ABSTRACT
Ruminococcus bromii is a dominant member of the human gut microbiota that plays a key role in releasing energy from dietary starches that escape digestion by host enzymes via its exceptional activity against particulate “resistant” starches. Genomic analysis of R. bromii shows that it is highly specialized, with 15 of its 21 glycoside hydrolases belonging to one family (GH13). We found that amylase activity in R. bromii is expressed constitutively, with the activity seen during growth with fructose as an energy source being similar to that seen with starch as an energy source. Six GH13 amylases that carry signal peptides were detected by proteomic analysis in R. bromii cultures. Four of these enzymes are among 26 R. bromii proteins predicted to carry dockerin modules, with one, Amy4, also carrying a cohesin module. Since cohesin-dockerin interactions are known to mediate the formation of protein complexes in cellulolytic ruminococci, the binding interactions of four cohesins and 11 dockerins from R. bromii were investigated after overexpressing them as recombinant fusion proteins. Dockerins possessed by the enzymes Amy4 and Amy9 are predicted to bind a cohesin present in protein scaffoldin 2 (Sca2), which resembles the ScaE cell wall-anchoring protein of a cellulolytic relative, R. flavefaciens. Further complexes are predicted between the dockerin-carrying amylases Amy4, Amy9, Amy10, and Amy12 and two other cohesin-carrying proteins, while Amy4 has the ability to autoaggregate, as its dockerin can recognize its own cohesin. This organization of starch-degrading enzymes is unprecedented and provides the first example of cohesin-dockerin interactions being involved in an amylolytic system, which we refer to as an “amylosome.”
IMPORTANCE
Fermentation of dietary nondigestible carbohydrates by the human colonic microbiota supplies much of the energy that supports microbial growth in the intestine. This activity has important consequences for health via modulation of microbiota composition and the physiological and nutritional effects of microbial metabolites, including the supply of energy to the host from short-chain fatty acids. Recent evidence indicates that certain human colonic bacteria play keystone roles in degrading nondigestible substrates, with the dominant but little-studied species Ruminococcus bromii displaying an exceptional ability to degrade dietary resistant starches (i.e., dietary starches that escape digestion by host enzymes in the upper gastrointestinal tract because of protection provided by other polymers, particle structure, retrogradation, or chemical cross-linking). In this report, we reveal the unique organization of the amylolytic enzyme system of R. bromii that involves cohesin-dockerin interactions between component proteins. While dockerins and cohesins are fundamental to the organization of cellulosomal enzyme systems of cellulolytic ruminococci, their contribution to organization of amylases has not previously been recognized and may help to explain the starch-degrading abilities of R. bromii.
INTRODUCTION
The impact of the human intestinal microbiota upon health is increasingly recognized (1, 2). The dense microbial community within the large intestine depends mainly on the fermentation of nondigestible carbohydrates as its source of energy. For many diets, the single largest source of fermentable carbohydrate entering the colon is estimated to be resistant starch (RS) (3), which is defined as dietary starch that escapes digestion by host enzymes in the upper gastrointestinal tract because of protection provided by other polymers (RS1), particle structure (RS2), retrogradation (RS3), or chemical cross-linking (RS4) (4). Supplementation of diets with resistant starch can confer health benefits, especially in reducing insulin resistance and in protection against colorectal cancer, that are considered to be mediated mainly by microbial fermentation products (5, 6).
The only starch-degrading enzyme systems from human gut symbionts to have been studied in any detail are those of Bacteroides thetaiotaomicron and Eubacterium rectale. B. thetaiotaomicron relies on a “sequestration” system, encoded by the sus gene cluster, in which outer membrane Sus proteins mediate the binding and transport of partial hydrolysis products of starch into the periplasm, where they are processed further (7–10). The Firmicutes species E. rectale A1-86 and related Roseburia spp. appear to rely on a large extracellular amylase that is anchored to the cell wall, together with membrane-associated binding proteins and hydrolases that are upregulated by growth on starch (11–13). Although they are able to degrade soluble starches, these species do not show significant ability to degrade and utilize raw particulate starches or even resistant starches that have been pretreated by boiling (14). In contrast, recent investigations have strongly implicated relatives of another Firmicutes species, Ruminococcus bromii, as an important keystone species in the breakdown of resistant starch in the human large intestine. The populations of this group of bacteria detected in fecal samples are stimulated in individuals given diets enriched in RS2 or RS3 (15–17), while individuals lacking R. bromii apparently ferment RS3 inefficiently (16). R. bromii is a specialized amylolytic bacterium belonging to the Ruminococcaceae, a family of Firmicutes that is better known for the ability of certain rumen species to degrade cellulose (18). R. bromii shows high degradative activity against raw or boiled RS2 and RS3 resistant starches in comparison with other amylolytic human intestinal bacteria such as B. thetaiotaomicron, E. rectale, and Bifidobacterium adolescentis (14, 19). Indeed, even nongrowing R. bromii cells were found to stimulate the growth of those other amylolytic human gut bacteria by releasing soluble sugar from resistant starches (14).
In this investigation, we use genomics, proteomics, and protein-protein interaction studies to reveal the presence of unique enzyme systems in R. bromii that are likely to explain its exceptional ability to degrade starches and starch particles. In particular, we demonstrate for the first time the involvement of cohesin (Coh)-dockerin interactions, previously shown to be of importance mainly in lignocellulose-degrading enzyme systems, in the organization of microbial starch-degrading enzyme systems.
RESULTS
GHs of Ruminococcus bromii L2-63.
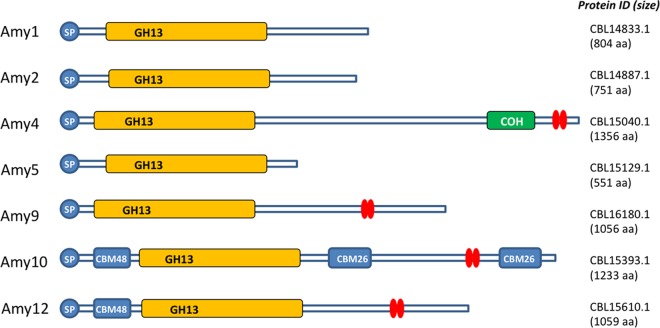
The genome of R. bromii L2-63 encodes only 21 glycoside hydrolases (GHs), which compares with much larger numbers of GH enzymes (50 to 150) in other glycan-utilizing human colonic Firmicutes and up to 350 in Bacteroides spp. (20, 21). Of the 21 R. bromii GH enzymes, 15 belong to GH13, a hydrolase family dedicated largely to the degradation of starch, while the single GH31 and GH77 enzymes may also play significant roles in starch degradation (see Table S1 and Fig. S1 in the supplemental material), thus indicating a high degree of nutritional specialization. The remaining four GH enzymes comprise three lysozymes (two GH23 and one GH25) and one β-glucosidase (GH3). N-terminal signal peptides (SP) are evident in seven of the GH13s, which are therefore likely to be secreted (Fig. 1).
FIG 1 .
Major extracellular amylases of R. bromii L2-63. Modular organization is shown for seven predicted gene products that carry GH13 catalytic modules and signal peptides (SP). The cohesin (COH) of Amy4, the CBMs from family 26 or family 48, and the dockerins (red double ellipses) are shown schematically. aa, amino acids.
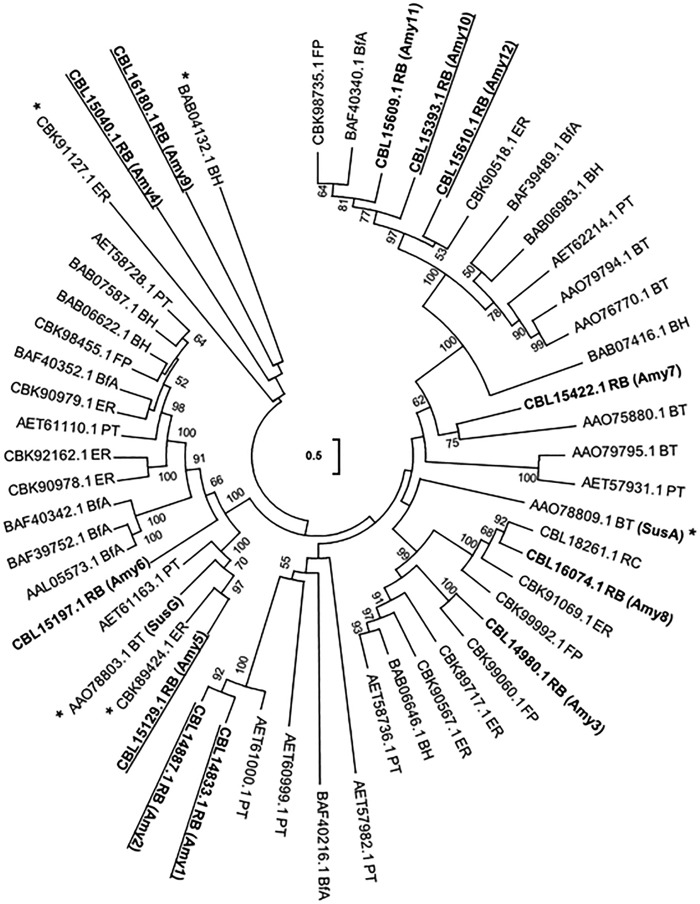
The relationships of R. bromii GH13 sequences were explored using BLASTP searches and phylogenetic comparisons that included three other prominent amylolytic species, B. thetaiotaomicron, E. rectale, and B. adolescentis, and two related nonamylolytic members of the Ruminococcaceae, Ruminococcus champanellensis and Faecalibacterium prausnitzii, from the human colon (Fig. 2). Among the R. bromii GH13 enzymes that carry SPs, two (Amy10 and Amy 12) group with GH13 subfamily 14, which includes pullulanases (22), and one (Amy5) groups with E. rectale α-amylase EUR_01860 (13), while Amy1, Amy2, Amy4, and Amy9 are not closely related to GH13s from the three other amylolytic human gut species. Amy4 is, however, distantly related to an enzyme from Bacillus halodurans that belongs to GH13 subfamily 19 and is known to release maltohexaose from amylose (22, 23) (Fig. 2). The closest relatives of Amy1 and Amy2 were found in the genome of the soil bacterium Paenibacillus terrae. The remaining enzymes that lack SPs grouped with a variety of other subfamilies (Fig. 2). Amy11 and Amy12 are encoded by adjacent genes within the genome, as are Amy8 and Amy15, whereas the genes encoding the remaining 11 GH13 enzymes are unlinked.
FIG 2 .
Phylogenetic tree comparing GH13 enzymes from Ruminococcus bromii and seven other bacterial genomes. R. bromii L2-63 (RB) GH13 sequences (labeled Amy1, Amy2, Amy3, etc.) are compared with those from other human gut species (the starch-utilizing strains Eubacterium rectale A1-86 [ER], Bacteroides thetaiotaomicron VPI-5482 [BT], and Bifidobacterium adolescentis ATCC15703 [BfA] and two other human colonic Ruminococcaceae species, Ruminococcus champanellensis 18P13 [RC] and Faecalibacterium prausnitzii L2-6 [FP]) and also with those from two non-gut species, Bacillus halodurans C-125 (BH) and Paenibacillus terrae HPL-003 (PT), that were known from the results of BLASTp queries of the NCBI database to possess proteins that were the closest matches to R. bromii Amy4. R. bromii sequences that are predicted to possess signal peptides are underlined. Sequences with functions concerned with glycogen or trehalose metabolism (including R. bromii L2-63 Amy 13, Amy14, and Amy15) that are predicted by KEGG GH annotation have been omitted from the tree. Sequences marked with an asterisk (*) encode enzymes that have been experimentally characterized. Bootstrap values, expressed as a percentage of 1,000 replications, are given at the branching nodes. This tree is unrooted and was constructed using the maximum-likelihood method. The scale bar (center of tree) refers to the number of amino acid differences per position.
Dockerin and cohesin modules and carbohydrate-binding modules (CBMs) in Ruminococcus bromii gene products.
Large numbers of proteins carrying dockerin modules have been reported in Ruminococcus species that are involved in degrading plant fiber (18, 24, 25). Notably, the rumen cellulolytic species R. flavefaviens FD1 encodes over 220 such proteins (18, 24), and those dockerins play a key role in the assembly of a cellulosome complex in this species via specific interactions with cohesin modules (26, 27). It was therefore of interest to discover that the genome of R. bromii L2-63 encodes 26 clearly identifiable dockerins, all but one of which are present in proteins that carry N-terminal signal peptides. Four of these dockerin-containing proteins (Amy4, Amy9, Amy10, and Amy12) are associated with GH13 catalytic modules involved in starch breakdown (Fig. 1). Of the remainder, 10 are associated with putative membrane proteins of unknown function, four with peptidase- or protease-related modules, and one with a putative terpene cyclase (see Fig. S2 in the supplemental material).
In addition, we found four predicted gene products (designated scaffoldin 1 [Sca1], Sca2, Sca3, and Sca4) that include cohesin modules (Fig. 3). Two of these have features that are of particular interest. Sca1 corresponds to the 1,356-amino-acid amylase precursor Amy4 (CBL15040) and carries a cohesin (Coh1) followed by a dockerin at its C terminus. Sca2 (534 amino acids; CBL15370) has a cohesin (Coh2) together with a putative sortase signal motif at its C terminus, reminiscent of the ScaE protein that has been shown to mediate anchoring of the cellulosome complex to the cell surface in R. flavefaciens 17 (28). Sca3 carries four repeat “X25” domains. We were therefore interested in determining whether the cohesins found in these four R. bromii proteins could interact with dockerin-containing proteins to help anchor enzymes to the cell surface and to form enzyme complexes, as described below. The R. bromii genome also encodes eight carbohydrate-binding modules (CBMs) that are associated with GH13 enzymes. Amy10 carries two CBM26 modules and one CBM48 module (Fig. 1), while each of five other enzymes carries a single CBM48 module; the CBM26 and CBM48 modules are typically involved in binding to starch (http://www.cazy.org).
FIG 3 .
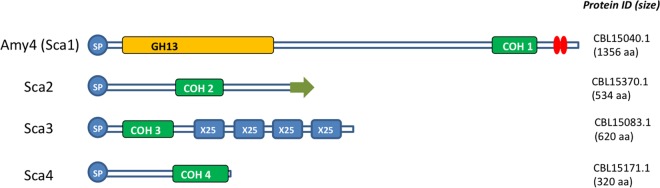
Schematic representation of cohesin-carrying proteins of Ruminococcus bromii L2-63. The four proteins are designated scaffoldins. Scaffoldin 1 (Sca1) contains a GH13 amylase module and is synonymous with the amylase Amy4. Sca2 carries a predicted C-terminal sortase signal (indicated by an arrow). The X25 domains in Sca3 show some similarity to starch-specific CBMs found in the Bacteroides thetaiotaomicron proteins SusE and SusF (10).
Growth and native starch-degrading activity of Ruminococcus bromii.
R. bromii strains grow poorly on media lacking rumen fluid, indicating that they have complex growth requirements (14, 29) and fail to grow in the rumen fluid-free medium YCFA (30), which supports the growth of many other human colonic anaerobes. YCFA medium was therefore modified here to include additional filter-sterilized vitamins that enable good growth of R. bromii L2-63 (see Materials and Methods); we refer to this basal medium as RUM medium. As shown in Table 1, similar amylase activities were detected for mid-exponential-growth-phase cells grown in RUM medium with fructose or with soluble starch as the energy source, showing that amylase activity is expressed constitutively in this bacterium. Most (53% to 75%) of the amylase activity was detected in the cell pellet rather than in the culture supernatant, indicating that it was cell associated. R. bromii enzyme preparations from stationary-phase cultures were active against both raw and preboiled starches, but activity was greater against the boiled starches, especially in the case of potato starch (Table 2).
TABLE 1 .
Impact of growth substrate on amylase activity in R. bromii L2-63a
| Growth substrate | Culture OD650 ± SD | Amylase activity (units/mg cell protein ± SD)b |
|
|---|---|---|---|
| Culture supernatant | Cell pellet | ||
| 0.2% fructose | 0.52 ± 0.03 | 0.022 ± 0.007 | 0.047 ± 0.025 |
| 0.2% potato starch | 0.49 ± 0.01 | 0.027 ± 0.009 | 0.059 ± 0.039 |
Data were determined in cultures growing exponentially on RUM medium and are presented as means and SDs of the results of three replicate experiments.
1 unit = 1 μM glucose/min.
TABLE 2 .
Activity of R. bromii L2-63 native amylases against different starchesa
| Type of starch | Total culture activity (%) under indicated condition |
|
|---|---|---|
| Boiled | Untreated | |
| Potato starch (Sigma, S2004) | 100 | 6.2 |
| High-amylose corn starch (Sigma, S4180) | 44.1 | 18.8 |
| Corn starch (Sigma, S9679) | 83.0 | 24.6 |
| Novelose 330 (National Starch) | 55.8 | 27.3 |
Enzyme preparations from stationary-phase cultures grown for 48 h in RUM medium with boiled 0.2% Novelose 330 as the energy source were used. Data are shown as percentages of the activity obtained for boiled potato starch, based on the means of the results of triplicate assays (SD were less than 1% of the mean in all cases) combining data from supernatant, washings and residual pellet fractions.
Proteomic analysis of prominent cell pellet-associated and supernatant proteins from starch-grown R. bromii cultures led to the detection of six proteins carrying GH13 modules (Table 3; see also Fig. S3 and S4 in the supplemental material). Five proteins carrying GH13 modules (identified as Amy1, Amy2, Amy4, Amy9, and Amy12) were detected among 23 prominent spots analyzed using two-dimensional (2D) gel separations of proteins from the culture supernatant. Amy4, Amy2, and Amy9, together with Amy10, were also identified in the cell pellet fraction from the 23 analyzed spots (Table 3). Thus, six of the seven predicted extracellular GH13 enzymes that carry signal peptides, and all four of dockerin-carrying enzymes Amy4, Amy9, Amy10, and Amy12, were detected among the major proteins expressed by R. bromii (Fig. 1).
TABLE 3 .
Major proteins identified in R. bromii L2-63a
| R. bromii protein detected | Protein length (aa) | Cell pellet bit score (% coverage) | Culture supernatant bit score (% coverage) | Protein ID |
|---|---|---|---|---|
| Glycosidases | ||||
| Amy 4 | 1,356 | 759 (13)b | 1,591 (25) | CBL15040.1 |
| Amy 1 | 804 | 409 (11) | CBL14833.1 | |
| Amy 2 | 751 | 510 | 1,150 | CBL14887.1 |
| Amy 9 | 1,056 | 250 (4) | 1,135 (21) | CBL16180.1 |
| Pullulanases, type I | ||||
| Amy 10 | 1,233 | 1,190 (19) | CBL15393.1 | |
| Amy 12 | 1,059 | 552 (9) | CBL15610.1 | |
| Hypothetical | 548 | 113 | CBL14834.1 | |
| Pyruvate, phosphate dikinase | 875 | 1,201 (28) | 1,291 (29) | CBL14812.1 |
| Hypothetical | 630 | 389 (18) | 449 (16) | CBL14592.1 |
| Dockerin type I repeat | 734 | 63 | CBL15687.1 | |
| Chaperone (DnaK) | 718 | 96 (4) | 270 (10) | CBL15021.1 |
| Archaea/vacuole-type H+-ATPase subunit A | 584 | 788 (25) | CBL15964.1 | |
| Chaperonin GroL | 542 | 308 (11) | CBL15709.1 | |
| Aconitase | 643 | 257 (9) | 782 (27) | CBL15613.1 |
| Putative uncharacterized | 572 | 211 (11) | CBL14592.1 | |
| Hypothetical | 1,495 | 1,247 (19) | CBL15066.1 | |
| Carbamoyl-phosphate synthase large subunit | 1,350 | 187 (4) | CBL14589.1 | |
| Pyruvate-flavodoxin oxidoreductase | 1,186 | 162 (3) | WP_021883784.1 | |
| Translocase subunit SecA | 955 | 564 (14) | CBL15065.1 | |
| Isoleucyl-tRNA synthetase | 921 | 337 | CBL15771.1 | |
| Acetaldehyde dehydrogenase/alcohol dehydrogenase AdhE | 873 | 640 (16) | CBL14797.1 |
Cultures were grown on RUM medium containing 0.2% soluble starch for 48 h. Proteomic analysis and the preparation of cell pellet and culture supernatants are described in Materials and Methods. Data presented are based on analysis of the most prominent spots (molecular mass, >60 kDa) recovered from two SDS gel separations of cell pellet (23 spots) and supernatant (23 spots) preparations. aa, amino acid; ID, identifier.
The scores and percent coverage values shown represent the top hits against an R. bromii-encoded protein for a given protein spot in the cell pellet or supernatant fractions. Several of these proteins were detected in multiple spots recovered from 2D gels, reflecting charge variants.
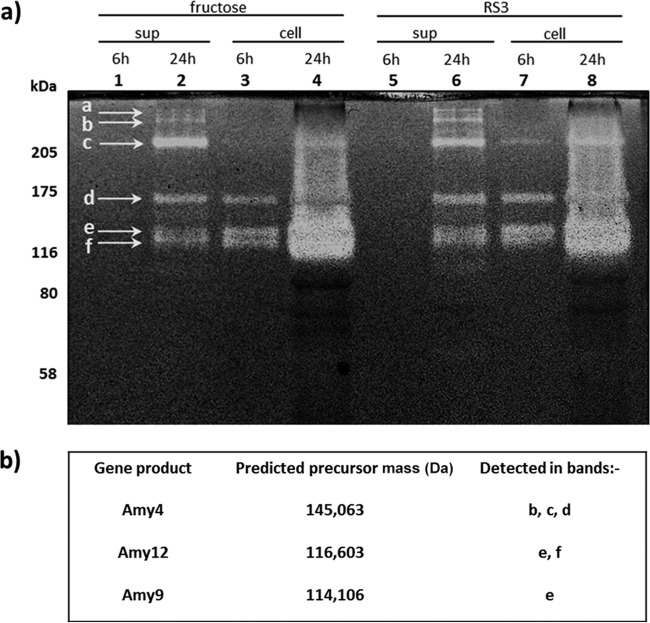
Zymogram analysis of one-dimensional gel separations, involving pretreatment of the native protein fractions at 60°C followed by renaturation (11), was also used to compare the major amylases produced by R. bromii L2-63 grown on RUM medium with fructose or boiled RS3 starch as an energy source. As for total amylase assayable activity (Table 1), no difference between fructose- and starch-grown cultures was evident in activity profiles, further confirming that expression of these major amylases is constitutive (Fig. 4). Major activity bands were excised from the SDS gel and sequenced by liquid chromatography tandem mass spectrometry (LC MS/MS). Five bands were found to contain amylases; these corresponded to three dockerin-containing starch-degrading enzymes (two α-amylases, Amy4 [CBL15040] and Amy9 [CBL16180], and a pullulanase, Amy12 [CBL15610]) among the four discussed above. While the positions of Amy9 and Amy12 were consistent with their predicted molecular sizes, Amy4 sequences were detected in at least two activity bands of higher molecular mass (>250 kDa).
FIG 4 .
Detection of major R. bromii amylases by zymogram analysis and sequencing. (a) Zymogram showing activity of amylases against RS3 for R. bromii L2-63 cells grown for 24 h on 0.2% fructose or 0.2% RS3. Values on the left correspond to the molecular masses determined by staining the gel with Coomassie blue, prior to staining the gel with iodine to visualize clear zones of amylase activity. sup, supernatant proteins; cell, cell-associated proteins. Bands a to f, visible active bands. (b) Identification of amylolytic enzymes from excised bands by LC-MS/MS. In addition, a homologue of a Cna (collagen adhesion)-type protein was detected in band a, a hypothetical protein (RBR_05030) in band b, RNA polymerase subunit B in band d, and a hypothetical protein (RBR_07100) in band e.
Dockerin-cohesin interactions in Ruminococcus bromii.
Individual dockerin and cohesin modules were expressed as recombinant products in Escherichia coli as Xyn (dockerin) or CBM (cohesin) fusion proteins. The resulting recombinant chimeric proteins were purified to allow investigation of interactions between them using an array approach and enzyme-linked immunosorbent assay (ELISA) analysis. By those means, we analyzed interactions among 11 dockerins (including the four that are found in proteins with GH13 modules) and four cohesins (see Materials and Methods). The cohesin from Amy4 (Coh1) bound the dockerins of Amy9 and Amy10 as well as the dockerin of Amy4 itself (Table 4 and Fig. 5). The demonstration that Amy4-Coh1 recognizes its own C-terminal dockerin is intriguing, as this suggests the presence of a mechanism that could result in multimerization. Such cohesin-dockerin pairs that presumably result in multimerization are known in other microbial systems (24, 31, 32). Sca2-Coh2 bound strongly to two of the dockerins associated with GH13 modules (Amy4 and Amy9) and to three other dockerins, including one with a peptidase module and one containing leucine-rich-repeat (LRR)-type sequences (Table 4; see also Fig. S2 in the supplemental material). LRR sequences are also present in multiple dockerin-carrying gene products in R. flavefaciens (24) and are thought to be involved in a variety of protein-protein interactions in other bacteria, including interactions with host proteins (33). The cohesins present in two other gene products (Sca3 and Sca4) bound all four of the dockerins found in GH13 enzymes.
TABLE 4 .
Interactions of recombinant R. bromii dockerin and cohesin modules determined by a microarray approacha
| Dockerin | Interaction |
Associated domain(s) | |||
|---|---|---|---|---|---|
| Coh1 (Amy4) | Coh2 (Sca2) | Coh3 (Sca3) | Coh4 (Sca4) | ||
| Amy9 | +++++ | +++ | +++++ | +++++ | GH13 |
| CBL16032.1 | +++ | ++ | +++++ | +++++ | Peptidase |
| CBL14720.1 | +++++ | +++ | +++++ | +++++ | 2× LRR |
| Amy4 | ++++ | +++ | +++++ | +++++ | GH13, Coh1 |
| Amy10 | +++++ | + | +++++ | +++++ | GH13, CBM26, CBM48 |
| CBL14834.1 | +++++ | +++ | +++++ | +++++ | |
| CBL14836.1 | − | − | − | + | Cysteine protease |
| CBL15647.1 | − | − | − | − | |
| Amy12 | − | − | ++ | +++++ | GH13, CBM48 |
| CBL15625.1 | − | − | − | − | |
| CBL16049.1 | − | − | − | − | |
Proteins were expressed as recombinant products (CBM fusions [cohesins] and XYN fusions [dockerins]) in E. coli (see Materials and Methods). LRR, = leucine-rich repeat. +++++, very strong; ++++, strong; +++, moderate; ++, weak; +, very weak; −, none.
FIG 5 .
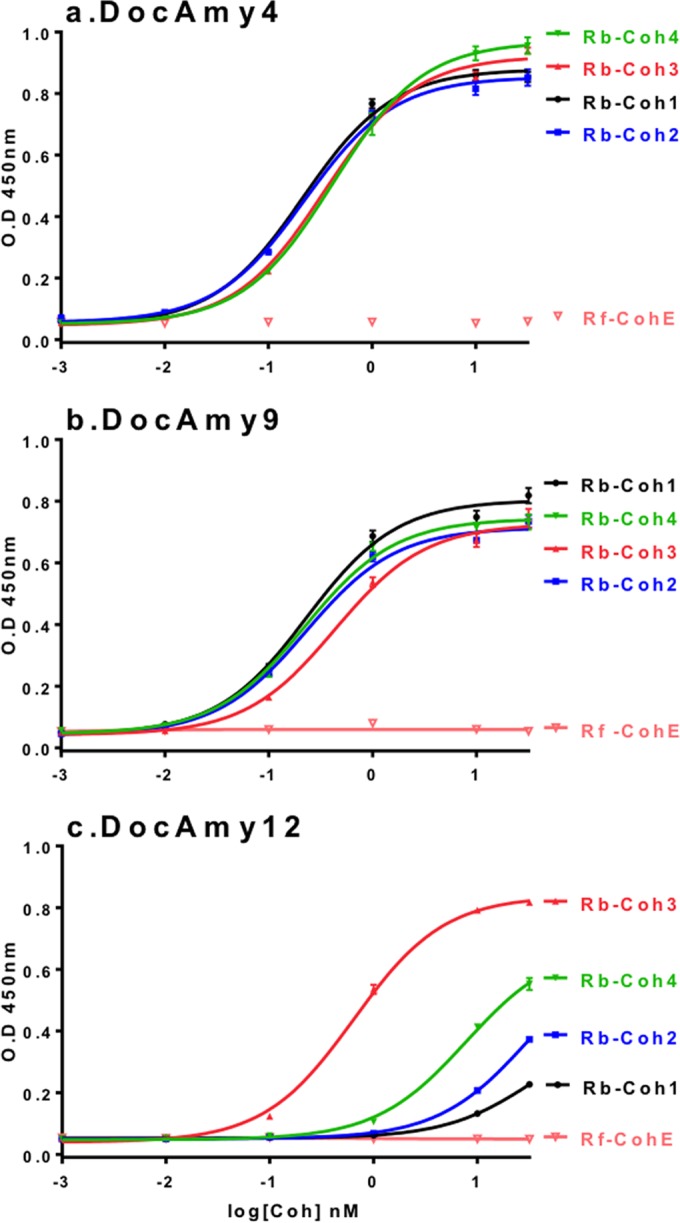
Interactions of recombinant dockerins and cohesins from R. bromii L2-63. Selected cohesin-dockerin interactions were examined by ELISA experiments. (a and b) Amy4 (a) and Amy9 (b) dockerins interact strongly with all four cohesins of R. bromii. (c) Strong interaction of the Amy12 dockerin with Coh3, moderate interaction with Coh4, and negligible interaction with Coh1 and Coh2. CohE from R. flavefaciens FD1 was included in the experiment as a negative control. Error bars indicate the standard deviations from the means of the results determined for triplicate samples from one experiment.
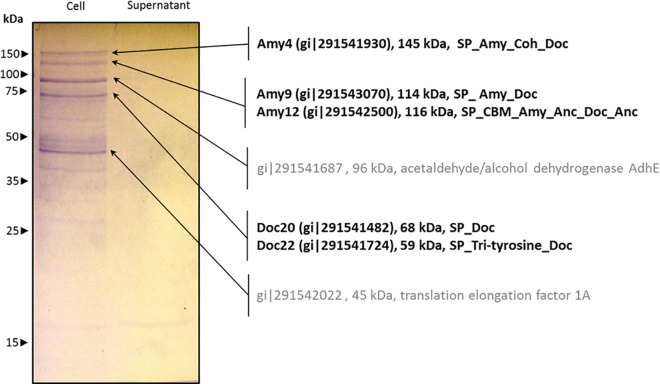
His6-tagged recombinant Amy4-Coh1 was also used to investigate interactions with native R. bromii proteins. A pulldown experiment using Coh1 detected the Amy9 and Amy12 proteins, two dockerin-containing membrane proteins of unknown function, and the Amy4 amylase itself (Fig. 6). This confirms that the Amy4 cohesin recognizes several dockerins present in native R. bromii proteins, including Amy9 and Amy4. The recovery of Amy12 was not predicted from the studies performed with isolated recombinant dockerin modules, and this might imply that there are other binding mechanisms involving Sca3 or Sca4 or perhaps involving substrate binding. Otherwise, the results obtained with native R. bromii proteins agreed well with the interactions seen between the recombinant dockerins and cohesins (Table 4).
FIG 6 .
Identification of native R. bromii proteins that interact with overexpressed Amy4-Coh1. Cell-associated or cell-free supernatant proteins of R. bromii L2-63 grown with boiled Novelose RS3 starch as the energy source were incubated with His6-tagged Coh1 at 37°C as described in Materials and Methods. Proteins binding to Coh1 were recovered and separated by SDS-PAGE, followed by proteomic analysis of individual bands. Protein identifiers (ID), molecular-mass values, and conserved modules for identified proteins are shown on the right of the figure. Numbers on the left indicate sizes and positions of molecular mass markers. SP, signal peptide; Amy, amylase; Coh, cohesin; Doc, dockerin; Anc, cell wall surface anchor; CBM, carbohydrate-binding module; Tri-tyrosine, triple-tyrosine motif.
DISCUSSION
Firmicutes bacteria belonging to the Ruminococcaceae represent 10% to 25% of the microbiota of healthy individuals, according to the results of molecular surveys (34). Certain of these bacteria appear to play important roles in the degradation of insoluble substrates in the human large intestine (21). Ruminococcus relatives were the only bacterial group to be found significantly associated with the fiber fraction of human fecal samples on the basis of 16S rRNA sequence analysis, accounting for 12.2% of fiber-associated but only 3.3% of liquid-phase sequences (35). Furthermore, key roles in the degradation of insoluble substrates have been ascribed recently to two species. The newly defined species R. champanellensis is the only human intestinal bacterium so far reported to degrade microcrystalline cellulose (25, 36) and has been detected mainly in methanogenic individuals (37). A related species from the human colon, “R. bicirculans,” shows a more specialized capacity to utilize plant cell wall polysaccharides, including β-glucans, but is unable to degrade cellulose or xylan (38). R. bromii, on the other hand, is a remarkably specialized amylolytic bacterium that has no ability to degrade plant cell wall polysaccharides but shows very high degradative activity against resistant starches (14).
Large numbers of dockerin modules are found in proteins from cellulolytic Ruminococcus species that have been shown to produce cellulosomes, notably, R. flavefaciens (24). Interactions of dockerins with cohesin modules present in scaffolding proteins are responsible for the organization of the members of a diverse set of plant cell wall-degrading enzymes into cellulosome complexes in R. flavefaciens (26, 39, 40) and in other Clostridium-related cellulolytic bacteria (27). A cellulosome complex has also been discovered recently in the human colonic species R. champanellensis (25). In contrast, the noncellulolytic human colonic species R. bicirculans carries only a single dockerin and a single cohesin that are associated with a GH73 enzyme (38); in general, there are few examples of dockerins and cohesins playing a role in protein complexes from noncellulolytic species. It was therefore of some surprise to detect 26 dockerin- and four cohesin-carrying proteins in the genome of the specialist starch-degrader R. bromii L2-63. We have shown here that some of these modules are involved in the organization of the major extracellular amylases of R. bromii into multiprotein complexes, or “amylosomes.”
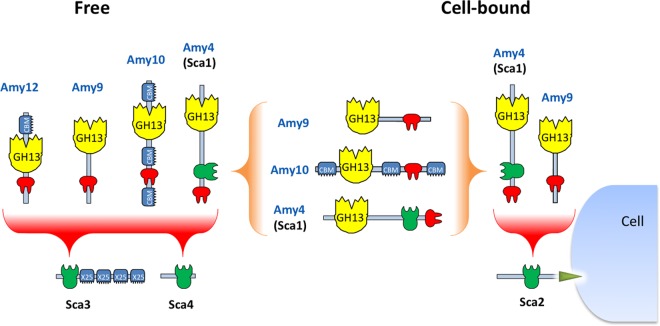
On the basis of the evidence presented here, the possible arrangements of the major extracellular R. bromii amylases were revealed and are summarized in Fig. 7. The Sca2 protein carries a C-terminal motif that predicts attachment to the cell surface via a sortase-mediated mechanism, as previously demonstrated for the ScaEs of R. flavefaciens and R. champanellensis (25, 26). The Amy9 and Amy4 amylases have the potential to link directly to Sca2 via its C-terminal dockerin. The Sca2 cohesin was also able to bind dockerins from proteins other than amylases, including peptidases, suggesting that it has a more general role as a cell surface-anchoring mechanism. The Amy4 enzyme is unusual in that it carries a cohesin and a dockerin in addition to a catalytic module (Fig. 7). Even more unusual, however, is that the Amy4 cohesin recognizes the dockerin of Amy4 itself; this suggests that Amy4 should be able to form multimeric assemblages. The Amy4 cohesin also provides a partner for the dockerins from Amy10 and Amy9, suggesting that minicomplexes involving two or more amylases are formed. The frequency with which the Amy4 cohesin engages the dockerin of another Amy4 protein, as opposed to the dockerins from Amy9 and Amy10, presumably depends on the relative abundances of the Amy9, Amy10, and Amy4 dockerins and their relative affinities for the Amy4 cohesin. The roles of the other two cohesin-carrying proteins, Sca3 and Sca4, are not yet clear, but both were able to bind dockerins from all four dockerin-carrying GH13 enzymes. The presence of four X25 modules on Sca3 (Fig. 3) is particularly intriguing, since similar modules bind starch in cell surface proteins SusE and SusF of B. thetaiotaomicron and have also been observed in a pullulanase of Bacillus acidopullulyticus (10, 41). Dockerin-bearing amylases, i.e., Amy4, Amy9, Amy10, and Amy12, would then be equipped with strong starch-binding properties, through their interaction with Sca3. Binding to starch is also likely to be mediated by the CBM48 modules present in Amy9 and Amy10 when present in the complex. Amy10 also carries two CBM26 modules, which are thought to have a particularly important role in binding to raw starches (23). With regard to enzyme specificity, the predicted pullulanase activity of Amy10 and Amy12 is likely to be complemented by the distinct actions of Amy4 and Amy9, although the specificities of the R. bromii enzymes have yet to be determined experimentally. Amy10 and Amy12 also contain additional modules of unknown function that might possibly mediate cell surface attachment. Future work will explore whether other strains of R. bromii, notably, the ATCC27255 type strain, which shows activity on RS as well as growth characteristics similar to those seen with R. bromii L2-63 (14), produce a similar type of amylosome complex; however, such work depends on genome sequencing, which is currently available only for R. bromii L2-63.
FIG 7 .
Potential dockerin-mediated interactions of R. bromii amylases with cohesin-carrying proteins and with each other. On the basis of observed interactions with recombinant cohesins and dockerins (Table 4 and Fig. 5), we can predict that the Amy4 and Amy9 enzymes are likely to bind to the cell surface via the Sca2 scaffoldin protein. Further complexes are likely to form between the Amy4, Amy9, and Amy10 proteins and between Amy4, Amy9, Amy10, and Amy12 and the Sca3 and Sca4 proteins. Binding of the enzymes to Sca3 would presumably confer starch-binding features to the resultant complex. Binding of dockerin-bearing amylases Amy9 and Amy10 to the cohesin of Amy4 would result in the formation of multienzyme complexes that can be part of a cell-bound or cell-free system. Amy4 also has the potential to self-aggregate through interactions between its own cohesin and dockerin.
R. bromii L2-63 apparently produces its major amylases constitutively since we found no significant difference between cultures grown on fructose and those grown on starch in either enzyme assay or zymogram analysis results. This is in marked contrast to the substrate-inducible amylase systems of other human colonic anaerobes such as B. thetaiotaomicron (8), E. rectale (13) and Roseburia spp. (11, 12) and provides further evidence that R. bromii is extremely specialized in its utilization of carbohydrate substrates.
In conclusion, we have presented evidence that four of the major extracellular starch-degrading enzymes in R. bromii are attached to the cell surface and/or assembled into complexes via cohesin-dockerin interactions. This provides the first example of the involvement of dockerin and cohesins, best known from their roles in the cellulosomes that are responsible for lignocellulose breakdown (27), in a starch-degrading enzyme system, and we therefore refer to the complexes formed as “amylosomes.” It seems likely that this organization will help to explain the exceptional degradative activity shown by R. bromii against particulate starches (14).
MATERIALS AND METHODS
Sequence analysis and phylogeny of GH13 enzymes.
The R. bromii L2-63 genome was sequenced by the Pathogen Genomics group at the Wellcome Trust Sanger Institute (United Kingdom) as part of the EU MetaHit project (http://www.sanger.ac.uk/resources/downloads/bacteria/metahit/). Prediction of cohesin and dockerin modular sequences was performed using the BLASTP and tBLASTn algorithm (42), employing known dockerin and cohesin sequences as queries. Analysis of carbohydrate-active enzymes (CAZymes) was performed using the CAZy database (http://www.cazy.org), with reference also to the KEGG database (43). Related GH13 protein sequences were detected using BLASTp. The results were filtered to exclude all matches with E values of >1e-10, sequence identity of <35%, or bit scores of <200, and the remaining best-hit annotations were assigned to the CAZy database sequences. The sequences were then aligned by ClustalW (44) and used to construct a maximum-likelihood phylogenetic tree, using MEGA6.0 software (45).
Growth medium for R. bromii.
Isolation of R. bromii L2-63 from a human fecal sample using a rumen-fluid-based medium was described by Ze et al. (14). Semidefined RUM medium, which was developed for the present work as a modification of YCFA medium (30), consists of (per 100 ml) Casitone (1 g), yeast extract (0.25 g), NaHCO3 (0.4 g), resazurin (0.1 mg), biotin (1 µg), cobalamin (1 µg), p-aminobenzoic acid (3 µg), folic acid (5 µg), pyridoxamine (15 µg), K2HPO4 (0.045 g), KH2PO4 (0.045 g), NaCl (0.09 g), (NH4)2SO4 (0.09 g), MgSO4 ⋅ 7H2O (0.009 g), and CaCl2 (0.009 g) and the short-chain fatty acids (final concentrations) acetate (33 mM), propionate (9 mM), and isobutyrate, isovalerate, and valerate (1 mM each). Cysteine (0.1 g/100 ml) was added to the medium following boiling and was dispensed into Hungate tubes while the tubes were flushed with CO2. After autoclaving, filter-sterilized solutions were added to give a final concentration of thiamine and riboflavin of 0.05 µg·ml−1 (each), pantothenate and nicotinamide of 1 µg·ml−1, pantethine of 50 µg·ml−1, and tetrahydrofolic acid of 0.1 µg·ml−1. Additional trace minerals FeSO4 ⋅ 7H2O (final concentration, 0.4 µg·ml−1), ZnSO4 ⋅ 7H2O (20 ng·ml−1), MnCl2 ⋅ 4H2O (6 ng·ml−1), H3BO3 (60 ng·ml−1), CoCl2 ⋅ 6H2O (40 ng·ml−1), CuCl2 ⋅ 2H2O (2 ng·ml−1), NiCl2 ⋅ 6H2O (4 ng·ml−1), NaMoO4 ⋅ 2H2O (6 ng·ml−1), and NaSeO3 (15 ng·ml−1) and EDTA (1 µg·ml−1) were also included here, but we have now shown that these are not required for growth of R. bromii L2-63. Carbohydrate or other energy sources were added as required, and the final pH of the medium was adjusted to 6.8 ± 0.2. With 0.2% fructose as the energy source, culture optical densities (ODs) and growth rates for R. bromii L2-63 on RUM medium were very similar to those obtained on rumen fluid medium (14).
Proteomic analysis.
Proteomic analysis was performed on culture supernatants and cell pellets obtained from triplicate biological experiments. R. bromii L2-63 was grown anaerobically in 150 ml of RUM medium supplemented with 0.5% soluble potato starch (Sigma) at 37°C to an OD at 650 nm (OD650) of between 0.7 and 0.8. After centrifugation at 9,000 × g, at 4°C, the culture supernatant was dialyzed (four times with 4 liters of distilled water, at 4°C), freeze-dried, and resuspended in 1.5 ml of resuspension Tris buffer (50 mM Tris [pH 8.8], 10% glycerol, 0.1% Triton X-100) supplemented with 1× protease inhibitor cocktail (P8465; Sigma). The cell pellet was washed three times with 1.5 ml phosphate-buffered saline (PBS) and, following one freeze-thaw cycle, resuspended in 1.5 ml of resuspension Tris buffer with protease inhibitor. Cell lysis was achieved by adding 1.2 g of 1-mm-diameter zirconia beads (BioSpec Products, OK, USA) and beating the cells twice on an MP FastPrep-24 bead beater for 30 s at 6.0 m/s. Nonsolubilized debris was removed by centrifugation at 10,000 × g for 5 min. Culture supernatant and cellular fractions were stored at −70°C until further analysis. Protein concentrations of the supernatant and cellular fractions were measured using Bradford reagent (Sigma Aldrich, Dorset, United Kingdom). Aliquots of 350 µg of protein were precipitated in 25% trichloroacetic acid (TCA)–20 mM dithiothreitol (DTT) for 1 h on ice, followed by centrifugation at 10,000 × g for 10 min at 4°C. Pellets were washed four times with 1 ml ice-cold acetone containing 20 mM DTT. After removal of the acetone, the protein pellets were resuspended in Rabilloud buffer (7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.5% biolite Ampholite; pH 3 to 10) and 200 to 250 µg of protein was separated on two-dimensional gels using 17-cm-long immobilized pH gradient (IPG) strips (pH 4 to 7) as described elsewhere (46). Gel images were analyzed with PDQuest Advanced 8.0.1 (BioRad, Hertfordshire, United Kingdom). The densest spots with an apparent molecular mass of between 49 and 145 kDa were excised from the gels manually and subjected to trypsinization and protein identification by the use of Nano LC MS/MS, followed by analysis of the total current ion data using the MASCOT search engine as described previously (46).
Investigation of native amylolytic activity.
The starch-mediated inducibility of amylase activity was investigated in cultures of R. bromii L2-63 grown in 100 ml RUM medium supplemented with 0.2% soluble starch or 0.2% fructose and incubated anaerobically at 37°C to the exponential phase (OD650 of between 0.45 and 0.55) in three independent experiments. Cells were collected and extracellular/cell-associated fractions were obtained as described above. Amylase activity was determined with 0.1% boiled potato starch substrate (Sigma Aldrich, Dorset, United Kingdom) as the substrate from triplicate incubations in PBS (pH 7) with CaCl2 (0.1 mM) at 37°C by measuring the release of reducing sugars (47). Amylase preparations used to investigate activity against different starches (Table 2) were obtained from stationary-phase cultures grown in RUM medium containing 0.2% boiled Novelose 330 for 48 h.
Activity of enzymes in polyacrylamide gels (zymogram analysis).
Amylolytic enzymes of R. bromii were also analyzed using starch zymograms (11). R. bromii was grown on RUM medium containing 0.2% Novelose (filter sterilized and preboiled for 10 min) (RS3) or 0.2% fructose and incubated at 37°C for up to 48 h. The cells were then harvested following centrifugation at 14,000 × g for 10 min at 4°C. Culture supernatant was collected, dialyzed for 24 h with distilled H2O (3 changes), frozen overnight at −20°C, and then lyophilized. The cell pellet was washed twice and resuspended in 50 mM sodium phosphate buffer (pH 6.5) before storage at −20°C. Cellular and supernatant protein samples were denatured by heating at 60°C for 20 min and then subjected to SDS-PAGE with 0.2% RS3 (Novelose 330) incorporated into the separating gel. After the separation was performed, the gel was washed twice with 200 ml of washing solution (10 mM Tris-HCl [pH 7.5], 2 mM DTT, 20% isopropanol) at room temperature. The proteins were then renatured by rocking the gel in 200 ml of renaturing solution (50 mM Tris-HCl [pH 6.8], 2 mM DTT, 1 mM EDTA) overnight at 4°C. The renatured gel was transferred to 200 ml of sodium phosphate buffer (50 mM, pH 6.8), soaked for 1 h at 4°C, and then incubated at 37°C for 4 h. The gel was subjected to Coomassie staining to record the position of protein ladders (Novagen; EMD Millipore, MA, USA). The gel was neutralized by washing in Tris-HCl solution (0.1 M, pH 8.0) for at least 4 h with several solution changes in the first hour. After neutralization, the gel was stained with 20% Lugol’s solution (Sigma) until clear zones of starch hydrolysis were visible.
Recombinant CBM-cohesin and xylanase-dockerin fusion products.
Cloning of CBM-fused cohesins and xylanase-fused dockerins was performed as described by Ben David et al. (25). Procedures for expression in E. coli BL21(DE3) and purification of the recombinant proteins are described in the same reference.
CBM-based microarray.
Microarray methods were as described in Ben David et al. (25). CBM-cohesin samples were diluted in Tris-buffered saline (TBS) (pH 7.4) to final concentrations of 9, 3, 1, 0.3, and 0.1 µM and printed onto cellulose-coated glass slides (type GSRC-1; Advanced Microdevices Pvt. Ltd., Ambala Cantt, India) by the use of a Microbiology Grid 610 Microarrayer (Digilab, Inc., Marlborough, MA). The printed microarrays were blocked by incubating the slides in blocking buffer (1% bovine serum albumin–TBS–10 mM CaCl2–0.05% Tween 20) at room temperature for 30 min. Afterward, chosen Xyn-Doc samples (3 nM in blocking buffer) were incubated with the slide at room temperature for 30 min followed by 3 washing steps (5 min each) in washing buffer (TBS–10 mM CaCl2–0.05% Tween 20). Fluorescent staining was accomplished by adding Cy3-labeled anti-Xyn and Cy5-labeled anti-CBM (diluted 1:1,000) in blocking buffer for 30 min. The probed slides were again washed 3 times, air dried, and scanned for fluorescence signals using a Typhoon 9400 variable-mode imager (GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
ELISA.
The ELISA procedure was performed as described earlier (25). The coating step was performed with 15 nM Xyn-Doc proteins. A concentration gradient (0.01 to 1,000 nM) of CBM-Coh was then applied to the coated Maxisorp 96-well plate (Greiner Bio-One, Belgium).
Interactions of Coh1 with native R. bromii proteins.
The interaction of the cohesin module of Amy4 (Coh1) with proteins of R. bromii L2-63 was evaluated using a pulldown assay. The R. bromii L2-63 Coh1 module was cloned into pIVEX (5 Prime, Hamburg, Germany) and His6-tagged (N-terminal) versions of the product were overexpressed and purified in vitro. Coh1 was overexpressed using a cell-free RTS 100 E. coli HY overexpression system (5 Prime, Hamburg, Germany). His6-tagged proteins were bound to Talon Dynabeads (Invitrogen, Oslo, Norway) following the recommendations of the manufacturer. The beads with affinity-bound Coh1 modules were washed once, and then 500 µl of a lysate of R. bromii L2-63 cells in PBS was allowed to interact with them. After incubation for 20 min at room temperature (RT), the beads were washed 3 times and then the proteins were eluted as recommended by the manufacturer. The eluted proteins were separated on a 12% SDS-PAGE gel and subjected to Coomassie staining. The observed bands were excised from the gel and subjected to LC-MS/MS identification as described above.
SUPPLEMENTAL MATERIAL
Comparison of GH complements of R. bromii (heat map). Download
Predicted R. bromii proteins carrying dockerin modules. Download
Proteome of the cell pellet fraction from R. bromii L-63 cultures grown on RUM medium supplemented with 0.2% potato starch. Download
Proteome of the supernatant fraction of R. bromii L2-63 cultures grown on RUM medium supplemented with 0.2% potato starch. Download
Glycoside hydrolase families in the R. bromii genome compared with those of three other amylolytic species and two nonamylolytic Ruminococcus species from the human colon.
GH13 enzymes encoded by the R. bromii L2-63 genome.
ACKNOWLEDGMENTS
We acknowledge support from BBSRC grant no. BB/L009951/1, from the Scottish government Food, Land and People program, and from the Society for Applied Microbiology. E.A.B. is supported by a grant (no. 1349/13) from the Israel Science Foundation (ISF), Jerusalem, Israel, and by a grant from the United States-Israel Binational Science Foundation (BSF). E.A.B. is the incumbent of the Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Thanks are due to Fergus Nicol for proteomic analysis and to Auriane Bernard for enzyme assays on stationary-phase cultures. We also thank Julian Parkhill and Keith Turner (Wellcome Trust Sanger Institute, Cambridge, United Kingdom) for making the R. bromii L2-63 genome sequence available for analysis.
Footnotes
Citation Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM, Bayer EA, Flint HJ. 2015. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. mBio 6(5):e01058-15. doi:10.1128/mBio.01058-15.
REFERENCES
- 1.Sekirov I, Russell SL, Caetano M, Antunes L, Finlay BB. 2010. Gut microbiota in health and disease. Physiol Rev 90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Flint HJ, Scott KP, Louis P, Duncan SH, Louis P. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 3.Cummings JH, Macfarlane GT. 1991. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 4.Englyst HN, Kingman SM, Cummings JH. 1992. Classification and measurement of nutritionally important starch fractions. Eur J Clin Nutr 46(Suppl 2):S33–S50. [PubMed] [Google Scholar]
- 5.Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. 2005. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr 82:559–567. [DOI] [PubMed] [Google Scholar]
- 6.Toden S, Bird AR, Topping DL, Conlon MA. 2007. Dose-dependent reduction of dietary protein-induced colonocyte DNA damage by resistant starch in rats correlates more highly with caecal butyrate than with other short chain fatty acids. Cancer Biol Ther 6:253–258. doi: 10.4161/cbt.6.2.3627. [DOI] [PubMed] [Google Scholar]
- 7.Reeves AR, Wang GR, Salyers AA. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol 179:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 10.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. 2012. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J Biol Chem 287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsay AG, Scott KP, Martin JC, Rincon MT, Flint HJ. 2006. Cell-associated α-amylases of butyrate-producing firmicute bacteria from the human colon. Microbiology 152:3281–3290. doi: 10.1099/mic.0.29233-0. [DOI] [PubMed] [Google Scholar]
- 12.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, Mayer CD, Young P, Rucklidge G, Ramsay AG, Flint HJ. 2011. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc Natl Acad Sci U S A 108(Suppl 1):4672–4679. doi: 10.1073/pnas.1000091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cockburn DW, Orlovsky NI, Foley MH, Kwiatkowski KJ, Bahr CM, Maynard M, Demeler B, Koropatkin NM. 2015. Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol Microbiol 95:209–230. doi: 10.1111/mmi.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abell GC, Cooke CM, Bennett CN, Conlon MA, McOrist AL. 2008. Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. FEMS Microbiol Ecol 66:505–515. doi: 10.1111/j.1574-6941.2008.00527.x. [DOI] [PubMed] [Google Scholar]
- 16.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dassa B, Borovok I, Ruimy-Israeli V, Lamed R, Flint HJ, Duncan SH, Henrissat B, Coutinho P, Morrison M, Mosoni P, Yeoman CJ, White BA, Bayer EA. 2014. Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One 9:e99221. doi: 10.1371/journal.pone.0099221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ze X, Le Mougen F, Duncan SH, Louis P, Flint HJ. 2013. Some are more equal than others: the role of keystone species in the degradation of recalcitrant substrates. Gut Microbes 4:236–240. doi: 10.4161/gmic.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. [DOI] [PubMed] [Google Scholar]
- 21.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B. 2006. Dividing the large glycosyl hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng Design Sel 19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 23.Boraston AB, Healey M, Klassen J, Ficko-Blean E, Lammerts van Bueren A, Law V. 2006. A structural and functional analysis of α-glucan recognition by family 25 and 26 carbohydrate binding modules reveals a conserved mode of starch recognition. J Biol Chem 281:587–598. doi: 10.1074/jbc.M509958200. [DOI] [PubMed] [Google Scholar]
- 24.Rincon MT, Dassa FHJ, Travis AJ, Jindou S, Borovok I, Lamed R, Bayer EA, Henrissat B, Coutinho PM, Antonopoulos DA, Berg Miller ME, White BA. 2010. Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium Ruminococcus flavefaciens FD1. PLoS One 5:e12476. doi: 10.1371/journal.pone.0012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben David Y, Dassa B, Borovok I, Lamed R, Koropatkin NM, Martens EC, White BA, Bernalier-Donadille A, Duncan SH, Flint HJ, Bayer EA, Moraïs S. 2015. Ruminococcal cellulosome systems from rumen to human. Environ Microbiol. doi: 10.1111/1462-2920.12868. [DOI] [PubMed] [Google Scholar]
- 26.Rincon MT, Ding S-Y, McCrae SI, Martin JC, Aurilia V, Lamed R, Shoham Y, Bayer EA, Flint HJ. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J Bacteriol 185:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayer EA, Lamed R, White BA, Flint HJ. 2008. From cellulosomes to cellulosomics. Chem Rec 8:364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- 28.Rincon MT, Cepeljnik T, Martin JC, Lamed R, Barak Y, Bayer EA, Flint HJ. 2005. Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the bacterial cell wall. J Bacteriol 187:7569–7578. doi: 10.1128/JB.187.22.7569-7578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbeck JL, Bryant MP. 1974. Nutritional features of the intestinal anaerobe Ruminococcus bromii. J Appl Microbiol 28:1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. 2012. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 78:420–428. doi: 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayer EA, Coutinho PM, Henrissat B. 1999. Cellulosome-like sequences in Archaeoglobus fulgidus: an enigmatic vestige of cohesin and dockerin domains. FEBS Lett 463:277–280. doi: 10.1016/S0014-5793(99)01634-8. [DOI] [PubMed] [Google Scholar]
- 32.Voronov-Goldman M, Lamed R, Noach I, Borovok I, Kwiat M, Rosenheck S, Shimon LJ, Bayer EA, Frolow F. 2011. Non-cellulosomal cohesin from the hyper-thermophilic archaeon Archaeoglobus fulgidus. Proteins 79:50–60. doi: 10.1002/prot.22857. [DOI] [PubMed] [Google Scholar]
- 33.Waldemarsson J, Areschoug T, Lindahl G, Johnsson E. 2006. The streptococcal Bir and Sir proteins define a family of surface proteins with leucine-rich repeats: camouflaging by other surface proteins. J Bacteriol 188:378–388. doi: 10.1128/JB.188.2.378-388.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lay C, Sutren M, Violaine Rochet V, Saunier K, Doré J, Rigottier-Gois L. 2005. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ Microbiol 7:933–946. doi: 10.1111/j.1462-2920.2005.00763.x. [DOI] [PubMed] [Google Scholar]
- 35.Walker AW, Duncan SH, Harmsen HJ, Holtrop G, Welling GW, Flint HJ. 2008. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol 10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- 36.Chassard C, Delmas E, Robert C, Bernalier-Donadille A. 2010. The cellulose-degrading microbial community of the human gut varies according to the presence or absence of methanogens. FEMS Microbiol Ecol 74:205–213. doi: 10.1111/j.1574-6941.2010.00941.x. [DOI] [PubMed] [Google Scholar]
- 37.Robert C, Bernalier-Donadille A. 2003. The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbiol Ecol 46:81–89. doi: 10.1016/S0168-6496(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 38.Wegmann U, Louis P, Goesmann A, Henrissat B, Duncan SH, Flint HJ. 2014. Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (“Ruminococcus bicirculans”) reveals two chromosomes and a selective capacity to utilize plant glucans. Environ Microbiol 16:2879–2890. doi: 10.1111/1462-2920.12217. [DOI] [PubMed] [Google Scholar]
- 39.Ding SY, Rincon MT, Lamed R, Martin JC, McCrae SI, Aurilia V, Shoham Y, Bayer EA, Flint HJ. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J Bacteriol 183:1945–1953. doi: 10.1128/JB.183.6.1945-1953.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White BA, Lamed R, Bayer EA, Flint HJ. 2014. Biomass utilization by gut microbiomes. Annu Rev Microbiol 68:279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 41.Turkenburg JP, Brzozowski AM, Svendsen A, Borchert TV, Davies GJ, Wilson KS. 2009. Structure of a pullulanase from Bacillus acidopullulyticus. Proteins 76:516–519. doi: 10.1002/prot.22416. [DOI] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vodovnik M, Duncan SH, Reid MD, Cantlay L, Turner K, Parkhill J, Lamed R, Yeoman CJ, Miller ME, White BA, Bayer EA, Marinšek-Logar R, Flint HJ. 2013. Expression of cellulosome components and type IV pili within the extracellular proteome of Ruminococcus flavefaciens 007. PLoS One 8:e65333. doi: 10.1371/journal.pone.0065333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lever M. 1977. Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAHBAH)—effect of bismuth on reaction. Anal Biochem 81:21–27. doi: 10.1016/0003-2697(77)90594-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of GH complements of R. bromii (heat map). Download
Predicted R. bromii proteins carrying dockerin modules. Download
Proteome of the cell pellet fraction from R. bromii L-63 cultures grown on RUM medium supplemented with 0.2% potato starch. Download
Proteome of the supernatant fraction of R. bromii L2-63 cultures grown on RUM medium supplemented with 0.2% potato starch. Download
Glycoside hydrolase families in the R. bromii genome compared with those of three other amylolytic species and two nonamylolytic Ruminococcus species from the human colon.
GH13 enzymes encoded by the R. bromii L2-63 genome.