Abstract
The pathophysiology of IDH mutations in tumorigenesis is increasingly described, yet the prognostic significance of IDH1 and IDH2 mutations in AML remains controversial. The primary objective of this study was to define the natural history and prognosis of patients with AML and IDH1 or IDH2 mutations and provide historical survival expectations. A total of 826 patients treated from 2010 to 2014 at a single institution were evaluated, including 167 patients (20%) with AML and IDH1 or IDH2 mutations. Median age was 62 years (range 18–92). There were 59 IDH1-R132, 83 IDH2-R140, and 23 IDH2-R172 mutations. Clinicopathologic characteristics associated with IDH-mutations included older age, less frequent therapy-related status, and increased incidence of intermediate-risk cytogenetics, FLT3-ITD mutations, and NPM1 mutations. Remission rates (CR/CRi) by AML treatment status were: induction, 68%; Salvage-1 (S1), 42%; and Salvage-2 and beyond (S2+), 27%. No difference in response was identified by IDH mutation status. Similarly, overall survival (OS) was not dependent on IDH status within any cohort. The median OS was 15.4 months in induction, 8.7 months in S1, and 4.8 months in S2+. This analysis defines the clinical outcome associated with IDH-mutations in both the front-line and salvage AML treatment settings, and confirms that response rate and OS for both IDH-mutated and IDH wild-type AML patients is comparable. This provides contemporary data to be used for comparison with results of novel investigational (e.g., selective IDH inhibitor) strategies.
Introduction
Since the discovery of mutations within the genes for the isocitrate dehydrogenase (IDH) enzymes IDH1 and IDH2 among patients with acute myeloid leukemia (AML), much insight has been gained regarding the incidence and unique pathophysiology of these mutations. The recurring trio of pathogenic IDH mutations in AML patients, IDH1-R132, IDH2-R172, and IDH2-R140, occur within the conserved active site and lead to loss of the expected Krebs Cycle reaction of isocitrate to alpha-ketoglutarate (αKG), while promoting a reverse reaction reducing αKG to the onco-metabolite 2-hydroxyglutarate (2HG) [1]. 2HG accumulation is purported to block normal cellular differentiation and promote tumorigenesis through competitive inhibition of αKG-dependent enzymatic activities, such as TET2-dependent DNA hydroxymethylation, histone demethylation, and HIF-1α activation [2–4]. Supporting evidence demonstrates AML with IDH mutations are specifically characterized by a distinct and globally hypermethylated DNA signature leading to impaired hematopoietic differentiation, which can be reversed with small molecule IDH inhibition [5–7].
IDH1/2 mutations are noted in ~20% in AML, including 6–16% for IDH1 and 8–19% for IDH2 mutations [8]. IDH1/2 mutations are identified more frequently in older patients, those with diploid or other intermediate-risk cytogenetics, and frequently co-occur with FLT3-ITD and NPM1 mutations, while being nearly mutually exclusive with both TET2 and WT1 mutations [9–13].
Despite the recent insights into the distinct pathophysiology of IDH mutants, the prognostic significance of IDH mutations remains controversial [14]. IDH1/2 mutations in conjunction with NPM1-mutant and FLT3-ITD negative molecular status have been associated with particularly favorable outcome in otherwise intermediate-risk AML [11,15,16], while other large studies have identified worse OS with IDH mutations including the NPM1-mutated IDH-mutated subset of patients [9,17–20], and other studies report a lack of prognostic significance [21–25].
The purpose of this analysis is to better define the clinical characteristics, natural history, and prognosis of AML patients with IDH-mutations, and to define the outcomes of IDH-mutated patients. This can be used as a reference expectation against which novel approaches and treatment strategies may be evaluated.
Methods
Eligible patients comprised all adults with a diagnosis of AML treated at M.D. Anderson Cancer Center (Houston, TX) from January 2010 to December 2014. Newly diagnosed and previously treated patients were eligible. A total of 826 patients with known IDH1 and IDH2 status and who received treatment at MD Anderson were included; these included 167 patients with IDH mutations. Details of treatments received are provided in Supporting Information Table 1.
From January 2010 to September 2012, molecular analysis was performed using polymerase chain reaction (PCR) amplification followed by Sanger sequencing using previously described methodology and PCR primers from Integrated DNA Technologies [26]. Beginning in September 2012, IDH molecular testing was performed within our institutional next-generation sequencing (NGS) hematologic malignancy platform within our CLIA-certified molecular diagnostics laboratory.
Patient characteristics are summarized using median (range) for continuous variables and frequency (percentage) for categorical variables. Categorical variables were compared for significance using the χ2 or Fisher’s exact test, and continuous variables were analyzed using the Wilcoxon Rank-Sum test. Statistical analyses were conducted in SAS 9.0 and significance was defined as a P value of <0.05. Overall survival (OS) was measured as the time from presentation to date of death or date of last follow-up (censored), and was calculated by Kaplan–Meier method using the log-rank test. Informed consent was obtained following institutional guidelines and in accordance with the Declaration of Helsinki.
Results
A total of 826 patients with known IDH1 and IDH2 status were evaluated, including 167 patients (20% of cohort) with IDH1 or IDH2 mutations. Patients with IDH-mutations included 59 (7%) with IDH1-R132, 83 (10%) with IDH2-R140 mutations, and 23 (3%) with IDH2-R172 mutations. Due to the low frequency of IDH2-R172 mutations, patients with IDH2-R140 and IDH2-R172 mutations were analyzed together as IDH2 mutants. In two patients, both an IDH1-R132 and IDH2-R140 mutation were identified concurrently. Within this cohort, 562 (68%) patients presented at the time of AML diagnosis for induction treatment, 120 (15%) patients at the time of salvage-1 (S1) and 144 (17%) patients at the time of salvage-2 or beyond (S2+). Detailed clinical and disease-specific characteristics by mutational status are shown in Table I. The median age for all patients was 62 years (range 18–92); 373 patients (45%) were ≥65 years of age.
TABLE I.
Clinical Characteristics and Patient Outcome of Study Cohort
| Characteristic | IDH wild-type [n = 659] | IDH-mutated [n = 167] | P-value | IDH1 mutated [n = 59] | IDH2-mutated [n = 106] | P-value |
|---|---|---|---|---|---|---|
| Median age (range) | 61 (18–92) | 67 (22–90) | <0.0005 | 67 (45–83) | 66 (22–90) | 0.31 |
| Male sex (%) | 376 (57) | 94 (56) | 0.857 | 32 (52) | 62 (58) | 0.45 |
| WBC count (×109/L) | 5.2 (0.2–363) | 3.6 (0.2–227.1) | 0.18 | 2.8 (0.4–164.7) | 4.8 (0.2–227.1) | 0.11 |
| ANC (×109/L) | 1.1 (0–103.7) | 0.42 (0–24.2) | <0.0005 | 0.3 (0–11.0) | 0.7 (0–24.2) | 0.008 |
| PLT count (×109/L) | 44 (2–568) | 55 (1–547) | 0.025 | 59 (1–553) | 55 (4–547) | 0.98 |
| BM Blasts (%) | 41 (0–98) | 60 (2–99) | <0.0005 | 67 (1–98) | 57 (1–99) | 0.28 |
| PB Blasts (%) | 14 (0–99) | 30 (0–99) | <0.0005 | 30 (0–99) | 29 (0–99) | 0.76 |
| LDHa | 760 (7–17,486) | 624 (142–42,000) | 0.003 | 602 (323–6,639) | 626 (142–42,000) | 0.41 |
| Therapy-related | 112 (17) | 13 (8) | 0.003 | 5 (8) | 8 (8) | 0.80 |
| Cytogeneticsb | <0.0005 | 0.15 | ||||
| Favorable | 58 (9) | 1 (2) | 1 (2) | 0 (0) | ||
| Intermediate | 350 (53) | 129 (77) | 43 (70) | 86 (81) | ||
| Unfavorable | 227 (34) | 32 (19) | 16 (26) | 16 (15) | ||
| Indeterminate | 24 (4) | 5 (3) | 1 (2) | 4 (4) | ||
| FLT3-ITD mutations (%) | 126 (19) | 45 (27) | 0.035 | 12 (21) | 32 (30) | 0.20 |
| NPM1 mutations (%) | 112 (17) | 55 (33) | <0.0005 | 23 (38) | 32 (30) | 0.37 |
IDH indicates isocitrate dehydrogenase; WBC, white blood cell; ANC, absolute neutrophil count; PLT, platelet; BM, bone marrow; PB, peripheral blood; LDH; lactate dehydrogenase; FLT3-ITD, FLT3 internal tandem duplication; NPM1, nucleophosmin.
Institutional normal reference range for LDH is 313 to 618 IU/L.
ECOG/SWOG classification system.
Compared with IDH wild-type patients, IDH1 and IDH2 mutated patients were older (median age 67 years vs. 61 years, P <0.0005), had a higher platelet count (median 55 ×109/L vs. 44 ×109/L, P = 0.025) and increased bone marrow blast percentage at presentation (60% vs. 41%, P <0.0005) (Table I). While total white blood cell (WBC) count was similar for both IDH-mutated and wild-type patients, the peripheral blast percentage was significantly higher in IDH-mutants compared to IDH wild-type (30% vs. 14%, P <0.0005), while the absolute neutrophil count (ANC) of IDH-mutated patients was significantly lower (0.42 × 109/L vs. 1.1 × 109/L, P <0.0005). IDH1-mutated patients had a significantly lower ANC than the IDH2-mutated patients (P = 0.008), this was the only clinicopathologic characteristic evaluated that differed between IDH1 and IDH2-mutated patients in our cohort (Table I). Additionally, IDH-mutated patients were more likely to have intermediate-risk cytogenetics (77% vs. 53%, P <0.0005), less likely to have therapy-related AML (8% vs. 17%, P = 0.003) and IDH-mutations were more frequently associated with the concurrent presence of FLT3-ITD mutations (27% vs. 19%, P = 0.035) and NPM1 mutations (33% vs. 17%, P <0.0005).
As expected, remission rates differed significantly based on AML treatment (salvage) status. Overall complete remission and complete remission with incomplete count recovery (CR/CRi) rate was 68% for AML induction, 42% for S1, and 27% for S2+ patients. Within each stage of treatment, response rate was not impacted by IDH mutational status (Table II). Treatment strategies received were heterogeneous and were dependent on factors such as patient age, performance status, comorbidities, and therapies received prior to institutional referral in applicable patients, and are detailed in Supporting Information Table 1. In the (n = 219) patients less than 60 years of age receiving induction AML therapy, the CR/CRi rate was 83% (n = 183); in the (n = 343) patients ≥60 years of age, the CR/CRi rate was 53% (n = 182), and response by age group was not affected by the presence or absence of IDH mutations.
TABLE II.
Response to Treatment by Salvage Status and IDH Mutation Status
| n | CR/CRi | P-value | Median OS | P-value | |
|---|---|---|---|---|---|
| Induction | 562 | 382 (68%) | 0.33 | 15.4 mo | 0.59 |
| IDH1+ | 37 | 24 (65) | 13.0 mo | ||
| IDH2+ | 69 | 42 (61) | 15.7 mo | ||
| IDH wild-type | 456 | 316 (69) | 15.3 mo | ||
| Salvage-1 | 120 | 50 (42%) | 0.76 | 8.7 mo | 0.44 |
| IDH1+ | 9 | 4 (40) | 5.9 mo | ||
| IDH2+ | 18 | 9 (50) | 11.1 mo | ||
| IDH wild-type | 92 | 37 (41) | 7.7 mo | ||
| ≥ Salvage-2 | 144 | 40 (27%) | 0.76 | 4.8 mo | 0.16 |
| IDH1+ | 14 | 5 (36) | 4.0 mo | ||
| IDH2+ | 19 | 5 (26) | 5.9 mo | ||
| IDH wild-type | 111 | 30 (27) | 4.8 mo |
IDH indicates isocitrate dehydrogenase; CR, complete remission; CRi, complete remission with incomplete count recovery; OS, overall survival.
With regards to hypomethylating agent (HMA) therapy in AML patients with IDH mutations, we evaluated specifically the (n = 175) patients receiving induction AML therapy with a HMA-based regimen. This included 48 IDH1/2-mutated patients and 127 IDH wild-type patients, with a median age of 75 (range 37–92). In this HMA-based induction treatment cohort, there was no impact of IDH-mutations on the overall response rate (45% vs. 58%, P = 0.13) or overall survival (9.5 vs. 10.3 months, P = 0.8) in this elderly induction treatment cohort (Supporting Information Fig. 1).
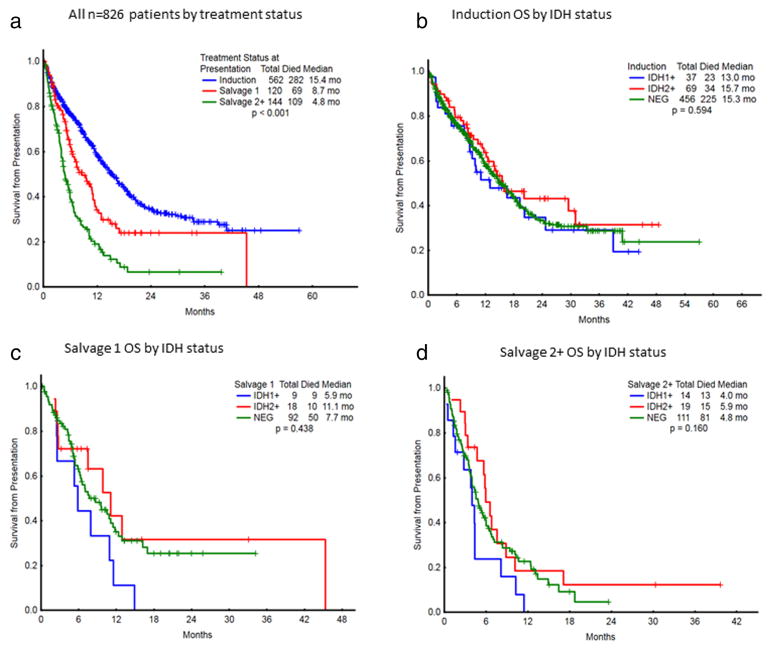
Overall survival (OS) by IDH mutational status and treatment status is presented in Table II and Fig. 1. There were no statistically significant differences in OS based on the presence of IDH1 or IDH2 mutations for any treatment cohort, either in the induction setting, or in either salvage setting. As no significant survival differences were identified in IDH2-mutant patients based on the presence of an IDH2-R140 versus IDH2-R172 mutation, IDH2-mutant patients were analyzed together due to the low frequency of IDH2-R172 mutations. In newly-diagnosed patients, median OS was 13.0 months for IDH1-mutated, 15.7 months for IDH2-mutated, and 15.3 months for IDH wild-type patients (P = 0.59). Median OS in the S1 setting was 5.9 months for IDH1-mutated, 11.1 months for IDH2-mutated, and 7.7 months for IDH wild-type patients (P = 0.44). For patients treated at the time of Salvage-2 and beyond, median OS was 4.0 months for IDH1-mutated, 5.9 months for IDH2-mutated, and 4.8 months for IDH wild-type (P = 0.16) (Fig. 2).
Figure 1.
(a) Overall survival of all n = 826 patients by treatment status. (b) Induction OS by IDH status. (c) Salvage 1 OS by IDH status. (d) Salvage 2+OS by IDH status.
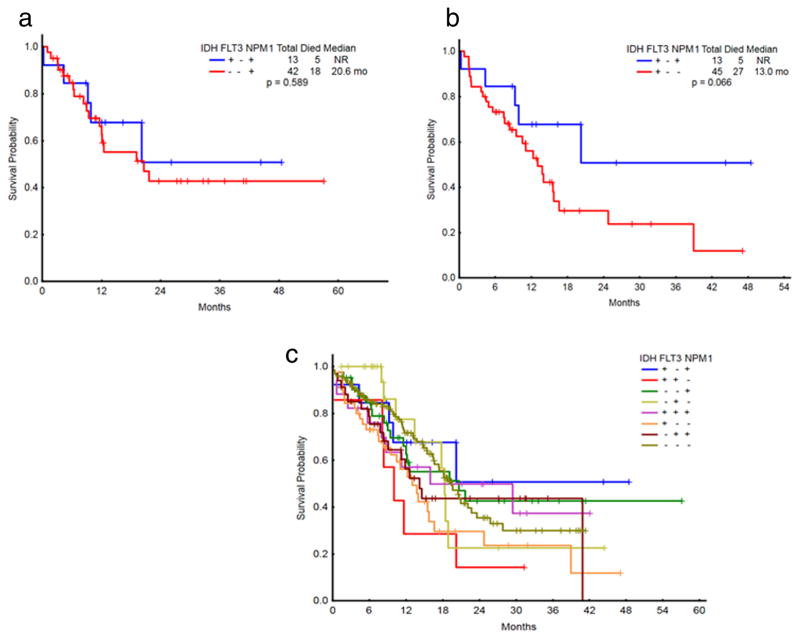
Figure 2.
(a) OS of newly diagnosed AML patients with intermediate-risk cytogenetics by molecular status (IDH+/NPM1+compared to NPM1+alone). (b) OS of newly diagnosed AML patients with intermediate-risk cytogenetics by molecular status (IDH+/NPM1+compared to IDH+alone). (c) OS of newly diagnosed AML patients with intermediate-risk cytogenetics by molecular status.
We then analyzed the 310 patients with intermediate-risk cytogenetics [27] presenting at diagnosis, in order to evaluate the impact of FLT3-ITD, NPM1, and IDH1/2 molecular combinations on OS within this molecularly defined induction subgroup (Fig. 2). Patients with intermediate-risk cytogenetics and NPM1 mutations alone (n = 42) had a median OS of 20.6 mo and a 2-yr OS of 43%, which was not unlike the outcome of intermediate-risk patients without FLT3-ITD and with concurrent NPM1 and IDH mutations (n = 13, 2-yr OS of 51%, with median OS not yet reached, P = 0.59, Fig. 2a). This compares with a median OS of 13.0 months and 2-yr OS of 30% in the IDH-mutant only intermediate-risk cohort (P = 0.067). When further evaluating only those patients in the front-line intermediate-risk cohort with age restricted to less than 60 (n = 122), the 5 patients with concurrent NPM1 and IDH mutations were noted to have 2-year OS of 100% [median follow-up time of 16 months], which was not significantly different from the outcome of IDH-only mutations (n = 10, 2-yr OS 60%, P = 0.10) and NPM1-only mutations (n = 16, 2-yr OS 58%, P = 0.10) (Supporting Information Fig. 1).
Discussion
The primary aim of the current analysis was to define the prognostic significance of IDH1 and IDH2 mutations in patients with AML. While several studies have reported on the incidence and prognosis of IDH mutations in patients with AML, available data is conflicting and the significance of IDH mutations on AML outcome has been unclear. This question is of primary importance given the recent development of novel therapeutic agents targeting mutant IDH1 or IDH2 with promising preliminary clinical results, with objective responses seen in ~50% of AML patients treated in the relapsed/refractory setting with IDH1 or IDH2 inhibitors [28,29]. Importantly, our study provides a useful baseline expectation for patient outcomes, against which investigational strategies can be assessed.
We confirm that IDH-mutated patients have distinctive clinicopathologic characteristics including older age, increased incidence of FLT3-ITD and NPM1 mutations, intermediate-risk cytogenetics, higher platelet count, and increased bone marrow blast percentage at diagnosis [14]. We additionally identify increased circulating blasts and decreased ANC in IDH-mutated patients, and less frequent occurrence of IDH-mutations in the setting of therapy-related AML.
We were unable to confirm a uniquely improved OS of patients with AML with FLT3-ITD negative, NPM1-mutated, and IDH1/2-mutated intermediate-risk disease. In the study by Patel et al., the 21 patients with this molecular phenotype had a 3-year OS of 89%, compared with 31% in the FLT3-ITD negative, NPM1-mutated, and IDH wild-type population [11]. This compares to 2-year OS of 51% and 43% in our cohort equivalents. This disparity is likely in part accounted for by difference in patient age between studies, with exclusively patients <60 years in the former study, and a median age of 62 years in our current study population. Notably, when we analyzed only those front-line induction AML patients with intermediate-risk cytogenetics and FLT3-ITD negative, NPM1-mutated, and IDH wild-type status who were also <60 years of age, the five corresponding patients were noted to have 100% OS with a median follow-up time of 16 months, which due to the small numbers of younger induction patients in our cohort was not of statistical significance.
Most importantly, we describe the natural history of patients with IDH-mutated AML, which to the best of our knowledge is one of the largest studies of clinical outcomes by IDH status and salvage treatment status to date. We confirm that response rate and OS for IDH-mutated is not dissimilar to IDH wild-type AML at all treatment junctures, and provide a useful reference for comparison for future studies in this patient population. Lastly, we enthusiastically note the improved outcomes in AML salvage therapy among all AML patients compared to historical controls, particularly including historical analyses from our institution [30,31]. Compared to an OS of less than 2 months for S2+ AML two decades ago, current S2+ median survival has more than tripled, due to continued gains in supportive care, participation in promising clinical trials, and available effective salvage therapies.
Supplementary Material
Acknowledgments
Contract grant sponsor: MD Anderson Cancer Center Support Grant (CCSG); Contract grant number: CA016672.
Contract grant sponsors: Jeanne F. Shelby Scholarship Fund, R. Lee Clark Fellow award.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Conflict of interest: SA is an employee of Agios Pharmaceuticals. HK has received research funding from Agios, and both CDD and HK have served on Agios advisory committees. The remaining authors declare no competing interests.
Authors Contributions
SA is an employee of Agios Pharmaceuticals. HK has received research funding from Agios, and both CDD and HK have served on Agios advisory committees. The remaining authors declare no competing interests. CDD and HK designed the research, performed the research, analyzed the data, and wrote the paper. MB and SP analyzed the data and performed statistical analysis. MR and KPP performed the research and contributed vital analytical tools. FR, SA, MK, KT, TK, GGM, JC analyzed the data and wrote the paper.
References
- 1.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxygluta-rate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. Epub 2010/02/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. Epub 2010/06/02. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wise DR, Ward PS, Shay JE, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. Epub 2011/11/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. Epub 2012/02/22. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. Epub 2010/12/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kernytsky A, Wang F, Hansen E, et al. IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood. 2015;125:296–303. doi: 10.1182/blood-2013-10-533604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Im AP, Sehgal AR, Carroll MP, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: Associations with prognosis and potential treatment strategies. Leukemia. 2014;28:1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. Epub 2010/06/23. eng. [DOI] [PubMed] [Google Scholar]
- 10.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. Epub 2010/04/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. Epub 2012/03/16. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rampal R, Alkalin A, Madzo J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou KG, Jiang LJ, Shang Z, et al. Potential application of IDH1 and IDH2 mutations as prognostic indicators in non-promyelocytic acute myeloid leukemia: A meta-analysis. Leuk Lymphoma. 2012;53:2423–2429. doi: 10.3109/10428194.2012.695359. [DOI] [PubMed] [Google Scholar]
- 14.DiNardo CD, Propert KJ, Loren AW, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013;121:4917–4924. doi: 10.1182/blood-2013-03-493197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green CL, Evans CM, Zhao L, et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118:409–412. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- 16.Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: Integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011 Jul 28;118:1069–1076. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 17.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. Epub 2009/08/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravandi F, Patel K, Luthra R, et al. Prognostic significance of alterations in IDH enzyme isoforms in patients with AML treated with high-dose cytarabine and idarubicin. Cancer. 2012;118:2665–2673. doi: 10.1002/cncr.26580. Epub 2011/10/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbas S, Lugthart S, Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood. 2010;116:2122–2126. doi: 10.1182/blood-2009-11-250878. Epub 2010/06/12. eng. [DOI] [PubMed] [Google Scholar]
- 20.Nomdedeu J, Hoyos M, Carricondo M, et al. Adverse impact of IDH1 and IDH2 mutations in primary AML: Experience of the Spanish CETLAM group. Leuk Res. 2012;36:990–997. doi: 10.1016/j.leukres.2012.03.019. Epub 2012/04/24. eng. [DOI] [PubMed] [Google Scholar]
- 21.Chotirat S, Thongnoppakhun W, Promsuwicha O, et al. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:5. doi: 10.1186/1756-8722-5-5. Epub 2012/03/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, Zhu YM, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 23.Kosmider O, Gelsi-Boyer V, Slama L, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–1096. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- 24.Wagner K, Damm F, Gohring G, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. Epub 2010/04/07. eng. [DOI] [PubMed] [Google Scholar]
- 25.Thol F, Damm F, Wagner K, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116:614–616. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- 26.Patel KP, Ravandi F, Ma D, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: Frequency and clinicopathologic features. Am J Clin Pathol. 2011;135:35–45. doi: 10.1309/AJCPD7NR2RMNQDVF. Epub 2010/12/22. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–4083. Epub 2000/12/09. eng. [PubMed] [Google Scholar]
- 28.Stein EM, Altman JK, Collins R, et al. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant metabolic enzyme, induces durable remissions in a phase I study in patients with IDH2 mutation positive advanced hematologic malignancies. Blood. 2014;124:115. (Oral ASH Abstract) [Google Scholar]
- 29.Yaqub F. Inhibition of mutant IDH1 in acute myeloid leukaemia. Lancet Oncol. 2015;16:e9. doi: 10.1016/S1470-2045(14)71140-4. [DOI] [PubMed] [Google Scholar]
- 30.Giles F, O’Brien S, Cortes J, et al. Outcome of patients with acute myelogenous leukemia after second salvage therapy. Cancer. 2005;104:547–554. doi: 10.1002/cncr.21187. [DOI] [PubMed] [Google Scholar]
- 31.Pemmaraju N, Kantarjian H, Garcia-Manero G, et al. Improving outcomes for patients with acute myeloid leukemia in first relapse: A single center experience. Am J Hematol. 2015;90:27–30. doi: 10.1002/ajh.23858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.