Abstract
Cholangiocarcinoma (CCA) is the most common malignant heterogeneous polygenetic carcinoma with a high incidence in Asia. Most patients would die within 1 year after diagnosis and the 5 year survival rate is less than 10-20% worldwide. Single nucleotide polymorphisms (SNPs) in genes regulate telomere maintenance, mitosis, and inflammation, and may help predict individual susceptibility to certain drugs, environmental factor, and risks to particular diseases. The gene-gene interaction and the regulation of SNPs have not been assessed extensively in CCA. According to our previous study, the GRB2-associated-binding protein (Gab1) gene rs3805246 (X2=5.015, P=0.025, OR=0.531, 95% CI 0.304-0.928) and epidermal growth factor receptor (EGFR) gene rs2007000 (X2=7.934, P=0.005, OR=2.148, 95% CI 1.255-3.675) presented significant difference between CCA patients and controls. This study conducted a population-based analysis using 225 CCA cases (153 biliary tract cancer patients and 72 gall bladder cancer patients) to assess the association between SNPs and progression of CCA patients, including the overall survival and the prognosis analysis. Results showed that an increased susceptibility of BTC was significantly associated with SNP loci distribution frequency in EGFR rs2107000 (X2=7.934, P=0.005, OR=2.148, 95% CI 1.255-3.675). Furthermore, multivariate factor regression analysis represented cholelithiasis medical history of BTC patients can be an effective evaluation criteria of BTC susceptibility in early stage. This study also assessed the relationship between these genotypic polymorphisms and clinicopathologic data, including tumor differentiation stage and overall survival. This is the first study identifying that EGFR polymorphisms are associated with BTC and EGFR rs2017000 polymorphisms may be an important survival predictor in BTC patients.
Keywords: Cholangiocarcinoma, molecular marker, SNPs
Introduction
Cholangiocarcinoma (CCA) is a malignant neoplasm arising from the biliary epithelium that constitutes approximately 2% of all reported cancer and accounts for about 3% of all gastrointestinal malignancies [1]. CCA may arise from every portion of the biliary system and can be anatomically classified as intrahepatic or extrahepatic CCA which can be called as biliary tract cancer (BTC). Epidemiological data show that the incidence and prevalence of CCA are increasing within decades worldwide. According to a conservative estimation, 15 million people were infected worldwide in 2004, of which over 85% was from China. Data from the population-based cancer registry in Shanghai indicated that the incidence of CCA has increased more rapidly than other malignancies in China [2]. Currently, the therapeutic modalities for patient with CCA include surgery (most efficient way), radiation therapy, chemotherapy (e.g., 5-FU alone or in combination with methotrexate, leucovorin, cisplatin, mitomycin C, or interferon alpha (IFN-α), chemoradiation therapy, photodynamic therapy, liver transplantation, and palliative therapy [3,4]. Even with a successful surgery, prognoses of patients with CCA are still very poor, with an average 5-year survival rate of 5%-10% [5,6].
Cancer prognosis and survival rate are associated with individual health status, heterogeneity and disease phenotype. CCA is a multi-gene involved hereditary cancer. Many risk factors affect disease status of CCA, including age, genetics and life style [7]. In addition, several chronic inflammation diseases such as bile duct stones, hepatitis, and parasitic infections, increase the risk of CCA [8]. To identify specific genes involved in the cancer development is crucial for novel therapeutics development, cancer prevention and early diagnosis [9]. Several studies focusing on association of SNPs and prognosis prediction have been developed recently [10-12]. The high-throughput SNPs technique is the most amenable method to deal with large-scale analysis in human genome-wide level. Indeed, studies of target SNP selection for genotype-phenotype association have met some success in genes and genomic regions [9,13]. Genome Wide Association Studies (GWAS) have identified four SNPs (PBX1, RORα, NTN1, and SYT6) to be associated with prognosis in patients with early-onset breast cancer [14]. Moreover, the prognosis of bladder cancer is highly correlated with G26071 mutation in EGFR gene [15]. Also, SNP loci T393 of GNAS1 encoding Gas protein has been used as a clinical prognosis evaluation marker in several cancers, including bladder cancer, colorectal cancer and larynx squamous cell carcinoma [16-18].
Many important biological processes, such as cell growth and survival, organ morphogenesis, neovascularization, and tissue repair and regeneration, are regulated by receptor tyrosine kinase (RTK) activation. In general, RTK activity is strictly regulated in normal cell; but dysregulation or constitutive activation of RTK has been found in wide range of cancers [19]. Signaling transduction pathway of EGFR is the major explored RTK pathway which involves in tumor proliferation, differentiation, apoptosis and metastasis, and Gab1 is one of the downstream element of the EGFR pathway [20,21]. Gab1 is a docking protein that recruits phosphoinositide 3-kinase (PI3K) and other effector proteins in response to the activation of many RTKs. As one of the transmembrane glycoproteins, EGFR is a member of epidermal growth factor family. In hilar CCA, down-regulation of Gab1 inhibits cell proliferation and migration [22]. The primary mechanism of EGF-induced stimulation of sub-stream molecular signaling pathway via Gab1 has been demonstrated in mouse embryonic fibroblast cells [23]. KRAS mutation and increased level of EGFR might benefit from dual target RTKs inhibitor in CCA therapy [6]. This study focused on the association between CCA development and molecular mechanism of RTKs pathway, and further detected the corresponding genomic distribution of SNPs to CCA differentiation, survival and prognosis, to explore new genetic markers which can be used to reflect the prognosis of CCA.
Material and method
Study population
Participants of this case-control study came from the Shengjing hospital of China medical university, Republic of China. 225 BTC patients were tested with computed tomography (CT) and magnetic resonance cholangiopancreatography (MRCP) examination for the confirmation of disease status and a written informed consent was received from each patient. Patients were divided into two groups, bile duct carcinoma (BDC, N=153) and gall bladder cancer (GBC, N=72), and a confirmed diagnosis of cholangiocarcinoma based on clinical and histopathological evaluation was received from each patient. In addition, both BDC and GBC patients were categorized into low and high differentiation stage by pathological examination. This study was approved by the relevant Institutional Review Boards for human research in China.
SNP discovery and genotyping in Gab1, EGFR and EGF
Single-nucleotide polymorphisms (SNPs) were selected base on Tag SNP and their putative functional significance. Tag SNPs approach searched the Han Chinese data according to the disequilibrium information from the international HapMap project. The following criteria were used to determine tagging SNPs: (1) a minor allele frequency (MAF) > 5%, and (2) linkage disequilibrium (LD) of r2 > 0.8 between SNPs marker over the genome. Three Taq SNPs of Gab1, the rs3828512, rs3805236 and rs300919, were chosen for further genotyping analysis. Based on the reports in previous publications, NCBI database and Japanese Single Nucleotide Polymorphisms (http://snp.ims.u-tokyo.ac.jp), rs4444903 of EGF and rs1140475, rs2017000, rs884419 of EGFR were also selected, respectively, for further analysis.
Isolating genomic DNA from whole blood
Human blood samples were collected from all of the 225 patients. Whole genomic DNA was extracted from patients’ blood using the Aidlab Genomic DNA Extraction Kit (Aidlab Co., Beijing, China) following to the manufacturer’s instruction.
DNA extraction from PPFE samples
DNA isolation was performed using QiAmp DNA FFPE Tissue kit (Qiagen, Courtaboeuf, France). To extract DNA from formalin fixed paraffin embedded (FFPE) tissue, eight 5-8 μm thick serial sections were cut from each parafilm block and were stored in Eppendorf vials. Parafilm was dewaxed with xylene (10 sec) and centrifuged twice at 14000 rpm for 2 min to wash 100% ethanol from the pellet. Pellet was suspended in 180 μl tissue lysis buffer (ALT buffer, Qiagen) and 20 μl proteinase K. After gently mixed, the sample was incubated at 56°C for 1 hr and agitated at 90°C for 1 hr. Tissue DNA was extracted with MinElute column (Qiagen) according to the manufacture’s instruction. DNA was diluted to a final concentration of 20 ng/μl for TaqMan PCR.
TaqMan PCR
TaqMan allelic discrimination assay was used to detect the presence of eight SNPs loci in BTC. The alleles-specific probes were labeled with the fluorescence reporter dye FAM and VIC at their 5’-end. Reactions were performed in a 5 µl solution with 900 nM specific primers, 200 nM specific TaqMan probes, 10 ng DNA using 384-well plates and TaqMan Genotyping Master Mix (Applied Biosystem) as previously described [24] using ABI Prism® sequencing detection system (ABI7900, Applied Biosystem). Data were analyzed using ABI7900 SDS software and all of the tests were performed in a triplicated manner of experiment design.
Statistical analysis
All statistical analysis was conducted by Statistical Packages for Social Sciences 13.0 (SPSS 13.0). Description statistics between BDC and GBC groups were calculated using Student t test (numeric variables) and Chi-square test (categorical variables). HaploView V. 4.0 was used to assess Hardy-Weinberg equilibrium and linkage disequilibrium [25]. Fisher’s extract test was applied when the subject number was less than 5 per test. The distribution and allele frequency were identified by Chi-square test. Odds ratios (ORs) and 95% confidence interval (CIs) were estimated using unconditional logistic regression to determine the magnitude and statistical significance of associations. The statistical software UNPHASED 3.0.7 (Frank Dudbridge, MRC Biostatistics Unit Cambridge, UK) was applied to assess gene-gene interaction and allele-disease associations. Furthermore, the Kaplan-Meier Survival curve analysis (using Log-rank test) was used to study the association of the identified potential risk factor with patient survival rate (3 year).
Results
Results of Genotyping of SNPs of Gab1, EGFR and EGF and the Hardy-Weinberg equilibrium analysis were conducted in our previous study. In this study, we focused on the association between SNPs and prognosis of BTC. According to the differentiation status of prognostic pathological results, BTC patients were divided into three groups, the highly differentiated group (HD), medium differentiated group (MD) and low differentiated group (LD). However, all of the genetic frequency distribution data were not statistically significant (P > 0.05) (Table 1).
Table 1.
Comparison of genotype frequency distribution of SNPs and differentiation status of BTC
| Gene | SNPs | Total | HD | MD | LD | X2 | P-value | |
|
|
||||||||
| n | 225 | 132 | 71 | 22 | ||||
|
| ||||||||
| Gab1 | rs3828512 | AA | 101 | 60 | 32 | 9 | ||
| AG | 105 | 62 | 33 | 10 | ||||
| GG | 19 | 10 | 6 | 3 | ||||
| 0.918 | 0.922 | |||||||
| rs3805236 | AA | 123 | 75 | 35 | 13 | |||
| AG | 88 | 52 | 29 | 7 | ||||
| GG | 14 | 5 | 7 | 2 | ||||
| 3.979 | 0.409 | |||||||
| rs300919 | CC | 119 | 73 | 34 | 12 | |||
| TC | 91 | 53 | 31 | 7 | ||||
| TT | 15 | 6 | 6 | 3 | ||||
| 3.915 | 0.418 | |||||||
| rs3805246 | GG | 71 | 45 | 22 | 4 | |||
| GA | 119 | 66 | 40 | 13 | ||||
| AA | 35 | 21 | 9 | 5 | ||||
| 3.168 | 0.530 | |||||||
| EGFR | rs1140475 | CC | 189 | 112 | 61 | 16 | ||
| TC | 33 | 18 | 10 | 5 | ||||
| TT | 3 | 2 | 0 | 1 | ||||
| 4.144 | 0.387 | |||||||
|
| ||||||||
| Gene | SNPs | Total | HD | MD | LD | X2 | P-value | |
|
| ||||||||
| rs2017000 | GG | 89 | 56 | 30 | 3 | |||
| GA | 110 | 63 | 32 | 15 | ||||
| AA | 26 | 13 | 9 | 4 | ||||
| 7.311 | 0.120 | |||||||
| rs884419 | AA | 65 | 39 | 21 | 5 | |||
| GA | 117 | 70 | 33 | 14 | ||||
| GG | 43 | 23 | 17 | 3 | ||||
| 2.746 | 0.601 | |||||||
| EGF | rs4444903 | GG | 89 | 55 | 27 | 7 | ||
| GA | 99 | 57 | 32 | 10 | ||||
| AA | 37 | 20 | 12 | 5 | ||||
| 1.244 | 0.871 | |||||||
HD: highly differentiated group; MD: medium differentiated group; LD: low differentiated group.
We also divided the BTC group into BDC and GBC subgroups to see if there was any difference between differentiation status and SNPs of Gab1, EGFR and EGF. Unfortunately, no significant difference was observed within these two subgroups (Tables 2 and 3). This study further conducted a lifetime analysis for BTC group using Kaplan-Meier estimation and Log-rank test to compare the survival difference of each SNP among BTC patients. Results showed that the difference of median survival time of BTC patients with different genotypes had statistically significance (P=0.021). The median survival time of patients with A/A genotype of rs2017000 in EGFR was significantly lower than patients with G/G and G/G genotypes (A/A: 11 month; 95% CI 4.421-17.579; G/G: 18 month; 95% CI 15.851-20.149; G/A: 19 month; 95% CI 17.557-20.443) (Figure 1; Table 4). After controlling other confounders, such as gender, age, disease history of biliary stone, differentiation status of BTC and SNP genotypes through multivariate Cox regression analysis, the effect of A/A genotype of rs2017000 in EGFR was still significantly lower than the other two genotypes (Table 5).
Table 2.
Comparison of genotype frequency distribution of SNPs and differentiation status of BDC and GBC
| Gene | SNPs | Total | HD | MD | LD | X2 | P-value | |
|
|
||||||||
| n | 153 | 92 | 45 | 16 | ||||
|
| ||||||||
| Gab1 | rs3828512 | AA | 70 | 43 | 21 | 6 | ||
| AG | 71 | 43 | 20 | 8 | ||||
| GG | 12 | 6 | 4 | 2 | ||||
| 1.060 | 0.901 | |||||||
| rs3805236 | AA | 82 | 50 | 22 | 10 | |||
| AG | 60 | 38 | 18 | 4 | ||||
| GG | 11 | 4 | 5 | 2 | ||||
| 3.990 | 0.407 | |||||||
| rs300919 | CC | 79 | 50 | 21 | 8 | |||
| TC | 63 | 37 | 20 | 6 | ||||
| TT | 11 | 5 | 4 | 2 | ||||
| 1.747 | 0.782 | |||||||
| rs3805246 | GG | 49 | 31 | 15 | 3 | |||
| GA | 81 | 46 | 25 | 10 | ||||
| AA | 23 | 15 | 5 | 3 | ||||
| 2.176 | 0.703 | |||||||
| EGFR | rs1140475 | CC | 128 | 78 | 39 | 11 | ||
| TC | 23 | 13 | 6 | 4 | ||||
| TT | 2 | 1 | 0 | 1 | ||||
| 5.294 | 0.258 | |||||||
|
| ||||||||
| Gene | SNPs | Total | HD | MD | LD | X2 | P-value | |
|
| ||||||||
| rs2017000 | GG | 61 | 42 | 17 | 2 | |||
| GA | 74 | 40 | 23 | 11 | ||||
| AA | 18 | 10 | 5 | 3 | ||||
| 6.469 | 0.167 | |||||||
| rs884419 | AA | 45 | 26 | 15 | 4 | |||
| GA | 79 | 49 | 21 | 9 | ||||
| GG | 29 | 17 | 9 | 3 | ||||
| 0.748 | 0.945 | |||||||
| EGF | rs4444903 | GG | 61 | 38 | 17 | 6 | ||
| GA | 68 | 39 | 21 | 8 | ||||
| AA | 24 | 15 | 7 | 2 | ||||
| 0.494 | 0.974 | |||||||
Table 3.
Comparison of genotype frequency distribution of SNPs and differentiation status of GBC
| Gene | SNPs | Total | HD | MD | LD | X2 | P-value | |
|
|
||||||||
| n | 72 | 40 | 26 | 6 | ||||
|
| ||||||||
| Gab1 | rs3828512 | AA | 31 | 17 | 11 | 3 | ||
| AG | 34 | 19 | 13 | 2 | ||||
| GG | 7 | 4 | 2 | 1 | ||||
| 0.773 | 0.942 | |||||||
| rs3805236 | AA | 41 | 25 | 13 | 3 | |||
| AG | 28 | 14 | 11 | 3 | ||||
| GG | 3 | 1 | 2 | 0 | ||||
| 2.204 | 0.698 | |||||||
| rs300919 | CC | 40 | 23 | 13 | 4 | |||
| TC | 28 | 16 | 11 | 1 | ||||
| TT | 4 | 1 | 2 | 1 | ||||
| 3.377 | 0.497 | |||||||
| rs3805246 | GG | 22 | 14 | 7 | 1 | |||
| GA | 38 | 20 | 15 | 3 | ||||
| AA | 12 | 6 | 4 | 2 | ||||
| 2.028 | 0.731 | |||||||
| EGFR | rs1140475 | CC | 61 | 34 | 22 | 5 | ||
| TC | 10 | 5 | 4 | 1 | ||||
| TT | 1 | 1 | 0 | 0 | ||||
| 0.933 | 0.920 | |||||||
|
| ||||||||
| Gene | SNPs | Total | HD | MD | LD | X2 | P-value | |
|
| ||||||||
| rs2017000 | GG | 28 | 14 | 13 | 1 | |||
| GA | 36 | 23 | 9 | 4 | ||||
| AA | 8 | 3 | 4 | 1 | ||||
| 4.820 | 0.306 | |||||||
| rs884419 | AA | 20 | 13 | 6 | 1 | |||
| GA | 38 | 21 | 12 | 5 | ||||
| GG | 14 | 6 | 8 | 0 | ||||
| 5.361 | 0.252 | |||||||
| EGF | rs4444903 | GG | 28 | 17 | 10 | 1 | ||
| GA | 31 | 18 | 11 | 2 | ||||
| AA | 13 | 5 | 5 | 3 | ||||
| 5.162 | 0.271 | |||||||
Figure 1.
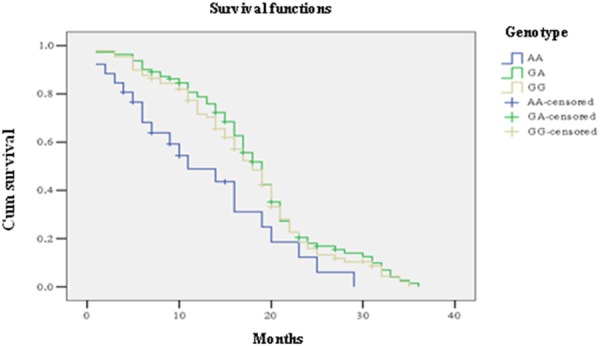
Kaplan-Meier’s 3 year survival curve of 206 BTC patients grouped by their genotype for SNP EGFR rs2017000. The corresponding median survival time of bile duct patients in GG genotype (median: 18 month; 95% CI 15.851-20.149) and GA type (median: 19 month; 95% CI 17.557-20.443) was significant higher than in AA type (median: 11 month; 95% CI 4.421-15.579).
Table 4.
Association between each genotyping frequency of SNP and prognosis of BTC patients
| Gene | SNPs | Total (n) | Followed (n) | Lost (n) | Median survival time | Log rank | |||
|
| |||||||||
| Month | 95% CI | X2 | P-value | ||||||
|
| |||||||||
| Gab1 | rs3828512 | AA | 101 | 95 | 6 | 18 | 16.010-21.394 | ||
| AG | 105 | 97 | 8 | 17 | 15.032-19.736 | ||||
| GG | 19 | 14 | 5 | 17 | 14.909-20.369 | ||||
| 1.236 | 0.896 | ||||||||
| rs3805236 | AA | 123 | 112 | 11 | 18 | 15.749-22.004 | |||
| AG | 88 | 82 | 6 | 17 | 13.595-20.337 | ||||
| GG | 14 | 12 | 2 | 18 | 15.396-21.002 | ||||
| 3.990 | 0.458 | ||||||||
| rs300919 | CC | 119 | 111 | 8 | 17 | 14.547-20.379 | |||
| TC | 91 | 83 | 8 | 17 | 13.382-22.098 | ||||
| TT | 15 | 12 | 3 | 17 | 14.759-19.980 | ||||
| 1.170 | 0.967 | ||||||||
| rs3805246 | GG | 71 | 68 | 3 | 19 | 15.345-23.117 | |||
| GA | 119 | 109 | 10 | 17 | 14.958-19.756 | ||||
| AA | 35 | 29 | 6 | 18 | 15.327-20.015 | ||||
| 2.340 | 0.753 | ||||||||
| EGFR | rs1140475 | CC | 189 | 177 | 12 | 19 | 16.333-22.584 | ||
| TC | 33 | 26 | 7 | 18 | 15.308-21.132 | ||||
| TT | 3 | 3 | 0 | 18 | 15.988-21.050 | ||||
| 2.736 | 0.664 | ||||||||
|
| |||||||||
| Gene | SNPs | Total (n) | Followed (n) | Lost (n) | Median survival time | Log rank | |||
|
| |||||||||
| Month | 95% CI | X2 | P-value | ||||||
|
| |||||||||
| rs2017000 | GG | 89 | 82 | 7 | 18 | 15.851-20.149 | |||
| GA | 110 | 103 | 7 | 19 | 17.557-20.443 | ||||
| AA | 26 | 21 | 5 | 11 | 4.421-17.579 | ||||
| 7.760 | 0.021 | ||||||||
| rs884419 | AA | 65 | 60 | 5 | 18 | 15.721-20.225 | |||
| GA | 117 | 106 | 11 | 17 | 14.336-20.001 | ||||
| GG | GG | 43 | 40 | 3 | 17 | 15.878-19.546 | |||
| 0.987 | 0.836 | ||||||||
| EGF | rs4444903 | GG | 89 | 84 | 5 | 17 | 14.993-21.346 | ||
| GA | 99 | 93 | 6 | 18 | 16.086-20.754 | ||||
| AA | 37 | 29 | 8 | 16 | 13.467-19.585 | ||||
| 2.082 | 0.685 | ||||||||
Table 5.
Multivariate Cox regression of potential factors affecting BTC prognosis
| Factor | β value | P value | OR | 95% CI |
|---|---|---|---|---|
| A/A in rs2017000 | 0.814 | 0.002 | 1.921 | 1.141-2.594 |
| Gender | 1.107 | 0.157 | 1.043 | 0.511-3.372 |
| Age | -0.201 | 0.482 | 1.047 | 0.807-1.421 |
| Disease history of BS | 0.311 | 0.001 | 2.781 | 2.274-5.941 |
| Differentiation status | 1.296 | 0.002 | 1.664 | 1.259-3.082 |
BS: biliary stone.
RTK related downstream signaling molecular activation to regulate many key process including cell growth, survival, organ morphogenesis, neovascularization, tissue repair and regeneration. Our previous study assessed the genome association of CCA revealed no significant SNPs frequency distribution difference in Gab1 and EGF that associated to CCA susceptibility. Particularly, patients with A/A in Gab1 rs3805246 and G/G+G/A in EGFR rs2017000 simultaneously had significantly higher chance to have CCA. Current study evaluated Gab1, EGFR and EGF SNPs allele frequency distribution according two subtypes and three differentiation stage (low, middle or high) of CCA. Result revealed no one of these SNPs displayed a significant genotype distribution whatever in gallbladder cancer or biliary tract cancer (data not shown). The selecting 8 loci of Gab1 rs3828512, rs3805236, rs300919 and rs3805246, EGFR rs1140475, rs2017000 and rs884419 and EGF rs444903 presented no association to the low, middle and high grade CCA differentiation status were using multivariate logistic regression model in SPSS 13.0.
To evaluate the association of risk factors (gender, age, tumor differentiation stage, and gallstone history) with patients 3 year overall survival, done by Kaplan-Meier survival curve analysis (Table 5). Kaplan-Meier assay showed CCA patient carry EGFR rs2017000 A/A genotype represented poor prognosis and overall survival time (median: 11 months; 95% CI 4.421-17.579) compared to G/G (median: 18 month; 95% CI 15.851-20.149) and G/A (median: 19 month; 95% CI 17.557-20.443) genotype, is graphically in Figure 1. Therefore, a multivariate analysis was required in order to exclude the effect of risk factors for CCA. In the multivariate survival analysis using regression model, dependent variable as the EGFR rs2017000 A/A genotype (P=0.002, OR=1.921) and cholelithiasis medical history (P=0.001, OR=2.781) was significantly associated with CCA susceptibility (Table 5). However, this association was not observed in gender and age (P > 0.05).
Discussion
Recent years, various analytical methodologies for SNPs markers screening provide a comprehensive way for analyzing human genome and identifying potential mutated genes and genomic regions contributing to the disease phenotype. T/T genotype of rs887569 in EZH2 gene was indicated to be correlated with longer overall survival in CCA patients [26]. Among 198 CCA patients from northeastern Thailand, the G/G genotype of rs6726395 in NRF2 gene was found to be associated with longer survival with a hazard ratio of 0.54 (95% CI: 0.31-0.94) [27]. CCA is a biliary epithelium originated malignant neoplasm which is considered as a complicated multi-gene involved hereditary disease which is interacted by both environmental and genetic factors [28]. Generally, CCA is detected and diagnosed clinically at an incorporative stage and a majority of them show poor prognoses even after surgical resection [29]. The clinical course and outcome of CCA cancer varies among people, but the effect of genetic variability on prognosis is poorly uderstood. Therefore, the predictive marker identification of CCA aims to evaluate patient’s prognosis and overall outcome, which is very meaningful for judging the probability of cancer recurrence after standard treatment.
Recent studies showed that the receptor tyrosine kinases (RTKs) pathway plays an important role in the pathogenesis, development and survival prognosis in various human malignant tumors [30,31]. For example, a study which recruited 129 breast cancer patients indicates that one of the RTK family, the MET, is a prognostic factor for disease-specific survival in breast cancer patients receiving neo-adjuvant chemotherapy [32]. Genome-wide analysis of ICC suggests that RTKs are associated with ICC and RTK inhibitors which have a novel therapeutic potential to improve ICC [33]. Result of this study showed that Gab1, EGFR and EGFR SNPs genotypes have no correlation to BTC differentiation status. Moreover, same result was also identified in Gab1, EGFR and EGF genotype compared to the two subtype of BTC (BDC group and GBC group).
Up to date, the cancer risk which is associated with individual’s SNP has been documented in many studies. A genome-wide analysis of breast cancer identifies SNPs in five genes associated with susceptibility, including TNRC9, FGFR2, MAP3K1, H19 and LSP1 [34]. Another genome-wide association study (GWAS) recruited 19,091 cases and 20,606 controls of East-Asian descendant, including Chinese, Korean, and Japanese women, provided strong evidence for a novel breast cancer susceptibility locus represented by rs9485372, near the TAB2 gene (6q25.1), and identifies two possible susceptibility related loci located in the ESR1 gene (11q24.3) [35]. This study explored A/A genotype of rs2017000 in EGFR strongly correlated with shorter overall survival time than the other two genotypes. EGFR heterodimers have been shown to increase cell motility underlying downstream signaling pathway activation. In addition, rs2017000 locus is located in the intron of exon 21 of EGFR gene, and exon 21 is a tyrosin kinase domain, which was found to be associated with ovarian cancer and lung cancer development [21,36,37]. Several conditions of liver or bile duct may cause BTC, such as parasitic infections, primary sclerosis cholangitis, biliary-duct cysts, hepatolithiasis and toxins [38-40]. In this study, patients with cholelithiasis medical history showed higher BTC susceptibility in the multivariate factor regression analysis. Thus, the cholelithiasis may act as a BTC marker in early diagnosis. Although choleithiasis has not been demonstrated to be clearly associated with BTC, pancreaticobiliary maljunction (PBM) with bile duct dialation, cholelithiasis and cholecystectomy are associated exclusively with EH-BTC other than gallbladder cancer [41].
In summary, in this clinical study, we provided convincing evidence for the significant association between disease susceptibility and SNPs of rs2017000 in EGFR. Our study also suggested that BTC patients carrying genetic allele EGFR rs2017000 G/G+G/A genotype might have longer overall survival time and better prognosis with optimal clinical treatment. Using multivariate regression analysis controlling for potential confounders, effects of EGFR rs2017000 was still significant on the association with BTC.
Disclosure of conflict of interest
None.
References
- 1.Charbel H, Al-Kawas FH. Cholangiocarcinoma: epidemiology, risk factors, pathogenesis, and diagnosis. Curr Gastroenterol Rep. 2011;13:182–187. doi: 10.1007/s11894-011-0178-8. [DOI] [PubMed] [Google Scholar]
- 2.Jin F, Devesa SS, Zheng W, Blot WJ, Fraumeni JF Jr, Gao YT. Cancer incidence trends in urban Shanghai, 1972-1989. Int J Cancer. 1993;53:764–770. doi: 10.1002/ijc.2910530510. [DOI] [PubMed] [Google Scholar]
- 3.Wijaya I, Abdullah M. Diagnosis and treatment update: cholangiocarcinoma. Acta Medica Indonesiana. 2011;43:212–215. [PubMed] [Google Scholar]
- 4.Yoon JH, Gores GJ. Diagnosis, Staging, and Treatment of Cholangiocarcinoma. Curr Treat Options Gastroenterol. 2003;6:105–112. doi: 10.1007/s11938-003-0011-z. [DOI] [PubMed] [Google Scholar]
- 5.Anderson CD, Chari RS, Delbeke D, Rice MH, Pinson CW. The role of PET imaging in the evaluation of cholangiocarcinoma and gallbladder carcinoma. Gastroenterology. 2003;124:A827–A827. doi: 10.1016/j.gassur.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts LR, Factor VM, Thorgeirsson SS. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142:1021–1031. e15. doi: 10.1053/j.gastro.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee BS, Cha BH, Park EC, Roh J. Risk factors for perihilar cholangiocarcinoma: A hospital-based case-control study. Liver Int. 2015;35:1048–1053. doi: 10.1111/liv.12618. [DOI] [PubMed] [Google Scholar]
- 8.de Martel C, Plummer M, Franceschi S. Cholangiocarcinoma: descriptive epidemiology and risk factors. Gastroenterol Clin Biol. 2010;34:173–180. doi: 10.1016/j.gcb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Engle LJ, Simpson CL, Landers JE. Using high-throughput SNP technologies to study cancer. Oncogene. 2006;25:1594–1601. doi: 10.1038/sj.onc.1209368. [DOI] [PubMed] [Google Scholar]
- 10.Lin CY, Chen JY, Lin TN, Jeng WJ, Huang CH, Huang CW, Chang SW, Sheen IS. IL28B SNP rs12979860 is a critical predictor for on-treatment and sustained virologic response in patients with hepatitis C virus genotype-1 infection. PLoS One. 2011;6:e18322. doi: 10.1371/journal.pone.0018322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan XL, Moyer AM, Fridley BL, Schaid DJ, Niu N, Batzler AJ, Jenkins GD, Abo RP, Li L, Cunningham JM, Sun Z, Yang P, Wang L. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17:5801–5811. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Sohn I, Son DS, Kim DH, Ahn T, Jung SH. Prediction of a time-to-event trait using genome wide SNP data. BMC Bioinformatics. 2013;14:58. doi: 10.1186/1471-2105-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wjst M. Target SNP selection in complex disease association studies. BMC Bioinformatics. 2004;5:92. doi: 10.1186/1471-2105-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rafiq S, Tapper W, Collins A, Khan S, Politopoulos I, Gerty S, Blomqvist C, Couch FJ, Nevanlinna H, Liu J, Eccles D. Identification of inherited genetic variations influencing prognosis in early-onset breast cancer. Cancer Res. 2013;73:1883–1891. doi: 10.1158/0008-5472.CAN-12-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naji F, Attaleb M, Laantri N, Benchakroun N, El Gueddari B, Benider A, Azeddoug H, Ennaji MM, El Mzibri M, Khyatti M. Identification of G2607A mutation in EGFR gene with a significative rate in Moroccan patients with nasopharyngeal carcinoma. Cell Mol Biol. 2010;56(Suppl):OL1442–1446. [PubMed] [Google Scholar]
- 16.Frey UH, Eisenhardt A, Lummen G, Rubben H, Jockel KH, Schmid KW, Siffert W. The T393C polymorphism of the G alpha s gene (GNAS1) is a novel prognostic marker in bladder cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:871–877. doi: 10.1158/1055-9965.EPI-04-0720. [DOI] [PubMed] [Google Scholar]
- 17.Frey UH, Alakus H, Wohlschlaeger J, Schmitz KJ, Winde G, van Calker HG, Jockel KH, Siffert W, Schmid KW. GNAS1 T393C polymorphism and survival in patients with sporadic colorectal cancer. Clin Cancer Res. 2005;11:5071–5077. doi: 10.1158/1078-0432.CCR-05-0472. [DOI] [PubMed] [Google Scholar]
- 18.Lehnerdt GF, Franz P, Winterhoff S, Bankfalvi A, Grehl S, Lang S, Schmid KW, Siffert W, Jahnke K, Frey UH. The GNAS1 T393C polymorphism predicts survival in patients with advanced squamous cell carcinoma of the larynx. Laryngoscope. 2008;118:2172–2176. doi: 10.1097/MLG.0b013e318185793ds. [DOI] [PubMed] [Google Scholar]
- 19.Ito F. Foreword. Target therapy for cancer: anti-cancer drugs targeting growth-factor signaling molecules. Biol Pharm Bull. 2011;34:1773. doi: 10.1248/bpb.34.1773. [DOI] [PubMed] [Google Scholar]
- 20.Van den Eynde M, Baurain JF, Mazzeo F, Machiels JP. Epidermal growth factor receptor targeted therapies for solid tumours. Acta Clin Belg. 2011;66:10–17. doi: 10.2143/ACB.66.1.2062508. [DOI] [PubMed] [Google Scholar]
- 21.Dova L, Pentheroudakis G, Georgiou I, Malamou-Mitsi V, Vartholomatos G, Fountzilas G, Kolaitis N, Kitsiou E, Pavlidis N. Global profiling of EGFR gene mutation, amplification, regulation and tissue protein expression in unknown primary carcinomas: to target or not to target? Clin Exp Metastasis. 2007;24:79–86. doi: 10.1007/s10585-007-9055-0. [DOI] [PubMed] [Google Scholar]
- 22.Sang H, Li T, Li H, Liu J. Down-regulation of Gab1 inhibits cell proliferation and migration in hilar cholangiocarcinoma. PLoS One. 2013;8:e81347. doi: 10.1371/journal.pone.0081347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattoon DR, Lamothe B, Lax I, Schlessinger J. The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol. 2004;2:24. doi: 10.1186/1741-7007-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Paolicchi E, Pacetti P, Giovannetti E, Mambrini A, Orlandi M, Crea F, Romani AA, Tartarini R, Danesi R, Peters GJ, Cantore M. A single nucleotide polymorphism in EZH2 predicts overall survival rate in patients with cholangiocarcinoma. Oncol Lett. 2013;6:1487–1491. doi: 10.3892/ol.2013.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khunluck T, Kukongviriyapan V, Puapairoj A, Khuntikeo N, Senggunprai L, Zeekpudsa P, Prawan A. Association of NRF2 polymorphism with cholangiocarcinoma prognosis in Thai patients. Asian Pac J Cancer Prev. 2014;15:299–304. doi: 10.7314/apjcp.2014.15.1.299. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava K, Srivastava A, Sharma KL, Mittal B. Candidate gene studies in gallbladder cancer: a systematic review and meta-analysis. Mutat Res. 2011;728:67–79. doi: 10.1016/j.mrrev.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakanuma Y, Sato Y, Harada K, Sasaki M, Xu J, Ikeda H. Pathological classification of intrahepatic cholangiocarcinoma based on a new concept. World J Hepatol. 2010;2:419–427. doi: 10.4254/wjh.v2.i12.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y, Guo JJ, Liu YM, Wu XL. microRNA-34a Inhibits the Growth, Invasion and Metastasis of Gastric Cancer by Targeting PDGFR and MET Expression. Biosci Rep. 2014;34:e00112. doi: 10.1042/BSR20140020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Rothlin CV, Leighton JA, Ghosh S. Tyro3, Axl, and Mertk Receptor Signaling in Inflammatory Bowel Disease and Colitis-associated Cancer. Inflamm Bowel Dis. 2014;20:1472–1480. doi: 10.1097/MIB.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh YW, Lee HJ, Ahn JH, Lee JW, Gong G. MET expression is associated with disease-specific survival in breast cancer patients in the neoadjuvant setting. Pathol Res Pract. 2014;210:494–500. doi: 10.1016/j.prp.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Andersen JB, Thorgeirsson SS. Genetic profiling of intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol. 2012;28:266–272. doi: 10.1097/MOG.0b013e3283523c7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R SEARCH collaborators; Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schürmann P, Dörk T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X kConFab; AOCS Management Group. Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long J, Cai Q, Sung H, Shi J, Zhang B, Choi JY, Wen W, Delahanty RJ, Lu W, Gao YT, Shen H, Park SK, Chen K, Shen CY, Ren Z, Haiman CA, Matsuo K, Kim MK, Khoo US, Iwasaki M, Zheng Y, Xiang YB, Gu K, Rothman N, Wang W, Hu Z, Liu Y, Yoo KY, Noh DY, Han BG, Lee MH, Zheng H, Zhang L, Wu PE, Shieh YL, Chan SY, Wang S, Xie X, Kim SW, Henderson BE, Le Marchand L, Ito H, Kasuga Y, Ahn SH, Kang HS, Chan KY, Iwata H, Tsugane S, Li C, Shu XO, Kang DH, Zheng W. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8:e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang GC, Lin JY, Wang Z, Zhou Q, Xu CR, Zhu JQ, Wang K, Yang XN, Chen G, Yang JJ, Huang YJ, Liao RQ, Wu YL. Epidermal growth factor receptor double activating mutations involving both exons 19 and 21 exist in Chinese non-small cell lung cancer patients. Clin Oncol. 2007;19:499–506. doi: 10.1016/j.clon.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Schilder RJ, Sill MW, Chen X, Darcy KM, Decesare SL, Lewandowski G, Lee RB, Arciero CA, Wu H, Godwin AK. Phase II study of gefitinib in patients with relapsed or persistent ovarian or primary peritoneal carcinoma and evaluation of epidermal growth factor receptor mutations and immunohistochemical expression: a Gynecologic Oncology Group Study. Clin Cancer Res. 2005;11:5539–5548. doi: 10.1158/1078-0432.CCR-05-0462. [DOI] [PubMed] [Google Scholar]
- 38.Plieskatt JL, Deenonpoe R, Mulvenna JP, Krause L, Sripa B, Bethony JM, Brindley PJ. Infection with the carcinogenic liver fluke Opisthorchis viverrini modifies intestinal and biliary microbiome. FASEB J. 2013;27:4572–4584. doi: 10.1096/fj.13-232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joo I, Lee JM. Imaging bile duct tumors: pathologic concepts, classification, and early tumor detection. Abdom Imaging. 2013;38:1334–1350. doi: 10.1007/s00261-013-0027-3. [DOI] [PubMed] [Google Scholar]
- 40.Jablonska B. Biliary cysts: etiology, diagnosis and management. World J Gastroenterol. 2012;18:4801–4810. doi: 10.3748/wjg.v18.i35.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bragazzi MC, Cardinale V, Carpino G, Venere R, Semeraro R, Gentile R, Gaudio E, Alvaro D. Cholangiocarcinoma: Epidemiology and risk factors. Translation Gastrointestinal Cancer. 2012;1:9. [Google Scholar]


