Abstract
Purpose
TUNEL assay is widely used to evaluate cell death. Quantification of TUNEL-positive (TUNEL+) cells in tissue sections is usually performed manually, ideally by two masked observers. This process is time consuming, prone to measurement errors, and not entirely reproducible. In this paper, we describe an automated quantification approach to address these difficulties.
Methods
We developed an ImageJ macro to quantitate cell death by TUNEL assay in retinal cross-section images. The script was coded using IJ1 programming language. To validate this tool, we selected a dataset of TUNEL assay digital images, calculated layer area and cell count manually (done by two observers), and compared measurements between observers and macro results.
Results
The automated macro segmented outer nuclear layer (ONL) and inner nuclear layer (INL) successfully. Automated TUNEL+ cell counts were in-between counts of inexperienced and experienced observers. The intraobserver coefficient of variation (COV) ranged from 13.09% to 25.20%. The COV between both observers was 51.11 ± 25.83% for the ONL and 56.07 ± 24.03% for the INL. Comparing observers' results with macro results, COV was 23.37 ± 15.97% for the ONL and 23.44 ± 18.56% for the INL.
Conclusions
We developed and validated an ImageJ macro that can be used as an accurate and precise quantitative tool for retina researchers to achieve repeatable, unbiased, fast, and accurate cell death quantitation. We believe that this standardized measurement tool could be advantageous to compare results across different research groups, as it is freely available as open source.
Keywords: cell death, inner nuclear layer, NMDA, outer nuclear layer, retina, retinal detachment, TUNEL assay
In this study, we developed and validated an automated ImageJ macro to quantitate cell death by TUNEL assay. The macro segmented retinal layers successfully and quantitated TUNEL-positive cells close to observer values. We believe this tool can be useful for improving cell death quantitation.
In recent years, research of degenerative retinal diseases has been focused on understanding cell death and ways of interfering with it. Several experimental animal models of diabetic retinopathy, retinitis pigmentosa, retinal detachment, and glaucoma, have revealed that cell death in the retina can occur through different modalities, including apoptosis and necroptosis among others, and encompasses multiple inducers and pathways and displays different morphologic characteristics.1–5 From the many tools used to characterize and quantify cell death, the terminal deoxynucleotidyl transferase (dUTP) nick end labeling (TUNEL) assay is likely the most widespread method. This assay detects fragmented or nicked DNA by means of a terminal deoxynucleotidyl transferase enzyme, which incorporates fluorescent-labeled dUTPs in damaged nucleic acid regions. First described by Gavrieli et al.,6 this assay has been a sensitive method for detecting apoptosis.7 However, as TUNEL assay detects DNA fragments regardless of the induced cell death pathway, it cannot distinguish between apoptotic or other forms of programmed cell death.3,8–10 Despite its limitations, TUNEL assay remains the most widespread method used to screen for any form of programmed cell death, to date.
In order to quantitate TUNEL assay results in tissue sections, manual counting of TUNEL-positive (TUNEL+) cells, ideally by two masked observers is performed.11 Observers count TUNEL+ cells and measure the respective retinal area, and results are expressed as either TUNEL+ cells/area or TUNEL+ cells/total cells. However, this process is time consuming, prone to measurement errors, and not entirely reproducible. Therefore, an automated approach is of interest to address these difficulties. To the best of our knowledge, there is no automated cell death quantitation method available, capable of segmenting retinal layers and counting individual TUNEL+ cells in the retina without performance variability.
The purpose of this study was to introduce a novel ImageJ macro for the automated detection of cell death in retinal cross-sections. In addition, we studied differences in grading between an experienced and an inexperienced observer. Because the introduction of a new measurement method requires that it be parallel with the gold standard, or commonly accepted method, we validated this macro by comparing its performance relative to that of two observers. We found that the ImageJ macro can achieve a fast and accurate retinal cell death quantitation.
Materials and Methods
Fluorescence Microscopy Image Database
Digital images of TUNEL-stained retinal cross-sections were obtained from the Angiogenesis Laboratory fluorescence microscopy database. In order to quantitate TUNEL+ cells on the outer nuclear layer (ONL), we randomly selected 90 images from an experimental murine retinal detachment model which induces photoreceptor cell death by subretinal injection of sodium hyaluronate.12 For experimental comparison between groups, we selected 30 images from an experimental group (mammalian sterile 20-like kinase 2 [MST2] knock-out mice) and compared them to a control group (C57BL/6 mice), as previously published.13 To assess inner nuclear layer (INL) TUNEL+ cells, we randomly selected 90 images from an experimental N-methyl-d-aspartic acid (NMDA)-induced excitotoxicity murine model that uses two different drug doses (10 and 100 nM). Eyes from these experimental models were enucleated, and 7- to 10-μm thin sections were cut using a model CM1850 cryostat (Leica Microsystems, Bannockburn, IL, USA). TUNEL staining was performed with ApopTag fluorescein direct in situ apoptosis detection kit (Millipore, Bedford, MA, USA), according to manufacturer's instructions. Prior to being mounted, cell nuclei were stained with Quinolinium, 4-[3-(3-methyl-2(3H)-benzothiazolylidene)-1-propenyl]-1-[3-(trimethylammonio)propyl]-, di-iodide (TO-PRO 3; Life Technologies, Grand Island, NY, USA).
Because we sought to build an assorted and representative sample, we included images from the aforesaid models acquired by multiple independent researchers, which used different acquisition protocols. These images were acquired using fluorescence microscopy (Axio Imager model M2; Carl Zeiss, Inc., Thornwood, NY, USA) with a 20×/0.8 numerical aperture air objective, as we considered this magnification was the most suitable to assess a reasonably large area without missing details in cell morphology. Image exclusion criteria were poor TUNEL staining quality, uneven image focus, ONL or INL layers not distinguishable, retina cross-section not centered on the image frame, or significant shadowing. Finally, selected digital images were exported as 24-bit red-green-blue (RGB) images in an uncompressed tagged image file format (TIFF) for further analysis.
ImageJ Macro Script Programming
To develop an automated method to quantitate cell death by TUNEL assay in retinal cross-section images, we designed a macro for the ImageJ platform (version 1.49, http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA) and coded the script using IJ1 programming language on Fiji image processing software (http://fiji.sc/Fiji, in the public domain). After TIFF files importation, the most relevant image processing steps are detailed as follows.
Image Scale Setup.
The image spatial scale (pixel-to-mm2) was acquired from the microscope bundled software (Zen version 1.1; Carl Zeiss, Inc.) in order to express results in mm2. This scale value can be easily obtained from image metadata.
ONL and INL Segmentation.
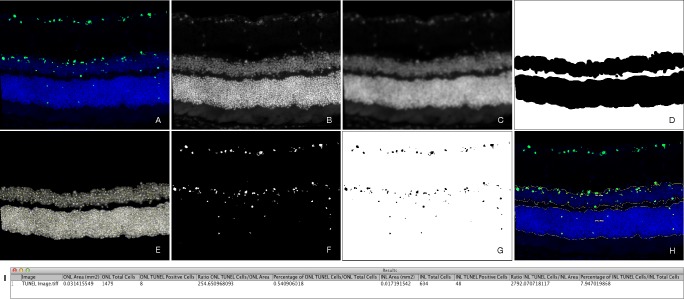
From the native RGB digital image (Fig. 1A), an 8-bit blue channel was extracted, and a Gaussian blur filter was applied (Figs. 1B, 1C) in order to reduce noise and detail.14 After contrast enhancement, we used Tsai moment-preserving thresholding method to obtain a binary image.15 Following several erosion iterations to achieve a fitted segmentation, outlier pixels were removed to a specific radius. The largest area in the image was presumed to be the ONL area. Then, the segmented ONL region of interest was subtracted from the processed blue channel. The INL area was segmented following the same approach as that used for the ONL area (Fig. 1D).
Figure 1.
Segmentation of ONL, INL, and quantitation of TUNEL-positive cells by ImageJ macro. From the native RGB image (A), an 8-bit blue channel was extracted (B), and a Gaussian blur filter was applied (C). Using the Tsai moment-preserving thresholding method, we segmented ONL and INL layers (D) and, by determining the local maxima corresponding to cell nuclei, we identified individual cells at these layers (E). The 8-bit green channel was extracted, and background noise was subtracted (F). Using the Tsai moment-preserving thresholding method and binary watershed segmentation, we counted TUNEL+ cells (G). A JPEG image overlay was automatically created and exported for visual assessment of the quantitation (H). Results from the macro were reported (I) as area (mm2), total cells (count), TUNEL+ cells (count), TUNEL+ cells-to-area (count/mm2) ratio, and percentage of TUNEL+ cells over total cells (%), for both ONL and INL.
ONL and INL Total Cells.
Outer nuclear layer and INL regions of interest were used to determine specific areas where total cells were counted. By determining the local maxima corresponding to cell nuclei, individual cells at these layers were identified and recorded (Fig. 1E).
ONL and INL TUNEL+ Cells.
From the native RGB digital image, an 8-bit green channel was extracted, and background noise was subtracted, in order to achieve a cleaner image (Fig. 1F). Following that, we applied Tsai moment-preserving thresholding method to obtain a binary image and used binary watershed segmentation to separate any contiguous cells (Fig. 1G). This filter first calculates the Euclidian distance map and finds the ultimate eroded points. Following, it dilates each of the latter as far as possible, either until the edge of the particle is reached, or the edge touches a region of another. Individual cells were counted according to specific size and circularity.
After image processing, the macro generated a report with 10 quantitative outcome variables (Fig. 1I), which included (1) ONL area (mm2); (2) ONL total cells (count); (3) ONL TUNEL+ cells (count); (4) ONL TUNEL+ cells to ONL area (count/mm2) ratio; (5) percentage of ONL TUNEL+ cells/ONL total cells (%); (6) INL area (mm2); (7) INL total cells (count); (8) INL TUNEL+ cells (count); (9) ratio of INL TUNEL+ cells to INL area (count/mm2); and (10) percentage of INL TUNEL+ cells/INL total cells (%). Additionally, for a qualitative analysis of the script performance, a JPEG (Joint Photographic Experts Group) image overlay was automatically created and exported for visual assessment of segmented areas and quantitated TUNEL+ cells (Fig. 1H).
Script Training, Validation, and Statistical Analysis
To calibrate and train the macro to experimental observer values, we first defined specific criteria to count TUNEL+ cells, including fluorescent signal colocalization between green and blue channels and nuclei-like size and shape, and if TUNEL+ cells appeared clumped, cell number was counted according to clump size. Most important, we encouraged observers to keep the same criteria for all images. Initially, images were assessed by an independent observer (BD) who manually segmented ONL and INL areas and counted TUNEL+ cells by using ImageJ (version 1.48). Script parameters were fine tuned to these values. Following that, 2 masked observers, 1 experienced (PT) and 1 inexperienced observer (BT), assessed the image dataset using the same approach. Observers were given duplicate images within the set, without their knowledge, in order to assess measurement reproducibility in a blind manner. Outcome variables were recorded as was time required to quantitate each image.
Statistical analysis was performed with Prism version 6 software (GraphPad, San Diego, CA, USA) and JMP Pro software version 11.2.0 (SAS, Carey, NC, USA). Normality was assessed with Shapiro-Wilk test. Statistical significance for differences between groups was determined with Wilcoxon signed-rank test for matched pairs and 1-way ANOVA with Tukey post hoc correction for multiple comparisons. For correlation analysis, values were fitted to linear regression, and results were expressed as Spearman ρ correlation coefficient and R2 goodness of fit for the model. Intra- and interobserver agreement analyses werre achieved by plotting the mean differences between measurements relative to their means, using Bland-Altman plot method.16 Coefficients of variation (COV) were calculated as differences between measurements over relative to their means: [(A − B)/([A + B]/2) × 100].17 A and B represent either first and second measurement in intraobserver agreement, or different observers in interobserver agreement. Results were expressed as means ± standard deviations (SD). A P value of <0.05 was considered statistically significant.
Results
All images that matched our selection criteria and were included in the dataset were processed by the ImageJ macro, with accurate segmentation of ONL and INL areas and TUNEL+ cell count, as seen in the exported JPEG image overlay (Supplementary Fig. S1B). Additionally, all defined variables were quantitated and reported.
ImageJ Macro and Observers' Performance For Area and TUNEL-Positive Cell Density
Results from observers and macro measurements can be seen in the Table. ONL area measured by the automated macro (0.028 mm2) was similar to that measured by the inexperienced observer (0.028 mm2, P = 0.958) and 10% less than that measured by the experienced observer (0.030 mm2, P = 0.019). No statistically significant differences were observed in TUNEL+ cell counts and ratio of TUNEL+ cells to area between the observers and macro. INL area measured by the macro (0.016 mm2) was similar to that of the inexperienced observer (0.018 mm2, P = 0.286) and 10% less than that of the experienced observer (0.018 mm2, P = 0.009). No statistically significant differences were observed in TUNEL+ cell counts and ratio of TUNEL+ cells to area between the experienced observer and macro. In contrast, significant differences were seen between the variables of the inexperienced observer and those of the the macro (P < 0.001).
Table.
Outcome Variables for Observers and ImageJ Macro
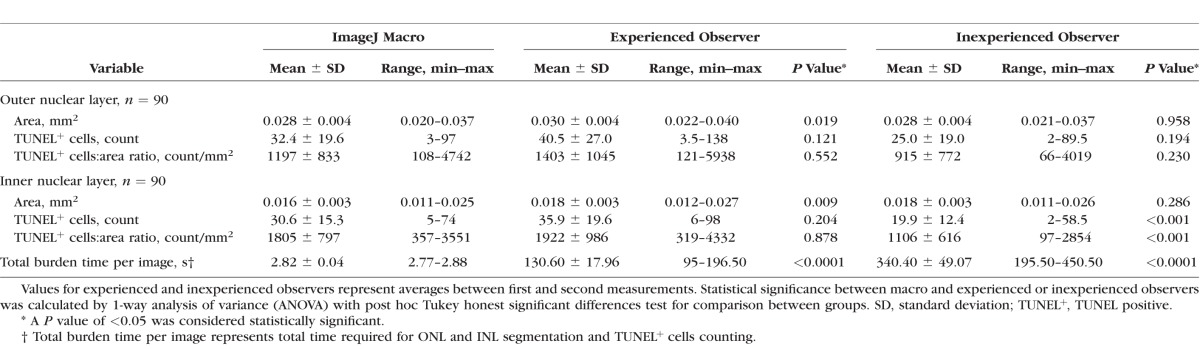
We calculated observers' total burden time per image by adding the time needed for ONL and INL segmentation and TUNEL+ cell counting. On average, an experienced observer needed 2.16 minutes per image, whereas the inexperienced observer needed more than 5.66 minutes. Macro needed less than 3 seconds per image with batch processing (using 2.8-GHz Core i7 iMac model computer; Apple Co., Cupertino, CA, USA), which was 60- and 160-fold faster than the experienced and inexperienced observers, respectively (Supplementary Fig. S4).
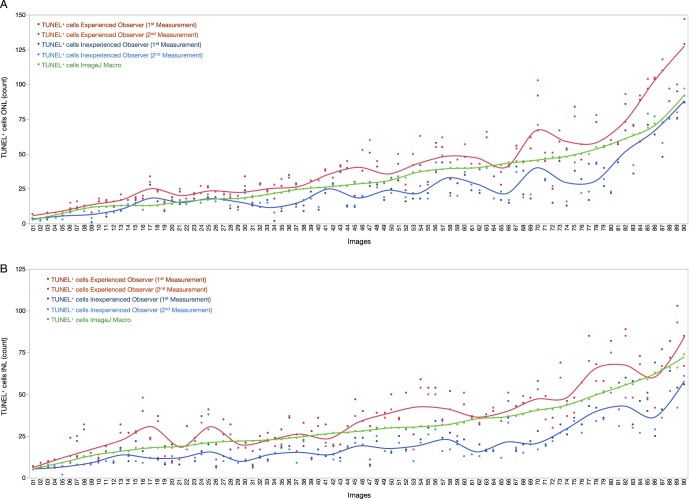
Next, ONL and INL TUNEL+ cell values were plotted for each image, observers, and automated macro. TUNEL+ cell values for the macro were located between the lower values of the inexperienced observer and the higher counts of the experienced observer, with only few exceptions corresponding to images with significant background noise, due to lack of postprocessing in the native image (Figs. 2A, 2B).
Figure 2.
TUNEL-positive cell counts for the macro and mean values of inexperienced and experienced observers. Cell counts for the ImageJ macro and first and second measurements from observers plotted for every image, for ONL (A) and INL (B) datasets. A colored trendline represents mean variations for each observer. Macro TUNEL+ counts are located between observer's measurements.
Intraobserver Agreement and Correlation
Since repeatability is a crucial element for any measurement method, we first sought to determine intraobserver variability. To answer this question, observers quantitated 180 images within the dataset, twice, in a masked fashion. Values from the first and second measurements were compared for each image. We then plotted their differences relative to the means, using Bland-Altman plots. The COV of TUNEL+ cell counts between these two measurements for the ONL dataset was 15.21 ± 19.69% for the inexperienced observer and 13.09 ± 13.84% for the experienced observer (Supplementary Table S1; Supplementary Figs. S2A, S2B). For the INL dataset, the COV was 16.41 ± 15.39% for the inexperienced observer and 25.20 ± 21.82% for the experienced observer (Supplementary Table S1; Supplementary Figs. S2B, S3A). Spearman ρ correlation coefficient between first and second measurements from the inexperienced observer was 0.95 in ONL images and 0.92 in INL images (Supplementary Table S1; Supplementary Figs. S2C–S3C). This coefficient was 0.96 in ONL images and 0.93 in INL images for the experienced observer (Supplementary Table S1, Supplementary Figs. S2D–S3D).
Interobserver Agreement and Correlation
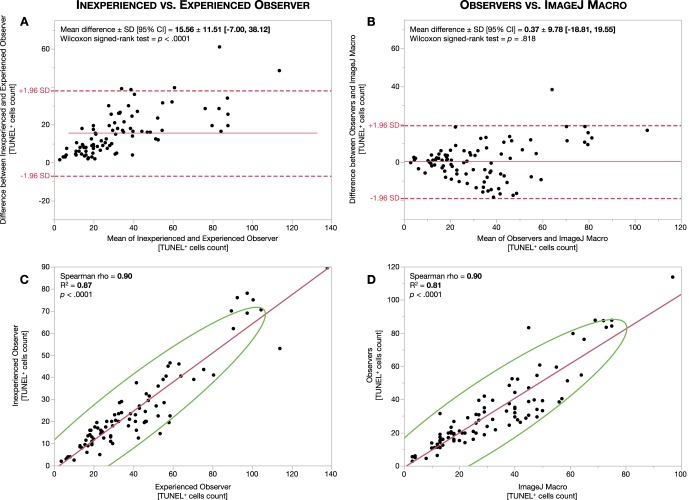
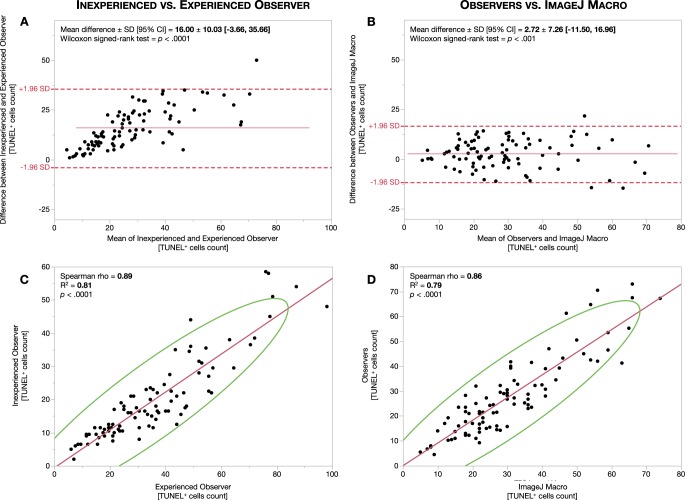
To determine interobserver variability, we averaged the first and second measurements of each observer and obtained their respective mean TUNEL+ cell count. COV of variation of TUNEL+ cell counts between the two observers was 51.11 ± 25.83% for the ONL dataset (Supplementary Table S1; Fig. 3A) and 56.07 ± 24.03% for the INL dataset (Supplementary Table S1; Fig. 4A). Following, we sought to determine whether the ImageJ macro would agree with the observers. We used the limits of agreement between these two observers to define the range to which the macro TUNEL+ cell counts should fit. Of note, these ranges were estimated for the whole dataset and should not be extrapolated or predicted as per image. Compared to the COV for the observers, the COV for the macro was 23.37 ± 15.97% for the ONL (Supplementary Table S1; Fig. 3B) and 23.44 ± 18.56% for the INL (Supplementary Table S1; Fig. 4B). These results can be visualized in Bland-Altman plots, as the values for the macro relative to those for the observers presented a smaller bias value and a more grouped and narrower confidence interval than that existing between observers. In addition, Spearman ρ coefficient between observers was 0.90 for the ONL and 0.89 for the INL and 0.90 for the ONL and 0.86 for the INL between observers and macro (Supplementary Table S1; Figs. 3C, 3D, 4C, 4D).
Figure 3.
Outer nuclear layer inter-observer agreement and correlation. Bland-Altman plots for ONL measurement agreement between observers (A) and between observers and ImageJ macro (B) are shown. Red lines represent mean differences or bias. Red dashed lines represent 95% limit of agreement (±1.96 SD). Correlation analysis between observers (C) and between observers and ImageJ macro (D). Red lines represent the fitted linear regression trendline. Green lines represent the 95% confidence interval area.
Figure 4.
Inner nuclear layer interobserver agreement and correlation. Bland-Altman plots for INL measurement agreement between observers (A) and between observers and ImageJ macro (B). Red lines represent mean differences or bias. Red dashed lines represent the 95% limit of agreement (±1.96 SD). Correlation analysis between observers (C) and between observers and ImageJ macro (D). Red lines represent the fitted linear regression trendline. Green lines represent the 95% confidence interval area.
Experimental Testing
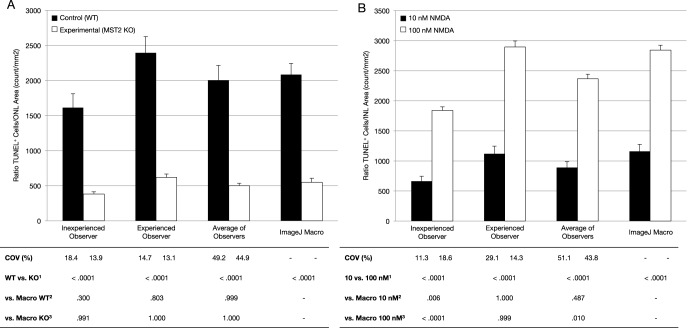
To confirm its validation and to further test performance, we sought to determine whether the macro was able to detect statistically significant differences, if present, between experimental groups. We randomly selected 30 images from a control group and 30 from an experimental group and compared results between observers and macro.
In the ONL dataset (Fig. 5A), all observers and macro were able to detect a statistically significant difference between the experimental group and their respective control (P < 0.0001). No statistical differences were observed in control and experimental groups between macro and average of all observers. However, there were small but statistically significant differences in the control values obtained by the different observers (P = 0.004) but no statistical significant differences obtained by the different observers in the experimental group (P = 0.943).
Figure 5.
Experimental testing of ImageJ macro. From the ONL dataset, 30 images from a WT control group and 30 from an experimental (MST2 knock-out) group were selected (A). From the INL dataset, 30 images from a 10 nM NMDA group and 30 from a 100 nM NMDA group were selected (B). Columns represent mean and standard errors of the mean values for the TUNEL+ cells-to-area per observer ratio. 1Wilcoxon signed rank test between groups within the same observer. 2One-way ANOVA with Tukey post hoc correction for comparison between all control groups or 10 nM against the macro. 3One-way ANOVA with Tukey post hoc correction for comparison between all experimental groups or 100 nM against the macro.
In the INL dataset (Fig. 5B), all observers and macro were able to detect a statistically significant difference between the two experimental groups (P < 0.0001). No statistical differences were observed for both 10 and 100 nM groups between the experienced observer and macro. There was though a statistically significant difference in the absolute values obtained for the 10 nM group (P = 0.018) and 100 nM group (P < 0.0001) between the two observers.
Discussion
In this study, we developed an ImageJ macro that was able to segment ONL and INL retinal layers and count TUNEL+ cells accurately. The reported parameters were able to fit within the established limits of agreement and were highly correlated with the observers' measurements. In addition, the macro was able to detect statistically significant differences between experimental groups. Collectively, these results validate the macro as a measurement tool for automated cell death quantitation in the retina. We speculate that the advent of this tool can aid in achieving accurate, repeatable, fast, and unbiased measurements.
A moderate degree of variability was observed between repeated measurements in both the inexperienced and experienced observers, as expected. Collectively, both observers presented good reproducibility as shown by bias value and range of limits of agreement. Bland-Altman plots for the inexperienced observer displayed a lower bias value and narrower limits of agreement compared to the experienced observer, most likely due to the time invested in the analysis and higher TUNEL+ cell counts by the experienced observer. Still, both of the observers were able to detect statistically significant differences among experimental groups, and the magnitude of these difference was higher for the experienced observer. In contrast, Spearman correlation coefficient for both of the observers showed similar values between groups. As Bland and Altman previously described, statistical hypothesis tests and correlation coefficients are not suitable statistical tests for comparing the agreement of two measurement methods. The former only determines whether the means of groups are equal or not. The latter measures the linear dependence between two variables, hence a paralleled change in the measurement magnitude will not affect the correlation coefficient but will certainly affect agreement.16,18 We suggest that, for the purpose of validating new measurement techniques, Bland-Altman plots and COV should be calculated, and a limit of agreement should be determined beforehand.
The ImageJ macro was able to detect layers and count TUNEL+ cells within the limits of agreement determined by both observers. The coefficients of variation between macro and average of observers were approximately half the value of those between observers, for both ONL and INL dataset. These values are supported by the narrower limits of agreement between macro and average of observers than between both observers, as seen in Bland-Altman plots. These premises argue in favor of the high level of accuracy of this measurement tool.16 On a side note, TUNEL+ cell count variability was increased in INL images with more TUNEL+ cells, probably owing to the presence of cells of different sizes19 and staining patterns. Nonetheless, because there is no absolute reference value in TUNEL+ cell counting, we believe it is crucial to follow consistent criteria when counting cells, in order to detect consistent differences between groups.
The burden time to analyze these images is significant. On average, an experienced observer needed more than 2 minutes per image, whereas an inexperienced observer required more than 5 minutes. In contrast, the macro required less than 3 seconds for each image analysis, when executed in batch mode. This advantage opens up the possibility of assessing a larger area of retina. We calculated that the time required to manually count two images per retina would still be higher than analyzing the entire retina with eight to nine images using ImageJ macro, even considering increased acquisition time. The ability to examine larger retina areas in the same amount of time will result in higher experimental objectivity.
While estimating expected sample size for each experimental group, the variability both of the biological model and the measurement tool should be taken into account. If the measurement tool used to quantitate outcome variables has significant variability, it increases the overall variability as bias and limits of agreement are also subject to sampling variation.20 In essence, higher variability reduces the ability to detect statistical significance, increasing required sample size. Therefore, using an automated measurement tool can reduce bias and sample size.
Image Acquisition, Postprocessing Recommendations, and Limitations
To challenge the macro versatility, we included images taken by multiple observers using different acquisition parameters. However, we recommend a standardized acquisition and postprocessing approach customized for each researcher and equipment used and testing the macro prior to experimental analysis. Images should ideally be centered, with ONL and INL layers not merged, without shadows or uneven focus, to ensure an appropriate macro performance. Image background noise should be reduced as much as possible.
Study Limitations.
Still, several limitations should be considered. First, macro results were compared against two observers. However, these observers performed with good reproducibility. Second, the macro assumes the largest nuclei area in the image to be the ONL area. This should be considered when assessing images from experimental neurodegenerative models with significant ONL/INL thinning. Finally, digital images should be acquired with a 20×/0.8 numerical aperture objective, as this is the chosen working magnification of the script for the aforementioned reasons. We are currently working on new macro versions to broaden the type of images suitable for automated analysis.
In summary, the developed and validated ImageJ macro can be an accurate and precise quantitative tool with which retina researchers can achieve repeatable, unbiased, fast, and above all, accurate cell death quantitation. We believe this standardized measurement tool could be advantageous to compare results across different research groups. The macro will be freely available to all as open source.
Supplementary Material
Acknowledgments
Supported by Bayer Healthcare Global Ophthalmology Awards Program (GOAP; DEM); the Yeatts Family Foundation (DGV, JWM); Loefflers family foundation (JWM,DGV); a 2013 Macula Society Research Grant award (DGV); a Bausch & Lomb Vitreoretinal Fellowship (HM); a Physician Scientist Award (DGV) and an unrestricted grant (JWM) from the Research to Prevent Blindness Foundation; NEI Grant R21EY023079-01A1 (DGV); and NEI Grant EY014104 (MEEI Core Grant). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Please visit http://imagej.net/TUNEL_Cell_Counter to download the macro and original image dataset. We added a detailed tutorial, setup instructions, and suggestions to improve performance.
Disclosure: D.E. Maidana, Bayer Healthcare (F); P. Tsoka, None; B. Tian, None; B. Dib, None; H. Matsumoto, None; K. Kataoka, None; H. Lin, None; J.W. Miller, None; D.G. Vavvas, None
References
- 1. Cuenca N,, Fernández-Sánchez L,, Campello L,, et al. Cellular responses following retinal injuries and therapeutic approaches for neurodegenerative diseases. Prog Retin Eye Res. 2014; 43: 17–75. [DOI] [PubMed] [Google Scholar]
- 2. Murakami Y,, Matsumoto H,, Roh M,, et al. Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsRNA-induced retinal degeneration. Cell Death Differ. 2014; 21 : 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murakami Y,, Matsumoto H,, Roh M. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci U S A. 2012; 109 : 14598–14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson TV,, DeKorver NW,, Levasseur VA,, et al. Identification of retinal ganglion cell neuroprotection conferred by platelet-derived growth factor through analysis of the mesenchymal stem cell secretome. Brain. 2014; 137 (pt 2) : 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galluzzi L,, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008; 135 : 1161–1163. [DOI] [PubMed] [Google Scholar]
- 6. Gavrieli Y,, Sherman Y,, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992; 119 : 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liming P,, Bradley CJ,, Liu JJ. The correlativity analysis of six methods of detecting apoptosis. Chin Med Sci J. 1999; 14 : 145–151. [PubMed] [Google Scholar]
- 8. Grasl-Kraupp B,, Ruttkay-Nedecky B,, Koudelka H,, Bukowska K,, Bursch W,, Schulte-Hermann R. In situ detection of fragmented DNA (TUNEL assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: a cautionary note. Hepatology. 1995; 21 : 1465–1468. [DOI] [PubMed] [Google Scholar]
- 9. Pasparakis M,, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015; 517 : 311–320. [DOI] [PubMed] [Google Scholar]
- 10. Trichonas G,, Manola A,, Morizane Y,, et al. A novel nonradioactive method to evaluate vascular barrier breakdown and leakage. Invest Ophthalmol Vis Sci. 2010; 13 : 317–323. [DOI] [PubMed] [Google Scholar]
- 11. Negoescu A,, Guillermet C,, Lorimier P,, Brambilla E,, Labat-Moleur F. Importance of DNA fragmentation in apoptosis with regard to TUNEL specificity. Biomed Pharmacother. 1998; 52 : 252–258. [DOI] [PubMed] [Google Scholar]
- 12. Matsumoto H,, Miller JW,, Vavvas DG. Retinal detachment model in rodents by subretinal injection of sodium hyaluronate. J Vis Exp. 2013; 79: 50660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsumoto H,, Murakami Y,, Kataoka K,, et al. Mammalian STE20-like kinase 2, not kinase 1, mediates photoreceptor cell death during retinal detachment. Cell Death Dis. 2014; 5 : e1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lunscher WH,, Beddoes MP. Optimal edge detector design I: parameter selection and noise effects. IEEE Trans Pattern Anal Mach Intell. 1986; 8 : 164–177. [DOI] [PubMed] [Google Scholar]
- 15. Tsai W-H. Moment-preserving thresholding: a new approach. Comput Vis Graph Image Process. 1985; 29 : 377–393. [Google Scholar]
- 16. Bland JM,, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986; 1 : 307–310. [PubMed] [Google Scholar]
- 17. Filippi M,, Horsfield MA,, Rovaris M,, et al. Intraobserver and interobserver variability in schemes for estimating volume of brain lesions on MR images in multiple sclerosis. AJNR Am J Neuroradiol. 1998; 19 : 239–244. [PMC free article] [PubMed] [Google Scholar]
- 18. Bland JM,, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995; 346 : 1085–1087. [DOI] [PubMed] [Google Scholar]
- 19. Chan TF,, Chiu K,, Lok CKM, et al. Morphometric analyses of retinal sections. J Vis Exp. 2012; 60: 3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bland JM,, Altman DG. Agreed statistics: measurement method comparison. Anesthesiology. 2012; 116 : 182–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.