Abstract
Insecticides impose extreme selective pressures on populations of target pests and so insecticide resistance loci of these species may provide the footprints of ‘selective sweeps'. To lay the foundation for future genome-wide scans for selective sweeps and inform genome-wide association study designs, we set out to characterize some of the baseline population genomic parameters of one of the most damaging insect pests in agriculture worldwide, Helicoverpa armigera. To this end, we surveyed nine Z-linked loci in three Australian H. armigera populations. We find that estimates of π are in the higher range among other insects and linkage disequilibrium decays over short distances. One of the surveyed loci, a cytochrome P450, shows an unusual haplotype configuration with a divergent allele at high frequency that led us to investigate the possibility of an adaptive introgression around this locus.
Introduction
New genomic technologies allow population genetic studies to move beyond questions of migration and population structure generally to those that identify loci within the genome that exhibit extreme gene flow or population structure or other signs that may be interpreted as selection. One strategy to identify potential insecticide-resistance loci is to seek genomic regions that appear to exhibit the characteristics of positive selection such as extended linkage disequilibrium (LD), reduced nucleotide variation and increased proportions of rare variants in the frequency spectra (Nielsen, 2005). These parameters are expected to vary between different populations and different regions in the genome due to the interplay between drift, recombination, mutation and selection. Consequently, some inquiry into what constitutes the baseline population genomics parameters of a species is required before deviations from neutral expectations can be detected.
A genome-wide survey of molecular variation within the model lepidopteran, Bombyx mori, reported that LD decayed over very short distances, with the implication that selective sweeps would be limited to small regions (Xia et al., 2009). Signals of selection were detected at 1041 regions of which 354 were protein-coding genes. These were deemed good candidates for domestication genes, including those involved in silk production, as there has been recent strong selection for such traits. It is reasonable to propose that, in pesticide-resistant organisms where extremely strong selection is exerted on natural populations, similar approaches may identify new candidate resistance genes.
Helicoverpa armigera is a significant lepidopteran pest of agriculture throughout Africa, Asia, Europe and Australia. High polyphagy coupled with an ability to rapidly evolve resistance to insecticides make it responsible for damage to crops estimated at >US$2 billion annually. Resistance to insecticide sprays in H. armigera drove the introduction of insecticidal transgenic cotton to Australia and Asia. The recent incursion of H. armigera into Brazil (Tay et al., 2013) also threatens agricultural productivity in the New World. Population genomics approaches can characterize past and present population structure throughout the species range and identify adaptive loci such as those that confer resistance to insecticides.
Helicoverpa is a well-defined genus within the heliothine subfamily of noctuid moths where its monophyly is strongly supported by morphology and molecular characterization (Matthews, 1999; Cho et al., 2008). Within the genus, however, relationships between species are less clear often due to morphological similarities. For instance, crop damage by H. armigera in Australia is sometimes misattributed to H. punctigera and vice versa (Zalucki et al., 1986). H. armigera and its New World counterpart, H. zea were once thought to constitute one cosmopolitan species but Hardwick (1965) placed them into separate species groups when he distinguished five species groups on the basis of penis structure: armigera, gelotopoeon, hawaiiensis, punctigera, and zea. Subsequent data from immunological assays and mitochondrial DNA sequence analyses suggested that H. zea is more closely related to H. armigera than some other species within the zea group such as H. assulta (Mitter et al., 1993; Behere et al., 2007). Hybridization between H. armigera and H. assulta and between H. armigera and H. zea is possible in the laboratory (Laster and Hardee, 1995; Wang and Dong, 2001). H. armigera and H. assulta are sympatric, whereas the geographical distributions of H. zea and H. armigera were not thought to overlap in the field until the recent incursion of H. armigera into Brazil.
Previous molecular population genetic studies of H. armigera used various markers and usually aimed to characterize population structure rather than identify loci under selection. An allozyme study of 12 H. armigera populations dispersed throughout Australia suggested limited population structure (FST =0.01; Daly and Gregg, 1985). Similarly, analysis of mitochondrial sequences reveals minimal differentiation among global samples with most of the variation distributed throughout the species range (Behere et al., 2007). A survey of eight microsatellite loci revealed that FST among Australian populations was 0.003 after the high frequency of null alleles (>10%) was taken into account (Endersby et al., 2007). Microsatellite loci in lepidopterans have also been associated with transposable elements causing whole loci to vary in copy number (Zhang, 2004), which motivated the development of exon-primed intron-crossing (EPIC) markers with primers that bind to conserved exon sequences, reducing the frequency of null alleles and allowing the characterization of the more variable intronic sequences (Tay et al., 2008). EPIC markers also have the advantage of applicability across related species.
Here we examine nucleotide diversity and LD at nine Z-linked EPIC markers in H. armigera. H. armigera follows a ZZ/ZW sex determination with the female being the heterogametic sex. The focus on Z-linked loci meant that we were able to Sanger-sequence amplicons directly from females and thereby (i) prevent insertion/deletion heterozygosity from confounding sequence traces and (ii) measure the extent of LD directly without having to infer gametic phase. This enabled us to quantify the extent of genomic nucleotide diversity within H. armigera and determine whether LD declines in H. armigera over short distances as it is reported to decline in B. mori. In doing this, we identified a locus exhibiting unusual patterns of nucleotide diversity. In order to determine whether a selective sweep was the best model to describe the patterns in this locus, we sequenced two additional flanking loci, sequenced the gene in related species and characterized the global distribution of the putative sweep haplotype.
Materials and methods
To examine baseline nucleotide diversity in H. armigera, we chose EPIC markers from the Z chromosome. Loci were chosen according to the following criteria: they were likely to be located on the Z chromosome, likely to be dispersed across the Z chromosome, we were confident in their gene models, their introns were of a size appropriate for reliable PCR amplification, and had flanking exon sequence conservation. We chose to examine Apt and Tpi because they had been reported to be Z-linked in Lepidoptera species (Jiggins et al., 2005; Yasukochi et al., 2006; d'Alencon et al., 2010). Two P450 genes were chosen but neither were expected to have a role in insecticide resistance as their orthologs have roles in development (Willingham and Keil, 2004; Feyereisen, 2005). The other loci were chosen without regard to gene ontology.
Development of Z-linked EPIC markers
B. mori Z-linked proteins were selected from the silkworm genome database (http://www.silkdb.org/cgi-bin/silkgo/index.pl) and cross-checked against Genbank accessions to obtain more detailed annotations. Protein sequences were checked against the B. mori genome to ensure that they were single copy and subsequently used to identify orthologs in the H. armigera contig database (Helicoverpa Genome Consortium, unpublished) under the protein2genome model in Exonerate (Slater and Birney, 2005). Other criteria were to avoid loci that mapped to the ends of contigs (because sequence quality could be compromised) and regions containing repeat sequences.
A total of nine B. mori proteins with a BLASTX score of at least 200 were shortlisted so as to include all five scaffolds of the B. mori Z chromosome (nscaf1690, nscaf2210, nscaf2734, nscaf3040 and nscaf3068). EPIC markers were designed to span at least one intron with product sizes ranging from 600 to 1200 bp with exon sequences that were at least 50 bp away from either end of an intron. The loci down3 (downstream 3 kb) and up3 (upstream 3 kb) were subsequently included after the patterns of variation around Cyp303a1 were observed although they did not return any matches to known protein-coding sequences.
Twenty families were generated from single-pair matings of a laboratory-maintained colony to follow the markers through a pedigree. F1 individuals were sexed as pupae and re-assessed as adults. This colony was initiated from field samples collected in the vicinity of Toowomba, Australia (27º34′S, 151º57′E) in 2002 but has since been subjected to multiple injections of another laboratory-maintained strain, GR, to counteract the effects of inbreeding depression (Mahon et al., 2008). A separate set of pedigrees was used to ascertain Z-linkage of the Cyp303a1 alleles as our colony had fixed for the Ins200 allele. These families were derived from field samples collected in the MacIntyre Valley and obtained as ethanol-preserved moths (parents) and pupae (offspring). DNA extractions were carried out using a standard phenol–chloroform procedure. Scoring was performed by visualizing PCR products on agarose gels.
Samples
A total of 199 H. armigera (26 Australian and 173 non-Australian), 5 H. assulta and 20 H. punctigera DNA samples were obtained from G Behere; data on the collection of these samples is outlined in Behere et al. (2007). The Australian H. armigera data set consisted of 16 females from Dalmore, Victoria (38º11′S, 145º25′E) and 10 females from Orbost, Victoria (37º42′S, 148º27′E), both of which come from samples from the work by Behere et al. (2007) and a new collection of 112 females from MacIntyre Valley, Queensland (28º32′S, 150º18′E). The three collection sites are all temperate agricultural regions. However, they are currently classified into distinct bioregions (of which there are 89 in Australia; http://www.environment.gov.au/land/nrs/science/ibra). The Orbost and Dalmore samples were collected off corn and the MacIntyre Valley population off cotton. The Victorian female samples were identified by inferring hemizygosity from Sanger sequencing of Z-linked loci, that is, if overlapping traces (indicating heterozygosity for small indel polymorphisms) were present in the chromatograms, the sample would be designated 'male'. The MacIntyre Valley samples were collected as eggs from the field in 2010 and laboratory reared; sex was determined directly from visual inspection. The Victorian and MacIntyre Valley collections thus represent spatially and temporally separated populations of Australian H. armigera. For the genotyping of Cyp303a1 in non-Australian samples, the data set consisted of 35 individuals from Burkina Faso, 40 individuals from Uganda, 32 individuals from China, 12 individuals from Pakistan and 54 individuals from India. The sexes of these individuals were not known.
Sequencing reactions and quality checks
Cycling conditions varied slightly depending on the targets but were generally 35–40 cycles of 94 °C for 30 s, 60–65 °C for 30 s and 72 °C for 1.5 min. All PCR reactions were carried out using NEB Standard Taq polymerase and buffer (catalog number M0273). The final concentration of reagents was 0.025U/μl polymerase, 1x buffer, 200 μM dNTPs, 0.3 μM forward primer and 0.3 μM reverse primer. Sanger-sequencing of PCR products was performed on an ABI3730XL system (Macrogen, Korea).
Sequence quality checks were carried out using Sequencher 4.72 (Gene Codes, Ann Arbor, MI, USA), and sequences were manually edited to match the consensus-by-majority sequence if the base confidence was <40%. This approach was adopted to remove polymorphisms likely to be introduced by sequencing errors, especially for single-nucleotide indels occurring in a homopolymeric run. The disadvantage is that true polymorphisms occurring at low frequency are potentially discarded, but the preference was to adopt a conservative estimate of polymorphism given our expectations of a high-diversity genome.
Sequence diversity and nucleotide divergence
For H. punctigera and H. zea sequences, a repository was available for BLAST searches and accessing contigs (Helicoverpa Genome Consortium, unpublished). In the case of H. assulta where no such database could be interrogated, PCR reactions were carried out with H. armigera primers and Sanger-sequenced; orthology is assumed because only a single specific product was amplified. Multiple sequence alignment was performed using Seaview 4.0 (Gouy et al., 2010) and ClustalX (Larkin et al., 2007). Interspecies alignments were carried out in a two-step process: first, by defining each intraspecies alignment as a profile and, second, by aligning the profiles using the profile alignment option in ClustalX. Maximum-likelihood trees were constructed using PhyML under a GTR model, with support for clades based on 100 bootstrap replicates. Analyses of polymorphism and LD were carried out using DnaSP 5.10.01 (Librado and Rozas, 2009), with alignment files indicated as haploid Z chromosome. Estimates of polymorphism and divergence presented are uncorrected with respect to models of DNA evolution. Population differentiation was evaluated using an unbiased estimator of FST proposed by Hudson et al. (1992).
Linkage disequilibrium
LD was estimated as the square of the correlation coefficient, r2, for each pair of single-nucleotide polymorphisms using only parsimony-informative sites; sites segregating for three or four nucleotides and all indel polymorphisms were ignored. The statistical significance of each pairwise comparison was evaluated using Fisher's exact test and the χ2 test followed by Bonferroni correction for multiple testing. The number of significant pairwise comparisons as evaluated by Fisher's exact test (Supplementary Table S1) is more conservative, but we wanted to relax these constraints given our hypothesis of a low-LD genome; hence LD heatmaps were plotted using the outcomes of the χ2 test, which tended to evaluate a higher number of results as significant. The heatmap for Cyp303a1 and its flanking regions (3 kb upstream and downstream) was plotted by concatenating the sequences of up3, Cyp303a1 and down3 in individuals (five insertion and six deletion alleles) where data were available for all three loci. Heatmaps were visualized using the LDheatmap package in R (http://www.r-project.org/).
Decay of LD over physical distance was modeled on the expectations of Hill and Weir (1988) and implemented with the nonlinear least-squares function in R.
Coalescent simulations
A Monte Carlo program, msms (Ewing and Hermisson, 2010) was used to generate samples evolving under a neutral infinite-sites model based on the coalescent process, assuming a large and constant population size. All simulations were performed using the sample size n and number of segregating sites S as minimal input parameters. The value of n=63 was chosen to reflect the allele frequencies in field populations while maximizing the number of sequenced alleles in the analyses, that is, 44 Ins200 and 19 Del200 alleles (defined below). The value of S=80 was obtained from empirical data (Table 1). The recombination parameter C was estimated using two methods: the number of minimum recombination events (Rm) using the method of Hudson and Kaplan (1985), and R from Hudson (1987), which is based on the variance of the average number of differences between pairs of sequences in a sample.
Table 1. Nucleotide diversity and Tajima's D for nine loci surveyed in this study.
| Locus | n | No. of sites (bp)a | Sb | π | Tajima's D | |||
|---|---|---|---|---|---|---|---|---|
| Apt (55) | Dalmore (8) | 751–772 | 26 | 81 | 0.01 | 0.02 | 0.57 | −0.99 |
| Orbost (5) | 17 | 0.01 | 1.48 | |||||
| M. Valley (41) | 80 | 0.02 | −1.05 | |||||
| Cycle (21) | M. Valley (20) | 824 | 40 | 0.01 | −0.02 | |||
| Cyp303a1 (83)c | Dalmore (14) | 470–515 | 58 | 80 | 0.05 | 0.05 | 1.39 | 1.77 |
| Orbost (10) | 50 | 0.05 | 2.27* | |||||
| M. Valley (56) | 76 | 0.05 | 1.85 | |||||
| Cyp305b1 (22) | M. Valley (21) | 649 | 56 | 0.02 | −0.30 | |||
| Period (36) | Dalmore (13) | 336–511 | 36 | 54 | 0.03 | 0.02 | −0.79 | −1.57 |
| Orbost (5) | 29 | 0.03 | −0.71 | |||||
| M. Valley (17) | 52 | 0.03 | −1.67 | |||||
| Phc (36) | Dalmore (12) | 482–534 | 64 | 94 | 0.04 | 0.04 | −0.47 | −0.48 |
| Orbost (7) | 61 | 0.04 | −0.39 | |||||
| M. Valley (16) | 77 | 0.04 | −0.19 | |||||
| SCAP (12) | M. Valley (11) | 840 | 104 | 0.03 | −0.79 | |||
| Tc (16) | M. Valley (15) | 817 | 60 | 0.02 | 0.43 | |||
| Tpi (33) | Dalmore (11) | 514–544 | 100 | 125 | 0.07 | 0.06 | 0.08 | 0.04 |
| Orbost (2) | 35 | 0.06 | NA | |||||
| M. Valley (19) | 115 | 0.06 | −0.24 | |||||
Where estimates are presented in two columns under a single heading, the left column represents estimates for an individual population while the right column represents estimates after pooling sequences of all three populations. Figures in brackets after the locus name represent the total number of sequences surveyed, including the reference strain. Tajima's D for the Orbost population of Tpi is not available as a minimum of four sequences are required. *P<0.05.
The number of sites is presented as a range due to the differing subsets of indel polymorphisms present in different populations. As gapped sites are excluded from this analysis, the lower boundary represents the number of sites considered when alleles from all three populations are pooled.
Number of segregating sites, including singletons.
Includes two sequences from a laboratory-maintained colony.
For the simulations under a single-locus selection model, effective population sizes (Ne) between 105 and 107 and an allele frequency of 0.3 were used with the SF switch, with time t set to 0 to represent selection occurring up to the present time. The number of data sets in Table 3 (D) were chosen from simulations that resulted in the highest probability (typically Ne=107) so as to maximize the available data for subsequent analyses. Two values of the selection coefficient, s, were tested: the first representing weak-to-modest levels of selection (s=0.01) and the second representing a strong positive selection (s=0.1). The effect of the beneficial allele on the heterozygote was set to be half of that of the homozygote, that is, -SAA 2Nes -SAa 1Nes.
Results
Development of Z-linked EPIC markers
The development of nine H. armigera Z-linked markers in this study (Supplementary Table S2) was informed by previous reports of synteny in lepidopterans (Jiggins et al., 2005; Yasukochi et al., 2006; d'Alencon et al., 2010). Apt and Tpi have been established as Z-linked loci in multiple species, whereas the other loci were chosen because they were single-copy sequences that had 1:1 orthologs on B. mori Z-linked genes. Sex-limited inheritance of PCR amplicon size variation across pedigrees confirmed Z-linkage for Cyp303a1, Phc and Period (Supplementary Figures S1 and S2). For the remaining four loci, direct sequencing of amplicons was carried out on female samples without pedigree analyses. The absence of overlapping traces in the chromatograms indicated that the sequences were hemizygous and confirmed that these four loci were also on the Z chromosome.
Sequence diversity
We initially characterized five loci for which Z-linkage was determined by pedigree analyses (Apt, Cyp303a1, Period, Phc and Tpi) in two Victorian population samples described by Behere et al. (2007). Consistent with mitochondrial DNA analyses by Behere et al. (2007), we found no evidence for structure between these two populations (FST<0.06 at all loci examined). These initial results prompted us to expand the data set by obtaining an additional Australian population. To avoid the redundancy of work associated when scoring males (see Materials and methods section), we obtained 112 adult females from McIntyre Valley (on the border of New South Wales and Queensland) and scored them at all nine Z-linked loci. There was no evidence for population structure between the McIntyre Valley samples and the two Victorian populations (Supplementary Table S3), which suggested that LD analyses could be conducted on alleles pooled from all three populations (see below).
Levels of nucleotide diversity across all loci and the three Australian collection sites were high (694 single-nucleotide polymorphisms in <6 kb of sequence) and did not differ substantially between collection sites for any locus (Table 1). However, π values differed up to sixfold across loci (0.01–0.06 nucleotide differences per site) while indel variation differed by up to sevenfold across loci (0.002–0.014 indel events per site; Supplementary Table S4). Haplotype diversities were in the range of 0.7–1 for each locus per location, and we did not observe significant geographic structuring of haplotypes. Six of the nine loci had a negative Tajima's D, indicating an elevated number of rare variants in the samples although statistically the values were non-significant (consistent with the neutral model). The most notable feature of the frequency spectrum analysis was that Cyp303a1 had a highly positive Tajima's D. This was also true when each population was looked at individually, although only the Orbost population crossed the standard significance threshold.
Linkage disequilibrium
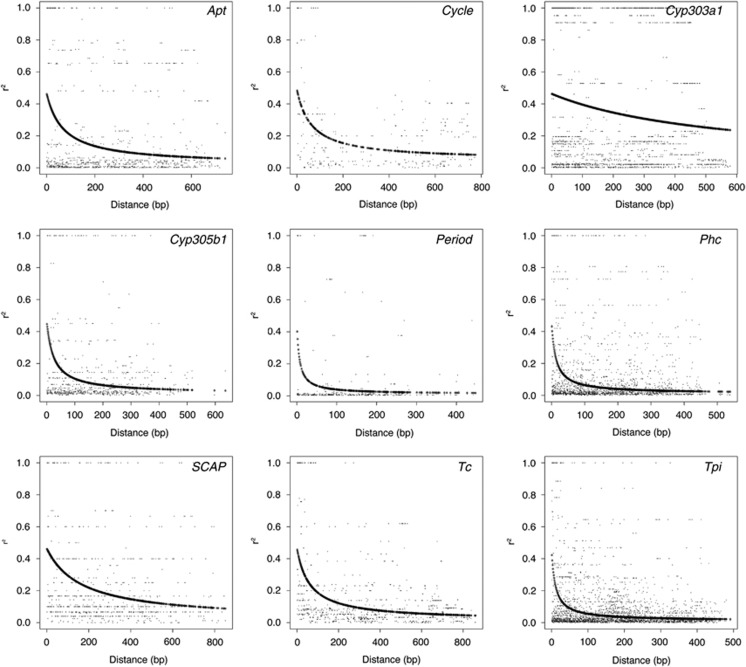
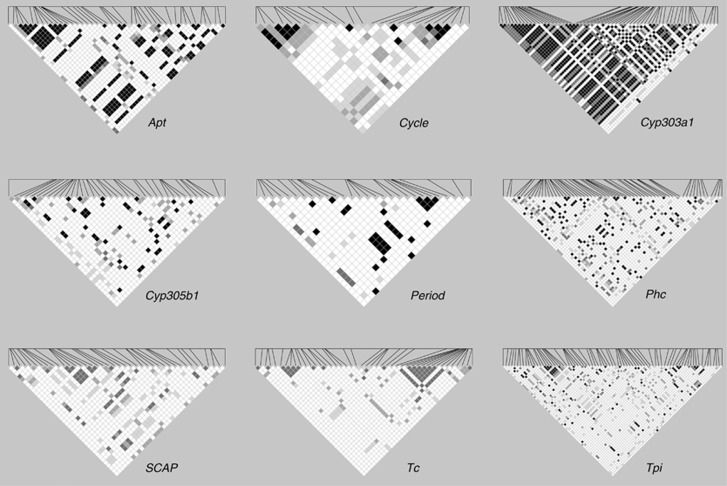
LD was calculated after pooling alleles from all three populations to maximize the sample sizes and thereby increase the power to detect significant associations. The level of LD in H. armigera was generally very low and of a similar magnitude to that seen in B. mori. LD was found to halve within 200 bps at each locus with the exception of Cyp303a1, whereby the distance at which r2 reached half its maximal estimated value was beyond the size of the 600/800 bp sequenced region (Figure 1). The paucity of LD at Phc and Tpi is striking given the total number of comparisons involved (Figure 2, Supplementary Table S1) as there are 63 and 77 parsimony-informative sites in Phc and Tpi, respectively.
Figure 1.
Plots of r2 over physical distance in base pairs, with curves showing the decay of LD modeled on the expectations of Hill and Weir (1988). The approximate distance at which E(r2) decays to 0.2 (approximately half the maximum estimated value across all nine loci) for each decay curve is 104, 122, >579, 25, 7, 14, 186, 76 and 11 bp for Apt, Cycle, Cyp303a1, Cyp305b1, Period, Phc, SCAP, Tc and Tpi, respectively. Maximum values of E(r2) ranged from 0.39 to 0.51. The number of alleles sampled for each locus is reported in Table 1.
Figure 2.
LD heatmaps for each of the nine loci plotted as significance of the r2 value in pairwise comparisons of segregating sites. Only bi-allelic sites are included. In all cases, the physical distance between the first and last site does not exceed 1 kb. Shading indicates significance level with black: P<0.001 (significant by Bonferroni), 80% grey: P<0.001, 50% grey: 0.001<P<0.01, 20% grey: 0.01<P<0.05, white: not significant.
Signals of selection at Cyp303a1?
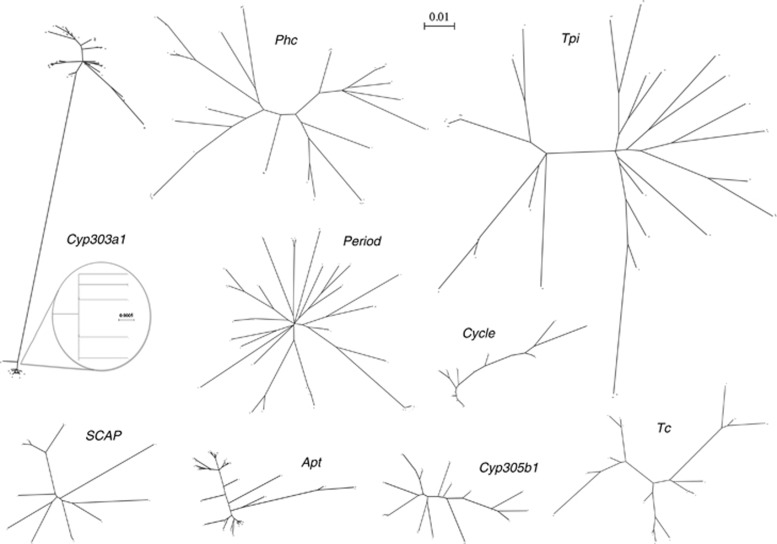
The positive Tajima's D values and the excessive LD at Cyp303a1 can be further understood by the allelic network of this locus relative to that of the other loci (Figure 3). An unrooted maximum-likelihood tree reveals an anomalous long branch separating two Cyp303a1 haplogroups that we will refer to as Del200 and Ins200 because a diagnostic feature of the two haplogroups is a 200-bp indel. The other surveyed loci exhibit more gradations in their phylogenies. Tpi does have a long internal branch but that can be attributed to a single stretch of 25 nucleotides containing five fixed differences. Even after exclusion of the 200-bp indel in Cyp303a1, the long internal branch is still apparent (Supplementary Figure S3) as there are 31 other fixed differences that are interspersed throughout the sequence alignment. The extent of divergence between the Ins200 and Del200 haplogroups raised concerns as to whether they represented two allelic types or paralogs. However, we confirmed that the indel polymorphism segregated in an allelic Z-linked manner where female offspring always presented only one copy of the locus, inherited from the male parent (Supplementary Figure S2).
Figure 3.
Unrooted maximum-likelihood trees for each of the nine loci. Each tip represents an allele. A long branch separates the two clades representing the Ins200 (above) and Del200 (below) alleles at Cyp303a1. Inset: The clade containing the Del200 alleles has very short branch lengths, with 32 out of 39 individuals carrying the same haplotype.
A total of 44 Ins200 and 39 Del200 alleles were sequenced so that the level of variation within haplogroups could be compared with the divergence between haplogroups (Table 2). The Del200 haplogroup contained very short branch lengths and was dominated by a single haplotype (32 of the 39 individuals). Within the Del200 haplogroup, there were very low levels of variation, evident in the small number of segregating sites and haplotypes compared with an equivalent number of Ins200 alleles. No indel polymorphisms were observed within the Del200 haplogroup and Tajima's D was significantly negative. In contrast, the Ins200 haplogroup had levels of nucleotide and indel diversity of a similar magnitude to that of other loci. The π value of the Ins200 haplogroup was 20 times that of the Del200 haplogroup, and multiple smaller indels were present within the Ins200 haplogroup. The divergence between haplogroups exceeded levels of nucleotide diversity at all other loci examined in this study.
Table 2. Comparison of the Cyp303a1 Del200 and Ins200 haplogroups.
| Statistic | Del200 | Ins200 |
|---|---|---|
| Number of sequences, n | 39 | 44 |
| Number of sites, excluding gaps | 523 | 681 |
| Segregating sites (including singletons), S | 9 | 63 |
| Parsimony informative sites | 1 | 45 |
| Number of haplotypes, excluding gaps | 7 | 32 |
| Number of indel events, I | 0 | 17 |
| Nucleotide diversity per site, π | 0.001 | 0.019 |
| Tajima's D | −2.23** | −0.43 |
| Between haplogroups | ||
| Number of fixed differences | 31 | |
| Nucleotide divergence | 0.09 | |
**P<0.01.
To assess whether evolution at the Cyp303a1 locus was compatible with the neutral model, two tests were carried out using coalescent simulations. The first test addressed the likelihood of obtaining i identical alleles from a sample of size n given the diversity of the sample (Hudson et al., 1994). By analyzing the length of the amplicons, the frequency of the Del200 haplogroup was observed to be approximately 28% in all three populations. The preceding haplotypic analysis indicated that 32/39 or 82% of alleles would fall under a single haplotype, hence the value of i was determined to be 14 (0.28 × 0.82 × 63) for a sample size of n=63. Simulated data sets were generated under three scenarios: (i) no recombination, (ii) C=Rm, the minimum number of recombination events which underestimates the total number of recombination events (Hudson and Kaplan, 1985), and (iii) C=R estimated from the method of Hudson (1987). The recombination parameter, C, was estimated using the Ins200 haplogroup data. The first two scenarios are conservative, yet the probability of obtaining a subset of identical alleles does not exceed 12%. Under the third scenario, which includes a modest amount of recombination, the probability is significant enough to reject the neutral model (P<0.013; Table 3 (A)).
Table 3. Coalescent simulations with and without selection.
|
Recombination parameter |
|||
|---|---|---|---|
| C=0 | C=Rm | C=R | |
| No selection | |||
| (A) Number of data sets | 10 000 | 10 000 | 10 000 |
| P (i⩾14) | 0.120 | 0.064 | 0.013 |
| (B) Number of data sets | 1200 | 643 | 126 |
| P (d<5∪d>30) | 0.10 | <0.01 | <0.01 |
| Selection coefficient, s=0.01 | |||
| (C) Number of data sets | 1000 | 1000 | 1000 |
| P (i⩾14) | >0.90 | >0.90 | >0.85 |
| (D) Number of data sets | 976 | 974 | 954 |
| P (d<5∪d>30) | 0.05 | <0.01 | <0.001 |
| Selection coefficient, s=0.1 | |||
| (E) Number of data sets | 1000 | 1000 | 1000 |
| P (i⩾14) | >0.95 | >0.95 | >0.95 |
| (F) Number of data sets | 977 | 968 | 962 |
| P (d<5∪d>30) | 0.04 | <0.01 | <0.001 |
Probability of observing (A, C, E) a minimum of i identical sequences using a threshold value determined from empirical observations of the frequency of the Del200 haplogroup and (B, D, F) a major haplogroup that is highly divergent from all other alleles in the population whereby the pairwise distance, d, between the major allele and all other sequences in a data set is either <5 (representing variants within the Del200 haplogroup) or >30 (representing the 31 fixed differences between the Ins200 and Del200 haplogroups). All data sets were simulated using the parameters n=63 and S=80. For simulations with selection, Ne ranged from 105 to 107and the SF option with t=0 and f=0.3 was used (Ewing and Hermisson, 2010). In addition to a no recombination scenario, two estimates of the recombination parameter, C, were included: the minimum number of recombination events, Rm=5 (Hudson, 1987), and the estimator based on the variance of the average number of differences between pairs of sequences, R=21.7 (Hudson and Kaplan, 1985).
The second test addressed the likelihood of observing a major haplogroup that is highly divergent from all other alleles in the population. The test was only conducted on data sets that fulfilled the criteria of the first test (i⩾14) and was implemented as follows: the pairwise distances (d) between the major haplotype and all others in the data set were calculated. Haplotypes that diverged at <5 substitutions represent variants in one haplogroup (corresponding to the Del200 haplogroup) while those containing >30 substitutions (fixed differences) represent haplotypes from other haplogroups (akin to the Ins200 haplogroup). Data sets containing any values of 5<d<30 are considered unlike our observed data set, that is, are not divided into diverged haplogroups. The simulations conducted in the absence of recombination recovered diverged haplogroups (like the observed data set) 10% of the time, whereas those with even the minimal level of recombination recovered the diverged haplogroups <1% of the time (Table 3 (B)). These simulations demonstrate that, under a strict neutral model of coalescence, it is highly unlikely to observe a haplotype network divided into divergent haplogroups with one containing limited diversity.
To assess whether selection was the most parsimonious explanation for both the low diversity of the Del200 haplogroup and the divergence between the two haplogroups, the two tests outlined above were carried out under two models incorporating the selection coefficient parameter, s (Table 3 (C–F)). Using an effective population size (Ne) between 105 and 107, allele frequency (f) of 0.3 and allowing selection to occur to the present time (t=0), the likelihood of obtaining 14 identical alleles out of 63 was >85% in all the scenarios tested (Table 3 (C and E)). However, the likelihood of obtaining a long internal branch fell to <1% when even a minimal amount of recombination was allowed, irrespective of the strength of selection (Table 3 (D and F)). In the absence of recombination, the diverged haplogroups were recovered approximately 5% of the time. These simulations suggest that positive selection in and of itself is insufficient to account for the patterns observed in our data set, and some secondary mechanism affecting recombination may have accompanied the selective event.
To further investigate whether selection affected Cyp303a1 and the Del200 haplogroup in particular, the frequency of the deletion allele outside of Australia was examined using PCR amplicon length analysis. The deletion haplogroup was not detected in any samples from India (n=54), Pakistan (n=12), Burkino Faso (n=35) or Uganda (n=40). However, three deletion alleles were present in the Chinese population (n=32) of which one was confirmed by Sanger sequencing. This Chinese Del200 allele differed from the major Del200 haplotype found in Australia at a single site. A low frequency of novel amplicon lengths were found in these global samples but most were approximately 800 bps, which is typical of the Ins200 haplogroup. Thus this locus contrasts dramatically with the multiple markers from multiple studies that exhibit low FST in global populations (Daly and Gregg, 1985; Nibouche et al., 1998; Zhou et al., 2000; Behere et al., 2007).
We postulated that the target of selection resulting in the patterns observed in the Del200 haplogroup may not be within the sequence of the EPIC amplicon we surveyed but another polymorphism that is in LD with it. To determine whether there were any amino-acid changes at Cyp303a1 that could be the variant targeted by selection, the complete coding region was sequenced from two Ins200 and two Del200 alleles of a laboratory-maintained colony. We did not detect any non-synonymous substitutions that could discriminate the two alleles, although there were four synonymous polymorphisms segregating.
To determine whether the polymorphism targeted by selection could be limited to the Cyp303a1 locus, adjacent regions 3 kb upstream and downstream of Cyp303a1 were sequenced from a subset of the field samples (5:7 and 11:12 Ins200:Del200 alleles, respectively). An LD heatmap illustrates that approximately 300 bp of the 3′ end of the upstream region is in LD with the indel (Supplementary Figure S4). Levels of diversity at this locus were similar to that of other loci with π=0.05 and π(i)=0.010 and Tajima's D was slightly negative (−0.29) but not statistically significant. In contrast, the downstream region (down3) exhibited very low diversity (π=0.004) with no indels and had a significantly negative Tajima's D (−1.95, P<0.05). Thus selection may indeed be acting downstream of Cyp303a1.
Divergence from other species
The lack of nucleotide polymorphism in the downstream locus is consistent with the idea that there has been a selective sweep in the vicinity, and the implication is that this downstream locus is closer to the target of selection. However, an alternate hypothesis is that strong purifying selection independent of the effect seen at Cyp303a1 acts upon this downstream locus, even though it appears to be non-coding (that is, sequences are constrained because any change alters an important function). The divergence between closely related species could help us discriminate between these two hypotheses. If such constraint is acting on the sequence 3 kb downstream of Cyp303a1, then it may have been acting since the divergence of the species. Table 4 shows the divergences between species for the nine loci and the region downstream of Cyp303a1. As expected from the species phylogeny, H. armigera sequences showed the greatest divergence with H. punctigera followed by H. assulta and then H. zea. H. zea appeared very similar to H. armigera. The down3 locus exhibited values that were similar to those of other loci, which rejects the hypothesis that the lack of diversity observed in H. armigera could reflect excessive purifying selection at that sequence over the period encompassing the divergence of these species. Rather, the lack of diversity despite divergence favours a model of recent positive selection affecting loci in this region.
Table 4. Nucleotide divergence between H. armigera and H. assulta, H. punctigera and H. zea.
| Locus | Nucleotide divergence, Dxy, between H. armigera and | ||
|---|---|---|---|
| H. assulta | H. punctigera | H. zea | |
| Apt | 0.06 | 0.08 | 0.04 |
| Cycle | 0.06 | 0.06 | 0.03 |
| Cyp303a1 | 0.11 | 0.18 | 0.12 |
| Cyp305b1 | 0.04 | 0.10 | 0.04 |
| down3 | 0.07 | 0.13 | 0.05 |
| Period | 0.05 | 0.08 | 0.03 |
| Phc | 0.07 | 0.11 | 0.03 |
| SCAP | 0.10 | 0.19 | 0.04 |
| Tc | 0.09 | 0.12 | 0.05 |
| Tpi | 0.07 | 0.11 | 0.07 |
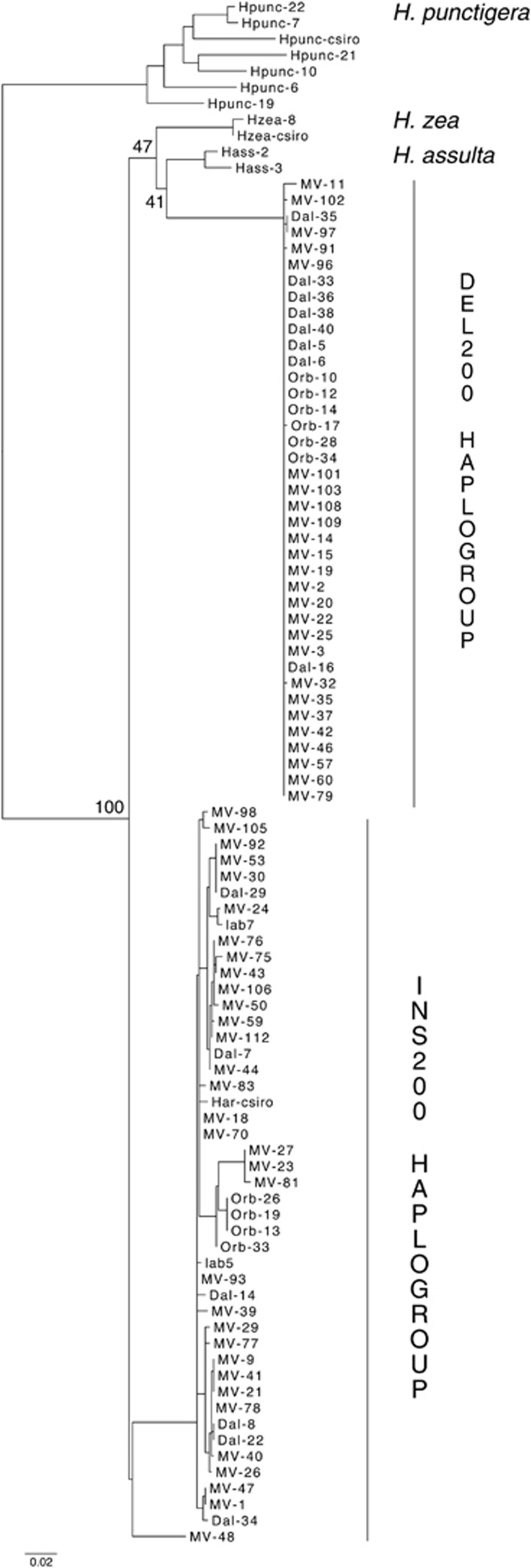
Furthermore, rather than showing selective constraint, the Cyp303a1 amplicon shows high divergence relative to the values observed at other loci in H. assulta and H. zea. The divergence across the intron suggests overlapping indels during the evolution of this locus, making alignments difficult. For instance, the H. punctigera sequence shared one characteristic of the deletion variant in that it was lacking the 200-bp insert. However, it also contained a 100-bp deletion in a region common to both subgroups and various smaller indel polymorphisms in the regions flanking the H. armigera 200-bp indel. The H. assulta sequence had an 800-bp insertion that incorporated the 200-bp H. armigera insertion. The H. zea sequence incorporated parts of the H. armigera insertion. A maximum-likelihood tree of the sequenced region across H. armigera, H. assulta, H. punctigera and H. zea (Figure 4) shows low bootstrap support of the relationships between species, except for the placement of H. punctigera as the outgroup. We also scored the state of the H. punctigera, H. assulta and H. zea sequences where the fixed differences between the Ins200 and Del200 haplogroups occurred (Supplementary Table S5). Although the three outgroup sequences superficially have more states in common with the insertion variant, there is no clear indication that the insertion is the ancestral state.
Figure 4.
Maximum-likelihood tree of Cyp303a1 sequenced region from H. armigera, H. assulta, H. punctigera and H. zea. For clarity, only bootstrap values pertaining to the relationships between species are shown.
Discussion
The levels of diversity we observe on the Z chromosome of H. armigera (average π=0.03) are high relative to genome-wide estimates of diversity in other insects, which are generally high among that of other taxa (Leffler et al., 2012). In Aedes aegypti, Anopheles funestus and Anopheles gambiae for instance, nucleotide diversity in noncoding regions is approximately 0.01; in Drosophila melanogaster it is ~0.01 and in D. simulans it is ~0.02 (Morlais and Severson, 2003; Wondji et al., 2007; Langley et al., 2012; O'Loughlin et al., 2014). It is worth noting that, if the mutation rates differ between the sexes, then nucleotide diversity at neutral loci can differ between sex chromosomes and autosomes (Vicoso and Charlesworth, 2006). In Lepidopterans where males are the homogametic sex, a Z chromosome spends 2/3 of its evolutionary time in males, whereas an autosome only spends half of its time in this potentially more mutagenic sex. Furthermore, positive selection could cause the Z chromosome to evolve faster as recessive alleles are exposed in females (Vicoso and Charlesworth, 2006). Therefore, it is possible that the nucleotide diversity we observe on the Z of H. armigera may be elevated relative to the genome-wide value. If estimates from silk moths are used as a guide (Sackton et al. 2014), the autosomal diversity will be approximately 60% of that Z chromosomes (that is, ~0.02), and that would not alter our conclusion that H. armigera exhibits high levels of nucleotide diversity.
The frequency of insertions and deletions is also higher in H. armigera (π(i)=0.005) relative to D. melanogaster where π(i) is <0.003 for intergenic and intronic regions (Ometto et al., 2005). A pragmatic consequence of such a high indel frequency is that direct sequencing of EPIC PCRs in this species may be problematic at autosomal loci (and Z loci in males) because the sequence trace at each frequently spaced indel (every 200 bps) will feature two overlapping sequences (observed as double peaks on the sequence chromatograms) that may be hard to disentangle. At a theoretical level, we are left with the question of whether the high nucleotide diversity (π≈θ) in this species is due to a large effective population size or a high mutation rate (θ=4Neμ).
This study also reveals that the H. armigera genome displays remarkably limited LD. For eight of the nine loci characterized herein, r2 drops to half its estimated maximal value within 200 bps, and this is low relative to that of the other lepidopterans so far characterized and that of other insects (Supplementary Table S6). Regardless of the reasons for the different levels of LD in different species, it creates an important design consideration for future population genomic studies in these insects. For instance, a rigorous genome-wide association study in H. armigera would need such a high marker density that whole-genome sequencing might be preferable to technologies that genotype 'tag' single-nucleotide polymorphisms. The high levels of nucleotide diversity coupled with rapid decay of LD also mean that genotype imputation approaches will be limited. Another challenge of allele-rich architecture is genome assembly itself because alleles may be confused as paralogs. However, an advantage of a low-LD, high-diversity genome should be easier identification of causal variants in genome-wide association studies or selective sweep studies.
This study supports previous findings of little population subdivision in Australian H. armigera (Daly and Gregg, 1985; Endersby et al., 2007). Low FST values at multiple loci sampled from spatially and temporally different populations suggest extensive gene flow. The paucity of LD is consistent with this scenario—if the three populations were genetically differentiated, we would expect a modest degree of significant associations due to 'admixture' from the pooling of alleles. The presence of a single haplotype that appears to have recently arisen to similar intermediate frequencies (the Del200 haplotype) in geographically separated samples is parsimoniously explained by extensive gene flow in Australia.
The divergence data reported here also suggests that H. zea are not substantially diverged from H. armigera. For instance, divergence between H. armigera and H. zea at the Phc locus (0.03) was less than that observed between some H. armigera alleles (π=0.04). This is consistent with the origin of H. zea from within an ancestral H. armigera population as proposed by Mallet et al. (1993) and affirmed by Behere et al. (2007).
A footprint of a selective sweep?
The Cyp303a1 locus exhibits multiple patterns that are aberrant relative to the other loci surveyed here and that are inconsistent with neutral expectations. Among these is extended LD and an unusual frequency spectrum of polymorphisms. These patterns can be attributed to the occurrence of two divergent haplogroups, one of which seems to have recently arisen to high frequency in Australian populations as it exhibits very little allelic diversity despite it being at 28% frequency. The coalescent simulations performed here show that such patterns are extremely unlikely in a neutral model, particularly when so much recombination is observed in the H. armigera genome. As discussed below, in order to see such patterns, the extent of recombination among the sampled alleles must have been distorted by the influence of selection, a molecular mechanism limiting the site of recombination at meiosis, and/or population demographics.
A subset of highly similar alleles within a set of diverged alleles has been reported at particular loci in other species and is often attributed to partial selective sweeps—the model is that a favourable variant has increased in frequency at such a rate that recombination has not had time to occur, enabling nearby variants to 'hitchhike' to intermediate frequency (Hudson et al., 1994; Schlenke and Begun, 2004; Sanchez-Gracia and Rozas, 2007). In a similar manner, the lack of variation in our deletion haplotype is inconsistent with a scenario of neutral polymorphism at intermediate frequency or the scenario of an old polymorphism maintained through balancing selection. Instead, the selective sweep model is supported by the Del200 haplogroup displaying a skewed frequency spectrum (Tajima's D is significantly negative), which can be interpreted as recent positive selection resulting in an intermediate frequency of the polymorphism. Furthermore, the pattern of polymorphism in a noncoding region 3 kb downstream of this locus also shows a significantly negative Tajima's D value. However, there is no clear bifurcation into haplogroups at this downstream locus. This could be explained by a hard sweep focussed on a variant closer to the down3 locus, purging variation at it; the Cyp303a1 intron that is further away from this selective pressure thus sits on the 'shoulder' of the sweep where a limited amount of recombination has prevented fixation of the Del200 haplogroup.
A second explanation for the patterns observed at Cyp303a1 is that recombination is suppressed in the Del200 haplogroup because of an uncharacterized molecular feature such as an inversion or perhaps the 200-bp indel itself has prevented exchange between chromosomes at prophase I of meiosis. This would explain the second extraordinary feature of the Cyp303a1 genealogy—the accumulation of so many divergent sites between the Ins200 and the Del200 haplogroups (31 fixed differences). Recombination would be unimpeded among the Ins200 alleles but they could not recombine with alleles from the Del200 haplogroup. However, such recombination suppression would not explain the Tajima's D test results among the Del200 alleles or the down3 locus. The molecular explanation for the lack of recombination would therefore still need to be accompanied by a secondary selection event.
A third way that recombination could be distorted is if our samples were influenced by demographic events such that alleles were not sampled from a population where random mating had been occurring throughout the history of their coalescence. We have already noted that most of the data presented here are consistent with previous suggestions of little population structure in H. armigera. However, the population structure at the Cyp303a1 locus is exceptional in that the Del200 haplogroup is present at high frequency in all Australian samples yet does not occur in African, Indian or Pakistani populations and is at very low frequency in the Chinese population we surveyed. These data support the model that the Del200 haplogroup arose in Australia and has increased to its current frequency of 28% due to positive selection and has spread to China. The alternate model, separating the originating country (for example, China) from the sweep to high frequency, implies that the selective agent driving the sweep is geographically limited to Australia; this is a more complex and therefore less likely scenario.
We have not surveyed the other Z-linked loci outside Australia, yet mitochondrial DNA, allozyme, microsatellite and EPIC PCR analyses do not suggest that Australia houses particularly divergent or isolated alleles (Daly and Gregg, 1985; Nibouche et al., 1998; Behere et al., 2007; Endersby et al., 2007; Tay et al., 2008). So if there is no evidence of a reservoir of diverged alleles in Australian H. armigera, where does the Del200 haplogroup come from? One possibility is that it was introduced via introgression from a related species. This notion is appealing because it explains the high level of divergence between the two haplogroups. The term 'comet allele' has been proposed to describe haplotypes that have introgressed across species or sub-species boundaries—similar to comets, they have 'dipped' into this system from another lineage (Staubach et al., 2012). There are precedents for such events in other species such as that described by Brand et al. (2013) where D. simulans alleles have entered the D. sechellia genome in an adaptive process. The divergence between the two haplogroups is as great as between H. armigera and H. assulta at other loci. Given the observed frequencies of the Del200 haplotype in our Asian and African populations, an Australian origin appears most likely and is consistent with the hypothesized radiation of heliothines on this continent (Matthews, 1999). Our data do not provide evidence for a source population from H. assulta, H. punctigera or H. zea. However, population structures and levels of diversity in H. punctigera and H. assulta are not as well characterized—the provenance of the H. armigera divergent allele could be a cryptic race or isolated population of either species given their overlapping ranges. Alternatively, it could be from another species not characterized here such as Helicoverpa hardwicki, Helicoverpa prepodes and members of the genus Australothis and Heliocheilus, which are endemic to Australia (Matthews, 1999; Cho et al., 2008).
Thus to explain the patterns we see at the Cyp303a1 locus, we are left with two alternate hypotheses both of which involve a selective sweep. In the first, our coalescent simulations suggest that the high level of diversity within H. armigera coupled with a molecular-based suppression of recombination in the Del200 haplogroup may have allowed the emergence of a highly divergent and recently adaptive allele. Alternatively, an adaptive introgression of this locus from another species would explain the sweep of a highly diverged allele through Australian populations.
Finally, we note that the selective agent believed to be driving the patterns in the genealogy of Cyp303a1 is unknown. Given that Cyp303a1 is a cytochrome P450 gene, insecticides could be candidates because genes in the P450 multigene family are frequently associated with insecticide resistance (Feyereisen, 2005). However, Cyp303a1 is a strict (1:1) ortholog to a Drosophila gene that has been functionally characterized as being essential for mechanosensation and chemosensation and is expressed only in the sensory bristles (Willingham and Keil, 2004). The occurrence of 1:1 orthology across this taxonomic distance is notable given the multiple gene gain and loss events commonly observed in multigene families and supports the grouping of Cyp303a1 with the developmental rather than detoxification class of P450s. If Cyp303a1 is the target of selection, the causal variant would have to be a regulatory mutation as there are no amino-acid differences between the Ins200 and Del200 haplotypes, and there was no copy number variation detected at this locus. If insecticide selection is acting on the function of the Cyp303a1 locus, then a sensing function may be more likely than a detoxifying one. The second possibility is that the target of selection is another gene and Cyp303a1 is merely a 'hitchhiker', although analysis of the contig on which Cyp303a1 is located has not revealed any genes in the region extending 10 kb downstream (unpublished data).
In conclusion, this study established that the H. armigera genome exhibits high levels of nucleotide diversity within populations and generally high levels of recombination and gene flow yet we discovered an instance where deviations from these trends suggest that footprints of selection can be detected. Genome-wide scans for signals of selection are a complementary approach to genome-wide association study in identifying candidate genes for phenotypes of interest. Evaluating the role of demographic processes in shaping genome architecture remains a major challenge, and new tests for identifying selection will need to accommodate more complex scenarios potentially including introgression from other species.
Data archiving
The sequences for Cyp303a1 have been submitted to GenBank (accession numbers KR709083-5). All other sequences are available in the Dryad repository under the doi:10.5061/dryad.123qg.
Acknowledgments
We thank the leaders of the Helicoverpa Genome Consortium for permission to use the data ahead of publication, Ganesh Behere and Wee Tek Tay for providing access to their frozen collections and David Clarke and Robert Good for bioinformatic assistance. SVS was supported by a University of Melbourne Research Scholarship and the CSIRO.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Behere G, Tay W, Russell D, Heckel D, Appleton B, Kranthi K et al. (2007). Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea. BMC Evol Biol 7: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Kingan SB, Wu L, Garrigan D. (2013). A selective sweep across species boundaries in Drosophila. Mol Biol Evol 30: 2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Mitchell A, Mitter C, Regier J, Matthews M, Robertson R. (2008). Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Syst Entomol 33: 581–594. [Google Scholar]
- d'Alencon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S et al. (2010). Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci 107: 7680–7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JC, Gregg P. (1985). Genetic variation in Heliothis in Australia: species identification and gene flow in the two pest species H. armigera (Hübner) and H. punctigera Wallengren (Lepidoptera: Noctuidae). Bull Entomol Res 75: 169–184. [Google Scholar]
- Endersby NM, Hoffmann AA, McKechnie SW, Weeks AR. (2007). Is there genetic structure in populations of Helicoverpa armigera from Australia? Entomol Exp Appl 122: 253–263. [Google Scholar]
- Ewing G, Hermisson J. (2010). MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26: 2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyereisen R. (2005). Insect cytochrome P450. Compr Mol Insect Sci 4: 1–77. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Hardwick DF. (1965). The corn earworm complex. Memoirs Entomol Soc Can 97: 5–247. [Google Scholar]
- Harris C, Rousset F, Morlais I, Fontenille D, Cohuet A. (2010). Low linkage disequilibrium in wild Anopheles gambiae s.l. populations. BMC Genet 11: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Weir BS. (1988). Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33: 54–78. [DOI] [PubMed] [Google Scholar]
- Hudson RR. (1987). Estimating the recombination parameter of a finite population model without selection. Genet Res 50: 245–250. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ. (1994). Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. (1992). Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Mavarez J, Beltran M, McMillan WO, Johnston JS, Bermingham E. (2005). A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics 171: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE et al. (2012). Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Laster ML, Hardee DD. (1995). Intermating compatibility between North American Helicoverpa zea and Heliothis armigera (Lepidoptera: Noctuidae) from Russia. J Econ Entomol 88: 77–80. [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, Venkat A et al. (2012). Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol 10: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Mahon RJ, Olsen KM, Downes S. (2008) Isolations of Cry2Ab resistance in Australian populations of Helicoverpa armigera (Lepidoptera: Noctuidae) are allelic J Econ Entomol 101: 909–914. [DOI] [PubMed]
- Mallet J, Korman A, Heckel DG, King P. (1993). Biochemical genetics of Heliothis and Helicoverpa (Lepidoptera: Noctuidae) and evidence for a founder event in Helicoverpa zea. Ann Entomol Soc Am 86: 189–197. [Google Scholar]
- Matthews M. (1999) Heliothine Moths of Australia: A Guide to Pest Bollworms and Related Noctuid Groups. Monographs on Australian Lepidoptera, vol. 7, CSIRO Publishing: Melbourne, Australia. ISBN 0643063056. [Google Scholar]
- Mitter C, Poole RW, Matthews M. (1993). Biosystematics of the heliothinae (lepidoptera: noctuidae). Ann Rev Entomol 38: 207–225. [Google Scholar]
- Morlais I, Severson DW. (2003). Intraspecific DNA variation in nuclear genes of the mosquito Aedes aegypti. Insect Mol Biol 12: 631–639. [DOI] [PubMed] [Google Scholar]
- Nibouche S, Bues R, Toubon JF, Poitout S. (1998). Allozyme polymorphism in the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae): comparison of African and European populations. Heredity 80: 438–445. [Google Scholar]
- Nielsen R. (2005). Molecular signatures of natural selection. Annu Rev Genet 39: 197–218. [DOI] [PubMed] [Google Scholar]
- O'Loughlin SM, Magesa S, Mbogo C, Mosha F, Midega J, Lomas S et al. (2014). Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories. Mol Biol Evol 31: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto L, Stephan W, De Lorenzo D. (2005). Insertion/deletion and nucleotide polymorphism data reveal constraints in Drosophila melanogaster introns and intergenic regions. Genetics 169: 1521–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Corbett-Detig RB, Nagaraju J, Vaishna L, Arunkumar KP, Hartl DL. (2014). Positive selection drives faster-Z evolution in silkmoths. Evolution 68: 2331–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gracia A, Rozas J. (2007). Unusual pattern of nucleotide sequence variation at the OS-E and OS-F genomic regions of Drosophila simulans. Genetics 175: 1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. (2004). Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G, Birney E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubach F, Lorenc A, Messer PW, Tang K, Petrov DA, Tautz D. (2012). Genome patterns of selection and introgression of haplotypes in natural populations of the house mouse (Mus musculus). PLoS Genet 8: e1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay W, Behere G, Heckel D, Lee S, Batterham P. (2008). Exon-primed intron-crossing (EPIC) PCR markers of Helicoverpa armigera (Lepidoptera: Noctuidae). Bull Entomol Res 98: 509–518. [DOI] [PubMed] [Google Scholar]
- Tay WT, Soria MF, Walsh T, Thomazoni D, Silvie P, Behere GT et al. (2013). A brave new world for an Old World pest: Helicoverpa armigera (Lepidoptera: Noctuidae) in Brazil. PLoS One 8: e80134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. (2006). Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet 7: 645–653. [DOI] [PubMed] [Google Scholar]
- Wang C, Dong J. (2001). Interspecific hybridization of Helicoverpa armigera and H. assulta (Lepidoptera: Noctuidae). Chinese Sci Bull 46: 489–491. [Google Scholar]
- Willingham AT, Keil T. (2004). A tissue specific cytochrome P450 required for the structure and function of Drosophila sensory organs. Mech Dev 121: 1289–1297. [DOI] [PubMed] [Google Scholar]
- Wondji CS, Hemingway J, Ranson H. (2007). Identification and analysis of single nucleotide polymorphisms (SNPs) in the mosquito Anopheles funestus, malaria vector. BMC Genomics 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z et al. (2009). Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi Y, Ashakumary LA, Baba K, Yoshido A, Sahara K. (2006). A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects. Genetics 173: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalucki M, Daglish G, Firempong S, Twine P. (1986). The biology and ecology of Heliothis armigera (Hubner) and Heliothis punctigera Wallengren (Lepidoptera, Noctuidae) in Australia: what do we know? Aust J Zool 34: 779–814. [Google Scholar]
- Zhang DX. (2004). Lepidopteran microsatellite DNA: redundant but promising. Trends Ecol Evol 19: 507–509. [DOI] [PubMed] [Google Scholar]
- Zhou X, Faktor O, Applebaum SW, Coll M. (2000). Population structure of the pestiferous moth Helicoverpa armigera in the Eastern Mediterranean using RAPD analysis. Heredity 85: 251–256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.