Better tools are needed for detecting Mycobacterium tuberculosis and defining the dynamic interactions of M. tuberculosis with its human host.
Abstract
The world is in need of more effective approaches to controlling tuberculosis. The development of improved control strategies has been hampered by deficiencies in the tools available for detecting Mycobacterium tuberculosis and defining the dynamic consequences of the interaction of M. tuberculosis with its human host. Key needs include a highly sensitive, specific nonsputum diagnostic; biomarkers predictive of responses to therapy; correlates of risk for disease development; and host response–independent markers of M. tuberculosis infection. Tools able to sensitively detect and quantify total body M. tuberculosis burden might well be transformative across many needed use cases. Here, we review the current state of the field, paying particular attention to needed changes in experimental paradigms that would facilitate the discovery, validation, and development of such tools.
Mycobacterium tuberculosis has been performing its deadly dance with Homo sapiens since before the duo emerged together from Africa (Comas et al., 2013). Tuberculosis (TB) continues to cause an outsized burden of morbidity and mortality. Currently, one third of the global population is thought to harbor latent infection, on the basis of sustained lymphocyte reactivity with M. tuberculosis proteins in the absence of symptoms. Those with latent infection have an ∼10% lifetime risk of developing active disease. TB claims ∼1.4 million lives annually, and often leaves disability behind in survivors (Murray et al., 2014; WHO, 2014a). These global numbers mask the profoundly inequitable distribution of this scourge of poverty, the burden of which falls heaviest on low- and middle-income countries (WHO, 2014a).
Approaches to decreasing TB morbidity and mortality, along with M. tuberculosis transmission, can be conceived as falling into three general categories, the first of which is treatment of disease. Effective treatment is central to any strategy for control. In addition to preventing morbidity and mortality, timely effective diagnosis and treatment of TB are critically important for decreasing transmission because pulmonary disease and transmission are tightly linked (Russell, 2011). Important barriers to effective treatment include the lack of availability of diagnostics that afford timely, effective ascertainment of TB, accurate tools to monitor responses to treatment, and the need for dramatically shorter courses of chemotherapy, which could potentially increase patient compliance and decrease the emergence of drug resistance.
The second approach is the prevention of disease. As preventing disease prevents morbidity, mortality, and transmission, this approach has obvious pride of place in terms of potential. It provides the underlying rationale for vaccination, as well as for secondary prophylaxis strategies. Unfortunately, the current barriers here are just as obvious. BCG, brought into the world some 90 yr ago, remains the only licensed vaccine for TB. Although efficacy is high against disseminated disease in infants (Trunz et al., 2006), a benefit that may well be provided by M. tuberculosis infection itself in older individuals (Karp et al., 2015), BCG’s efficacy in preventing pulmonary TB is highly variable, ranging from negative numbers to 80%, with the lowest efficacy seen in areas of high transmission and endemnicity (Colditz et al., 1994; Dye, 2013; Mangtani et al., 2014). Further, whereas the efficacy of secondary prophylaxis with the antimycobacterial drug isoniazid (focused on those at highest clinical risk, including those in the first year after infection, and those at life stages and/or with intercurrent conditions that predispose to disease progression) has been demonstrable in low transmission settings such as the global North, it has not been demonstrated in high transmission settings such as South Africa (Churchyard et al., 2014; Wood and Bekker, 2014).
A final approach to limiting TB morbidity and mortality is the prevention of infection. Raising the bar to the establishment of sustained infection—a term that will be used throughout this Perspective to refer to the sustained presence of viable M. tuberculosis bacilli, with or without ongoing replication and subclinical or clinical disease—has promise for preventing both disease and transmission. Successful infection likely involves a single airborne droplet nucleus capable of reaching the alveolus, containing a single—or at most a few—organisms (Mills et al., 1960; Riley et al., 1995). Abortive infection, defined operationally by evidence of robust T cell reactivity, followed by the reversion of such, is seen in longitudinal studies of humans (Shah et al., 2011) and guinea pigs infected by exposure to air vented from the rooms of humans with active TB disease (Dharmadhikari et al., 2011). Prevention of infection through better ventilation, and prevention of sustained infection through better nutrition, may well have played a role in decreasing M. tuberculosis transmission in urban North America in the early 20th century. BCG vaccination also appears to have some efficacy in preventing sustained infection in both adults and children (Soysal et al., 2005; Eisenhut et al., 2009; Barreto et al., 2011; Basu Roy et al., 2012; Chan et al., 2013; Roy et al., 2014). For this and other reasons (Karp et al., 2015), developing vaccines aimed at preventing sustained infection represents a promising approach. That said, such vaccines are not yet known, and practical ways of testing more general methods of raising the barrier to infection (e.g., improved ventilation and improved nutrition), and of translating such interventions to practice, remain to be devised.
In sum, current approaches for TB control remain insufficient. Further, as with the burden of TB itself, existing approaches also fail any reasonable equity agenda—whereas considerable success has been attained in much of the industrial North, these approaches have not led to control in endemic areas with high transmission pressures.
The development of novel approaches to TB control has been hampered by deficiencies in the available tools for detecting and quantifying the presence of M. tuberculosis in, and the dynamic consequences of the interactions of M. tuberculosis with, individual human hosts. Diagnosis of disease largely relies on direct detection of M. tuberculosis (via microscopy and/or culture) or M. tuberculosis DNA (via amplification techniques) in sputum. Detection of infection relies on evidence of sustained T cell reactivity with M. tuberculosis antigens (via tuberculin skin tests [TSTs] or IFN-γ release assays [IGRAs]) in peripheral blood. These tools provide neither an optimal foundation for TB control nor a clear path to the development of improved strategies. Fundamentally, the available tools fall well short of what is needed to reflect the underlying pathogenic complexities. These complexities deserve underscoring. Clinically silent, sustained infection actually represents a broad spectrum of responses, ranging from something close to sterilizing immunity to subclinical active disease. In turn, latency is itself part of a broader spectrum that encompasses active and fulminant disease (Barry et al., 2009). The overall spectrum is highly dynamic. Functional imaging and histological analysis of infected humans and nonhuman primates has shown that individual foci of infection can pursue diametrically opposite trajectories at the same time in the same lung (Russell et al., 2010; Lin et al., 2013, 2014). The response to therapeutic intervention appears to be just as complicated and dynamic (Russell et al., 2010; Lin et al., 2013). Improved tools that could provide actionable insight into these complexities have transformative potential.
TB diagnosis
Current diagnostics for pulmonary TB rely on sputum samples and suffer several shortcomings. To begin, detection of M. tuberculosis in airway secretions depends on the presence of necrotic foci of infection in contiguity with the airway. As a result, by the time diagnosis in sputum becomes possible, most patients will have had active disease for quite some time, often with considerable lung damage. The practical necessity for M. tuberculosis to be present in airway secretions underscores that diagnosis through sputum, perforce, only occurs after individuals are likely capable of transmitting M. tuberculosis to others.
Several alternative methods for diagnostic testing of sputum exist, but they all have shortcomings. A molecular test, the Xpert MTB/RIF test (Cepheid), is sensitive (98% in microscopy positive subjects; 72% in microscopy negative subjects) and specific (99%; Boehme et al., 2010), but requires instrumentation not broadly available in endemic settings. Microscopy is broadly available and highly specific, but lacks sensitivity, missing the diagnosis in over one third of patients seeking care (Davis et al., 2013). Mycobacterial culture is the gold standard for TB diagnosis, but it provides results after considerable delay. In some endemic settings, clinicians are disinclined to obtain sputum in the first place (Satyanarayana et al., 2015), and in these settings diagnosis and misdiagnosis proceeds from history, physical exam, and chest x-ray alone.
Other challenges for TB diagnostics include HIV infection, which markedly lowers the sensitivity of sputum diagnostic tests (Lawn and Wood, 2011), and age. In infants, sputum is sparse and/or difficult to obtain, and analysis of repetitive early morning gastric aspirates or induced sputa has sensitivities that remain problematic (Starke, 2003; Zar et al., 2005). Obviously, sputum diagnostics have no utility in extrapulmonary disease, the diagnosis of which relies on samples (tissue or cerebrospinal fluid) collected invasively.
For all of these reasons, there is need for the development of a highly sensitive and specific diagnostic test for TB that is able to rapidly identify the presence of active disease in endemic settings and is performable at reasonable cost on an easily obtainable nonsputum sample such as blood, urine, or breath. Such a diagnostic might be based on the quantitative detection of M. tuberculosis or its products. In this vein, it remains an open question whether mycobacterial burden has a direct correlation with an individual’s location along the spectrum from latent infection to active disease to effectively treated—a question that will remain open until accurate methods for quantifying mycobacterial burden are developed (vide infra). A diagnostic might also be based on quantification of biomarkers of the host’s state.
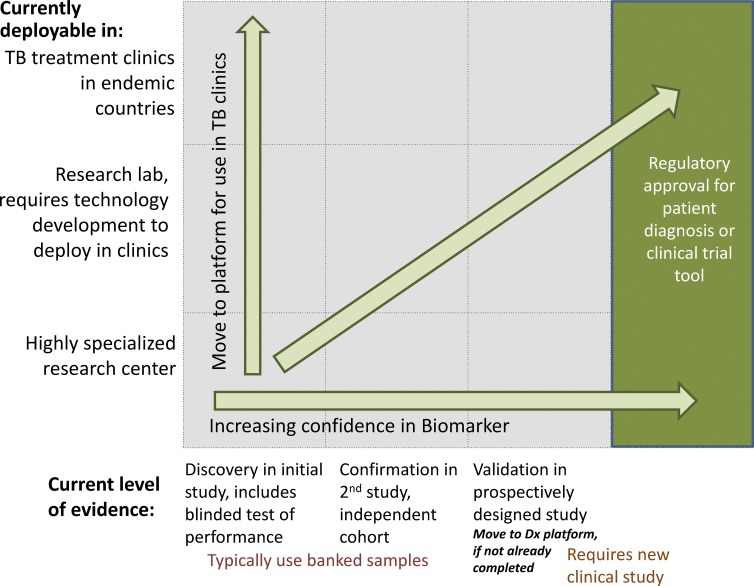
There are currently no validated nonsputum mycobacterial or host biomarkers for the diagnosis of pulmonary TB. A framework for the pathway for validating proposed biomarkers for specific use cases is presented in Fig. 1. Potential biomarkers must progress through sequential testing to confirm utility in an independent cohort, and then undergo validation in a prospective study. In addition, proposed biomarkers must move to a testing platform appropriate for the proposed use, which includes TB clinics in endemic countries—as is required for a nonsputum diagnostic. A large number of published studies declaring their goal to be diagnostic biomarker identification report the comparison of samples from patients with active disease to those from individuals with latent infection. Such study designs do not, however, directly address the appropriate use case, which is the differentiation of clinically symptomatic individuals with and without active TB (not to speak of the differentiation of those with minimally symptomatic active disease from those with latent infection). As such, the study designs standard in the field to date do not substantively move the field forward toward biomarker validation, and can only nominate hypotheses for follow up testing. The World Health Organization recently published a report of a consensus meeting on high priority target product profiles for new TB diagnostics that includes target product profiles (TPPs) for a rapid biomarker-based nonsputum diagnostic test, as well as for a community-based triage or referral test for identifying people suspected of having TB for whom confirmatory testing should follow (WHO, 2014b). The nonsputum diagnostic TPP is presented (with permission) in Table 1, with the purpose of providing a useful exemplar for those currently involved in biomarker research. The published TPPs outline the criteria a biomarker would likely need to meet, along with those of associated diagnostic testing platforms, to result in substantial impact. Progress toward meeting these TPPs is in brief summarized below. The TPP criterion for fast turnaround (<1 h) necessitates a focus on biomarkers that are present and can be directly measured in patient specimens; as such, cell stimulation and other culture-based methods are not discussed here.
Figure 1.
Framework for biomarker validation for new diagnostics and clinical trial tools.
Table 1.
TPP: A rapid biomarker-based non-sputum–based test for detecting TB
| Characteristic | Optimal requirements | Minimal requirements |
| Scope | ||
| Goal | To develop a rapid biomarker-based test that can diagnose pulmonary TB and optimally also extrapulmonary TB using non-sputum samples (for example, urine, blood, oral mucosal transudates, saliva, exhaled air) for the purpose of initiating TB treatment during the same clinical encounter or on the same day | |
| Target population | Target groups are adults and children including those who are HIV-positive and suspected of having active pulmonary TB or extrapulmonary TB in countries with a medium to high prevalence of TB as defined by WHO | |
| Target user of test | Health-care workers with a minimum of training | Trained microscopy technicians |
| Setting (level of the healthcare system) | Health posts without attached laboratories (that is, levels below microscopy centers) or higher levels of the health-care system | Primary health-care clinics with attached laboratories; peripheral microscopy centers or higher levels of the health-care system |
| Performance characteristics | ||
| Diagnostic sensitivity for pulmonary TB in adults | ≥98% for smear positive culture positive TB, ≥68% for smear negative culture positive TB (that is, similar to Xpert MTB/RIF assay) Overall pooled sensitivity should be ≥80% in adults with HIV infection | Overall ≥65% but should be >98% among patients with smear positive culture positive TB (that is, similar to smear microscopy) Overall pooled sensitivity should be better than smear microscopy in adults with HIV infection |
| Diagnostic sensitivity for extrapulmonary TB in adults | Ideally, should be ≥80% for all forms of microbiologically confirmed TB | No lower range of sensitivity was defined |
| Diagnostic sensitivity in children | Sensitivity for intrathoracic TB ≥66% for microbiologically confirmed TB (that is, similar to Xpert MTB/RIF) | No lower range of sensitivity was defined |
| Diagnostic specificity | At least as specific as Xpert MTB/RIF for pulmonary, extrapulmonary, and childhood TB (that is, 98% specificity compared against microbiological reference standard); test should distinguish between active TB and latent or past infection | |
| Operational characteristics | ||
| Sample type | Not invasive or minimally invasive, non-sputum samples | |
| Manual preparation of samples | Sample prep should be integrated or manual prep should not be required | Limited number of steps only; precise measuring should not be needed |
| Time to result | <20 min including time spent preparing sample | <1 h including time spent preparing sample |
| Instrument and power requirement | No instrument needed | Small, portable or hand-held instrument that can operate on battery or solar power |
| Maintenance and calibration | Disposable, no maintenance | Minimal maintenance required with automatic alert and remote calibration |
| Operating temperature and humidity level | +5°C to +50°C with 90% humidity | +5°C to +40°C with 70% humidity |
| Results capturing, documentation, data display | Instrument free test with ability to save test results using separate, attachable reader | Test menu must be simple to navigate; integrated screen, simple keypad or touch screen, ability to save results using instrument or separate reader |
| Internal quality control | Internal controls included for processing sample and detecting TB | Internal control only for processing sample |
| Price | ||
| Price of individual test | <US $4.00 | <US $6.00 |
In terms of the direct detection of M. tuberculosis or its products, detection of M. tuberculosis DNA in blood and urine from patients with pulmonary TB has better sensitivity than culture of these specimens, but remains well below required thresholds (Cannas et al., 2008; da Cruz et al., 2011; Theron et al., 2014). Among proteins and cell wall components, lipoarabinomannan (LAM) has received the most experimental attention. A commercially available urine LAM diagnostic test is available; however, poor sensitivity has led to limited use (Minion et al., 2011). There may be potential for improved sensitivity with other LAM assays (Mukundan et al., 2012; Chan et al., 2015; Hamasur et al., 2015). In HIV-positive patients, both M. tuberculosis DNA and LAM detection in urine hold promise due to the relatively high yield of identifying TB patients missed by sputum diagnostics, ease of collection, and nonspecific symptoms resulting from immune suppression (Aceti et al., 1999; Nakiyingi et al., 2014; Lawn et al., 2015). Analysis of antigen 85 has also been explored in blood and urine, with variable performance in small datasets (Bentley-Hibbert et al., 1999; Kashyap et al., 2007; Mukundan et al., 2012). Other proteins have been detectable in urine by mass spectrometry (Napolitano et al., 2008; Young et al., 2014). Overall, the ability to directly detect diverse M. tuberculosis products in blood or urine is promising. The generally observed low detection sensitivity may be improvable through advances in sample processing (Kashyap et al., 2007), reagent development, and/or the development of novel analytical platforms. It may also be possible to increase specificity, although problems with specificity may of course be caused by the presence of these same products in individuals with latent infection with M. tuberculosis and/or nontuberculous mycobacteria. However, these candidate biomarkers represent hypotheses that require evaluation in appropriately designed and powered studies.
On the host side, analysis of antibodies has not proved promising due to heterogeneity of the antibody response to M. tuberculosis (Kunnath-Velayudhan et al., 2010). Some traction has been achieved with transcriptomic profiling. Unbiased systems biology techniques revealed a type I IFN/inflammatory transcriptional signature in peripheral blood during pulmonary TB (Berry et al., 2010), a signature that was subsequently validated in a variety of different populations (Lesho et al., 2011; Lu et al., 2011; Maertzdorf et al., 2011, 2012; Bloom et al., 2013). Importantly, the signature has reasonable ability to differentiate TB from other respiratory and inflammatory diseases (Berry et al., 2010; Maertzdorf et al., 2012; Bloom et al., 2013; Kaforou et al., 2013). The signature also increases with disease activity and diminishes with treatment (Bloom et al., 2012). To date, however, sensitivity and specificity have not met minimum TPP requirements. Further, the complexities inherent in quantitative measurement of the expression levels of a large number of genes make this approach challenging as well as expensive.
Unbiased transcriptional profiling has also been applied to the development of a diagnostic test for childhood TB (Anderson et al., 2014). In children, diagnostic options are currently limited by the difficulty in obtaining samples for microbiological confirmation. As such, the validation of a new test is more difficult in the absence of a gold standard. In the aforementioned study, a transcriptional signature with sensitivity of 83% and specificity of 83% in culture-confirmed TB was reported, with a sensitivity estimated at 35–80% in children with possible or probable TB (i.e., without microbiological confirmation).
Beyond transcriptional signatures, host markers have generally shown significant overlap between symptomatic patients confirmed positive or negative for TB, leading to poor specificity for the markers tested to date. This has been true for serum micro-RNAs (Miotto et al., 2013; Zhang et al., 2013) and proteins (Phalane et al., 2013; Huang et al., 2014; Hur et al., 2015). Volatile organic compounds, likely arising from the host response, have been explored in breath. Using gas chromatography and mass spectroscopy for detection and quantification, a sensitivity of 84% and specificity 65% has been reported, but without an independent test of performance (Phillips et al., 2010). In sum, even highly complex gene signatures have, to date, failed to meet the high bar required for a diagnostic test. The consistent overlap between TB and non-TB patients across the majority of the host markers suggests the host response may indeed encompass a spectrum of activity that is variable within active TB and overlaps with latent TB infection as well as other illnesses. The exception may be pediatric patients, for whom receiver operator curves for host markers may actually improve on the accuracy of current diagnostic tests and algorithms. The complexity of reducing a high dimensional host signature into a practical test remains a key barrier to the application of this work in TB clinics in endemic settings.
In 2011, the Bill & Melinda Gates Foundation announced a Grand Challenge request for proposals to investigate biomarkers that could enable the development of a nonsputum-based diagnostic for TB. This resulted in investment in a diverse portfolio of projects investigating both pathogen and host biomarkers, including direct measurement of M. tuberculosis products in serum, plasma, and urine analysis of exosomes in blood as a potential enriched source of M. tuberculosis products, a targeted effort to characterize and develop new detection reagents for LAM, and broad efforts to profile host proteins, lipids, and metabolites for diagnostic potential. Data generated to date has underscored the aforementioned challenges, including the conclusion that antibody responses are unlikely to provide useful diagnostics for TB.
Monitoring therapeutic responses and assessing bacterial burden
Adequate tools for monitoring responses to existing TB therapeutics are also lacking. This poses a challenge for clinical management and imposes a barrier to the development of novel therapeutics. Similar to diagnostics, tools to monitor treatment must undergo prospective validation and translation to an appropriate testing platform, as depicted in Fig. 1. Standard therapy for treating active TB is for 6 mo, although data from clinical trials of shorter regimens suggest that 80–85% of patients can be successfully treated in 4 mo (Gillespie et al., 2014). However, whereas sputum culture conversion at 2 mo correlates with cure at the end of 6 mo of treatment and lack of relapse, the positive predictive value is low (Wallis, 2015). Moreover, there is no available biomarker that allows prediction of which individuals could be treated for 4 mo with a subsequent relapse rate equivalent to that of 6 mo therapy in nonstratified subjects. This has motivated a search for better biomarkers to identify individual patients who could be effectively treated with a shortened course.
There is a similar need for better biomarkers as surrogate endpoints in testing novel drugs and regimens, as the poor predictive power of sputum culture conversion to determine needed treatment duration, or to rank the relative potency of new regimens, necessitates follow up for at least 18 mo after randomization to measure the impact on relapse in the first year after finishing treatment. In addition to expense and risk, the reliance on sputum culture for drug development will potentially result in the discarding of novel agents with activity against the specific mycobacterial subpopulations most refractory to current drugs. The predictive link from sputum culture to patient outcome is likely weakened by the obvious bias toward identification of bacteria that have access to the airway. In addition to foci of infection in the lung that are occult with respect to sampling, M. tuberculosis can also be found outside the respiratory tract (Neyrolles et al., 2006; Das et al., 2013). There is also a clear bias toward the ascertainment of M. tuberculosis subpopulations that can be grown with standard culture methods, missing other viable populations (Garton et al., 2008; Mukamolova et al., 2010). Tools to quantify the total body burden of M. tuberculosis, both culturable and not, as well as the host response to any remaining mycobacteria, could enable more tailored individual treatment decisions as well as facilitate drug development. There is thus a clear need for the discovery and validation of biomarkers that can provide timely insights into therapeutic responses and cure, including methods for quantifying mycobacterial burden and biomarkers of the host response.
The development of more useful tools is made more challenging by the diverse use cases involved—including both clinical trials and routine clinical use and predictive biomarkers that allow stratification of patients and biomarkers that track response to treatment. For biomarkers to be used in clinical trials, a higher level of cost and complexity could be tolerated, but routine clinical use requires a field-deployable, low-cost test. These different use cases must be clearly defined during the research and development stages, and an appropriate test platform is required before moving to final validation and regulatory approval (Fig. 1). The vast majority of publications investigating treatment response biomarkers fail to articulate the intended use case and underlying TPP. Furthermore, most studies compare changes in proposed biomarkers over time during treatment without testing, or being powered to test, the correlation with patient outcome, i.e., relapse-free cure.
Opportunities to improve sputum analysis include the practical (reducing time and/or broadening application in low resource settings) and the more exploratory (providing information on bacterial populations that are currently unculturable). In the former vein, Friedrich et al. (2013) compared Xpert MTB/RIF results to smear microscopy and culture on solid and liquid media and found the Xpert MTB/RIF assay to have high sensitivity (97%) but poor specificity (49%) to identify culture positive specimens when Xpert is used as a binary readout. When using the quantitative measurement from the Xpert MTB/RIF assay, the change in quantitative sputum bacterial load correlated with smear grades, solid culture grades, and time to liquid culture positivity (all ρ = 0.73–0.74; P < 0.001), leaving the possibility that this quantitative data, if used in the analysis, may have additional utility in identifying patients who are cured (Friedrich et al., 2013). The Xpert MTB/RIF test detects DNA from dead as well as live mycobacteria. A recent study tested the value of adding propidium monoazide (which quenches PCR-mediated detection of DNA from dead mycobacteria) to allow for the specific detection of viable bacilli in sputum using Xpert MTB/RIF (Nikolayevskyy et al., 2015). Whereas a positive correlation (r = 0.61) with time to positivity of TB cultures on liquid media was reported, performance would need to be improved to allow practical use. Currently, results comparing DNA measurements with culture show relatively poor correlations. Further investigation will be needed to understand whether these correlations can be improved by accounting for live but unculturable bacteria in the sample. The contribution of such bacteria to patient outcomes, the need for continued treatment, and risk of relapse, must also be defined to understand the potential clinical utility of such analyses.
Beyond sputum, our fundamental lack of understanding of the total body burden and distribution of M. tuberculosis during latent infection, in active disease, and through the course of treatment should motivate studies to characterize total mycobacterial load. Such studies will need to start in fit-for-purpose animal models that display a broad spectrum of infection and disease states, such as cynomolgus macaques infected with low-dose Mtb (Capuano et al., 2003; Lin et al., 2012) and guinea pigs infected in natural transmission models (Dharmadhikari et al., 2011). Linked analysis of clinical specimens from well-characterized, longitudinal cohorts would be required to test hypotheses arising from such studies. Though challenging, progress in characterizing the total body M. tuberculosis burden will likely be critical for making progress in transforming the diagnosis, treatment, and prevention of TB.
On the host side, investigations into biomarkers that reflect TB burden remain at the hypothesis-generating stage. Higher baseline levels of antibodies to specific M. tuberculosis proteins were reported to correlate with slower sputum conversion, but antibody levels overlap with those seen in fast responders, hampering practical utility (Baumann et al., 2013). Plasma vascular endothelial growth factor concentrations at 2 wk have been positively correlated with time to sputum conversion (Riou et al., 2012). Decreases in frequencies of M. tuberculosis antigen-stimulated IFN-γ–secreting CD4+ T cells expressing CD38 and HLA-DR and Ki-67 were associated with resolution of sputum smear and culture positivity in a preliminary study (Adekambi et al., 2015). Detection of systemic heme oxygenase-1 (HO-1) levels showed some promise, with a larger number of patients who were successfully treated showing decreased HO-1 levels, and levels remaining unchanged in five patients who were culture positive at the end of 6 mo of treatment (Andrade et al., 2013). Nahid et al. (2014) measured hundreds of proteins in serum samples from 39 patients and identified a five-marker signature (IL-11, Rα, α2-Antiplasmin, PSME1, and SAA) that predicted conversion with an estimated 80 ± 11% sensitivity and 80 ± 7% specificity upon internal cross-validation. All of these studies require confirmation in independent cohorts and—most importantly—correlation with relapse-free cure.
High-resolution 3-dimensional imaging provides unique opportunities here (Chen et al., 2014). In a population of patients with multidrug-resistant TB undergoing 2 yr of therapy, quantitative, volumetric changes in the uptake of 2-deoxy-2-[18F]-fluro-D-glucose (FDG) at 2 and 6 mo, as quantified by positron emission tomography/computed tomography (PET/CT), was correlated with treatment outcome. Additional studies in drug-sensitive cohorts should provide additional data on the potential utility of such modalities in monitoring TB patients. M. tuberculosis-specific reporter molecules (Backus et al., 2011) compatible with PET/CT imaging hold potential for providing complementary data. The potential ability of the latter techniques to enable quantification of mycobacterial load in vivo is particularly exciting. Because of the requirement for specialized equipment, complexity, and cost, however, the use of such methods will likely be restricted to clinical trials.
Thus, although there are promising developments, significant work remains to be done to identify and validate therapeutic response biomarkers that can provide timely, practical, and actionable information on patient outcomes. To achieve this, longitudinal studies to characterize pathogen and host biomarkers, alongside imaging-based methods, will likely be required.
Prevention of disease and infection
TSTs and IGRAs provide presumptive evidence of sustained infection but are unable to differentiate between latent infection and active disease. More importantly, these assays provide no predictive power for the risk of progression to active disease. The identification of biomarkers that facilitate practical stratification of latently infected individuals for risk of progression to active disease could allow for rational targeting of secondary prophylactic interventions, both existing and novel. Furthermore, mechanistic insight into the biology of progression provided by such biomarkers holds promise for the development of novel host-directed therapeutic and/or preventive approaches. Finally, such stratification has promise for aiding TB vaccine development. Any such biomarker would need to undergo confirmation and prospective validation and a move to an appropriate detection platform for use in clinical trials or, more broadly, in endemic settings (Fig. 1). As recently reviewed (Karp et al., 2015), the TB vaccine enterprise is at something of a crossroads. Lacking animal models known to be predictive, or any validated correlate of protection, the field lacks a clearly rational way to up- or down-select vaccine candidates for clinical development. Moreover, as prevention of disease has been the standard test of efficacy, and only 10% of (immunocompetent) hosts with sustained infection develop TB over a lifetime, efficacy trial sizes are large and prohibitively costly. Dramatic decreases in trial size and length might be secured by targeting such trials to those at highest risk for progression to active disease. Progress made in identifying and validating such biomarkers, on the host side, is noted below.
Studies to identify biomarkers that provide a quantitative assessment of the risk of progression to active disease are difficult to perform due to the low rate of progression, leading to the need to prospectively collect samples from large populations over time. Validation similarly requires a large prospectively followed cohort. A recent study identified a gene expression signature comprising 16–21 signal genes and 10 reference genes—mirroring the type I IFN/inflammatory signature reported in cross-sectional studies of active TB noted above—that predictively (months prior) discriminates progressors from controls with a sensitivity and specificity of 67 and 81%, respectively (unpublished data; W. Hanekom, personal communication). Such performance holds out the possibility of identifying individuals at risk for TB well before they become symptomatic, although specificity may need to be improved and gene number decreased to make practical use of such a test.
Although it remains to be determined whether type I IFN activity is causally linked to progression and disease, biological plausibility is high (Dorhoi et al., 2014; Mayer-Barber et al., 2014; McNab et al., 2014; Redford et al., 2014). Indeed, this link may well underlie the early susceptibility to TB seen after HIV infection—itself associated with sustained increases in type I IFN production (Hyrcza et al., 2007; Byrnes et al., 2008). It might also contribute to the spring peak in TB diagnosis observed across latitudes in temperate climes (Willis et al., 2012), as a result of winter respiratory virus-induced type I IFNs amplifying the dynamic flux of granuloma activity in latent infection. Defining causality could be achieved either through testing whether pulmonary delivery of exogenous type I IFNs can induce reactivation in a fit-for-purpose animal model of latency (Lin et al., 2014) or, more directly, through testing whether monoclonal antibody–mediated blockade of type I IFN activity prevents progression to disease in individuals at high risk. If causal, how might this be exploited? For one, it suggests targets for host-directed therapy. It also suggests the potential for immunizing against other common pathogens that drive robust type I IFN activity as a strategy for preventing TB.
A further impediment to the effective prevention of TB is the lack of a validated correlate of vaccine-induced protection to aim for, given the lack of a vaccine with demonstrated efficacy. Moreover, there is growing realization in the field that the presumptive mechanistic correlate underlying TB vaccine development for the past decades is likely to be inadequate or incorrect (Karp et al., 2015). The nomination of novel mechanistic correlates of protection for vaccine candidates to aim at, deriving from a deeper understanding of TB immunopathogenesis, from the study of outlier populations (e.g., those with persistent exposure in the absence of apparent sustained infection), and/or from insights derived from the definition of correlates of risk has real promise (Karp et al., 2015). However, these biomarker issues are beyond the scope of the current Perspective.
Prevention of sustained infection appears to be a rational goal for novel vaccine strategies (Karp et al., 2015). Testing such vaccines with current tools is somewhat problematic. Sustained TST and/or IGRA positivity does not provide endpoints as firm as disease endpoints. The development of sensitive ways for detecting and quantifying low M. tuberculosis burdens, and/or the presence, sustained or not, of low levels M. tuberculosis infection, discussed above, would be a major advance for the development of vaccines aimed at preventing sustained infection.
The definition of correlates of protection is beyond the purview of this Perspective. It bears noting, however, that our available tools for identifying outlier individuals whose immunological interrogation might nominate a novel vaccine concept based on eliciting a type of uncommon immunity—highly exposed individuals without apparent sustained infection—have a major, illustrative limitation. The lack of demonstrable TST and/or IGRA reactivity may reflect a lack of infection, the development of a conventional T cell response with test reversion after the achievement of sterile protection, or the development of an uncommon, protective immune response that is not measurable by assays that depend on T cell reactivity with proteins.
Moving the field forward
There is a pressing need for new tools. That said, these needs are often not well served by the current study paradigms, which are generally not guided by the clarity provided by careful consideration of intended use case and the TPPs that derive from such. Under each use case, a detailed TPP provides the guidance needed to evaluate biomarker performance and utility against intended use. Targets within the TPP should, when possible, be informed by modeling the potential impact of a specific set of product attributes. This approach provides clear targets for researchers and product developers, and actionable results when targets are met. The articulation of clear use cases allows studies and clinical cohorts to be designed against the intended use, with the relevant control groups and key follow-up clinical data collected. Study size is often limited for early, exploratory biomarker projects. When moving to validation, appropriate power calculations are needed. It is noted that transformative progress may require funders to align behind a definitive approach to these challenges rather than enabling the proliferation of multiple, redundant, sub optimal studies.
Though publication of new biomarkers is common, refinement, validation and independent confirmation of such is not. Further, once discovered, the performance of proposed biomarkers is likely to benefit from focused efforts to improve the sensitivity and specificity and/or reduce the assay to manufacturability and practicality—bridging the gap between exploratory identification and reduction to practice by developers. A framework for these efforts is outlined in Fig. 1.
Acknowledgments
The authors thank C. Wilson, L. Stuart, K. Duncan, G. Kaplan, A. Kasmar, and W. Hannekom for helpful discussions.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- IGRA
- IFN-γ release assay
- LAM
- lipoarabinomannan
- TB
- tuberculosis
- TPP
- target product profile
- TST
- tuberculin skin test
References
- Aceti A., Zanetti S., Mura M.S., Sechi L.A., Turrini F., Saba F., Babudieri S., Mannu F., and Fadda G.. 1999. Identification of HIV patients with active pulmonary tuberculosis using urine based polymerase chain reaction assay. Thorax. 54:145–146. 10.1136/thx.54.2.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adekambi T., Ibegbu C.C., Cagle S., Kalokhe A.S., Wang Y.F., Hu Y., Day C.L., Ray S.M., and Rengarajan J.. 2015. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J. Clin. Invest. 125:1827–1838. 10.1172/JCI77990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.T., Kaforou M., Brent A.J., Wright V.J., Banwell C.M., Chagaluka G., Crampin A.C., Dockrell H.M., French N., Hamilton M.S., et al. KIDS TB Study Group. 2014. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med. 370:1712–1723. 10.1056/NEJMoa1303657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade B.B., Pavan Kumar N., Mayer-Barber K.D., Barber D.L., Sridhar R., Rekha V.V., Jawahar M.S., Nutman T.B., Sher A., and Babu S.. 2013. Plasma heme oxygenase-1 levels distinguish latent or successfully treated human tuberculosis from active disease. PLoS ONE. 8:e62618 10.1371/journal.pone.0062618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus K.M., Boshoff H.I., Barry C.S., Boutureira O., Patel M.K., D’Hooge F., Lee S.S., Via L.E., Tahlan K., Barry C.E. III, and Davis B.G.. 2011. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat. Chem. Biol. 7:228–235. 10.1038/nchembio.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto M.L., Pereira S.M., Pilger D., Cruz A.A., Cunha S.S., Sant’Anna C., Ichihara M.Y., Genser B., and Rodrigues L.C.. 2011. Evidence of an effect of BCG revaccination on incidence of tuberculosis in school-aged children in Brazil: second report of the BCG-REVAC cluster-randomised trial. Vaccine. 29:4875–4877. 10.1016/j.vaccine.2011.05.023 [DOI] [PubMed] [Google Scholar]
- Barry C.E. III, Boshoff H.I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R.J., and Young D.. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7:845–855. 10.1038/nrmicro2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy R., Sotgiu G., Altet-Gómez N., Tsolia M., Ruga E., Velizarova S., and Kampmann B.. 2012. Identifying predictors of interferon-γ release assay results in pediatric latent tuberculosis: a protective role of bacillus Calmette-Guerin?: a pTB-NET collaborative study. Am. J. Respir. Crit. Care Med. 186:378–384. 10.1164/rccm.201201-0026OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann R., Kaempfer S., Chegou N.N., Nene N.F., Veenstra H., Spallek R., Bolliger C.T., Lukey P.T., van Helden P.D., Singh M., and Walzl G.. 2013. Serodiagnostic markers for the prediction of the outcome of intensive phase tuberculosis therapy. Tuberculosis (Edinb.). 93:239–245. 10.1016/j.tube.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Bentley-Hibbert S.I., Quan X., Newman T., Huygen K., and Godfrey H.P.. 1999. Pathophysiology of antigen 85 in patients with active tuberculosis: antigen 85 circulates as complexes with fibronectin and immunoglobulin G. Infect. Immun. 67:581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M.P., Graham C.M., McNab F.W., Xu Z., Bloch S.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J., et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 466:973–977. 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom C.I., Graham C.M., Berry M.P., Wilkinson K.A., Oni T., Rozakeas F., Xu Z., Rossello-Urgell J., Chaussabel D., Banchereau J., et al. 2012. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS ONE. 7:e46191 10.1371/journal.pone.0046191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom C.I., Graham C.M., Berry M.P., Rozakeas F., Redford P.S., Wang Y., Xu Z., Wilkinson K.A., Wilkinson R.J., Kendrick Y., et al. 2013. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS ONE. 8:e70630 10.1371/journal.pone.0070630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme C.C., Nabeta P., Hillemann D., Nicol M.P., Shenai S., Krapp F., Allen J., Tahirli R., Blakemore R., Rustomjee R., et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015. 10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes A.A., Harris D.M., Atabani S.F., Sabundayo B.P., Langan S.J., Margolick J.B., and Karp C.L.. 2008. Immune activation and IL-12 production during acute/early HIV infection in the absence and presence of highly active, antiretroviral therapy. J. Leukoc. Biol. 84:1447–1453. 10.1189/jlb.0708438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannas A., Goletti D., Girardi E., Chiacchio T., Calvo L., Cuzzi G., Piacentini M., Melkonyan H., Umansky S.R., Lauria F.N., et al. 2008. Mycobacterium tuberculosis DNA detection in soluble fraction of urine from pulmonary tuberculosis patients. Int. J. Tuberc. Lung Dis. 12:146–151. [PubMed] [Google Scholar]
- Capuano S.V. III, Croix D.A., Pawar S., Zinovik A., Myers A., Lin P.L., Bissel S., Fuhrman C., Klein E., and Flynn J.L.. 2003. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect. Immun. 71:5831–5844. 10.1128/IAI.71.10.5831-5844.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.C., Yang C.H., Chang L.Y., Wang K.F., Kuo Y.C., Lin C.J., Lee S.W., Hsueh P.R., Fang C.T., and Huang L.M.. 2013. Lower prevalence of tuberculosis infection in BCG vaccinees: a cross-sectional study in adult prison inmates. Thorax. 68:263–268. 10.1136/thoraxjnl-2012-202208 [DOI] [PubMed] [Google Scholar]
- Chan C.E., Götze S., Seah G.T., Seeberger P.H., Tukvadze N., Wenk M.R., Hanson B.J., and MacAry P.A.. 2015. The diagnostic targeting of a carbohydrate virulence factor from M.tuberculosis. Sci Rep. 5:10281 10.1038/srep10281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.Y., Dodd L.E., Lee M., Paripati P., Hammoud D.A., Mountz J.M., Jeon D., Zia N., Zahiri H., Coleman M.T., et al. 2014. PET/CT imaging correlates with treatment outcome in patients with multidrug-resistant tuberculosis. Sci. Transl. Med. 6:ra166 10.1126/scitranslmed.3009501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchyard G.J., Fielding K.L., Lewis J.J., Coetzee L., Corbett E.L., Godfrey-Faussett P., Hayes R.J., Chaisson R.E., Grant A.D., and Thibela T.B.S.T.. Thibela TB Study Team. 2014. A trial of mass isoniazid preventive therapy for tuberculosis control. N. Engl. J. Med. 370:301–310. 10.1056/NEJMoa1214289 [DOI] [PubMed] [Google Scholar]
- Colditz G.A., Brewer T.F., Berkey C.S., Wilson M.E., Burdick E., Fineberg H.V., and Mosteller F.. 1994. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA. 271:698–702. 10.1001/jama.1994.03510330076038 [DOI] [PubMed] [Google Scholar]
- Comas I., Coscolla M., Luo T., Borrell S., Holt K.E., Kato-Maeda M., Parkhill J., Malla B., Berg S., Thwaites G., et al. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45:1176–1182. 10.1038/ng.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz H.L., de Albuquerque Montenegro R., de Araújo Lima J.F., da Rocha Poroca D., da Costa Lima J.F., Maria Lapa Montenegro L., Crovella S., and Charifker Schindler H.. 2011. Evaluation of a nested-PCR for mycobacterium tuberculosis detection in blood and urine samples. Braz. J. Microbiol. 42:321–329. 10.1590/S1517-83822011000100041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B., Kashino S.S., Pulu I., Kalita D., Swami V., Yeger H., Felsher D.W., and Campos-Neto A.. 2013. CD271+ bone marrow mesenchymal stem cells may provide a niche for dormant Mycobacterium tuberculosis. Sci. Transl. Med. 5:70ra13 10.1126/scitranslmed.3004912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.L., Cattamanchi A., Cuevas L.E., Hopewell P.C., and Steingart K.R.. 2013. Diagnostic accuracy of same-day microscopy versus standard microscopy for pulmonary tuberculosis: a systematic review and meta-analysis. Lancet Infect. Dis. 13:147–154. 10.1016/S1473-3099(12)70232-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmadhikari A.S., Basaraba R.J., Van Der Walt M.L., Weyer K., Mphahlele M., Venter K., Jensen P.A., First M.W., Parsons S., McMurray D.N., et al. 2011. Natural infection of guinea pigs exposed to patients with highly drug-resistant tuberculosis. Tuberculosis (Edinb.). 91:329–338. 10.1016/j.tube.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A., Yeremeev V., Nouailles G., Weiner J. III, Jörg S., Heinemann E., Oberbeck-Müller D., Knaul J.K., Vogelzang A., Reece S.T., et al. 2014. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol. 44:2380–2393. 10.1002/eji.201344219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C. 2013. Making wider use of the world’s most widely used vaccine: Bacille Calmette-Guerin revaccination reconsidered. J. R. Soc. Interface. 10:20130365 10.1098/rsif.2013.0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M., Paranjothy S., Abubakar I., Bracebridge S., Lilley M., Mulla R., Lack K., Chalkley D., and McEvoy M.. 2009. BCG vaccination reduces risk of infection with Mycobacterium tuberculosis as detected by gamma interferon release assay. Vaccine. 27:6116–6120. 10.1016/j.vaccine.2009.08.031 [DOI] [PubMed] [Google Scholar]
- Friedrich S.O., Rachow A., Saathoff E., Singh K., Mangu C.D., Dawson R., Phillips P.P., Venter A., Bateson A., Boehme C.C., et al. Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA). 2013. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir Med. 1:462–470. 10.1016/S2213-2600(13)70119-X [DOI] [PubMed] [Google Scholar]
- Garton N.J., Waddell S.J., Sherratt A.L., Lee S.M., Smith R.J., Senner C., Hinds J., Rajakumar K., Adegbola R.A., Besra G.S., et al. 2008. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5:e75 10.1371/journal.pmed.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie S.H., Crook A.M., McHugh T.D., Mendel C.M., Meredith S.K., Murray S.R., Pappas F., Phillips P.P., Nunn A.J., and Consortium R.E.. REMoxTB Consortium. 2014. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N. Engl. J. Med. 371:1577–1587. 10.1056/NEJMoa1407426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasur B., Bruchfeld J., van Helden P., Källenius G., and Svenson S.. 2015. A sensitive urinary lipoarabinomannan test for tuberculosis. PLoS ONE. 10:e0123457 10.1371/journal.pone.0123457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.L., Lee H.C., Yu C.W., Chen H.C., Wang C.C., Wu J.Y., and Lee C.C.. 2014. Value of procalcitonin in differentiating pulmonary tuberculosis from other pulmonary infections: a meta-analysis. Int. J. Tuberc. Lung Dis. 18:470–477. 10.5588/ijtld.13.0449 [DOI] [PubMed] [Google Scholar]
- Hur Y.G., Kang Y.A., Jang S.H., Hong J.Y., Kim A., Lee S.A., Kim Y., and Cho S.N.. 2015. Adjunctive biomarkers for improving diagnosis of tuberculosis and monitoring therapeutic effects. J. Infect. 70:346–355. 10.1016/j.jinf.2014.10.019 [DOI] [PubMed] [Google Scholar]
- Hyrcza M.D., Kovacs C., Loutfy M., Halpenny R., Heisler L., Yang S., Wilkins O., Ostrowski M., and Der S.D.. 2007. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 81:3477–3486. 10.1128/JVI.01552-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaforou M., Wright V.J., Oni T., French N., Anderson S.T., Bangani N., Banwell C.M., Brent A.J., Crampin A.C., Dockrell H.M., et al. 2013. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 10:e1001538 10.1371/journal.pmed.1001538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp C.L., Wilson C.B., and Stuart L.M.. 2015. Tuberculosis vaccines: barriers and prospects on the quest for a transformative tool. Immunol. Rev. 264:363–381. 10.1111/imr.12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap R.S., Rajan A.N., Ramteke S.S., Agrawal V.S., Kelkar S.S., Purohit H.J., Taori G.M., and Daginawala H.F.. 2007. Diagnosis of tuberculosis in an Indian population by an indirect ELISA protocol based on detection of Antigen 85 complex: a prospective cohort study. BMC Infect. Dis. 7:74 10.1186/1471-2334-7-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnath-Velayudhan S., Salamon H., Wang H.Y., Davidow A.L., Molina D.M., Huynh V.T., Cirillo D.M., Michel G., Talbot E.A., Perkins M.D., et al. 2010. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. USA. 107:14703–14708. 10.1073/pnas.1009080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn S.D., and Wood R.. 2011. Tuberculosis in antiretroviral treatment services in resource-limited settings: addressing the challenges of screening and diagnosis. J. Infect. Dis. 204(Suppl 4):S1159–S1167. 10.1093/infdis/jir411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn S.D., Kerkhoff A.D., Burton R., Schutz C., van Wyk G., Vogt M., Pahlana P., Nicol M.P., and Meintjes G.. 2015. Rapid microbiological screening for tuberculosis in HIV-positive patients on the first day of acute hospital admission by systematic testing of urine samples using Xpert MTB/RIF: a prospective cohort in South Africa. BMC Med. 13:192 10.1186/s12916-015-0432-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesho E., Forestiero F.J., Hirata M.H., Hirata R.D., Cecon L., Melo F.F., Paik S.H., Murata Y., Ferguson E.W., Wang Z., and Ooi G.T.. 2011. Transcriptional responses of host peripheral blood cells to tuberculosis infection. Tuberculosis (Edinb.). 91:390–399. 10.1016/j.tube.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Lin P.L., Dietrich J., Tan E., Abalos R.M., Burgos J., Bigbee C., Bigbee M., Milk L., Gideon H.P., Rodgers M., et al. 2012. The multistage vaccine H56 boosts the effects of BCG to protect cynomolgus macaques against active tuberculosis and reactivation of latent Mycobacterium tuberculosis infection. J. Clin. Invest. 122:303–314. 10.1172/JCI46252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.L., Coleman T., Carney J.P., Lopresti B.J., Tomko J., Fillmore D., Dartois V., Scanga C., Frye L.J., Janssen C., et al. 2013. Radiologic responses in cynomolgous macaques for assessing tuberculosis chemotherapy regimens. Antimicrob. Agents Chemother. 57:4237–4244. 10.1128/AAC.00277-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.L., Ford C.B., Coleman M.T., Myers A.J., Gawande R., Ioerger T., Sacchettini J., Fortune S.M., and Flynn J.L.. 2014. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat. Med. 20:75–79. 10.1038/nm.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Wu J., Wang H., Wang S., Diao N., Wang F., Gao Y., Chen J., Shao L., Weng X., et al. 2011. Novel biomarkers distinguishing active tuberculosis from latent infection identified by gene expression profile of peripheral blood mononuclear cells. PLoS ONE. 6:e24290 10.1371/journal.pone.0024290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Ota M., Repsilber D., Mollenkopf H.J., Weiner J., Hill P.C., and Kaufmann S.H.. 2011. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS ONE. 6:e26938 10.1371/journal.pone.0026938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Weiner J. III, Mollenkopf H.J., Bauer T., Prasse A., Müller-Quernheim J., Kaufmann S.H., and Kaufmann S.H.. TBornotTB Network. 2012. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. USA. 109:7853–7858. 10.1073/pnas.1121072109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangtani P., Abubakar I., Ariti C., Beynon R., Pimpin L., Fine P.E., Rodrigues L.C., Smith P.G., Lipman M., Whiting P.F., and Sterne J.A.. 2014. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin. Infect. Dis. 58:470–480. 10.1093/cid/cit790 [DOI] [PubMed] [Google Scholar]
- Mayer-Barber K.D., Andrade B.B., Oland S.D., Amaral E.P., Barber D.L., Gonzales J., Derrick S.C., Shi R., Kumar N.P., Wei W., et al. 2014. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 511:99–103. 10.1038/nature13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F.W., Ewbank J., Howes A., Moreira-Teixeira L., Martirosyan A., Ghilardi N., Saraiva M., and O’Garra A.. 2014. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-γ for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J. Immunol. 193:3600–3612. 10.4049/jimmunol.1401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C.C., O’Grady F., and Riley R.L.. 1960. Tuberculin conversion in the “naturally infected” guinea pig. Bull. Johns Hopkins Hosp. 106:36–45. [PubMed] [Google Scholar]
- Minion J., Leung E., Talbot E., Dheda K., Pai M., and Menzies D.. 2011. Diagnosing tuberculosis with urine lipoarabinomannan: systematic review and meta-analysis. Eur. Respir. J. 38:1398-1405. 10.1183/09031936.00025711 [DOI] [PubMed] [Google Scholar]
- Miotto P., Mwangoka G., Valente I.C., Norbis L., Sotgiu G., Bosu R., Ambrosi A., Codecasa L.R., Goletti D., Matteelli A., et al. 2013. miRNA signatures in sera of patients with active pulmonary tuberculosis. PLoS ONE. 8:e80149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K. 2011. WHO recommends against inaccurate tuberculosis tests. Lancet. 377:113–114. 10.1016/S0140-6736(11)60005-6 [DOI] [PubMed] [Google Scholar]
- Mukamolova G.V., Turapov O., Malkin J., Woltmann G., and Barer M.R.. 2010. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am. J. Respir. Crit. Care Med. 181:174–180. 10.1164/rccm.200905-0661OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan H., Kumar S., Price D.N., Ray S.M., Lee Y.J., Min S., Eum S., Kubicek-Sutherland J., Resnick J.M., Grace W.K., et al. 2012. Rapid detection of Mycobacterium tuberculosis biomarkers in a sandwich immunoassay format using a waveguide-based optical biosensor. Tuberculosis (Edinb.). 92:407–416. 10.1016/j.tube.2012.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C.J., Ortblad K.F., Guinovart C., Lim S.S., Wolock T.M., Roberts D.A., Dansereau E.A., Graetz N., Barber R.M., Brown J.C., et al. 2014. Global, regional, and national incidence and mortality for HIV, tuberculosis, and malaria during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 384:1005–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid P., Bliven-Sizemore E., Jarlsberg L.G., De Groote M.A., Johnson J.L., Muzanyi G., Engle M., Weiner M., Janjic N., Sterling D.G., and Ochsner U.A.. 2014. Aptamer-based proteomic signature of intensive phase treatment response in pulmonary tuberculosis. Tuberculosis (Edinb.). 94:187–196. 10.1016/j.tube.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakiyingi L., Moodley V.M., Manabe Y.C., Nicol M.P., Holshouser M., Armstrong D.T., Zemanay W., Sikhondze W., Mbabazi O., Nonyane B.A., et al. 2014. Diagnostic accuracy of a rapid urine lipoarabinomannan test for tuberculosis in HIV-infected adults. J. Acquir. Immune Defic. Syndr. 66:270–279. 10.1097/QAI.0000000000000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano D.R., Pollock N., Kashino S.S., Rodrigues V. Jr., and Campos-Neto A.. 2008. Identification of Mycobacterium tuberculosis ornithine carboamyltransferase in urine as a possible molecular marker of active pulmonary tuberculosis. Clin. Vaccine Immunol. 15:638–643. 10.1128/CVI.00010-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyrolles O., Hernández-Pando R., Pietri-Rouxel F., Fornès P., Tailleux L., Barrios Payán J.A., Pivert E., Bordat Y., Aguilar D., Prévost M.C., et al. 2006. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS ONE. 1:e43 10.1371/journal.pone.0000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolayevskyy V., Miotto P., Pimkina E., Balabanova Y., Kontsevaya I., Ignatyeva O., Ambrosi A., Skenders G., Ambrozaitis A., Kovalyov A., et al. 2015. Utility of propidium monoazide viability assay as a biomarker for a tuberculosis disease. Tuberculosis (Edinb.). 95:179–185. 10.1016/j.tube.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Phalane K.G., Kriel M., Loxton A.G., Menezes A., Stanley K., van der Spuy G.D., Walzl G., and Chegou N.N.. 2013. Differential expression of host biomarkers in saliva and serum samples from individuals with suspected pulmonary tuberculosis. Mediators Inflamm. 2013:981984 10.1155/2013/981984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M., Basa-Dalay V., Bothamley G., Cataneo R.N., Lam P.K., Natividad M.P., Schmitt P., and Wai J.. 2010. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb.). 90:145–151. 10.1016/j.tube.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Redford P.S., Mayer-Barber K.D., McNab F.W., Stavropoulos E., Wack A., Sher A., and O’Garra A.. 2014. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J. Infect. Dis. 209:270–274. 10.1093/infdis/jit424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R.L., Mills C.C., Nyka W., Weinstock N., Storey P.B., Sultan L.U., Riley M.C., and Wells W.F.. 1995. Aerial dissemination of pulmonary tuberculosis. A two-year study of contagion in a tuberculosis ward. 1959. Am. J. Epidemiol. 142:3–14. [DOI] [PubMed] [Google Scholar]
- Riou C., Perez Peixoto B., Roberts L., Ronacher K., Walzl G., Manca C., Rustomjee R., Mthiyane T., Fallows D., Gray C.M., and Kaplan G.. 2012. Effect of standard tuberculosis treatment on plasma cytokine levels in patients with active pulmonary tuberculosis. PLoS ONE. 7:e36886 10.1371/journal.pone.0036886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Eisenhut M., Harris R.J., Rodrigues L.C., Sridhar S., Habermann S., Snell L., Mangtani P., Adetifa I., Lalvani A., and Abubakar I.. 2014. Effect of BCG vaccination against Mycobacterium tuberculosis infection in children: systematic review and meta-analysis. BMJ. 349(aug04 5):g4643 10.1136/bmj.g4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.G. 2011. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol. Rev. 240:252–268. 10.1111/j.1600-065X.2010.00984.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.G., Barry C.E. III, and Flynn J.L.. 2010. Tuberculosis: what we don’t know can, and does, hurt us. Science. 328:852–856. 10.1126/science.1184784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana S., Subbaraman R., Shete P., Gore G., Das J., Cattamanchi A., Mayer K., Menzies D., Harries A.D., Hopewell P., and Pai M.. 2015. Quality of tuberculosis care in India: a systematic review. Int. J. Tuberc. Lung Dis. 19:751–763. 10.5588/ijtld.15.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Kasambira T.S., Adrian P.V., Madhi S.A., Martinson N.A., and Dorman S.E.. 2011. Longitudinal analysis of QuantiFERON-TB Gold In-Tube in children with adult household tuberculosis contact in South Africa: a prospective cohort study. PLoS ONE. 6:e26787 10.1371/journal.pone.0026787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soysal A., Millington K.A., Bakir M., Dosanjh D., Aslan Y., Deeks J.J., Efe S., Staveley I., Ewer K., and Lalvani A.. 2005. Effect of BCG vaccination on risk of Mycobacterium tuberculosis infection in children with household tuberculosis contact: a prospective community-based study. Lancet. 366:1443–1451. 10.1016/S0140-6736(05)67534-4 [DOI] [PubMed] [Google Scholar]
- Starke J.R. 2003. Pediatric tuberculosis: time for a new approach. Tuberculosis (Edinb.). 83:208–212. 10.1016/S1472-9792(02)00088-4 [DOI] [PubMed] [Google Scholar]
- Theron G., Peter J., Calligaro G., Meldau R., Hanrahan C., Khalfey H., Matinyenya B., Muchinga T., Smith L., Pandie S., et al. 2014. Determinants of PCR performance (Xpert MTB/RIF), including bacterial load and inhibition, for TB diagnosis using specimens from different body compartments. Sci Rep. 4:5658 10.1038/srep05658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunz B.B., Fine P., and Dye C.. 2006. Effect of BCG vaccination on childhood tuberculous meningitis and miliary tuberculosis worldwide: a meta-analysis and assessment of cost-effectiveness. Lancet. 367:1173–1180. 10.1016/S0140-6736(06)68507-3 [DOI] [PubMed] [Google Scholar]
- Wallis R.S. 2015. Sputum culture conversion as a tuberculosis biomarker: a glass half empty or half full? The Lancet. Respir. Med. 3:174–175. [DOI] [PubMed] [Google Scholar]
- WHO. 2014a. Global Tuberculosis Report 2014. http://www.who.int/tb/publications/global_report/en/
- WHO. 2014b. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. http://www.who.int/tb/publications/tpp_report/en/
- Willis M.D., Winston C.A., Heilig C.M., Cain K.P., Walter N.D., and Mac Kenzie W.R.. 2012. Seasonality of tuberculosis in the United States, 1993-2008. Clin. Infect. Dis. 54:1553–1560. 10.1093/cid/cis235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R., and Bekker L.G.. 2014. Isoniazid preventive therapy for tuberculosis in South Africa: an assessment of the local evidence base. S. Afr. Med. J. 104:174–177. 10.7196/samj.7968 [DOI] [PubMed] [Google Scholar]
- Young B.L., Mlamla Z., Gqamana P.P., Smit S., Roberts T., Peter J., Theron G., Govender U., Dheda K., and Blackburn J.. 2014. The identification of tuberculosis biomarkers in human urine samples. Eur. Respir. J. 43:1719-1729. 10.1183/09031936.00175113. [DOI] [PubMed] [Google Scholar]
- Zar H.J., Hanslo D., Apolles P., Swingler G., and Hussey G.. 2005. Induced sputum versus gastric lavage for microbiological confirmation of pulmonary tuberculosis in infants and young children: a prospective study. Lancet. 365:130–134. 10.1016/S0140-6736(05)17702-2 [DOI] [PubMed] [Google Scholar]
- Zhang X., Guo J., Fan S., Li Y., Wei L., Yang X., Jiang T., Chen Z., Wang C., Liu J., et al. 2013. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS ONE. 8:e81076 10.1371/journal.pone.0081076 [DOI] [PMC free article] [PubMed] [Google Scholar]