Abstract
Purpose of review
The circadian clock is an intricate biological timekeeper that is subject to fine-tuning mechanisms in order to maintain synchrony with the surrounding environment. One such mechanism is performed by the mammalian sirtuins that provide plasticity to the circadian clock by sensing cellular metabolic state. The sirtuins modulate the circadian epigenome and subsequent transcriptional control, and alterations to this organized system manifest in metabolic consequences, aging phenotypes and possibly cancer.
Recent findings
New information regarding sirtuin-dependent control of the circadian clock has emerged. In addition to SIRT1 and SIRT3, SIRT6 has been demonstrated as a critical regulator of circadian transcription that also serves as an interface with metabolic homeostasis. Also, new metabolic functions of SIRT1 have been described in the brain, which are critical to relay nutritional inputs to the central clock.
Summary
This review focuses on the link between the circadian clock and the sirtuins, with an emphasis on new findings. In addition, speculation on the possible connections at the physiological level will be made that could further link the clock to aging and cancer.
Keywords: Sirtuins, circadian clock, metabolism, nutrition, cancer, aging
Introduction
The circadian clock is a self-sustained biological pacemaker that operates with a periodicity of 24 hours, the purpose of which is to synchronize and maintain homeostasis of a number of physiological processes1, 2. Disruption in proper circadian timekeeping manifest in detrimental systemic effects and a number of clues from the clinic and laboratory suggest that these disturbances result in metabolic disruptions3, 4, cancer5, 6 and aging related phenotypes7–9. At the heart of the circadian molecular machinery are the core DNA-binding transcription factors, CLOCK and BMAL1, that drive the oscillation of ~10% of transcripts in the genome in a defined tissue-specific program 10, 11. CLOCK:BMAL1-dependent transcription of clock-controlled genes (CCGs) peaks during the day, while transcriptional feedback inhibition by the circadian repressors, Period (PER) and Cryptochrome (CRY) occurs at night 12, 13. In addition to the core transcriptional/translational feedback loop, regulation of circadian transcription is also subject to epigenetic modifications that are rhythmic over the day/night cycle 12, 14. An example of such histone modifications are mediated by the histone methyltransferases MLL115 and MLL316 on H3K4 that permit circadian gene expression, and intriguingly the tri-methylation of H3K4 is regulated by SIRT1 through cyclic deacetylation of MLL117.
The mammalian sirtuins modulate the circadian epigenome and provide specificity in transcriptional control. The sirtuins are an nicotinamide adenine dinucleotide (NAD+)-dependent family of histone deacetylases (HDACs), which are implicated in various physiological functions ranging from aging, maintenance of genome integrity, stress response to nutrient challenge, metabolic control and cancer 18–20. Remarkably, the seven mammalian sirtuins vary in their enzymatic activity (aside from their deacetylase function)21, biological targets, and cellular function 8, 22. The subcellular localization of the sirtuins is also varied: SIRT1 shuttles between the nucleus and cytoplasm, SIRT2 is cytoplasmic, SIRT3, SIRT4 and SIRT5 are mitochondrial, SIRT6 is nuclear and chromatin-bound, and SIRT7 is found largely in the nucleolus 19, 23.
Unsuspected Functions of the Sirtuins in Regulating the Clock
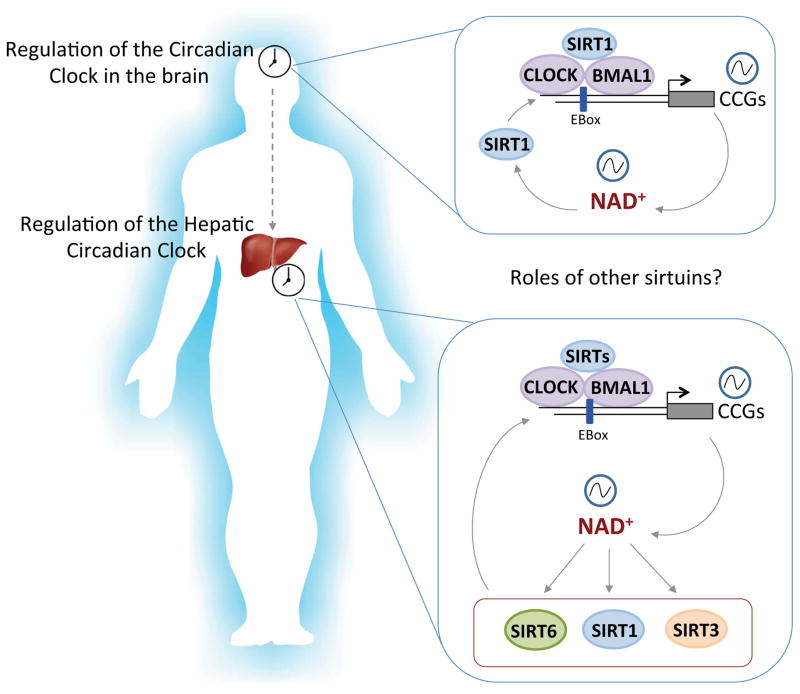
The mammalian sirtuins have been reported to regulate both the circadian clock in the brain as well as peripheral clocks, such as the liver (Figure 1). Work from a number of laboratories has shown that SIRT1 is involved in regulating circadian epigenetic control through H3K9 deacetylation, modulation of BMAL1 and PER2 acetylation state and stability, and subsequent control of circadian gene expression24, 25. Additionally, the circadian clock was previously shown to regulate mitochondrial oxygen consumption rate in an NAD+/SIRT3-dependent manner26. Given that levels of NAD+ are clock-controlled and oscillate over the circadian cycle27, 28, the question arises as to what other NAD+-dependent sirtuins are involved in clock regulation.
Figure 1. Sirtuin-dependent control of the circadian clock in the brain and periphery.
The core clock transcriptional machinery, directed by CLOCK and BMAL1, drives transcription of circadian gene expression, including nicotinamide phosphoribosyltransferase (Nampt), which subsequently results in oscillatory levels of NAD+. In the brain, NAD+-dependent SIRT1 activity is critical for circadian function both in the SCN and VMH. In the liver, SIRT1, SIRT6 and SIRT3 are involved in circadian transcription and metabolic regulation.
Suprachiasmatic nucleus, SCN; Ventromedial hypothalamus, VMH; Clock-controlled genes, CCGs.
Transcriptional control of SIRT6
In addition to the described roles of SIRT1 and SIRT3, SIRT6 was recently demonstrated to regulate the hepatic circadian clock. The defining feature of SIRT6 that distinguishes it from the other sirtuins is its constitutive chromatin localization 29, 30, that is maintained over the circadian cycle31. SIRT6 deacetylates H3K9 32, 33 and H3K56 34–36 which consequentially results in modulation of gene expression and telomere maintenance 37, 38. Strikingly, the genome-wide localization of SIRT6 is enriched at transcriptional start sites (TSS) of active genomic loci, that are also occupied by serine 5 phosphorylated RNA polymerase II 39. Collectively, these data suggest that SIRT6 functions as a transcriptional ‘marker’ that demarcates areas of the genome that are dynamically transcribed and subsequently silenced. Similarly, recent evidence supports a role for SIRT6 as an epigenetic safeguard for controlling proper cellular differentiation40, suggesting the HDAC function of SIRT6 is a critical control mechanism for proper transcription in multiple biological contexts. In further support of this concept, not only do SIRT6 and SIRT1 regulate unique sets of circadian genes in the liver, but SIRT6 is involved in modulating the proper chromatin recruitment of CLOCK:BMAL1 and SREBP1 and thereby regulating circadian gene expression31. This partitioned control of the nuclear sirtuins also results in differential jurisdiction of SIRT6 and SIRT1 in regulating circadian fatty acid metabolism, carbohydrates, peptides and cofactors in the liver31. The SIRT6-dependent regulation of circadian fatty acid synthesis, beta-oxidation and storage as triglycerides is also intriguing given that free fatty acids (FFA) are potent activators of SIRT6 HDAC activity in vitro41, suggesting a complex feedback regulation exists. These concepts underscore the unique ability of SIRT6 to dynamically modulate transcription in multiple contexts, one of which is the regulation of circadian gene expression in the liver.
Metabolic functions of SIRT1
Moreover, the complexity of sirtuin biology in regulating the clock is continually expanding as new roles for SIRT1 have recently emerged that dictate metabolic state. SIRT1 deacetylates and therefore modulates the circadian enzymatic activity of Acetyl-CoA Synthetase 1 (AceCS1), an enzyme involved in the production of acetyl-CoA from acetate. This results in the oscillation of acetyl-CoA over the circadian cycle and subsequently controls fatty acid elongation42. Because acetyl-coA is a key carbon donor in metabolism, this establishes the possibility of other biological processes that are dependent on acetyl-coA that could be linked to the clock and SIRT1. Also, the importance of non-transcriptional, metabolic and enzymatic feedback loops in circadian control43, which are accompanying mechanisms to the classic transcriptional/translational pathways, are critical in clock biological output.
Along a similar metabolic premise, SIRT1 was shown to play an important role in translating nutritional cues in the brain. In the ventromedial hypothalamus (VMH), SIRT1 was found to control circadian rodent behavior under specific conditions of light and food restriction, which also extends to effect circadian gene expression of the central clock in the suprachiasmatic nucleus (SCN)44. These results show that SIRT1 is a nutritional sensor in the VMH and is able to transmit metabolic information to the central clock that dictates systemic circadian behavior and rhythms. This data further demonstrates the plasticity of the clock system, that it is adaptive to its environment, and the importance of SIRT1 as a sensor that can transmit nutritional cues to alter transcriptional profiles.
The circadian clock has previously been implicated in insulin signaling. Insulin levels are dynamically rhythmic and genetic mutant models of the clock in rodents demonstrate the circadian effects on insulin and hepatic glucose production45–48. Similarly, SIRT1 has been reported to be involved in pancreatic beta cell secretion of insulin49 as well as a key regulator of insulin sensitivity50. The first preliminary data on circadian regulation of insulin sensitivity that is linked to SIRT1 was recently demonstrated in cultured hepatocytes, whereby insulin resistance induced by circadian misalignment was attenuated by pharmacological induction of SIRT1 activity51. This preliminary data is tantalizing, though requires further mechanistic insights in vivo to determine how critical SIRT1, or any other sirtuin are to the insulin pathway.
Circadian clock, sirtuins and cancer
In humans, circadian disruption which is prominent in shift workers puts them at increased risk for breast cancer52. In mice, an ablation of the central clock located in the SCN results in increased growth of tumor xenografts as compared to mice with an intact circadian pacemaker53. Also, the Per2m/m mice are highly sensitive to gamma-irradiation and exhibit increased levels of salivary gland hyperplasia54. The tumor suppressor p53 regulates Per2 expression by blocking CLOCK:BMAL1-dependent recruitment to the Per2 promoter and strikingly, p53-null mice exhibit a shorter circadian period and impaired photo-entrainment55. Most recently, the cancer/testis antigen PASD1, the expression of which is induced upon oncogenic transformation, was shown to interact with the clock complex and thereby repress circadian transcription56. Knockdown of PASD1 in human cancer cells was able to rescue the amplitude of circadian expression56, though what remains to be determined is the effect of PASD1 knockdown on tumorigenesis in vivo. Remarkably, these results indicate that loss of functional circadian transcription maybe a mechanism by which oncogenic transformation or tumor progression can occur. Yet, these results require additional experiments to determine the extent and molecular mechanisms by which the circadian clock is linked to cancer.
The connection between the circadian clock, the sirtuins and cancer has not been well established, yet a number of possible links can be made and will be discussed here. It is clear that SIRT1 does play a role in cancer, though this function seems to be context specific as SIRT1 has been reported to be a tumor suppressor and promoter. These ideas have been recently reviewed57, 58, and therefore we only focus on new data regarding the role of SIRT1 and cancer. Recent evidence elegantly illustrates that SIRT1 expression is elevated in leukemic stem cells, and a crosstalk exists between SIRT1 and MYC oncogenic signaling that is responsible for driving FLT3 receptor tyrosine kinase resistance in acute myeloid leukemia (AML)59. This data is interesting from a circadian perspective for two reasons. First, MYC is an E-Box binding transcription factor, similar to CLOCK and BMAL1, and the extent to which these two transcriptional programs overlap and share common gene targets is unknown. Therefore, SIRT1 could be playing a role in modulating more than one E-Box driven transcriptional pathway. Second, the role of the circadian clock, SIRT1 and MLL have been described15, 17, and it is likely that this transcriptional circuit could be highly relevant to leukemia. Indeed, recent evidence provides a mechanistic checks and balances system between the SIRT1 repressive chromatin complex and the MLL/DOT1L histone methyltransferase complex that is aberrantly expressed in some forms of leukemia. The DOT1L histone methyltransferase (required to for H3K79 dimethylation) cooperates with the MLL complex to aberrantly express a number of genes that drive tumorigenesis in leukemia driven by MLL rearrangement. This MLL-dependent transcription is counterbalanced by SIRT1 repressive deacetylation of H3K9 and subsequent methylation by SUV39H1 to restore gene expression profiles60. These results provide exciting new avenues for the need for pharmacological SIRT1-based intervention in MLL-rearranged leukemia.
Moreover, in human pancreatic ductal adenocarcinoma (PDAC) tumor specimens and matched normal pancreatic tissue, a marked deregulation of Sirt1 and a number of circadian genes (Bmal1, Per1-3 and Cry1-2) was observed, and an interesting correlation was seen whereby the expression of these genes was altered upon serum starvation in pancreatic cancer cell lines61. This data points to a nutritional sensing mechanism whereby SIRT1 activity can be altered, which may have beneficial therapeutic effects. SIRT6 has long been described as a key regulator of genome stability29, 36, and this concept agrees with data suggesting the role of SIRT6 as a tumor suppressor.
SIRT6 is a potent regulator of aerobic glycolysis in cancer cells, which is a key mechanism for energy production upon which cancer cells are reliant on for growth62. Loss of SIRT6 alone, without the activation of known oncogenes, results in tumor formation suggesting that this metabolic switch results in tumor initiation and most likely also drives tumor progression62. Though the direct link between SIRT6, cancer and the circadian clock has not been made, the SIRT6-dependent metabolic control mechanisms involved in oncogenic transformation are a tempting connection to the metabolic control exerted by the circadian clock. Indeed, a number of recent reports also show an association between metabolic state dictated by SIRT6 and different cancer types63–65.
Circadian clock, sirtuins and aging related pathologies
The role of SIRT1 in regulating the central circadian clock in the SCN has been linked to aging and recent reviews have covered this topic in detail66, 67. The brain-specific Sirt1−/− (BSKO) mice exhibit dampened circadian gene expression in the anterior hypothalamus (where the SCN is located), suggesting that SIRT1 positively regulates clock-controlled transcription68. These changes in gene expression result in altered circadian function, as the BSKO mice exhibit a lengthened circadian period and remarkably, at 5 months of age, the BSKO mice phenocopy the ‘aged’ phenotype observed in 22-month old WT animals68. This data suggests that loss of SIRT1 in the brain not only regulates the circadian clock but also accelerates the aging process, which is most likely mediated by NAD+. Changes in mitochondrial oxidative phosphorylation state were reported in aging and this was attributed to a decline in nuclear NAD+ levels and dependent on SIRT1 enzymatic activity69. Given that the circadian clock regulates oscillatory levels of NAD+ 27, 28, and that circadian transcription is dampened in the aging brain68, the decline in NAD+ levels during aging could be attributed to loss of clock function. Recent therapeutic strategies have been described using caloric restriction (CR) that could rescue the aging phenotype in cardiomyocytes70 and adipose tissue in a SIRT1-dependent manner71.
To date, the role of SIRT6 and aging has not been linked to the clock, but a number of clues suggest a probable connection exists. Genetic mouse models revealed that SIRT6 is involved in aging due to a number of reported factors: SIRT6 is implicated in regulation of genome stability29, DNA repair72, 73, telomere maintenance33, 34, and the dynamic regulation of stress and aging responsive genes32, 74, 75. Given the aging-dependent decline in NAD+ levels69 (that could be related to the loss of circadian function), it may be possible that SIRT6 activity would therefore decline with age and have many deleterious effects. For example, DNA repair has been linked to the circadian clock76–78 and to SIRT672, 73, and if this connection is related to aging is currently unknown. Also, male SIRT6 transgenic mice exhibit an extension in lifespan due to possible changes in the insulin-like growth factor 1 (IGF-1) signaling axis75. Interestingly, growth hormone (GH), which stimulates the production of IGF-1, is known to oscillate in a sleep and clock-dependent manner79, raising the possibility that the aging phenotype in SIRT6 transgenic animals could have a circadian connection. These possible connections between SIRT6, and even other mammalian sirtuins, in the context of clock control and aging require further exploration.
Concluding remarks
The circadian clock is a tightly regulated system that is essential to maintain organismal homeostasis in behavioral, metabolic, and endocrine rhythms. Yet, circadian time keeping is subject to numerous environmental cues that are needed to adjust our internal biological pacemaker to the environment, cellular metabolic state and even stress response. Therefore, the need for ‘metabolic sensors’ to modulate circadian rhythms is a necessity, and one class of these sensors are the mammalian sirtuins. To date, SIRT1, SIRT3 and now SIRT6 have been implicated in circadian control, both in regulating the central clock in the SCN as well as peripheral clocks. The accumulating data on the sirtuins demonstrates these HDACs are critical in the crosstalk between epigenetics, transcription, and metabolism. Our understanding of the extent to which the sirtuins regulate the clock is still incomplete, and possibly other SIRTs are also implicated in circadian control in the cytoplasm and nucleolus. Also, this review explores the possible connections linked to aging and cancer, yet we await more experimental evidence that will clarify the functions of the sirtuins in homeostatic control and contexts of disease state.
Key points.
The sirtuins serve as a fine-tuning mechanism for regulating circadian transcription and providing plasticity which is dependent on cellular metabolic state
The sirtuins and the clock therefore coordinate a critical crosstalk between epigenetics, transcription and metabolism
Deregulation of the sirtuin/clock axis could result in detrimental effects such as cancer and aging-related phenotypes
Acknowledgments
Special thanks to Paolo Sassone-Corsi for critical reading of the manuscript.
Financial Support and Sponsorship
S.M. was supported by NIH grant F32 GM097899 and by the UC Irvine Chao Family Cancer Center.
Footnotes
Conflicts of Interest
No conflicts of interest to declare.
References
- 1.Gamble KL, Berry R, Frank SJ, Young ME. Circadian clock control of endocrine factors. Nat Rev Endocrinol. 2014;10:466–75. doi: 10.1038/nrendo.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 3**.Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. This recent review highlights the recent advances in the clock field regarding the role of nutritional input on circadian metabolism. [DOI] [PubMed] [Google Scholar]
- 4.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–56. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 5.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 6.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 7.Froy O. Circadian rhythms, aging, and life span in mammals. Physiology (Bethesda) 2011;26:225–35. doi: 10.1152/physiol.00012.2011. [DOI] [PubMed] [Google Scholar]
- 8.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2013 doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–35. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010;13:1324–9. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–9. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 15.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–21. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valekunja UK, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A. 2013;110:1554–9. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilar-Arnal L, Katada S, Orozco-Solis R, Sassone-Corsi P. NAD(+)-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat Struct Mol Biol. 2015;22:312–8. doi: 10.1038/nsmb.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall JA, Dominy JE, Lee Y, Puigserver P. The sirtuin family's role in aging and age-associated pathologies. J Clin Invest. 2013;123:973–9. doi: 10.1172/JCI64094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi JE, Mostoslavsky R. Sirtuins, metabolism, and DNA repair. Curr Opin Genet Dev. 2014;26C:24–32. doi: 10.1016/j.gde.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldman JL, et al. Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation. Biochemistry. 2015;54:3037–50. doi: 10.1021/acs.biochem.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flick F, Luscher B. Regulation of sirtuin function by posttranslational modifications. Front Pharmacol. 2012;3:29. doi: 10.3389/fphar.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 25.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peek C, et al. Circadian Clock NAD+ Cycle Drives Mitochondrial Oxidative Metabolism in Mice. Science. 2013;341 doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 30.Tennen RI, Berber E, Chua KF. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131:185–92. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Masri S, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158:659–72. doi: 10.1016/j.cell.2014.06.050. This paper describes the newly defined role of SIRT6 in regulating circadian transcription and metabolism in the liver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–6. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–6. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–3. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toiber D, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–68. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tennen RI, Chua KF. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci. 2011;36:39–46. doi: 10.1016/j.tibs.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ram O, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–39. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Etchegaray JP, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol. 2015;17:545–57. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldman JL, Baeza J, Denu JM. Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. J Biol Chem. 2013;288:31350–6. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Sahar S, et al. Circadian Control of Fatty Acid Elongation by SIRT1-mediated Deacetylation of Acetyl-CoA Synthetase 1. J Biol Chem. 2014 doi: 10.1074/jbc.M113.537191. The authors define a new link between SIRT1, the clock and direct control of fatty acid metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–8. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Orozco-Solis R, Ramadori G, Coppari R, Sassone-Corsi P. SIRT1 Relays Nutritional Inputs to the Circadian Clock Through the Sf1 Neurons of the Ventromedial Hypothalamus. Endocrinology. 2015;156:2174–84. doi: 10.1210/en.2014-1805. A new role for SIRT1 and clock control in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010 doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Curr Biol. 2013;23:372–81. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–6. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bordone L, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun C, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–19. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Zhou B, et al. CLOCK/BMAL1 regulates circadian change of mouse hepatic insulin sensitivity by SIRT1. Hepatology. 2014;59:2196–206. doi: 10.1002/hep.26992. [DOI] [PubMed] [Google Scholar]
- 52.Schernhammer ES, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses' health study. J Natl Cancer Inst. 2001;93:1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 53.Filipski E, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–7. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 54.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 55.Miki T, Matsumoto T, Zhao Z, Lee CC. p53 regulates Period2 expression and the circadian clock. Nat Commun. 2013;4:2444. doi: 10.1038/ncomms3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Michael AK, et al. Cancer/Testis Antigen PASD1 Silences the Circadian Clock. Mol Cell. 2015;58:743–54. doi: 10.1016/j.molcel.2015.03.031. New insights on the role of the clock and cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Z, Fang D. The Roles of SIRT1 in Cancer. Genes Cancer. 2013;4:97–104. doi: 10.1177/1947601912475079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masri S, Kinouchi K, Sassone-Corsi P. Circadian clocks, epigenetics, and cancer. Curr Opin Oncol. 2015;27:50–6. doi: 10.1097/CCO.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, et al. SIRT1 Activation by a c-MYC Oncogenic Network Promotes the Maintenance and Drug Resistance of Human FLT3-ITD Acute Myeloid Leukemia Stem Cells. Cell Stem Cell. 2014;15:431–46. doi: 10.1016/j.stem.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen CW, et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med. 2015;21:335–43. doi: 10.1038/nm.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tavano F, et al. SIRT1 and circadian gene expression in pancreatic ductal adenocarcinoma: Effect of starvation. Chronobiol Int. 2015;32:497–512. doi: 10.3109/07420528.2014.1003351. [DOI] [PubMed] [Google Scholar]
- 62.Sebastian C, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–99. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu M, Seto E, Zhang J. E2F1 enhances glycolysis through suppressing Sirt6 transcription in cancer cells. Oncotarget. 2015;6:11252–63. doi: 10.18632/oncotarget.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choe M, et al. The RUNX2 Transcription Factor Negatively Regulates SIRT6 Expression to Alter Glucose Metabolism in Breast Cancer Cells. J Cell Biochem. 2015 doi: 10.1002/jcb.25171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim EJ, Juhnn YS. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway. J Biol Chem. 2015;290:9604–13. doi: 10.1074/jbc.M114.633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–71. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verdin E. The many faces of sirtuins: Coupling of NAD metabolism, sirtuins and lifespan. Nat Med. 2014;20:25–7. doi: 10.1038/nm.3447. [DOI] [PubMed] [Google Scholar]
- 68.Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–60. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gomes AP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–38. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu W, Zhou HF, Lin RB, Fu YC, Wang W. Shortterm calorie restriction activates SIRT14 and 7 in cardiomyocytes in vivo and in vitro. Mol Med Rep. 2014;9:1218–24. doi: 10.3892/mmr.2014.1944. [DOI] [PubMed] [Google Scholar]
- 71.Xu C, et al. Calorie Restriction Prevents Metabolic Aging Caused by Abnormal SIRT1 Function in Adipose Tissues. Diabetes. 2015;64:1576–90. doi: 10.2337/db14-1180. [DOI] [PubMed] [Google Scholar]
- 72.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–53. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–6. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kawahara TL, et al. Dynamic chromatin localization of Sirt6 shapes stress- and aging-related transcriptional networks. PLoS Genet. 2011;7:e1002153. doi: 10.1371/journal.pgen.1002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–21. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 76.Hirayama J, et al. Common light signaling pathways controlling DNA repair and circadian clock entrainment in zebrafish. Cell Cycle. 2009;8:2794–801. doi: 10.4161/cc.8.17.9447. [DOI] [PubMed] [Google Scholar]
- 77.Bee L, et al. Nucleotide excision repair efficiency in quiescent human fibroblasts is modulated by circadian clock. Nucleic Acids Res. 2015;43:2126–37. doi: 10.1093/nar/gkv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang TH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011;39:3176–87. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hastings M, O'Neill JS, Maywood ES. Circadian clocks: regulators of endocrine and metabolic rhythms. J Endocrinol. 2007;195:187–98. doi: 10.1677/JOE-07-0378. [DOI] [PubMed] [Google Scholar]