Abstract
Methylotrophic planktonic bacteria fulfill a particular role in the carbon cycle of lakes via the turnover of single-carbon compounds. We studied two planktonic freshwater lineages (LD28 and PRD01a001B) affiliated with Methylophilaceae (Betaproteobacteria) in Lake Zurich, Switzerland, by a combination of molecular and cultivation-based approaches. Their spatio-temporal distribution was monitored at high resolution (n=992 samples) for 4 consecutive years. LD28 methylotrophs constituted up to 11 × 107 cells l−1 with pronounced peaks in spring and autumn–winter, concomitant with blooms of primary producers. They were rare in the warm water layers during summer but abundant in the cold hypolimnion, hinting at psychrophilic growth. Members of the PRD01a001B lineage were generally less abundant but also had maxima in spring. More than 120 axenic strains from these so far uncultivated lineages were isolated from the pelagic zone by dilution to extinction. Phylogenetic analysis separated isolates into two distinct genotypes. Isolates grew slowly (μmax=0.4 d−1), were of conspicuously small size, and were indeed psychrophilic, with higher growth yield at low temperatures. Growth was enhanced upon addition of methanol and methylamine to sterile lake water. Genomic analyses of two strains confirmed a methylotrophic lifestyle with a reduced set of genes involved in C1 metabolism. The very small and streamlined genomes (1.36 and 1.75 Mb) shared several pathways with the marine OM43 lineage. As the closest described taxa (Methylotenera sp.) are only distantly related to either set of isolates, we propose a new genus with two species, that is, ‘Candidatus Methylopumilus planktonicus' (LD28) and ‘Candidatus Methylopumilus turicensis' (PRD01a001B).
Introduction
Pelagic freshwater bacteria are key factors in the biogeochemical processes of lakes. One particular role in the carbon cycle is fulfilled by methylotrophic bacteria that use single-carbon (C1) compounds such as methanol as sole sources of carbon and energy. Methanol is a main degradation product of pectin and lignin and is therefore mainly associated with plant growth and decay (Heikes et al., 2002; Vorholt, 2012). In aquatic systems, methanol and other C1 compounds (for example, methylated sulfur species and methylated amines) may also be linked to primary producers. C1 units can be either directly released by phytoplankton species or can result of bacterial breakdown of algal carbohydrates (Sieburth and Keller, 1989; Milne et al., 1995; Heikes et al., 2002). Other potential C1 sources include atmospheric deposition, allochthonous import (Heikes et al., 2002), or photochemical degradation of dissolved organic carbon (Dixon et al., 2011a; Mopper et al., 1991). Methanol is chemically stable and highly soluble in water because of its polarity (Dixon et al., 2011b; Heikes et al., 2002). So far, reports of in situ concentrations of methanol have been restricted to marine habitats with concentrations ranging in the nM scale (70–429 nmol l−1) and turnover rates as low as 1 day (range: 1–83 days) in very oligotrophic sites (Dixon et al., 2011a, b). An indication for high methylotrophic activity and an important role in the in situ biogeochemical cycling of C1 compounds was provided by the overrepresentation of methanol dehydrogenases (MDHs) affiliated with Methylophilaceae (Betaproteobacteria) in metatranscriptomic and metaproteomic surveys in freshwater and marine habitats (Sowell et al., 2011; Gifford et al., 2013; Georges et al., 2014; Hanson et al., 2014).
Aerobic methylotrophs can be found in various families of Proteobacteria, Verrucomicrobia, NC10, and Gram-positive microbes (Chistoserdova 2011). The family Methylophilaceae includes four formally described genera (Methylophilus, Methylovorus, Methylobacillus and Methylotenera) isolated from terrestrial environments or freshwater sediments (Yordy and Weaver, 1977; Jenkins et al., 1987; Govorukhina and Trotsenko, 1991; Kalyuzhnaya et al., 2006). Several representatives of these genera have been studied in detail and genome sequenced (Lapidus et al., 2011; Beck et al., 2014). Moreover, members of the planktonic marine lineage OM43 have been isolated via dilution to extinction methods and genome sequenced (Connon and Giovannoni, 2002; Giovannoni et al., 2008; Song et al., 2009; Huggett et al., 2012). OM43 have very small genomes (1.3 Mb) and a reduced set of genes encoding methylotrophic pathways (Giovannoni et al., 2008; Chistoserdova 2011; Halsey et al., 2011; Huggett et al., 2012). Members of this lineage are abundant in coastal oceans, especially during phytoplankton blooms (Sekar et al., 2004; Morris et al., 2006). Less is known about planktonic freshwater Methylophilaceae (Newton et al., 2011). Two ubiquitous species-like lineages (LD28 and PRD01a001B) are widely distributed and especially LD28 are highly abundant and seasonally persistent in the pelagic zone of lakes (Newton et al., 2011; Parveen et al., 2011; Salcher et al., 2011a; Eiler et al., 2012).
Here, we present an in-depth ecological assessment of members of the freshwater LD28 and PRD01a001B lineages in prealpine Lake Zurich, Switzerland. We used a multiphasic approach combining (i) high-resolution monitoring of their distribution, (ii) isolation of 127 pure cultures via dilution to extinction, (iii) genome sequencing of two representatives and (iv) laboratory experiments about their methylotrophic lifestyle and adaptation to cold water temperatures. Based on these analyses, we propose the establishment of a new genus with two species, that is, ‘Candidatus Methylopumilus planktonicus' and ‘Candidatus Methylopumilus turicensis'.
Materials and methods
Study site and sampling
Lake Zurich is a large, deep (136 m), prealpine, oligo-mesotrophic lake, characterized by persistent annual blooms of the toxic cyanobacterium Planktothrix rubescens (Posch et al., 2012). Water samples were taken every second week from 17 January 2008 to 14 December 2011 at the deepest part of the lake. Vertical profiles of temperature, conductivity, turbidity, oxygen and chlorophyll a content were recorded using a YSI multiprobe (Yellow Springs Instruments, model 6600, Yellow Springs, OH, USA) and a bbe fluoroprobe (TS-16-12, bbe Moldaenke GmbH, Schwentinental, Germany). The fluoroprobe was calibrated to distinguish different phytoplankton groups and P. rubescens according to the fluorescent spectra of their pigments (Beutler et al., 2002). Water samples were taken from 0, 5, 10, 20, 30, 40, 60, 80, 100 m depth, and additionally from the depth of recorded chlorophyll a maxima. Forty milliliters of water were fixed with formaldehyde (2% final concentration) for flow cytometric counts of bacteria, and 5–10 ml were fixed with paraformaldehyde (pH 7.4, 2% final concentration) for CARD-FISH (fluorescence in situ hybridization followed by catalyzed reporter deposition) analyses. Unfixed water samples for the isolation of bacteria and the preparation of media were transported to the laboratory within 15 min. Chemical parameters were determined in monthly intervals with standard techniques by the Zurich Water Supply Company (Zurich, Switzerland).
Abundances of microbes affiliated with LD28 and PRD01a001B
Total microbial abundances were determined by an inFlux V-GS cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA) equipped with an ultraviolet (355 nm) laser. All samples were stained with 4',6-diamidino-2-phenylindole (DAPI, 1 μg ml−1 final concentration), and weighted before and after measurements for sample volume quantification. Scatter plots of DAPI fluorescence vs 90° light scatter were analyzed with an in-house software (J Villiger, unpublished).
CARD-FISH with fluorescein-labeled tyramides was carried out as previously described (Sekar et al., 2003) with probes specifically detecting microbes affiliated with LD28 (probe LD28-1017; Salcher et al., 2011a) and a newly designed probe targeting the PRD01a001B lineage. Probe PRD-732 (5′-TCAGTATTAGTCCAGGGGGCTG-3′) and its competitor (5′-TCAGTATTAGSCCAGGGGGCTG-3′) was designed in ARB and tested following the workflow described in Salcher et al. (2011b). Highest stringency was obtained with 50% formamide in the hybridization buffer. CARD-FISH-stained samples were analyzed by fully automated high-throughput microscopy (Zeder and Pernthaler, 2009). Images were analyzed with the freely available image analysis software ACMEtool (technobiology.ch), and interfering autofluorescent cyanobacteria or debris particle were individually excluded from hybridized cells. At least 10 high-quality images and >1000 DAPI-stained bacteria were analyzed per sample. A lower limit for the quantification for FISH-stained cells (0.14% of all DAPI-stained particles) was applied as determined previously (Neuenschwander et al., 2015). Cell size estimations of LD28 bacteria were done on 12 samples (n=1225 cells) with the software LUCIA (Laboratory Imaging Prague, Prague, Czech Republic) following the workflow described in Posch et al. (2009). Samples corresponded to different LD28 population dynamics, that is, at times of population increase in spring (5 m depth, 09–24 April 2008), population decline in spring (5 m depth, 31 March–11 May 2009), population increase in autumn (5 m depth, 18 October–14 December 2011) and in the hypolimnion (80 m depth, 11 May 2009).
Isolation of planktonic bacteria affiliated with Methylophilaceae
A total of 46 isolation campaigns were carried out during 2010–2013. Unfixed water samples from 5 m depth were used for isolation of planktonic bacteria. Medium was mostly prepared from filtered and autoclaved lake water (0.2 μm pore sized polysulfone filters, Millipore, Billerica, MA, USA), amended with different types of carbon and nutrient sources (for details, see Supplementary Table S1). Two milliliter aliquots of medium were dispersed to 24-well plates (Thermo Fisher Scientific, Waltham, MA, USA) and inoculated with prefiltered (0.45 μm pore sized polycarbonate filters, Millipore) and diluted water samples. Total bacterial cell counts of inocula were determined by microscopy and samples were diluted to final inoculum sizes of approximately 1–10 cells well−1 (Supplementary Table S1). Twenty-four-well plates were incubated at close to in situ temperatures in a dark–light cycle (16:8 h) for 2–3 months. Growth of bacteria in individual wells was examined by pipetting 50 μl aliquots onto diagnostic glass slides (Thermo Fisher Scientific), which were air-dried at 45 °C, embedded into DAPI mix and inspected with a fluorescence microscope (Zeiss AxioImagerM.1, Carl Zeiss, Jena, Germany). Wells containing dense bacterial cultures were further split in 500 μl aliquots that were (i) propagated to fresh medium, (ii) amended with glycerol (30% final concentration) and stored at −80 °C, and (iii) pelleted by centrifugation (30 min, 16 000 × g) and used for DNA isolation, PCR and subsequent sequencing of the 16S ribosomal RNA genes. PCR was conducted with general bacterial primers GM3f and GM4r (Muyzer et al., 1995) and 1 μl template of either extracted DNA (GenElute Bacterial Genomic DNA Kit, Sigma-Aldrich, St Louis, MO, USA) or pelleted bacterial biomass subjected to 2–3 cycles of freeze thawing. PCR products were purified and sequenced with the ABI BigDye chemistry on an ABI 3130x Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) with primers GM3f, GM4r and GM1f (Muyzer et al., 1995). Partial sequences were assembled with the software DNA baser v3.5.0 (Heracle BioSoft, Lilienthal, Germany), aligned with the SINA web aligner (www.arb.silva.de) and imported into ARB (Ludwig et al., 2004) using the SILVA database SSU Ref 111 (Pruesse et al., 2007). A bootstrapped maximum likelihood tree (GTR-GAMMA model) of sequences of isolates affiliated with Methylophilaceae, their closest relatives and other described genera of Methylophilaceae was constructed on a dedicated web server (Stamatakis et al., 2005).
Growth characteristics and cell size of strain MMS-2-53
Strain MMS-2-53 was grown in triplicated setups of 40 ml sterile lake water amended with a mix of vitamins and amino acids (Supplementary Table S1) and different concentrations of methanol (0, 10 μM, 100 μM and 1 mM methanol) in Tissue Culture Flasks (25 cm2, TPP, Trasadingen, Switzerland) at 22 °C in the dark for 69 days. One milliliter subsamples were taken twice a week, fixed with formaldehyde and counted by flow cytometry (see above). Sizing of cells was conducted with a Zeiss AxioImagerM.1 microscope after DAPI staining and filtration (0.2 μm filters, Osmonics, Penang, Malaysia) with the software LUCIA (Posch et al., 2009). At least 100 individual cells per sample were subjected to image analysis (total n=4384).
Growth on methanol and methylamine and temperature preferences of strain MMS-2-53 were tested in triplicated setups of 40 ml sterile lake water amended with amino acids, vitamins and 1 mM methanol (variant A), or 1 mM methanol and 100 μM methylamine (variant B) in Tissue Culture Flasks (TPP) incubated in the dark at 6 °C, 13 °C, 18 °C and 22 °C for 6 weeks.
Genome sequencing and annotation of strains MMS-2-53 and MMS-10A-171
Whole-genome sequencing of strains MMS-2-53 and MMS-10A-171 was done after an initial multiple displacement amplification as cell yields and DNA concentrations were too low for direct sequencing. Unfixed cells of strain MMS-2-53 were sorted on a flow cytometer, lysed and amplified with either the Repli-G Single Cell Kit (Qiagen, Hilden, Germany; three parallel samples with 20 000–40 000 sorted cells) following the manufacturer's instructions or with a RepliPHI (epicenter, 14 samples with 5000 or 10 000 sorted cells) according to Dean et al. (2002) and Woyke et al. (2011). DNA extracts of strain MMS-10A-171 were amplified with the RepliPHI (11 samples). Multiple displacement amplification products were pooled for three and one individual libraries for strains MMS-2-53 and MMS-10A-171, respectively. 550-bp libraries were produced according to the manufacturer's instructions (TruSeq DNA PCR-Free LT Sample Preparation Kit, Illumina, San Diego, CA, USA) and 2 × 150-bp paired end sequences were generate on an Illumina MiSeq instrument (MCS versions 2.3.0.3 and 2.4.1.3, Illumina) with a 300-cycle MiSeq Reagent v2 kit (Illumina). Draft assemblies with A5-miseq (Coil et al., 2014) resulted in 6 and 38 contigs of strain MMS-2-53 and MMS-10A-171, respectively, whereof the largest 5 and 8 contigs (>5000 bp) were used for primer design. A total of 53 and 71 primers targeting contig ends and regions with a coverage of <10x were designed with the software Geneious 7.1.7 (www.geneious.com) and used for additional PCRs. Totals of 83 and 121 additional sanger sequences finally closed all gaps and poorly covered regions in the two genomes.
Gene calling and draft annotations were done with the Rapid Annotation using Subsystem Technology (RAST) server version 2.0 (Aziz et al., 2008) and the MicroScope web platform (Vallenet et al., 2013). Inconsistencies between the two methods and overlapping CDS were manually curated in ARTEMIS (Rutherford et al., 2000) and short CDS (<150 bp) without BLAST hits were discarded as artefacts. Homologies between genes of the two strains and their closest relatives, that is, methylophilales bacterium HTCC2181 (Giovannoni et al., 2008), Methylotenera mobilis JLW8, Methylotenera versatilis 301, Methylovorus glucosetrophus SIP3-4 (Lapidus et al., 2011), Methylophilus methylotrophus ATCC 53528 (NZ_ARJW00000000), Methylobacillus flagellatus KT (Chistoserdova et al., 2007) and a typical freshwater representative (Polynucleobacter necessarius subsp. asymbioticus QLW-P1DMWA-1; Meincke et al., 2012) were identified with the MicroScope web platform using protein identity cutoffs of ⩾30% and ⩾70% (minLrap ⩾0.8; maxLrap ⩾0). Reconstructions of methylotrophy pathways were modeled according to their closest relatives using comparative genomics tools of the MicroScope platform (Vallenet et al., 2013). Digital DNA–DNA hybridization of the two genomes and closely related strains was done via the Genome-to-Genome Distance Calculator using the GGDC 2.0 BLAST+ alignment method (Meier-Kolthoff et al., 2013). Annotated genome sequences have been deposited at EMBL (sequence accessions LN827929 and LN794158).
Statistical analyses
Before statistical analyses, data of relative abundances (% of DAPI) were arcsine transformed and cell numbers and physicochemical variables were log(x+1) transformed to better approximate a normal distribution. Redundancy analysis was used for determining the effects of environmental data on microbes affiliated with LD28 and PRD01a001B in Lake Zurich. The significance of added variables was tested by a Monte Carlo permutation test (500 permutations), and only variables with a highly significant correlation (P<0.0001) were included in the analysis. Significant differences in the growth of isolate MMS-2-53 according to different treatments were tested by one-way analysis of variances (Tukey post-hoc tests). All analyses were performed with the Microsoft EXCEL add-in program XLSTAT (www.xlstat.com).
Results
Spatio-temporal distribution of Methylophilaceae in Lake Zurich in relation to environmental parameters
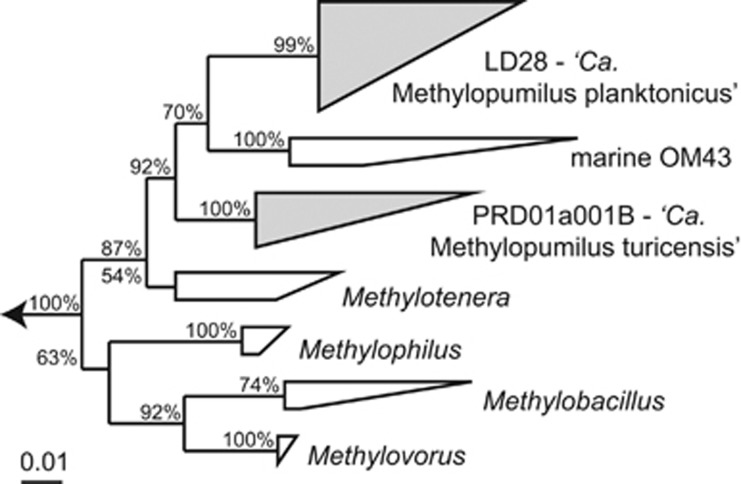
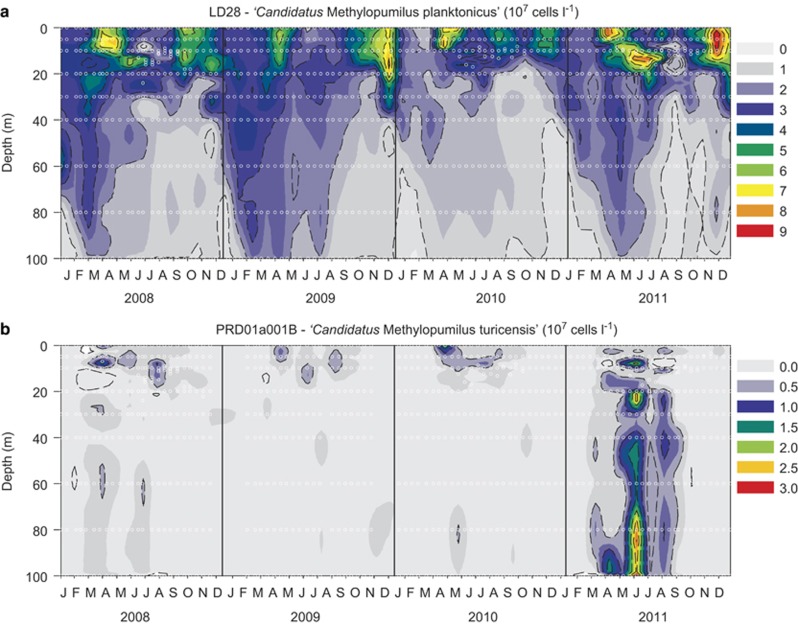
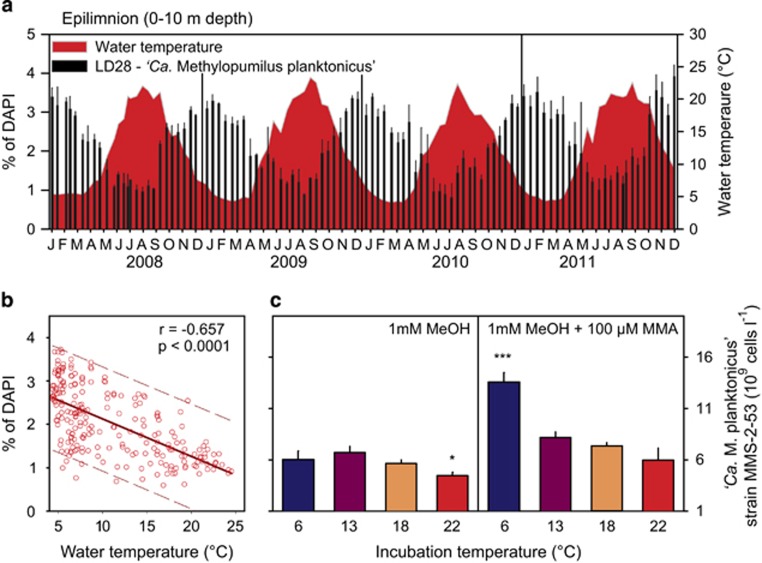
Microbes affiliated with LD28 (see Figure 1 for phylogenetic positioning) were present in all analyzed samples (n=992) in varying numbers. Two seasonal maxima (up to 11.3 × 107 cells l−1 or 4.1% of DAPI) in spring and autumn–winter were discernable in all four analyzed years (Figure 2a,Supplementary Figure S1). Bacteria of the PRD01a001B lineage were below the limit of detection (<0.14% of DAPI-stained cells) in most of our samples (n=642). However, a distinct maximum of up to 4% of DAPI-stained cells (3.2 × 107 cells l−1) was found in the hypolimnion in summer 2011 (Figure 2b, Supplementary Figure S1). This abundance peak lasted for several weeks between April and August 2011, but was absent in the other years, where maxima of 1% or 2.4 × 107 cells l−1 were restricted to the epilimnion in spring.
Figure 1.
Collapsed maximum likelihood tree of the 16S rDNA of Methylophilaceae. Lineages LD28 (‘Ca. Methylopumilus planktonicus) and PRD01a001B (‘Ca. Methylopumilus turicensis) are shown in gray. The scale bar at the bottom applies to 1% sequence divergence.
Figure 2.
Seasonality of abundances (107 cells l−1) of microbes affiliated with LD28 (‘Ca. Methylopumilus planktonicus'. (a) and PRD01a001B (‘Ca. Methylopumilus turicensis'. (b) in Lake Zurich, Switzerland, during the investigation period 2008–2011. Sampling dates and depths are displayed as white circles.
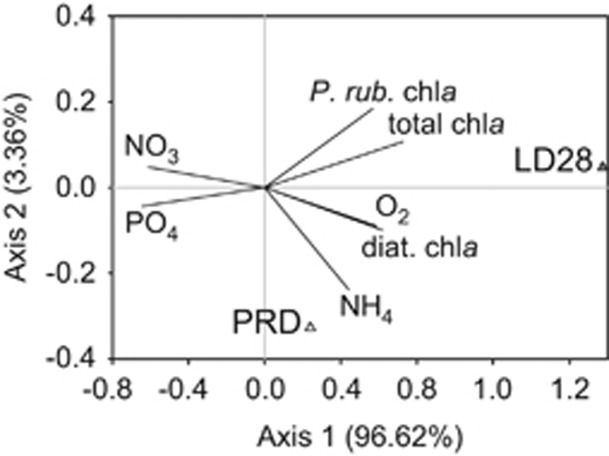
Redundancy analysis resulted in a significant explanation of variability in both lineages (Figure 3). Axis 1 correlated mostly with chlorophyll a concentrations (total chlorophyll a and chlorophyll a associated with the cyanobacterium Planktothrix rubescens and diatoms), nitrate, phosphate and ammonium, whereas the second axis separated chlorophyll a of P. rubescens from that of diatoms, as well as nitrate from phosphate and ammonium (see Supplementary Figure S2 for details of all environmental parameters). Population sizes of both lineages were clearly separated from each other; LD28 methylotrophs were more related to total and P. rubescens chlorophyll a (Pearson's correlation coefficient R=0.729 and R=0.611, respectively, P<0.0001), whereas PRD01a001B bacteria were more related to ammonium and diatom chlorophyll a (R=0.353 and R=0.309, respectively, P<0.0001). Nitrate and phosphate was negatively related to both methylotrophs (R=−0.618 and R=−0.233 for nitrate and R=−0.655 and R=−0.160 for phosphate, correlation to LD28 and PRD01a001B, respectively, P<0.0001).
Figure 3.
Redundancy analysis of environmental parameters explaining the variability in cell numbers of microbes affiliated with LD28 and PRD01a001B in Lake Zurich. LD28, abundances of LD28; PRD, abundances of PRD01a001B; total chla, total chlorophyll a; P.rub. chla, chlorophyll a associated with Planktothrix rubescens; diat. chla, chlorophyll a associated with diatoms; O2, oxygen concentrations; NH4, ammonium concentrations; NO3, nitrate concentrations; PO4, phosphate concentrations.
Isolation of Methylophilaceae from Lake Zurich
2077 of a total of 6507 inspected wells contained dense cultures, 788 of these appeared to be monocultures, and 115 and 12 of these were pure cultures affiliated with LD28 and PRD01a001B, respectively (Figure 1, Supplementary Figures S3, S4, Table S1). All isolates within each of the two lineages were very closely related to each other according to the 16S ribosomal RNA genes (98.6–100% and 99.7–100% sequence similarity for LD28 and PRD01a001B, respectively), but they were distantly related to the strains of the other genotype (95.2–96.5%) and to so far described taxa of Methylophilaceae (<95.6% Table 1). Both clades and the marine OM43 formed a monophyletic sister lineage to Methylotenera, with PRD01a001B branching more deeply from LD28 and OM43 (Figure 1, Supplementary Figures S3 and S4). One isolate of LD28 (strain MMS-2-53) and PRD01a001B (strain MMS-10A-171) were subjected to genome sequencing (Table 2). Digital whole-genome DNA–DNA hybridization of our isolates and genome sequenced Methylophilaceae resulted in very low similarities (<26.2% Table 1). Thus, these isolates, together with the marine OM43, likely represent a new genus within Methylophilaceae. We therefore propose to establish a new ‘Candidatus' genus with two new species, that is, ‘Candidatus Methylopumilus planktonicus' for isolates affiliated with LD28 (strain MMS-2-53) and ‘Candidatus Methylopumilus turicensis' for isolates affiliated with PRD01a001B (strain MMS-10A-171).
Table 1. 16S rDNA similarities, digital DNA–DNA hybridization, and proportions of shared protein-coding genes (⩾70% protein similarity) of the two strains and different members of Methylophilaceae.
|
16S rDNA |
DNA–DNA hyb. |
Shared genes (%) |
||||
|---|---|---|---|---|---|---|
| MMS-2-53 | MMS-10A-171 | MMS-2-53 | MMS-10A-171 | MMS-2-53 | MMS-10A-171 | |
| ‘Ca. M. planktonicus‘ MMS-2-53 | 96.5 | 22.5 | 12.8 | |||
| ‘Ca. M. turicensis' MMS-10A-171 | 96.5 | 22.5 | 12.4 | |||
| HTCC2181 (marine OM43) | 95.3 | 96.3 | 17.1 | 23.4 | 10.9 | 8.0 |
| Methylotenera mobilis JLW8 | 95.2 | 95.4 | 24.5 | 21.7 | 10.0 | 20.0 |
| Methylotenera versatilis 301 | 94.7 | 95.6 | 24.4 | 20.1 | 8.6 | 17.9 |
| Methylophilus methylotrophus ATCC 53528 | 93.7 | 93.8 | 21.3 | 19.9 | 8.4 | 14.2 |
| Methylobacillus flagellatus KT | 94.2 | 94.2 | 26.2 | 18.5 | 8.0 | 17.3 |
| Methylovorus glucosetrophus SIP3-4 | 93.9 | 94.4 | 25.1 | 20.2 | 8.4 | 19.8 |
Abbreviation: rDNA, ribosomal DNA.
Table 2. Genome statistics of strains MMS-2-53 and MMS-10A-171.
| ‘Ca. M. planktonicus' MMS-2-53 | ‘Ca. M. turicensis' MMS-10A-171 | |
|---|---|---|
| Genome size (bp) | 1 356 428 | 1 754 988 |
| Sequencing coverage | 481.6x | 191.6x |
| Organization | 1 chromosome | 1 chromosome |
| GC content (%) | 36.97 | 44.52 |
| Total genes | 1434 | 1806 |
| RNA genes | 45 | 49 |
| rRNA operons | 1 | 2 |
| Protein-coding genes | 1389 | 1757 |
| Mean-coding sequence length (bp) | 917.2 | 923.9 |
| Mean intergenic length (bp) | 69.8 | 83.2 |
| Protein-coding density (%) | 93.56 | 92.29 |
| EMBL sequence accession number | LN827929 | LN794158 |
Abbreviation: rRNA, ribosomal RNA.
Growth of strain MMS-2-53 with different concentrations of methanol
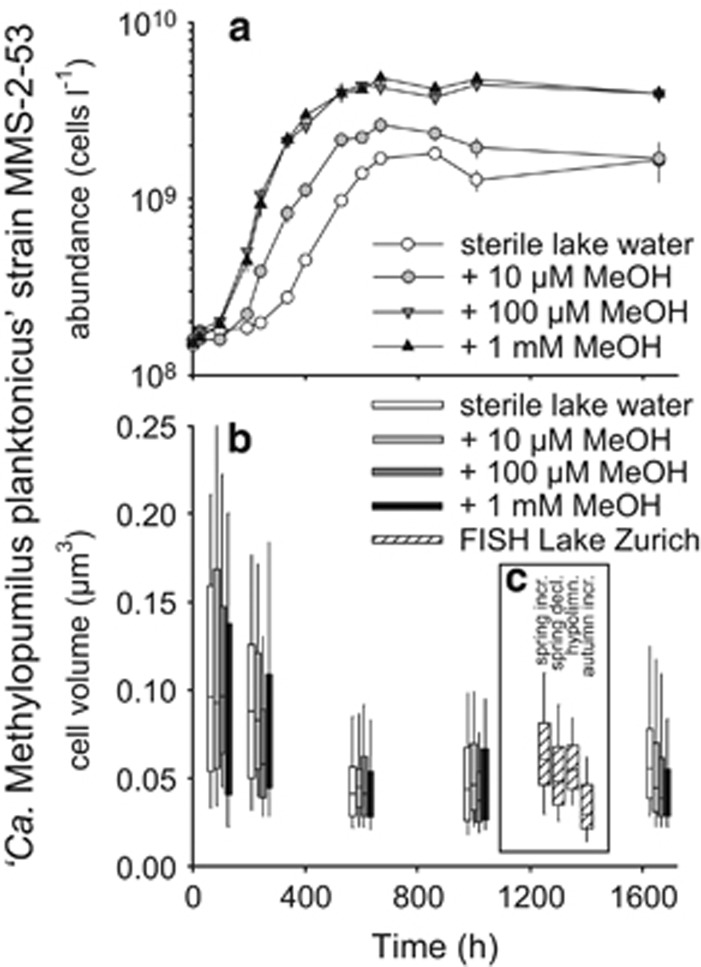
Cultivation of strain MMS-2-53 was only successful in sterile lake water, whereas no growth was visible on agar plates (neither prepared with lake water nor with conventional R2A, NSY or K-medium agar plates amended with methanol) or in conventional liquid media (R2, NSY, K-medium, inorganic basal medium amended with methanol). Strain MMS-2-53 reached maximal cell densities of 1.8 × 109 cells l−1 in sterile lake water after 36 days, corresponding to a maximal growth rate of μmax=0.16 d−1 (Figure 4a). Significantly higher growth yields (4.9 × 109 cells l−1) and growth rates (μmax=0.37 d−1) were achieved by the addition of methanol at different concentrations. Substrate saturation was apparently reached at 100 μM methanol, which yielded identical growth rates as the addition of 1 mM methanol. A precise calculation of Km or Ks was hampered by the unknown concentration of methanol in the lake water. The cell sizes of strain MMS-2-53 were very small (median=0.041 μm3, n=3552) during stationary growth phase (Figure 4b), however, volumes significantly increased during exponential phase (0.075 μm3, n=832). No significant differences in cell volume were recorded at different concentrations of methanol. Interestingly, cell volumes similar to those of stationary phase cultures were also obtained from CARD-FISH-stained preparations of LD28 microbes in Lake Zurich during population decline in the epilimnion in spring (0.048 μm3, n=416) and in the hypolimnion (0.055 μm3, n=83), whereas cells were even significantly smaller in autumn samples (0.03 μm3, n=327). LD28 growing to population maxima in the epilimnion in spring were larger (0.061 μm3, n=399) and in between the values obtained from cultures (Figure 4c).
Figure 4.
(a) Growth (cells l−1) of isolate MMS-2-53 (‘Ca. Methylopumilus planktonicus') in sterile lake water containing different amounts of methanol. (b, c) Cell volumes (μm3) of strain MMS-2-53 during growth and in situ in Lake Zurich at different depths and during different periods of the year (c, insert). spring incr., LD28 population increase in 5 m depth in spring; spring decl., population decline in 5 m depth in spring; hypolimn., LD28 in the hypolimnion (80 m depth); autumn incr., LD28 population increase in 5 m depth in autumn.
Effects of water temperature and additional C1 substrates
The relative abundances of LD28 microbes in Lake Zurich showed recurrent maxima in winter and were negatively related to water temperature in the upper water layers (epilimnion, 0–10 m depth, Figures 5a and b). To test for direct effects of water temperature and additional C1 substrates, we incubated strain MMS-2-53 with 1 mM methanol at different temperatures and in the presence or absence of 100 μM of methylamine (Figure 5c). There was only a slight effect of incubation temperatures on abundances when cells were grown with 1 mM methanol only (4.5–6.7 × 109 cells l−1). The addition of methylamine increased the cell yield by 1.2–2-fold and led to significantly higher cell numbers at 6 °C (12.8 × 109 cells l−1) when methylamine served as second C1 substrate.
Figure 5.
(a) Relative abundance of LD28 in the epilimnion (that is, 0–10 m depth) of Lake Zurich (% of DAPI) and water temperature (°C). (b) Correlation of relative abundances of LD28 to water temperature. (c) Abundance yield (109 cells l−1) of isolate MMS-2-53 grown for 6 weeks at different incubation temperatures and after addition of 100 μM methylamine.
Insight into the genomes of planktonic freshwater Methylophilaceae
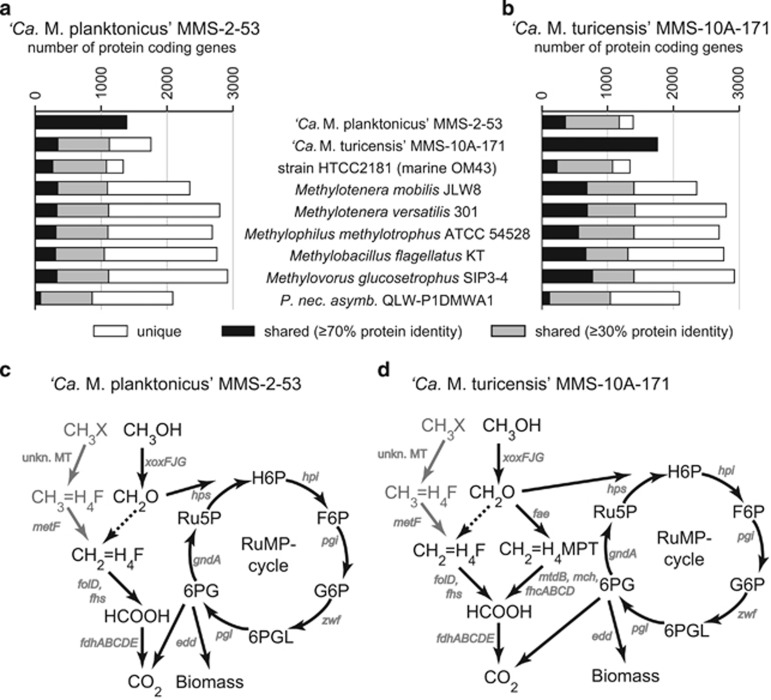
The sizes of the two newly sequenced complete genomes of strains MMS-2-53 and MMS-10A-171 were 1.36 and 1.75 Mb with a GC content of 37% and 44%, respectively (Table 2, Supplementary Figures S5 and S6). Only very low proportions of the 1389 and 1757 protein-coding genes were shared with other Methylophilaceae (<20%) at a protein similarity cutoff of ⩾70% (Figure 6a, Table 1). Nevertheless, we identified several striking similarities between our isolates and the marine OM43 lineage. First, both isolates encode genes for a special variant of rhodopsins (xanthorhodopsins), which are closely related to those in members of the OM43 lineage (protein similarities: 32.4–62.7%) and in Salinibacter ruber. None of the other so far sequenced Methylophilaceae possess rhodopsins. Both microbes were obligate methylotrophs, as genomic analyses identified an incomplete tricarboxylic acid cycle (the E1 and E2 subunits of the α-ketoglutarate dehydrogenase complex are missing), the biochemical basis of obligate methylotrophy. Moreover, no membrane transporters for sugars or amino acids were encoded in the genomes.
Figure 6.
(a, b) Number of protein-coding genes shared between ‘Ca. Methylopumilus planktonicus' MMS-2-53 (a) or ‘Ca. Methylopumilus turicensis' MMS-10A-171 (b) and genome sequenced members of Methylophilaceae and a typical freshwater microbe (Polynucleobacter necessarius subsp. asymbioticus QLW-P1DMWA-1). Protein identity cutoffs of ⩾30% and ⩾70% (minLrap⩾0.8) were applied. (c, d) Reconstruction of methylotrophic pathways of ‘Ca. Methylopumilus planktonicus' MMS-2-53 (c) and ‘Ca. Methylopumilus turicensis' MMS-10A-171 (d). Genes involved in methylotrophy are given in gray (see Supplementary Table S2 for more information). Dashed lines indicate spontaneous reactions, gray lines indicate possible pathways for CH3X oxidation. unkn. MT, unknown methyltransferase; CH3OH, methanol; CH3X, other C1 compounds, for example, methylamine; CH2O, formaldehyde; HCOOH, formate; H4F, tetrahydrofolate; H4MPT, tetrahydromethanopterin; H6P; hexulose-6-phosphate; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phospho-gluconate; Ru5P, ribulose-5-phosphate; RuMP cycle, ribulose monophosphate cycle.
Metabolic reconstructions of methylotrophic pathways, however, revealed several differences between the two strains and other Methylophilaceae (Figure 6b, Supplementary Table S2). Both isolates lacked genes encoding MDH but instead possessed genes xoxFJG, homologs of MDH, as well as PQQ biosynthesis genes (mxaRSACKL and pqqBCDEFG) that are required as a cofactor of MDH. Genes for a formaldehyde dehydrogenase were absent in both isolates, but all genes of the tetrahydrofolate (H4F) pathway for formaldehyde oxidation were present. The genomic organization of these genes involved in methanol and formaldehyde oxidation was highly similar to marine OM43. MMS-2-53 also lacked genes for the tetrahydromethanopterin (H4MPT) pathway of formaldehyde oxidation, while MMS-10A-171 encoded this pathway. Genes neither for the N-methylglutamate pathway nor for a methylamine dehydrogenase for methylamine oxidation were identified in both isolates. Formate is oxidized to CO2 with a formate dehydrogenase, with all genes organized serially in both strains (fdhABCDE). Further, both isolates encoded all genes of the ribulose monophosphate cycle and the Entner–Doudoroff pathway for the assimilation of carbon from C1 compounds.
Discussion
Phytoplankton as major sources of C1 substrates support the growth of planktonic freshwater Methylophilaceae
Our high-resolution sampling campaign in Lake Zurich (n=992 individual samples, Figure 2,Supplementary Figures S1 and S2) helped to identify potential environmental drivers for planktonic freshwater Methylophilaceae. The striking annually recurrent maxima of LD28 microbes in spring and autumn–winter seem to be mainly determined by the availability of C1 substrates released by phytoplankton. Seasonal maxima of these Methylophilaceae in late spring and autumn–winter of up to 3.5% of pyrosequencing reads were also reported from Lake Erken, Sweden (Eiler et al., 2012). However, the latter study found negative correlations to chlorophyll a, whereas we observed the opposite, which may be a consequence of the higher resolution of our data set (Figures 2 and 3). The same authors also reported an association of LD28 microbes with cyanobacterial blooms in several Swedish lakes (Eiler and Bertilsson, 2004), which is in accordance to our results from a single system (Figures 2 and 3). A positive correlation of LD28 to the chlorophyll a of the dominant primary producer of Lake Zurich, the cyanobacterium Planktothrix rubescens, has been reported before (Salcher et al., 2011a). Moreover, an operational taxonomic unit affiliated with LD28 was exclusively present in the depth layer of maximal P. rubescens densities in this lake (Van den Wyngaert et al., 2011). Methylophilaceae were also highly abundant in mesocosms containing the cyanobacterial toxin microcystin (Mou et al., 2013). Marine Methylophilaceae were enriched in seawater chemostats amended with diatom and cyanobacterial dissolved organic matter only after six generation times, but not after three, pointing at slow growth and a potential cross-feeding of C1 compounds that were released by other microbes (Landa et al., 2014). Slow growth is also in accordance with our experimental data from isolate MMS-2-53 (Figure 4). Interestingly, strain HTCC2181 of the marine OM43 lineage grows much faster than strain MMS-2-53 (Giovannoni et al., 2008; Halsey et al., 2011), although it shares several metabolic and genomic features with LD28 (for example, comparable cell yields, synergistic effects of a second C1 substrate, genome size, GC content and methylotrophic pathways).
Nutrient acquisition by LD28 and PRD01a001B
Both genotypes showed a negative correlation to nitrate and phosphate concentrations (Figure 3,Supplementary Figure S2), which is partly in agreement with earlier studies (Salcher et al., 2011a; Eiler et al., 2012). High-affinity phosphate transporters (Pst) for scavenging phosphorus at low concentrations were encoded in both genomes. Therefore, these microbes seem to be well adapted to phosphorus limitation. Some freshwater Methylophilaceae showed chemotaxis toward ammonium and phosphate (Dennis et al., 2013), although the taxonomic resolution in that study was not sufficient to discriminate between different lineages. We found a positive correlation to ammonium (Figure 3), as well as several genes encoding ammonium transporters in both genotypes. Thus, it seems likely that nitrogen demand is covered by ammonium uptake.
It is unclear what triggered the high abundances of microbes affiliated with PRD01a001B in summer 2011 (Figure 2, Supplementary Figure S1), but our observation nevertheless illustrates the importance of long-term time series for understanding the ecology and occurrence patterns of these slowly growing methylotrophs in lakes. Generally, the PRD01a001B lineage of Methylophilaceae is less explored than LD28, however, they were detected in a wide range of freshwater and oligosaline lakes (Wu et al., 2006).
Psychrophilic methylotrophs?
Negative correlations of planktonic freshwater Methylophilaceae and water temperature have been reported previously (Salcher et al., 2008, 2011a). However, so far it was not clear if these patterns were triggered by water temperature per se or by other co-varying factors such as higher nutrient availability in deeper water layers and during mixis events. Here, we experimentally demonstrate a relationship between incubation temperature and cell yield of strain MMS-2-53 (Figure 5). Although growth was also observed at higher temperatures (22 °C), bacteria reached significantly lower densities compared with 6 °C. Thus, we propose that these microbes are psychrophilic, that is, adapted to low temperatures. Interestingly, this psychrophilic aspect of growth seems to be enhanced by the availability of an additional C1 source, methylamine. Most of our strains were isolated during autumn–spring (65%), while in summer, when water temperatures reached >20 °C, we were less successful (Supplementary Table S1). However, further studies are needed to test for strain-specific temperature optima, as even closely related isolates can profoundly differ in their thermal adaptation (Hahn et al., 2014; Yung et al., 2014).
Genomic similarities and differences within Methylophilaceae
The very small genome size and low GC content of strain MMS-2-53 (Table 2, Supplementary Figure S5) is comparable to the streamlined genomes of marine methylotrophs HTCC2181 and HIMB624 of the OM43 clade (Giovannoni et al., 2008; Huggett et al., 2012). Strain MMS-10A-171 has a slightly larger genome, a higher GC content and shares more protein-coding genes with Methylotenera, Methylobacillus and Methylovorus (Table 2, Figure 6,Supplementary Figure S6). Both strains and members of the marine OM43 lineage encode similar types of rhodopsins, light driven proton pumps that are tuned to green light and thus, may be beneficial in productive conditions in lakes (Giovannoni et al., 2008). Moreover, our isolates were obligate methylotrophs, as already suggested earlier because LD28 bacteria did not incorporate amino acids or sugars in microautoradiography assays (Salcher et al., 2008, 2013). Both isolates possess genes encoding a special variant of MDH (xoxF) that most likely require the rare earth metal lanthanum as cofactor (Hibi et al., 2011; Beck et al., 2014). Methylotrophy is a very modular metabolic system; formaldehyde in particular can be oxidized via several different pathways, either by a formaldehyde dehydrogenase, via the H4F pathway or the H4MPT pathway (Chistoserdova 2011). Both isolates encode genes for the H4F pathway. Strain MMS-10A-171 also encodes genes for the H4MPT pathway, whereas it is absent in MMS-2-53 and in OM43 microbes (Giovannoni et al., 2008; Huggett et al., 2012; Figure 6b). Isolate MMS-2-53 grew to higher densities in the presence of methylamine as second C1 substrate, although we could not detect genes encoding methylamine oxidation (Figures 5 and 6). However, alternative routes for C1 substrate entry to the oxidative H4F pathway have been proposed for the marine OM43 (Halsey et al., 2011). One gene (metF) that may facilitate methylamine oxidation together with a so far unknown methyltransferase is present in both analyzed genomes (Figure 6, Supplementary Table S2); thus, these microbes may be able to oxidize also other C1 compounds than methanol. Methanol is mainly used by microbes for obtaining energy (up to 97% in oligotrophic regions of oceans) but can also contribute up to 50% to bacterial carbon demand (Dixon et al., 2011a, 2013). In the presence of other C1 compounds, methanol may thus be directly channeled to the ribulose monophosphate cycle for biomass production, whereas other C1 compounds may fuel energy demands. However, gene expression analyses are needed to verify this hypothesis and identify genes for methylamine oxidation.
Putative description of a new genus of Methylophilaceae
The low degree of sequence similarities on the level of 16S ribosomal RNA genes (<95.6%), digital whole-genome DNA–DNA hybridization (<26.2%) and shared protein-coding genes (<20%, Table 1) suggests that the here described isolates as well as the marine OM43 clade are not affiliated with any described genus of Methylophilaceae. Therefore, we propose a new provisional genus with two new species, that is, ‘Candidatus Methylopumilus planktonicus' for LD28 with MMS-2-53 as type strain and ‘Candidatus Methylopumilus turicensis' for PRD01a001B with MMS-10A-171 as type strain. Currently, additional analyses required for a formal species description, such as detailed metabolic analyses and cellular phospholipid fatty acids profiling, are hampered by the high amounts of biomass required for such assays. Moreover, we still face difficulties in composing a completely synthetic growth medium; so far, both isolates only grow well in sterile lake water.
Conclusion
We provide first insight in the ecology and unique lifestyle of an important group of so far uncultivated ubiquitous freshwater bacteria that occupy a distinct niche in the pelagic zone of lakes. This was achieved by combining high-resolution environmental monitoring, isolation of pure cultures, metabolic tests and genome sequencing of isolates. The two investigated strains remarkably differed in genome size (1.36 vs 1.76 Mb) and methylotrophic pathways (Figure 6,Table 2, Supplementary Table S2). It is conceivable that a common ancestor of our freshwater strains and the marine OM43 lineage already underwent gene streamlining (that is, loosing several genes encoding methylotrophic pathways), but also gained new genes via horizontal gene transfer (encoding, for example, rhodopsins). As genome reduction is more pronounced in strain MMS-2-53 than in MMS-10A-171, we believe that these microbes are ideal model organisms for future studies regarding gene loss and genome streamlining.
Acknowledgments
We thank captain E Loher for help during sampling and the Zurich Water Supply (WVZ) for the provision of limnochemical data. Illumina library preparation and sequencing was done at the Genetic Diversity Centre (GDC), ETH Zurich. This study was supported by the Swiss National Science Foundation (SNF, Project 31EE30-132771) as part of the European Science Foundation EUROCORES Programme EuroEEFG.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DAC, McTaggart TL, Setboonsarng U, Vorobev A, Kalyuzhnaya MG, Ivanova N et al. (2014). The expanded diversity of Methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLoS One 9: e102458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler M, Wiltshire K, Meyer B, Moldaenke C, Lüring C, Meyerhöfer M et al. (2002). A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynthesis Res 72: 39–53. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Lapidus A, Han C, Goodwin L, Saunders L, Brettin T et al. (2007). Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. J Bacteriol 189: 4020–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. (2011). Modularity of methylotrophy, revisited. Environ Microbiol 13: 2603–2622. [DOI] [PubMed] [Google Scholar]
- Coil D, Jospin G, Darling AE. (2014), A5-miseq: an updated pipeline to assemble microbial genomes from illumina miseq data. arXiv preprint1401.5130. [DOI] [PubMed]
- Connon SA, Giovannoni SJ. (2002). High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68: 3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang LH, Wu XH, Faruqi AF, Bray-Ward P et al. (2002). Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA 99: 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PG, Seymour J, Kumbun K, Tyson GW. (2013). Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J 7: 1661–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JL, Beale R, Nightingale PD. (2011. a). Rapid biological oxidation of methanol in the tropical Atlantic: significance as a microbial carbon source. Biogeosci Discuss 8: 3899–3921. [Google Scholar]
- Dixon JL, Beale R, Nightingale PD. (2011. b). Microbial methanol uptake in northeast Atlantic waters. ISME J 5: 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JL, Sargeant S, Nightingale PD, Colin Murrell J. (2013). Gradients in microbial methanol uptake: productive coastal upwelling waters to oligotrophic gyres in the Atlantic Ocean. ISME J 7: 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler A, Bertilsson S. (2004). Comparison of freshwater bacterial communities associated with cyanobacterial blooms in four swedish lakes. Environ Microbiol 6: 1228–1243. [DOI] [PubMed] [Google Scholar]
- Eiler A, Heinrich F, Bertilsson S. (2012). Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J 6: 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges AA, El-Swais H, Craig SE, Li WKW, Walsh DA. (2014). Metaproteomic analysis of a winter to spring succession in coastal northwest Atlantic Ocean microbial plankton. ISME J 8: 1301–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Booth M, Moran MA. (2013). Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J 7: 281–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782. [DOI] [PubMed] [Google Scholar]
- Govorukhina NI, Trotsenko YA. (1991). Methylovorus, a new genus of restricted facultatively methylotrophic bacteria. Int J Syst Bacteriol 41: 158–162. [Google Scholar]
- Hahn MW, Koll U, Jezberová J, Camacho A. (2014). Global phylogeography of pelagic Polynucleobacter bacteria: restricted geographic distribution of subgroups, isolation by distance, and influence of climate. Environ Microbiol 17: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey KH, Carter AE, Giovannoni SJ. (2011). Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ Microbiol 14: 630–640. [DOI] [PubMed] [Google Scholar]
- Hanson B, Hewson I, Madsen E. (2014). Metaproteomic survey of six aquatic habitats: discovering the identities of microbial populations active in biogeochemical cycling. Microb Ecol 67: 520–539. [DOI] [PubMed] [Google Scholar]
- Heikes BG, Chang W, Pilson MEQ, Swift E, Singh HB, Guenther A et al. (2002). Atmospheric methanol budget and ocean implication. Global Biogeochem Cycles 16: 1133. [Google Scholar]
- Hibi Y, Asai K, Arafuka H, Hamajima M, Iwama T, Kawai K. (2011). Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J Biosci Bioengineer 111: 547–549. [DOI] [PubMed] [Google Scholar]
- Huggett M, Hayakawa D, Rappe M. (2012). Genome sequence of strain HIMB624, a cultured representative from the OM43 clade of marine Betaproteobacteria. Stand Genomic Sci 6: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins O, Byrom D, Jones D. (1987). Methylophilus - a new genus of methanol-utilizing bacteria. Int J Syst Bacteriol 37: 446–448. [Google Scholar]
- Kalyuzhnaya MG, Bowerman S, Lara JC, Lidstrom ME, Chistoserdova L. (2006). Methylotenera mobilis gen. nov., sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int J Syst Evol Microbiol 56: 2819–2823. [DOI] [PubMed] [Google Scholar]
- Landa M, Cottrell MT, Kirchman DL, Kaiser K, Medeiros PM, Tremblay L et al. (2014). Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ Microbiol 16: 1668–1681. [DOI] [PubMed] [Google Scholar]
- Lapidus A, Clum A, LaButti K, Kaluzhnaya MG, Lim S, Beck DAC et al. (2011). Genomes of three methylotrophs from a single niche reveal the genetic and metabolic divergence of the Methylophilaceae. J Bacteriol 193: 3757–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucl Acid Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J, Auch A, Klenk H-P, Goker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meincke L, Copeland A, Lapidus A, Lucas S, Berry K, Glavina Del Rio T et al. (2012). Complete genome sequence of Polynucleobacter necessarius subsp. asymbioticus type strain (QLW-P1DMWA-1T). Stand Genomics 6: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne PJ, Riemer DD, Zika RG, Brand LE. (1995). Measurement of vertical distribution of isoprene in surface seawater, its chemical fate, and its emission from several phytoplankton monocultures. Marine Chem 48: 237–244. [Google Scholar]
- Mopper K, Zhou X, Kieber RJ, Kieber DJ, Sikorski RJ, Jones RD. (1991). Photochemical degradation of dissolved organic carbon and its impact on the oceanic carbon cycle. Nature 353: 60–62. [Google Scholar]
- Morris RM, Longnecker K, Giovannoni SJ. (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8: 1361–1370. [DOI] [PubMed] [Google Scholar]
- Mou X, Lu X, Jacob J, Sun S, Heath R. (2013). Metagenomic identification of bacterioplankton taxa and pathways involved in microcystin degradation in Lake Erie. PLoS One 8: e61890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Teske A, Wirsen C, Jannasch H. (1995). Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164: 165–172. [DOI] [PubMed] [Google Scholar]
- Neuenschwander SM, Pernthaler J, Posch T, Salcher MM. (2015). Seasonal growth potential of rare lake water bacteria suggest their disproportional contribution to carbon fluxes. Environ Microbiol 17: 781–795. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. (2011). A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol R 75: 14–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen B, Reveilliez J-P, Mary I, Ravet V, Bronner G, Mangot J-F et al. (2011). Diversity and dynamics of free-living and particle-associated Betaproteobacteria and Actinobacteria in relation to phytoplankton and zooplankton communities. FEMS Microbiol Ecol 77: 461–476. [DOI] [PubMed] [Google Scholar]
- Posch T, Franzoi J, Prader M, Salcher MM. (2009). New image analysis tool to study biomass and morphotypes of three major bacterioplankton groups in an alpine lake. Aquat Microb Ecol 54: 113–126. [Google Scholar]
- Posch T, Koster O, Salcher MM, Pernthaler J. (2012). Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nat Climate Change 2: 809–813. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acid Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Zeder M, Psenner R, Posch T. (2008). Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ Microbiol 10: 2074–2086. [DOI] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Frater N, Posch T. (2011. a). Vertical and longitudinal distribution patterns of differnt bacterioplankton populations in a canyon-shaped, deep prealpine lake. Limnol Oceanogr 56: 2027–2039. [Google Scholar]
- Salcher MM, Pernthaler J, Posch T. (2011. b). Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria 'that rule the waves' (LD12). ISME J 5: 1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher MM, Posch T, Pernthaler J. (2013). In situ substrate preferences of abundant bacterioplankton populations in a prealpine freshwater lake. ISME J 7: 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. (2003). An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol 69: 2928–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar R, Fuchs BM, Amann R, Pernthaler J. (2004). Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol 70: 6210–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth JM, Keller MD. (1989). Methylaminotrophic bacteria in xenic nanoalgal cultures: incidence, significance, and role of methylated algal osmoprotectants. Biol Oceanograph 6: 383–395. [Google Scholar]
- Song J, Oh H-M, Jang-Cheon C. (2009). Improved culturability of SAR11 strains in dilution-to-extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbiol Lett 295: 141–147. [DOI] [PubMed] [Google Scholar]
- Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF et al. (2011). Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5: 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. (2005). RAxML-II: a program for sequential, parallel and distributed inference of large phylogenetic. Concurr Comput-Pract Exp 17: 1705–1723. [Google Scholar]
- Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A et al. (2013). MicroScope - an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucl Acid Res 41: D636–D647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Wyngaert S, Salcher MM, Pernthaler J, Zeder M, Posch T. (2011). Quantitative dominance of seasonally persistent filamentous cyanobacteria (Planktothrix rubescens in the microbial assemblages of a temperate lake. Limnol Oceanogr 56: 97–109. [Google Scholar]
- Vorholt JA. (2012). Microbial life in the phyllosphere. Nat Rev Microbiol 10: 828–840. [DOI] [PubMed] [Google Scholar]
- Woyke T, Sczyrba A, Lee J, Rinke C, Tighe D, Clingenpeel S et al. (2011). Decontamination of MDA reagents for single cell whole genome amplification. PLoS One 6: e26161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zwart G, Schauer M, Kamst-van AMP, Hahn M. (2006). Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan plateau, China. Appl Environ Microbiol 72: 5478–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy JR, Weaver TL. (1977). Methylobacillus: a new genus of obligately methylotrophic bacteria. Int J Syst Bacteriol 27: 247–255. [Google Scholar]
- Yung C-M, Vereen MK, Herbert A, Davis KM, Yang J, Kantorowska A et al. (2014). Thermally adaptive tradeoffs in closely-related marine bacterial strains. Environ Microbiol: doi:10.1111/1462-2920.12714. [DOI] [PubMed]
- Zeder M, Pernthaler J. (2009). Multispot live-image autofocusing for high-throughput microscopy of fluorescently stained bacteria. Cytometry Part A 75A: 781–788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.