesis of poly-G oligonucleotide is problematic, because of the secondary conformations they form. Therefore it is quite possible that the poly-G preparations we used were not pure and also contained shorter poly-G oligonucleotides. Nevertheless, Fig.3 shows that the synthesis was apparently successful; reactivity to poly-G repeats increased when the number of guanines in the preparation increased. The prevalence of anti-G20 reactivity and the lack of anti-T20 in healthy human participants aroused our curiosity about the relative prevalence of runs of G20 and T20 in the human genome. This curiosity disclosed a greater than thousand-fold difference in the frequency of these two apparently uninteresting sequences. Before we discuss each of these unexpected findings, let us review the hypothesis behind the initial experiment.
Our interest in anti-DNA antibodies began some 15 years ago when we discovered that immunization of mice with a mouse monoclonal antibody that binds to the C-terminal domain of the p53 tumour-suppressor protein induced, via an anti-idiotypic network, antibodies both to p53 and to DNA.15 This finding could be explained by structural mimicry between damaged DNA and the monoclonal antibody to the C-terminal domain of p53, which binds damaged DNA; subsequently, we isolated a monoclonal antibody that mimicked the p53-binding site.4 Structural mimicry between the C-terminal domain of p53 and the monoclonal antibody was confirmed by the finding that both p53 and the antibody could bind the synthetic homo-oligonucleotide G20, but not A20, C20 or T20.4 The generation of a monoclonal mouse antibody that binds G20 indicates that mice are capable of making at least one such antibody. Our later development of an antigen microarray device for profiling antibody repertoires1–3 has enabled us to study autoantibody repertoires in SLE3 and other autoimmune diseases8 to large numbers of self-molecules. We therefore used this approach to learn if we might be able to define synthetic oligonucleotides bearing epitopes relevant to SLE; we predicted that only SLE patients who were positive for anti-DNA antibodies would harbour antibodies capable of binding to G20. We were surprised to discover that the healthy or sick humans we sampled, but not mice, bear IgG and IgM antibodies that bind to G20 (Figs1 and 2).
A number of points emerged regarding these antibodies: as anti-G20 antibodies are present in participants without detectable anti-dsDNA autoantibodies, we may conclude that such anti-G20 antibodies are distinct from the pathological anti-DNA antibodies found in patients with SLE or SSc. Apparently, the amounts of G20 in extracts of DNA used for dsDNA antibody testing are too few to detect these antibodies in participants who are free of pathological anti-DNA. Note that normal anti-G20 IgG and IgM binding was markedly reduced by shortening the poly-G20 chain or by small additions of other nucleotides, such as a single T placed at the 3′ end of a G16 chain; these findings argue for antibody binding to a unique conformation assumed by poly-G20. Indeed, it is known that poly-G sequences are prone to assume a specific conformation termed G-quadruplexes or G-tetrads, in which adjacent guanines form a four-stranded structure.16 Telomeres are also known to contain quadruplex conformations, which can down-regulate the action of telomerase enzymes affecting cell survival;17 for example, G quadruplex stabilization is thought to support the prolonged survival of cancer cells.18 G quadruplexes have also been found in the promoter regions of many proto-oncogenes.19–21 The presence of G quadruplexes may allow the direct binding of p53 when suppression of an oncogene is needed. It has been reported that some anti-DNA antibodies can enter living cells and their nuclei;22 further research must be done to learn whether normal anti-G20 antibodies might enter cells and bind to quadruplex structures in gene-promoters or in telomeres and so affect health or tumorigenesis.
The importance of conformation is also compatible with the observation that antibody binding to a poly-G molecule is markedly affected by the addition of a T to the 3′ but not to the 5′ end of the molecule (Fig.4). Note that the additions of a G to either the 3′ or the 5′ ends of a poly-T molecule enabled antibody binding (Fig.5); we do not yet know if the same antibodies are involved in binding to both the poly-G and the G-modified poly-T molecules.
Poly-G runs are also known to manifest unique physical properties such as transporting electrons23 and affecting charge;24 indeed, adjacent G bases appear to be critical in the electronic properties of telomeres.25 It remains to be determined whether anti-G20 antibodies can influence these properties.
Another unexpected observation of our present study is that most humans harbour IgM antibodies capable of binding to CpG repeats (Fig.6); CpG repeats are known to serve as sites for epigenetic regulation based on methylation of C.26 It would be important to learn whether the normal IgM anti-CpG antibodies we have detected here might exert any function in epigenesis. These antibodies might also exert an immune function; a subset of SLE patients appear to lack these IgM anti-CpG antibodies (Fig.6).
An immune function for IgM anti-G20 might also be considered as patients with PV appear to have reduced amounts of these antibodies (Fig.1). The present discovery of antibodies to G20, G-modified poly-T and CpG invite further research into their possible roles in health and disease.
No less intriguing are our findings regarding the prevalence and distribution of runs of G20 compared with T20 in the human genome (Tables1 and 2). Only a few genomes have been sequenced to the degree that allows us to draw conclusions with some degree of certainty – the human, the mouse and the fruit fly. The finding that the human, who has the most complex brain, has an extremely low number of poly-G repeats compared with the other species, and that those that overlap with genes are located predominantly in non-coding regions of genes expressed in the brain, suggests that a decrease in runs of G20 may exert some regulatory effect on the development or the function of the brain. It is conceivable that anti-G20 antibodies might play a role in the advancing complexity of brain function. The finding that p53 too binds G20 also suggests that the G20 motif might be worthy of study in neural development. Indeed it was found that p53 regulates renewing divisions in neural stem cells both during embryonic development and in adulthood, and also regulates neurite growth and axonal regeneration.27 Our observation that humans, compared with mice, uniquely feature both relatively reduced runs of poly-G and enhanced amounts of antibodies to G20 suggests the working hypothesis that there might be some association between these two apparently disparate phenomena; this remains to be further investigated. These musings, of course, bring us back to our seminal observation that G20 can serve as an epitope for binding both p53 and a monoclonal anti-DNA antibody.4 The connection between genomic G20 and its particular locations, p53, and prevalent antibodies to G20 might yet furnish novel perspectives on antibodies in development, as well as in health and disease.
Acknowledgments
This work was supported by the Open University of Israel grants (IG-0901 and IG-0902 to NS) and by a grant by the Laszlo N. Tauber Family Foundation to IRC and NS.
Disclosures
The authors declare that no competing interests exist.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. IgG reactivities to G20 compared with IgG reactivities to the other oligonucleotides in each human participant.
Figure S1. IgG Reactivity to G20 compared with all other oligonucleotides in healthy persons, scleroderma patients, systemic lupus erythematosus patients who are negative or positive for dsDNA.
Figure S2. IgM and IgG reactivities to G2T16 and T16G2 compared with T17.
Figure S2. The T20 to G20 ratio in the genomes of different organisms.
Table S1. The sensitivity of oligonucleotides whose reactivity was significantly increased or decreased in patients with systemic lupus erythematosus (positive predictive value ≥ 90%).
Appendix S1. List of oligonucleotides.
References
- Merbl Y, Zucker-Toledano M, Quintana FJ, Cohen IR. Newborn humans manifest autoantibodies to defined self molecules detected by antigen microarray informatics. J Clin Invest. 2007;117:712–8. doi: 10.1172/JCI29943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana FJ, Merbl Y, Sahar E, Domany E, Cohen IR. Antigen-chip technology for accessing global information about the state of the body. Lupus. 2006;15:428–30. doi: 10.1191/0961203306lu2328oa. [DOI] [PubMed] [Google Scholar]
- Fattal I, Shental N, Mevorach D, et al. An antibody profile of systemic lupus erythematosus detected by antigen microarray. Immunology. 2010;130:337–43. doi: 10.1111/j.1365-2567.2010.03245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkel J, Kam N, Erez N, Mimran A, Heifetz A, Eisenstein M, Rotter V, Cohen IR. Monoclonal antibody to a DNA-binding domain of p53 mimics charge structure of DNA: anti-idiotypes to the anti-p53 antibody are anti-DNA. Eur J Immunol. 2004;34:3623–32. doi: 10.1002/eji.200425371. [DOI] [PubMed] [Google Scholar]
- Arthritis Rheum 25 Arthritis Rheum 40 Criteria published by Tan EM 1982; 25: 1271, updated by Hochberg MC. 1997; 40: 1725.
- Preliminary criteria for the classification of systemic sclerosis (scleroderma). Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 1980;23:581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- Zagorodniuk I, Weltfriend S, Shtruminger L, Sprecher E, Kogan O, Pollack S, Bergman R. A comparison of anti-desmoglein antibodies and indirect immunofluorescence in the serodiagnosis of pemphigus vulgaris. Int J Dermatol. 2005;44:541–4. doi: 10.1111/j.1365-4632.2004.02541.x. [DOI] [PubMed] [Google Scholar]
- Fattal I, Shental N, Molad Y, et al. EBV antibodies mark SLE and scleroderma patients negative for anti-DNA. Immunology. 2014;141:276–85. doi: 10.1111/imm.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams http://bioinfogp.cnb.csic.es/tools/venny/
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths towards the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews BS, Eisenberg RA, Theofilopoulos AN, et al. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J Exp Med. 1978;148:1198–215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkel J, Erez-Alon N, Mimran A, Wolkowicz R, Harmelin A, Ruiz P, Rotter V, Cohen IR. Systemic lupus erythematosus in mice, spontaneous and induced, is associated with autoimmunity to the C-terminal domain of p53 that recognizes damaged DNA. Eur J Immunol. 2000;30:977–84. doi: 10.1002/(SICI)1521-4141(200004)30:4<977::AID-IMMU977>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Simonsson T. G-quadruplex DNA structures – variations on a theme. Biol Chem. 2001;382:621–8. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Lacroix L, Teulade-Fichou MP, et al. Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc Natl Acad Sci USA. 2001;98:3062–7. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xiang JF, Zhang H, Tang YL. Searching drug-like anti-cancer compound(s) based on G-quadruplex ligands. Curr Pharm Des. 2012;18:1973–83. doi: 10.2174/138161212799958369. [DOI] [PubMed] [Google Scholar]
- Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–72. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–8. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Lin S, Yuan G. Spectroscopic probing of recognition of the G-quadruplex in c-kit promoter by small-molecule natural products. Int J Biol Macromol. 2012;50:996–1001. doi: 10.1016/j.ijbiomac.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Yanase K, Smith RM, Puccetti A, Jarett L, Madaio MP. Receptor-mediated cellular entry of nuclear localizing anti-DNA antibodies via myosin 1. J Clin Invest. 1997;100:25–31. doi: 10.1172/JCI119517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhu J, Guo S, Li T, Li J, Wang E. Photoinduced electron transfer of DNA/Ag nanoclusters modulated by G-quadruplex/hemin complex for the construction of versatile biosensors. J Am Chem Soc. 2013;135:2403–6. doi: 10.1021/ja3089857. [DOI] [PubMed] [Google Scholar]
- Miura T, Thomas GJ., Jr Structure and dynamics of interstrand guanine association in quadruplex telomeric DNA. Biochemistry. 1995;34:9645–54. doi: 10.1021/bi00029a042. [DOI] [PubMed] [Google Scholar]
- Markus TZ, Daube SS, Naaman R. Cooperative effect in the electronic properties of human telomere sequence. J Phys Chem B. 2010;114:13897–903. doi: 10.1021/jp1064038. [DOI] [PubMed] [Google Scholar]
- Vinson C, Chatterjee R. CG methylation. Epigenomics. 2012;4:655–63. doi: 10.2217/epi.12.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Di Giovanni S. Gatekeeper between quiescence and differentiation: p53 in axonal outgrowth and neurogenesis. Int Rev Neurobiol. 2012;105:71–89. doi: 10.1016/B978-0-12-398309-1.00005-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. IgG reactivities to G20 compared with IgG reactivities to the other oligonucleotides in each human participant.
Figure S1. IgG Reactivity to G20 compared with all other oligonucleotides in healthy persons, scleroderma patients, systemic lupus erythematosus patients who are negative or positive for dsDNA.
Figure S2. IgM and IgG reactivities to G2T16 and T16G2 compared with T17.
Figure S2. The T20 to G20 ratio in the genomes of different organisms.
Table S1. The sensitivity of oligonucleotides whose reactivity was significantly increased or decreased in patients with systemic lupus erythematosus (positive predictive value ≥ 90%).
Appendix S1. List of oligonucleotides.


