Abstract
The over-collection and habitat destruction of natural Dendrobium populations for their commercial medicinal value has led to these plants being under severe threat of extinction. In addition, many Dendrobium plants are similarly shaped and easily confused during the absence of flowering stages. In the present study, we examined the application of the ITS2 region in barcoding and phylogenetic analyses of Dendrobium species (Orchidaceae). For barcoding, ITS2 regions of 43 samples in Dendrobium were amplified. In combination with sequences from GenBank, the sequences were aligned using Clustal W and genetic distances were computed using MEGA V5.1. The success rate of PCR amplification and sequencing was 100%. There was a significant divergence between the inter- and intra-specific genetic distances of ITS2 regions, while the presence of a barcoding gap was obvious. Based on the BLAST1, nearest distance and TaxonGAP methods, our results showed that the ITS2 regions could successfully identify the species of most Dendrobium samples examined; Second, we used ITS2 as a DNA marker to infer phylogenetic relationships of 64 Dendrobium species. The results showed that cluster analysis using the ITS2 region mainly supported the relationship between the species of Dendrobium established by traditional morphological methods and many previous molecular analyses. To sum up, the ITS2 region can not only be used as an efficient barcode to identify Dendrobium species, but also has the potential to contribute to the phylogenetic analysis of the genus Dendrobium.
Keywords: Dendrobium, ITS2, DNA barcode, species identification, genetic relationship
1. Introduction
Dendrobium Sw., one of the most important genera in the family Orchidaceae, comprising more than 1000 species [1,2,3,4], is mainly distributed in tropical and subtropical Asia, and Northern and Eastern Australia [5]. All these species are contained in the Conventions on International Trade in Endangered Species of Fauna and Flora (CITES). There are 74 Dendrobium species and two varieties in China, mainly distributed in the southern regions of the Tsinling Mountains [3]. Due to its pharmacological significance for human health, with reported effects including clearing away toxic materials accumulated in human tissues, enhancing the body’s immunity, reducing blood sugar levels, and prolonging life [6,7], Dendrobium has been one of the most well-known traditional herbal medicinal plants in China for centuries. For the commercial production of medicinal plants, Dendrobium plants are subjected to massive collection for trading in the medicine market in China. Nowadays, due to over-collection and habitat destruction, natural Dendrobium populations are under severe threat of extinction.
The accurate identification of Dendrobium species is critical to their safe utilization and genetic resource conservation. Traditional methods to identify Dendrobium species are based on phenotypic observations [8], while morphological characteristics are often affected by environmental and developmental factors [1,9,10]. Particularly, during the absence of flowering stages, the morphological characteristics of many Dendrobium species are extremely similar, rendering their differentiation very difficult and sometimes impossible [1,2,5,11]. Therefore, a simple and accurate identification method for Dendrobium plants is indispensable.
DNA barcoding is a new technique using a short and standardized DNA fragment to discriminate among species [12,13,14,15,16,17]. Irrespective of the morphological features of samples and the professional level of users, this technique should consistently identify a species [18]. In addition, some studies have reported that DNA barcoding was also applied for the identification of ancient archaeological samples [19,20] and for intraspecific and population studies [21]. In recent years, DNA barcoding has been generally improved as an efficient tool for species identification and has become a trend and area of recent interest for biology systematics and identification [13,14,22]. In plants, several regions of chloroplast DNA sequences (including matK, rbcL, psbA-trnH, and atpF-atpH spacer) and the internal transcribed spacer (ITS) region of the nuclear ribosomal DNA have been advocated as potential plant barcodes [13,23,24]. The internal transcribed spacer 2 (ITS2) is situated between the ribosomal genes 5.8S and 28S and probably has a function in the regulation of the transcription of active ribosomal subunits, as this spacer provides structural elements necessary for correct pre-rRNA processing [25,26,27]. Due to its valuable characteristics, including the availability of conserved regions for designing universal primers, the ease of its amplification, and sufficient variability to distinguish even closely related species, ITS2 was recently proposed as the standard barcode for authentication of medicinal plants [13,14,28,29,30,31,32]. In addition, it has been reported that ITS2 provides taxonomic signatures in systematic evolution [22,33,34] and can be used for rapid taxonomic classification [13,14,35].
The aim of the present study is to test whether ITS2 is a valuable marker for the application of barcoding in Dendrobium, and to apply ITS2 as a phylogenetic marker to infer genetic relationships among Dendrobium species.
2. Results
2.1. PCR Amplification, Success Rate and Sequence Characteristics
The amplification and sequence success rate of the ITS2 sequences from sampled specimens of Dendrobium species was 100%. The lengths of the ITS2 sequences used for the analyses were in the range of 243–258 bp, with an average of 248 bp. The mean GC content was 52.2%, with a range of 44.8%–64.7%. Therefore, the length and GC content of ITS2 sequences from the collected Dendrobium species were relatively variable.
2.2. Genetic Divergence within and between Species
When we used six metrics (average inter-specific distance, the minimum inter-specific distance, theta prime, average intra-specific distance, coalescent depth and theta) to estimate genetic divergences, the calculated results showed that ITS2 exhibited significant divergence at the inter-specific level (Table 1) to distinguish between closely related species. At the intra-specific level, relatively lower divergence was observed for all three corresponding metrics.
Table 1.
Analyses of inter-specific divergence and intra-specific variation of the ITS2 (Internal transcribed spacer 2) sequences in 364 samples of 64 Dendrobium species.
| Measurement | Kimura 2-Parameter (K2P) Value |
|---|---|
| All inter-specific distance | 0.182 ± 0.036 |
| Theta prime | 0.192 ± 0.036 |
| The minimum inter-specific distance | 0.185 ± 0.035 |
| All intra-specific distance | 0.007 ± 0.004 |
| Theta | 0.006 ± 0.003 |
| Coalescent depth | 0.014 ± 0.006 |
2.3. Assessment of Barcoding Gap
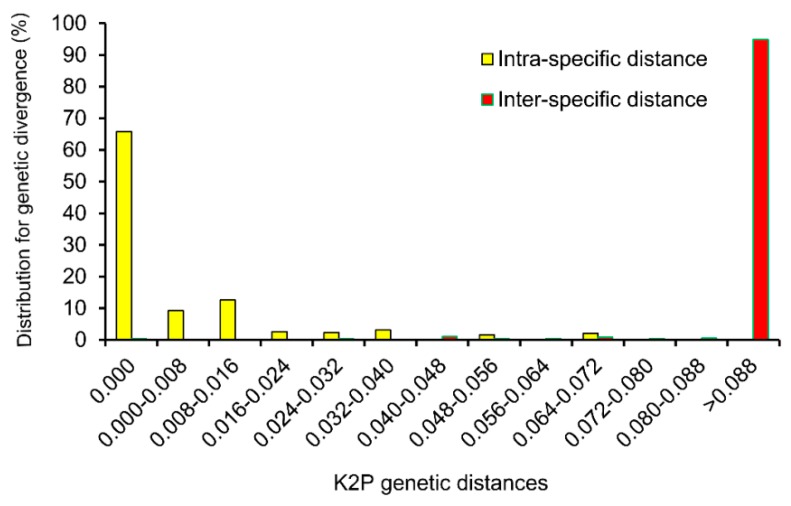
To examine intra- vs. inter-specific divergence, we investigated the distribution of genetic distance at a scale of 0.008 distance units. Only a slight overlap in inter-/intra-specific variation of ITS2 was found (Figure 1). The inter-specific distance was in the range of 0.000–0.415, equaled zero for only 0.38%, and the proportion of inter-specific genetic distance <0.072 was only 4.18%. The intra-specific distance ranged from 0.000 to 0.072, and most Dendrobium species with more than two samples in our study had a unique sequence (65.85%) in the ITS2 region. The results indicated an obvious barcoding gap between inter- and intra-specific divergence, and the ITS2 sequences could provide a useful way to authenticate different Dendrobium species.
Figure 1.
Relative distribution of inter-specific divergence between congeneric Dendrobium species and intra-specific variation in the ITS2 region using K2P genetic distance.
Wilcoxon two-sample tests also showed significant differences between the inter- and intra-specific divergences (p < 0.001). Therefore, ITS2 possessed intra- and inter-specific variation gaps.
2.4. Efficacy of ITS2 for Authentication
ITS2 possessed 85.9% and 82.8% identification success rates at the species level for BLAST1 and nearest genetic distance, respectively (Table 2). Thus, ITS2 region exhibited high identification efficiency.
Table 2.
Comparison of authentication efficiency for ITS2 using different methods.
| Methods of Identification | No. of Samples | No. of Species | Correct Identification (%) | Incorrect Identification (%) | Ambiguous Identification (%) |
|---|---|---|---|---|---|
| BLAST1 | 364 | 64 | 85.9 | 0 | 14.1 |
| Distance | 364 | 64 | 82.8 | 0 | 17.2 |
2.5. Evaluation of the Discriminatory Power of ITS2 Sequences
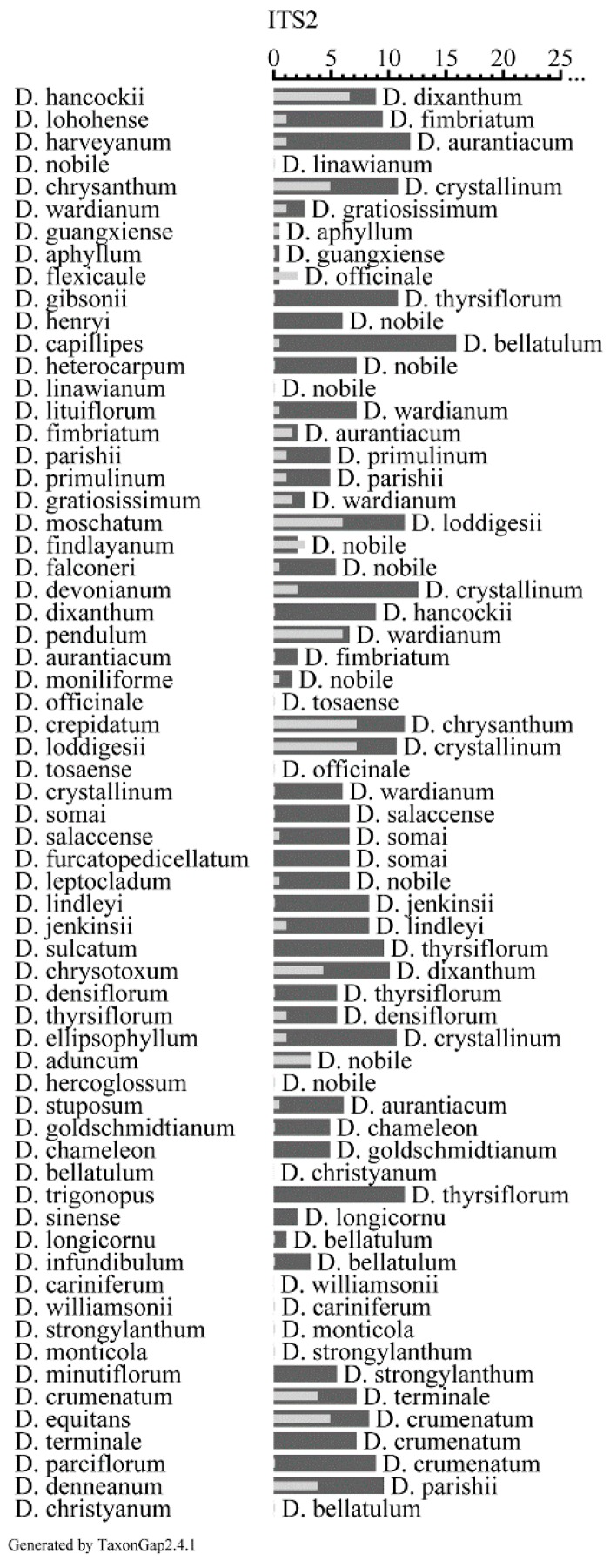
The discriminatory power of ITS2 sequences for collected samples was straightforwardly evaluated using software TaxonGap (Figure 2). As a result, over 79.7% of the species had larger inter- than intra-specific diversity; therefore, there were relatively clear species boundaries for ITS2 sequences. However, there were exceptions: 17.2% of the species (see dark grey bar, Figure 2) had identical sequences with their sister-species for D. nobile vs. D. linawianum, D. officinale vs. D. tosaense, D. hercoglossum vs. D. nobile, D. bellatulum vs. D. christyanum, D. cariniferum vs. D. williamsonii, and D. strongylanthum vs. D. monticola.
Figure 2.
The heterogeneity and separability for individual taxa of ITS2 based on 64 Dendrobium species by TaxonGap. The left side shows the complete list of Dendrobium species used in this study, including sequences we generated and those retrieved from GenBank (see Table 3 and Table S1). The right side depicts the within species heterogeneity (presented as light gray horizontal bar) and between-species separability (presented as dark grey horizontal bar) values (calculated by using similarity matrix for biomarker ITS2) with different OTUs (Operational Taxonomic Units) as matrix rows for ITS2. The names of the closest relatives (the taxon with the smallest separability) are listed at the right side of the dark grey bar.
2.6. Phylogenetic Analysis
According to the morphological classification of Dendrobium species reported previously [3,5,36], all Dendrobium species collected in this study (Table 3 and Table S1) belonged to 12 sections (sects.): Dendrobium, Grastidium, Chrysotoxae, Distichophyllum, Breviflores, Stuposa, Pedilonum, Formosae, Stachyobium, Crumenata, Aporum, and Strongyle. The neighbor-joining (NJ) phylogenetic tree constructed based on ITS2 sequences (Figure S1) grouped all the Dendrobium species into four main clusters, of which, however, only two (III, IV) are well supported considering bootstrap values.
Table 3.
Plant samples of Dendrobium used for analysis in this study with voucher number (No.) and GenBank Accession Number.
| Species Name | Voucher Number (No.) | Section | Collection Site | GenBank Accession No. |
|---|---|---|---|---|
| D. hancockii Rolfe | D-HZ001 | Dendrobium | Yunnan | KJ658296 |
| D. harveyanum Rchb. f. | D-HZ002 | Dendrobium | Yunnan | KJ658297 |
| D. nobile Lindl. | D-HZ003 | Dendrobium | Yunnan | KJ658298 |
| D. chrysanthum Wall. ex Lindl. | D-HZ004 | Dendrobium | Guangxi | KJ658299 |
| D. wardianum Warner | D-HZ005 | Dendrobium | Yunnan | KJ658300 |
| D. guangxiense S. J. Cheng et C. Z. Tang | D-HZ006 | Dendrobium | Yunnan | KJ658301 |
| D. guangxiense S. J. Cheng et C. Z. Tang | D-HZ007 | Dendrobium | Guangxi | KJ658302 |
| D. aphyllum (Rohb.) C. E. Fishcher | D-HZ008 | Dendrobium | Guangxi | KJ658303 |
| D. gibsonii Lindl. | D-HZ009 | Dendrobium | Yunnan | KJ658304 |
| D. gibsonii Lindl. | D-HZ010 | Dendrobium | Guangdong | KJ658305 |
| D. capillipes Rchb. | D-HZ011 | Dendrobium | Guangdong | KJ658306 |
| D. heterocarpum Lindl. | D-HZ012 | Dendrobium | Yunnan | KJ658307 |
| D. fimbriatum Hook. | D-HZ013 | Dendrobium | Guangxi | KJ658308 |
| D. primulinum Lindl. | D-HZ014 | Dendrobium | Yunnan | KJ658309 |
| D. falconeri Hook. | D-HZ015 | Dendrobium | Guangdong | KJ658310 |
| D. devonianum Paxt. | D-HZ016 | Dendrobium | Yunnan | KJ658311 |
| D. dixanthum Rchb. | D-HZ017 | Dendrobium | Zhejiang | KJ658312 |
| D. dixanthum Rchb. | D-HZ018 | Dendrobium | Yunnan | KJ658313 |
| D. pendulum Roxb. | D-HZ019 | Dendrobium | Yunnan | KJ658314 |
| D. moniliforme (Linn.) Sw. | D-HZ020 | Dendrobium | Yunnan | KJ658315 |
| D. officinale Kimura et Migo | D-HZ021 | Dendrobium | Yunnan | KJ658316 |
| D. officinale Kimura et Migo | D-HZ022 | Dendrobium | Guangxi | KJ658317 |
| D. officinale Kimura et Migo | D-HZ023 | Dendrobium | Zhejiang | KJ658318 |
| D. crepidatum Lindl. ex Paxt. | D-HZ024 | Dendrobium | Yunnan | KJ658319 |
| D. loddigesii Rolfe | D-HZ025 | Dendrobium | Yunnan | KJ658320 |
| D. crystallinum Tchb. F. | D-HZ026 | Dendrobium | Guangxi | KJ658321 |
| D. denneanum Kerr. | D-HZ041 | Dendrobium | Guangdong | KJ658336 |
| D. lindleyi Steudel | D-HZ027 | Chrysotoxae | Guangdong | KJ658322 |
| D. lindleyi Steudel | D-HZ028 | Chrysotoxae | Yunnan | KJ658323 |
| D. chrysotoxum Lindl. | D-HZ029 | Chrysotoxae | Yunnan | KJ658324 |
| D. densiflorum Lindl. | D-HZ030 | Chrysotoxae | Guangdong | KJ658325 |
| D. densiflorum Lindl. | D-HZ031 | Chrysotoxae | Guangxi | KJ658326 |
| D. thyrsiflorum Rchb. f. ex André | D-HZ032 | Chrysotoxae | Yunnan | KJ658327 |
| D. aduncum Wall. ex Lindl. | D-HZ033 | Breviflores | Gangdong | KJ658328 |
| D.hercoglossum Rehb. f. | D-HZ034 | Breviflores | Guizhou | KJ658329 |
| D. stuposum Lindl. | D-HZ035 | Stuposa | Yunnan | KJ658330 |
| D. stuposum Lindl. | D-HZ036 | Stuposa | Yunnan | KJ658331 |
| D. longicornu Lindl. | D-HZ037 | Formosae | Guangxi | KJ658332 |
| D. williamsonii Day et Rchb. F. | D-HZ038 | Formosae | Guangxi | KJ658333 |
| D. williamsonii Day et Rchb. F. | D-HZ039 | Formosae | Yunnan | KJ658334 |
| D. christyanum Rchb. F. | D-HZ042 | Formosae | Yunnan | KJ658337 |
| D. christyanum Rchb. F. | D-HZ043 | Formosae | Yunnan | KJ658338 |
| D. strongylanthum Rchb. F. | D-HZ040 | Stachyobium | Yunnan | KJ658335 |
Group I was the most complex, with 42 species, and was further divided into six subgroups. Among which, all species from sect. Dendrobium (33 species), Breviflores (two species), and Stuposa (one species) were included in Group I. Subgroup I-1 comprised 28 species, 25 from sect. Dendrobium and the remaining three from sect. Breviflores (D. aduncum and D. hercoglossum) and Grastidium (D. leptocladum). Subgroup I-2 included one species from sect. Chrysotoxae (D. chrysotoxum); while I-3 included six species from sect. Dendrobium and one from sect. Stuposa (D. stuposum). Subgroup I-4 contained two species from sect. Chrysotoxae (D. lindleyi and D. jenkinsii). Subgroup I-5 consisted of two species: one from sect. Dendrobium (D. gibsonii) and one from sect. Distichophyllum (D. ellipsophyllum). D. capillipes from sect. Dendrobium and D. trigonopus from sect. Formosae were assigned into subgroup I-6.
Group II comprised 16 species, and was further divided into four subgroups. Subgroup II-1 contained seven species from sect. Formosae. Subgroup II-2 included three species from sect. Chrysotoxae. Two species, D. goldschmidtianum and D. chameleon from sect. Pedilonum, constituted subgroup II-3. Compared with subgroup II-1, II-2 and II-3, subgroup II-4 was more complex, and comprised one species (D. terminale) from sect. Aporum, one species (D. parciflorum) from sect. Strongyle, and two species (D. crumenatum and D. equitans) from sect. Crumenata. All species (D. strongylanthum, D. monticola, and D. minutiflorum) from sect. Stachyobium constituted a separate group III. The species D. somai, D. salaccense and D. furcatopedicellatum from sect. Grastidium were distant from any other Dendrobium species, and were assigned into group IV (IV-1).
3. Discussion
A rapid and accurate method of species identification is essential to ensure the safe usage of drugs made from medicinal Dendrobium, and to preserve Dendrobium germplasm resources. To our knowledge, this is the first time that the ITS2 region has been utilized in identification of Dendrobium species using such a large sample size.
An ideal DNA barcode should possess high inter-specific but low intra-specific divergence in order to discriminate different species [13,37,38,39]. As in many previous studies [13,14,22,29], we found that ITS2 was a sufficiently variable DNA region among Dendrobium species for determination of genetic divergence, and also demonstrated a higher capability of successful discrimination (compared to 85.9% for BLAST1 method and 82.8% for nearest genetic distance method). For example, morphological traits of two species from sect. Breviflores (D. aduncum and D. hercoglossum) were very similar [3], but they could be accurately discriminated based on ITS2 regions. Similar satisfactory results were also obtained for discriminating D. thyrsiflorum and D. densiflorum, which are difficult to separate using morphological traits [3]. Therefore, we propose that ITS2 can be used for barcoding of Dendrobium species. However, it should be noted that some similar morphologic characteristics of Dendrobium species might make the classification of these species controversial. Hence, the taxonomic assignment of sequences from GenBank might not be accurate. If these factors were taken into account, the power of ITS2 in species discrimination might be estimated to be lower for Dendrobium.
However, ITS2 cannot easily solve all species determination problems in Dendrobium. For example, D. nobile and D. linawianum had identical ITS2 sequences, and another ITS2 sequence was common to D. officinale and D. tosaense. In addition, the number of Dendrobium that cannot be discriminated by ITS2 will probably increase with increasing species or sample set. Thus, other DNA barcodes might be worthwhile as complementary factors for discrimination of these species.
Recently, Dendrobium taxonomy is a global concern of biology systematics and is regarded as one of the most intricate challenges in Orchidaceae [4,5,11,40,41]. A number of molecular analyses have indicated that many morphological characters of the Dendrobium appear to be homoplasious, and several previously defined infrageneric taxa of Dendrobium are not monophyletic [4,5,11,42,43]. In the present study, we found that the ITS2 region was not only useful for DNA barcoding, but could also serve as a valuable standard phylogenetic marker for Dendrobium taxonomy. A dendrogram constructed with ITS2 data using the neighbor-joining (NJ) method indicated that the taxonomy of the infrageneric taxa of Dendrobium was complicated. As in previous studies [2,5,11,44], we also found that sect. Dendrobium was paraphyletic, and also strongly support the view that sects. Breviflores and Stuposa and D. chrysotoxum from sect. Chrysotoxae should be included in sect. Dendrobium [5]. D. ellipsophyllum from sect. Distichophyllum was grouped into I-5 together with D. gibsonii from sect. Dendrobium with middle support (bootstrap support (BS) = 68). Sect. Distichophyllum was reported to be a sister group of sect. Formosae in a previous study [5]. Since only one species of this section was tested in the present study, more sampling and more evidence is required to determine the relationship between species from sect. Distichophyllum and species from other sections. Xiang et al. showed that sect. Chrysotoxae was probably polyphyletic [5], and we obtained similar results that the collected species of sect. Chrysotoxae formed three groups (I-2, I-4, and II-2; Figure S1), and that D. lindleyi and D. jenkinsii, apart from other species from sect. Chrysotoxae, were grouped together (Figure S1, I-4) as Xiang et al. reported [5]. Some sections, such as sects. Stachyobium, Pedilonum and Formosae (except D. trigonopus), were found to be well supported as monophyletic (BS = 99, 99 and 96, respectively). Three sections, including sects. Aporum, Crumenata and Strongyle, formed the subgroup II-4 with strong support (BS = 97) as previously reported [5,38].
In addition, we found that D. trigonopus from sect. Formosae, unplaced species reported in previous studies [5,43], were grouped into subgroup I-6 together with D. capillipes with weak support (BS < 50). It appears that more sampling and more evidence are required to understand the evolutionary history of D. trigonopus. Differing from a previous study [5], we found that sect. Grastidium was not well supported as monophyletic. Apart from the other three species of sect. Grastidium, D. leptocladum was nested with species from sect. Dendrobium and grouped into subgroup I-1. Three species, D. somai, D. salaccense, and D. furcatopedicellatum from sect. Grastidium, were distant from any other Dendrobium species, and were assigned to subgroup IV-1, which may support the view that this section is poorly represented in mainland Asia [5].
Multiple copies of ITS (containing ITS1 and ITS2), one of the main factors that account for incomplete concerted evolution in plants, might cause the question whether the sequence obtained through PCR would be stable and representative, and might result in misleading phylogenetic inferences [14,29,45]. However, we think that the PCR-amplified copies could represent the dominant information of the repeated part of the ITS2 in plant genomes and, ITS2 can be effectively treated as a single locus as previous study [14]. In addition, there are many advantages for its widespread use, such as the levels of variations and multi-copy structure facilitating PCR amplification, even from herbarium specimens [29,46].
4. Materials and Methods
4.1. Plant Materials
A total of 364 samples of 64 species from genus Dendrobium were collected in this study. Among them, 43 specimens of 33 species sampled from the main distribution areas in China were used for sequencing (Table 3), whereas all other data (321 samples) was downloaded from GenBank (Table S1). The collected samples included D. nobile, D. officinale, D. fimbratum and D. chrysotoxum, which are listed in the Chinese Pharmacopoeia [6]. The species were verified and confirmed using the specimens stored in the herbarium of the Institute of Botany, Chinese Academy of Sciences, Beijing, China (http://www.nhpe.org). All corresponding voucher samples were deposited in the Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants, Hangzhou Normal University.
4.2. DNA Extraction, Amplification, and Sequencing
Fresh, young leaves of sampled specimens were randomly collected for genomic DNA isolation. The genomic DNA was isolated as described previously [47]. The ITS2 region was amplified using the following pair of universal primers [29]: ITS-2F, 5′-ATGCGATACTTGGTGTGAAT-3′; and ITS-3R, 5′-GACGCTTCTCCAGACTACAAT-3′. Primers were synthesized by Shanghai Sunny Biotechnology Co., Ltd. (Shanghai, China). PCR was conducted in 25 μL volumes containing 1× PCR Buffer, 2.5 mM Mg2+, 0.4 mM dNTPs, 0.5 μM of each primer, 1 U Taq DNA polymerase (TaKaRa Bio., Kyoto, Japan), and 30 ng genomic DNA template. The amplification was performed in a MJ Research PTC-100 thermal cycler (MJ Research, Waltham, MA, USA) with a PCR program: 94 °C for 4 min, followed by 35 cycles of 94 °C for 45 s, 56 °C for 45 s, 72 °C for 1.5 min, and a final extension at 72 °C for 10 min. The PCR products were sequenced by Shanghai Sunny Biotechnology Co., Ltd.
4.3. Data Analysis
The original sequences were assembled using CodonCode Aligner V3.0 (CodonCode Co., Centerville, MA, USA). The ITS2 sequences were subjected to Hidden Markov Model (HMM) [48] model analysis to remove the conserved 5.8S and 28S DNA sequences [49]. The sequences with less than 100 bp length and the possible contaminated sequences of fungi were discarded. The ITS2 sequences were aligned using Clustal W [50] and the genetic distances computed using MEGA 5.1 according to the Kimura 2-Parameter (K2P) model [51]. The average inter-specific distance, the minimum inter-specific distance and theta prime were used to represent inter-specific divergences using the K2P model [13,14,52,53]. The average intra-specific distance, coalescent depth and theta were calculated to evaluate the intra-specific variation [13,14,53]. The distributions of intra- versus inter-specific variability were compared using the DNA barcoding gaps (The difference between the intra- and inter-specific divergence values is named as “barcoding gap”) [13,14,53]. Wilcoxon two-sample tests were performed as indicated previously [13,23,24]. Two methods of species identification, including BLAST1 and the nearest distance method, were used to evaluate the species authentication efficacy [13,14,54]. In the BLAST1 method, all ITS2 sequences of Dendrobium species in this study were used as query sequences. BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search for the reference database for each query sequence. Correct identification means that the best BLAST hit of the query sequence is from the expected species; ambiguous identification means that the best BLAST hits for a query sequence were found to be those of several species including the expected species; and incorrect identification means that the best BLAST hit of the query sequence is not from the expected species [14]. In the nearest distance method, correct identification means that the hit in our database based on the smallest genetic distances is from the same species as that of the query; ambiguous identification means that several hits from our database were found to have the same smallest genetic distance to the query sequence; and incorrect identification means that the hit based on the smallest genetic distance is not from the expected species [14]. The discriminatory power of ITS2 sequences in species was calculated using TaxonGAP 2.4.1 software [55].
The phylogenetic analysis for collected Dendrobium species based on ITS2 sequences was performed using the neighbor-joining (NJ) method in MEGA 5.1. Bootstrap support (BS) values for individual clades were calculated by running 1000 bootstrap replicates of the data. Pholidota and Bulbophyllum are closely related to Dendrobium in Orchidaceae [5,56,57,58]. Four Pholidota species (P. chinensis, P. cantonensis, P. imbricata and P. carnea) and four Bulbophyllum species (B. orientale, B. inconspicuum, B. kwangtungense and B. omerandrum) were used as outgroups.
5. Conclusions
In summary, our study demonstrated that ITS2 might be a useful DNA barcode to identify Dendrobium species, and reconstruct the phylogeny of the genus Dendrobium. However, more Dendrobium species should be included in the future to verify whether the findings hold when even more closely related taxa are included. Our current work provided much useful genetic information about Dendrobium species, which will be useful for germplasm management and resource protection.
Acknowledgments
This study was supported in part by the National Natural Science Foundation of China (No. 30870180, 31470407), the Zhejiang Provincial Natural Science Foundation of China (LQ13H28006), and the Zhejiang Provincial Public Welfare Technology Applied Research Foundation of China (2014C32090).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/09/21975/s1.
Author Contributions
Conceived and designed the experiments: Shangguo Feng and Huizhong Wang; Performed the experiments: Shangguo Feng, Yan Jiang and Shang Wang; Collected samples: Huizhong Wang, Zhe Chen and Qicai Ying; Analyzed the data: Shangguo Feng, Shang Wang and Mengying Jiang; And drafted the manuscript: Shangguo Feng and Huizhong Wang.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wang H.Z., Feng S.G., Lu J.J., Shi N.N., Liu J.J. Phylogenetic study and molecular identification of 31 Dendrobium species using inter-simple sequence repeat (ISSR) markers. Sci. Hortic. 2009;122:440–447. doi: 10.1016/j.scienta.2009.06.005. [DOI] [Google Scholar]
- 2.Feng S.G., Lu J.J., Gao L., Liu J.J., Wang H.-Z. Molecular phylogeny analysis and species identification of Dendrobium (Orchidaceae) in China. Biochem. Genet. 2014;52:127–136. doi: 10.1007/s10528-013-9633-6. [DOI] [PubMed] [Google Scholar]
- 3.Tsi Z.H., Chen S.C., Luo Y.B., Zhu G.H. Flora Republicae Popularis Sinicae. Science Press; Beijing, China: 1999. Orchidaceae (3) [Google Scholar]
- 4.Adams P.B. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Bot. J. Linn. Soc. 2011;166:105–126. doi: 10.1111/j.1095-8339.2011.01141.x. [DOI] [Google Scholar]
- 5.Xiang X.G., Schuiteman A., Li D.Z., Huang W.C., Chung S.W., Li J.W., Zhou H.L., Jin W.T., Lai Y.J., Li Z.Y., et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 2013;69:950–960. doi: 10.1016/j.ympev.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Chinese Pharmmacopoeia Editorial Committee (CPEC) Theoretical and Applied Genetics. CPEC; Beijing, China: 2010. Pharmmacopoeia of the People’s Republic of China. [Google Scholar]
- 7.Bulpitt C.J., Li Y., Bulpitt P.F. The use of orchids in Chinese medicine. Genome Natl. Res. Counc. Can. 2007;100:558–563. doi: 10.1258/jrsm.100.12.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin W., Yao S. Cultivation and Appreciation of Noble Spring Orchid Cultivars. Guangdong Science and Technology Press; Guangzhou, China: 2006. [Google Scholar]
- 9.Ding G., Li X., Ding X., Qian L. Genetic diversity across natural populations of Dendrobium officinale, the endangered medicinal herb endemic to China, revealed by ISSR and RAPD markers. Genetika. 2009;45:375–382. doi: 10.1134/S1022795409030119. [DOI] [PubMed] [Google Scholar]
- 10.Lau D.T., Shaw P.C., Wang J., But P.P. Authentication of medicinal Dendrobium species by the internal transcribed spacer of ribosomal DNA. Planta Med. 2001;67:456–460. doi: 10.1055/s-2001-15818. [DOI] [PubMed] [Google Scholar]
- 11.Feng S., He R., Yang S., Chen Z., Jiang M., Lu J., Wang H. Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of Dendrobium species. Gene. 2015;567:182–188. doi: 10.1016/j.gene.2015.04.076. [DOI] [PubMed] [Google Scholar]
- 12.Hebert P.D.N., Penton E.H., Burns J.M., Janzen D.H., Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S., Yao H., Han J., Liu C., Song J., Shi L., Zhu Y., Ma X., Gao T., Pang X., et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao T., Yao H., Song J., Liu C., Zhu Y., Ma X., Pang X., Xu H., Chen S. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J. Ethnopharmacol. 2010;130:116–121. doi: 10.1016/j.jep.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Hajiahmadi Z., Talebi M., Sayed-Tabatabaei B.E. Studying genetic variability of Pomegranate (Punica granatum L.) based on chloroplast DNA and barcode genes. Mol. Biotechnol. 2013;55:249–259. doi: 10.1007/s12033-013-9676-2. [DOI] [PubMed] [Google Scholar]
- 16.Giudicelli G.C., Mader G., Brandao de Freitas L. Efficiency of ITS sequences for DNA barcoding in Passiflora (Passifloraceae) Int. J. Mol. Sci. 2015;16:7289–7303. doi: 10.3390/ijms16047289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Provan J., Gao L.M., Li D.Z. Sampling strategy and potential utility of indels for DNA barcoding of closely related plant species: A case study in taxus. Int. J. Mol. Sci. 2012;13:8740–8751. doi: 10.3390/ijms13078740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z.Y., Chen S.L. Identification of cortex herbs using the DNA barcode nrITS2. J. Nat. Med. Tokyo. 2013;67:296–302. doi: 10.1007/s11418-012-0681-8. [DOI] [PubMed] [Google Scholar]
- 19.Gismondi A., Rolfo M.F., Leonardi D., Rickards O., Canini A. Identification of ancient Olea europaea L. and Cornus mas L. seeds by DNA barcoding. Comptes Rendus Biol. 2012;335:472–479. doi: 10.1016/j.crvi.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Gismondi A., Leonardi D., Enei F., Canini A. Identification of plant remains in underwater archaeological areas by morphological analysis and DNA barcoding. Adv. Anthropol. 2013;3:240–248. doi: 10.4236/aa.2013.34034. [DOI] [Google Scholar]
- 21.Gismondi A., Fanali F., Labarga J., Caiola M., Canini A. Crocus sativus L. genomics and different DNA barcode applications. Plant Syst. Evol. 2013;299:1859–1863. doi: 10.1007/s00606-013-0841-7. [DOI] [Google Scholar]
- 22.Liu Z., Zeng X., Yang D., Ren G., Chu G., Yuan Z., Luo K., Xiao P., Chen S. Identification of medicinal vines by ITS2 using complementary discrimination methods. J. Ethnopharmacol. 2012;141:242–249. doi: 10.1016/j.jep.2012.01.057. [DOI] [PubMed] [Google Scholar]
- 23.Kress W.J., Erickson D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahaye R., van der Bank M., Bogarin D., Warner J., Pupulin F., Gigot G., Maurin O., Duthoit S., Barraclough T.G., Savolainen V. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. USA. 2008;105:2923–2928. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadjiolova K.V., Nicoloso M., Mazan S., Hadjiolov A.A., Bachellerie J.P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Nues R.W., Rientjes J.M.J., Morré S.A., Mollee E., Planta R.J., Venema J., Raué H.A. Evolutionarily conserved structural elements are critical for processing of Internal Transcribed Spacer 2 from Saccharomyces cerevisie Precursor Ribosomal RNA. J. Mol. Biol. 1995;250:24–36. doi: 10.1006/jmbi.1995.0355. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen A., Stothard J.R., Madsen H., Nalugwa A., Nyakaana S., Rollinson D. The ITS2 of the genus Bulinus: Novel secondary structure among freshwater snails and potential new taxonomic markers. Acta Trop. 2012;128:218–225. doi: 10.1016/j.actatropica.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Pang X., Song J., Zhu Y., Xu H., Huang L., Chen S. Applying plant DNA barcodes for Rosaceae species identification. Cladistics. 2011;27:165–170. doi: 10.1111/j.1096-0031.2010.00328.x. [DOI] [PubMed] [Google Scholar]
- 29.Yao H., Song J., Liu C., Luo K., Han J., Li Y., Pang X., Xu H., Zhu Y., Xiao P., et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao T., Yao H., Song J., Zhu Y., Liu C., Chen S. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evol. Biol. 2010;10:324. doi: 10.1186/1471-2148-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang X., Song J., Zhu Y., Xie C., Chen S. Using DNA barcoding to identify species within Euphorbiaceae. Planta Med. 2010;76:1784–1786. doi: 10.1055/s-0030-1249806. [DOI] [PubMed] [Google Scholar]
- 32.Gu W., Song J., Cao Y., Sun Q., Yao H., Wu Q., Chao J., Zhou J., Xue W., Duan J. Application of the ITS2 region for barcoding medicinal plants of Selaginellaceae in Pteridophyta. PLoS ONE. 2013;8:e67818. doi: 10.1371/journal.pone.0067818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman A.W. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003;19:370–375. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 34.Schultz J., Wolf M. ITS2 sequence-structure analysis in phylogenetics: A how-to manual for molecular systematics. Mol. Phylogenet. Evol. 2009;52:520–523. doi: 10.1016/j.ympev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Chiou S.J., Yen J.H., Fang C.L., Chen H.L., Lin T.Y. Authentication of medicinal herbs using PCR-amplified ITS2 with specific primers. Planta Med. 2007;73:1421–1426. doi: 10.1055/s-2007-990227. [DOI] [PubMed] [Google Scholar]
- 36.Schlechter R. In: The Orchidaceae of German New Guinea. Blaxell D.F., translator. The Australian Orchid Foundation; Melbourne, Australia: 1982. [Google Scholar]
- 37.Cowan R.S., Chase M.W., Kress W.J., Savolainen V. 300,000 species to identify: Problems, progress, and prospects in DNA barcoding of land plants. Taxon. 2006;55:611–616. doi: 10.2307/25065638. [DOI] [Google Scholar]
- 38.Li H.Q., Chen J.Y., Wang S., Xiong S.Z. Evaluation of six candidate DNA barcoding loci in Ficus (Moraceae) of China. Mol. Ecol. Resour. 2012;12:783–790. doi: 10.1111/j.1755-0998.2012.03147.x. [DOI] [PubMed] [Google Scholar]
- 39.Hebert P.D., Cywinska A., Ball S.L., deWaard J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yukawa T., Uehara K. Vegetative diversification and radiation in subtribe Dendrobiinae (Orchidaceae): Evidence from chloroplast DNA phylogeny and anatomical characters. Plant Syst. Evol. 1996;201:1–14. doi: 10.1007/BF00989048. [DOI] [Google Scholar]
- 41.Morris M.W., Steen W.L., Judd W.S. Vegetative anatomy and systematics of subtribe Dendrobiinae (Orchidaceae) Bot. J. Linn. Soc. 1996;120:89–144. doi: 10.1111/j.1095-8339.1996.tb00483.x. [DOI] [Google Scholar]
- 42.Burke J.M., Bayly M.J., Adams P.B., Ladiges P.Y. Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification. Aust. Syst. Bot. 2008;21:1–14. doi: 10.1071/SB07038. [DOI] [Google Scholar]
- 43.Schuiteman A. Dendrobium (Orchidaceae): To split or not split. Gard. Bull. Singap. 2011;63:245–257. [Google Scholar]
- 44.Clements M.A. Molecular phylogenetic systematics in Dendrobieae (Orchidaceae) Aliso. 2006;22:465–480. [Google Scholar]
- 45.Queiroz Cde S., Batista F.R., de Oliveira L.O. Evolution of the 5.8S nrDNA gene and internal transcribed spacers in Carapichea ipecacuanha (Rubiaceae) within a phylogeographic context. Mol. Phylogenet. Evol. 2011;59:293–302. doi: 10.1016/j.ympev.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Nieto Feliner G., Rossello J.A. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol. Phylogenet. Evol. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Feng S., Zhao H., Lu J., Liu J., Shen B., Wang H. Preliminary genetic linkage maps of Chinese herb Dendrobium nobile and D. moniliforme. J. Genet. 2013;92:205–212. doi: 10.1007/s12041-013-0246-y. [DOI] [PubMed] [Google Scholar]
- 48.Keller A., Schleicher T., Schultz J., Muller T., Dandekar T., Wolf M. 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–57. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Koetschan C., Hackl T., Muller T., Wolf M., Forster F., Schultz J. ITS2 Database IV: Interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Mol. Phylogenet. Evol. 2012;63:585–588. doi: 10.1016/j.ympev.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 50.Thompson J.D., Gibson T., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002 doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meier R., Zhang G., Ali F. The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Syst. Biol. 2008;57:809–813. doi: 10.1080/10635150802406343. [DOI] [PubMed] [Google Scholar]
- 53.Meyer C.P., Paulay G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross H.A., Murugan S., Li W.L. Testing the reliability of genetic methods of species identification via simulation. Syst. Biol. 2008;57:216–230. doi: 10.1080/10635150802032990. [DOI] [PubMed] [Google Scholar]
- 55.Slabbinck B., Dawyndt P., Martens M., de Vos P., de Baets B. TaxonGap: A visualization tool for intra-and inter-species variation among individual biomarkers. Bioinformatics. 2008;24:866–867. doi: 10.1093/bioinformatics/btn031. [DOI] [PubMed] [Google Scholar]
- 56.Ding X.Y., Wang Z.T., Xu H., Xu L.S., Zhou K.Y. Database establishment of the whole rDNA ITS region of Dendrobium species of “fengdou” and authentication by analysis of their sequences. Yao Xue Xue Bao. 2002;37:567–573. (In Chinese) [PubMed] [Google Scholar]
- 57.Yang L., Wang Z., Xu L. Simultaneous determination of phenols (bibenzyl, phenanthrene, and fluorenone) in Dendrobium species by high-performance liquid chromatography with diode array detection. J. Chromatogr. A. 2006;1104:230–237. doi: 10.1016/j.chroma.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Yao H., Song J.Y., Ma X.Y., Liu C., Li Y., Xu H.X., Han J.P., Duan L.S., Chen S.L. Identification of Dendrobium species by a candidate DNA barcode sequence: The chloroplast psbA-trnH intergenic region. Planta Med. 2009;75:667–669. doi: 10.1055/s-0029-1185385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.